Abstract
The Amphibacillus xylanus NADH oxidase, which catalyzes the reduction of oxygen to hydrogen peroxide with β-NADH, can also reduce hydrogen peroxide to water in the presence of free flavin adenine dinucleotide (FAD) or the small disulfide-containing Salmonella enterica AhpC protein. The enzyme has two disulfide bonds, Cys128-Cys131 and Cys337-Cys340, which can act as redox centers in addition to the enzyme-bound FAD (K. Ohnishi, Y. Niimura, M. Hidaka, H. Masaki, H. Suzuki, T. Uozumi, and T. Nishino, J. Biol. Chem. 270:5812–5817, 1995). The NADH-FAD reductase activity was directly dependent on the FAD concentration, with a second-order rate constant of approximately 2.0 × 106 M−1 s−1. Rapid-reaction studies showed that the reduction of free flavin occurred through enzyme-bound FAD, which was reduced by NADH. The peroxidase activity of NADH oxidase in the presence of FAD resulted from reduction of peroxide by free FADH2 reduced via enzyme-bound FAD. This peroxidase activity was markedly decreased in the presence of oxygen, since the free FADH2 is easily oxidized by oxygen, indicating that this enzyme system is unlikely to be functional in aerobic growing cells. The A. xylanus ahpC gene was cloned and overexpressed in Escherichia coli. When the NADH oxidase was coupled with A. xylanus AhpC, the peroxidase activity was not inhibited by oxygen. The Vmax values for hydrogen peroxide and cumene hydroperoxide reduction were both approximately 150 s−1. The Km values for hydrogen peroxide and cumene hydroperoxide were too low to allow accurate determination of their values. Both AhpC and NADH oxidase were induced under aerobic conditions, a clear indication that these proteins are involved in the removal of peroxides under aerobic growing conditions.
We have reported previously on a new group of facultatively anaerobic bacteria isolated from alkaline compost (17). Since the bacteria have unique phenotypic and chemotaxonomic characteristics (18), as well as bioenergetic properties (13), we named one of the isolate Amphibacillus xylanus (18). A. xylanus, which lacks a respiratory system and heme proteins, including catalase, grows well and has the same growth rate and cell yield under strictly anaerobic and aerobic conditions (18). This is due to the presence of anaerobic and aerobic pathways producing similar amounts of ATP (19). We purified NADH oxidase from aerobically grown A. xylanus (20) and found that hydrogen peroxide is formed during the reaction of the NADH oxidase. We have also found that the enzyme reduces hydrogen peroxide in the presence of additional free flavin adenine dinucleotide (FAD); this may be of physiological relevance because of the presence of 13 μM free FAD in aerobically growing cells. Such behavior indicates that the A. xylanus NADH oxidase has unique functional properties that are different from those of other known NADH oxidases (8, 10, 12, 27, 31–33).
The amino acid sequence of A. xylanus NADH oxidase exhibits up to 51.2% identity to the alkyl hydroperoxide reductase F-52a flavoprotein component (AhpF) from Salmonella enterica (36), but the ability of the latter to reduce hydrogen peroxide was not reported (11). We have shown that both enzymes reduce not only alkyl hydroperoxides but also hydrogen peroxide in the presence of the 22-kDa protein component (AhpC) of the alkyl hydroperoxide reductase from Salmonella enterica, and the turnover numbers of the reactions catalyzed by the NADH oxidase are extremely high compared with those of known peroxide-reducing enzymes (21). Complete reduction of the NADH oxidase by dithionite required 6 electron equivalents/mol of enzyme-bound flavin (25, 26). Such behavior indicated the presence of redox centers in addition to the FAD, and these were shown to be two disulfides, Cys128-Cys131 and Cys337-Cys340. To provide information about the relationship between such high turnover numbers and the three redox centers of the enzyme, the reductive half-reactions of the wild-type and the mutant (C337S/C340S) NADH oxidase were investigated and have been reported in a previous publication (22).
Hydrogen peroxide is presumed to be accumulated in aerobically growing cells of A. xylanus, which lack catalase and a respiratory chain (18), because the NADH oxidase should regenerate NAD+ from NADH formed in the aerobic pathway and produce hydrogen peroxide from oxygen. The ability to get rid of hydrogen peroxide must be high for the cells to be able to survive under aerobic conditions. In the presence of free FAD or the AhpC protein, the NADH oxidase shows scavenging activity for hydrogen peroxide (21, 23). In this study, we investigated these kinds of peroxide reductase activities, and we discuss them below in terms of their relevance to the aerobic metabolism of the bacteria.
MATERIALS AND METHODS
Bacterial growth.
A. xylanus Ep01 was grown aerobically at 39°C in glucose-containing medium as described previously (18). The cells were harvested by centrifugation after 7 h of cultivation (early stationary phase), washed with 50 mM sodium phosphate buffer (pH 7.0), and then stored at −80°C until use.
Enzymes and materials.
8-Dimethylaminoriboflavin (14) was kindly supplied by Kunio Matsui (Osaka International University for Women) and Sabu Kasai (Osaka City University) and converted to the FAD level with the FAD synthase complex from Brevibacterium ammoniagenes (35). 6-Hydroxy-riboflavin was a gift from Sandro Ghisla (University of Konstanz) and was converted similarly to FAD. Recombinant NADH oxidase and its mutants (C337S/C340S) were purified as described previously (26). Both the NADH oxidase and ahpC genes were cloned into the pTTQ18 vector and expressed in Escherichia coli JM109 by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration). Recombinant AhpC and NADH oxidase were purified in a manner similar to that used for recombinant NADH oxidase, since AhpC and NADH oxidase were separated by the first step of purification, a column of DE53 (2.6 by 33 cm) (25). The pools of the fractions containing AhpC and NADH oxidase after the DE53 purification were individually subjected to the next purification step as described previously (25).
Stopped-flow absorbance spectrophotometry.
The stopped-flow apparatus has been described previously (22). Anaerobiosis of the flow system was achieved by overnight equilibration with an anaerobic solution of protocatechuate-3,4-dioxygenase (a generous gift of David P. Ballou, University of Michigan). Rate constants were obtained by exponential fitting using the software Program A (developed by C.-J. Chiu, R. Chung, J. Dinverno, and D. P. Ballou), which permits the analysis of experimental data by exponential fitting based on the Marquardt algorithm (3).
Anaerobiosis for flavin reductase activity.
The enzyme solution (10 ml) containing 50 mM sodium phosphate buffer (pH 7.0), 0.5 mM EDTA, and enzyme (in the concentration range from 0.4 to 1.0 μM) was loaded into a tonometer. After anaerobiosis was established by repeated evacuation and equilibration with oxygen-free argon and equilibration at 25°C, the reaction was started by mixing the enzyme solution in the stopped-flow spectrophotometer with 20 to 200 μM FAD solutions containing different concentrations of NADH and monitored at 350 nm (NADH oxidation) and 450 nm (flavin reduction). The FAD solutions, containing 50 mM sodium phosphate buffer (pH 7.0), 0.5 mM EDTA, and 5 to 300 μM NADH (the concentrations were halved after mixing in the stopped-flow spectrophotometer), were made anaerobic by bubbling with oxygen-free argon at 25°C.
Anaerobiosis for reaction of reduced NADH oxidase with 6-hydroxy-FAD and 8-dimethylamino-FAD.
To 10 ml of ca. 20 μM NADH oxidase in the main compartment of a tonometer were added 233 μM xanthine, 2 μM benzyl viologen, and 2 μM methyl viologen. After anaerobiosis, a catalytic amount of milk xanthine oxidase was added from the side arm and the reduction was carried out at room temperature, taking more than 1 to 2 h. The reaction was started by mixing the enzyme solution with different concentrations of 6-hydroxy-FAD or 8-dimethylamino-FAD. The reaction with 6-hydroxy-FAD was monitored at 424 nm, and that with 8-dimethylamino-FAD was monitored at 520 nm.
NADH-dependent oxidase and peroxidase activities.
Oxygen consumption was determined polarographically at 25°C as described previously (23). The oxidase activity of NADH oxidase was measured spectrophotometrically at 340 nm and 25°C as described previously (25). Turnover studies of hydrogen peroxide or alkyl hydroperoxide reductase were carried out with a stopped-flow spectrophotometer (Hi-tech SF-61). We found that in the presence of Amphibacillus AhpC, the peroxide reductase activities were the same in air-saturated solution as those determined anaerobically (data not shown); therefore, we did not use an anaerobic stopped-flow assay system for the routine measurement of peroxide reductase activity. The enzyme-AhpC mixture (10 ml), containing 50 mM sodium phosphate buffer (pH 7.0), 0.5 mM EDTA, 300 mM ammonium sulfate, 1.12 μM enzyme, and 70.4 μM AhpC, was loaded into a tonometer. After equilibration at 25°C, the reaction was started by mixing with different NADH-peroxide mixtures, and the reaction was monitored at 340 nm. The NADH-peroxide mixtures contained 50 mM sodium phosphate buffer (pH 7.0), 0.5 mM EDTA, 0.1 to 1 mM hydrogen peroxide or cumene hydroperoxide, and 30 to 150 μM NADH at 25°C.
SOD activity and various peroxidase activities.
Superoxide dismutase (SOD) activity was determined by the ferricytochrome c-xanthine oxidase method (2). One enzyme unit of SOD was defined as that resulting in 50% inhibition of the detection reaction under the assay conditions. Each peroxidase activity was measured at both pH 7.0 (50 mM sodium phosphate buffer) and pH 8.5 (50 mM Tris buffer), because the internal pH of Amphibacillus, when incubated in the growth medium at pH 8.5 and 9.5, was 7.7 and 8.5, respectively. Other assays of peroxidases were performed at 39.5°C by methods used for: fatty acid peroxidase (4), cytochrome peroxidase (37), peroxidase (30), iodide peroxidase (1), glutathione peroxidase (16), chloride peroxidase (15), l-ascorbate peroxidase (34), and NADH peroxidase (23, 28).
Western blotting (immunoblotting).
Rabbit polyclonal anti-AhpC antibody and anti-NADH oxidase antibody from A. xylanus were prepared. Cell extracts from anaerobically and aerobically grown cultures were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane, which was soaked for 1 h at room temperature in blocking buffer (0.299 g of NaH2PO4 · 2H2O, 3.04 g of Na2HPO4 · 12H2O, and 8.77 g of NaCl in 1 liter [pH 7.2]) containing 5% (wt/vol) skim milk. The polyvinylidene difluoride membrane was incubated for 1 h at room temperature with anti-AhpC antibody in blocking buffer containing 5% (wt/vol) skim milk, washed five times with blocking buffer, incubated for 1 h with donkey anti-rabbit immunoglobulin G antibody conjugated with horseradish peroxidase, washed five times with blocking buffer, and then developed with a solution containing 50 mM Tris-HCl (pH 7.4), 0.6 mg of 3,3-diaminobenzidine tetrahydrochloride in 1 ml, 0.03% (wt/vol) cobalt chloride, and 0.03% (vol/vol) hydrogen peroxide.
Nucleotide sequence accession number.
The ahpC gene sequence reported has been entered into the DNA Data Bank of Japan under accession number AB018435.
RESULTS
Cloning, and expression of the ahpC gene from A. xylanus.
A partial open reading frame (ORF) homologous to the S. enterica ahpC structural gene has been found upstream of the A. xylanus NADH oxidase structural gene (5). The fragment was contained in a 9-kb EcoRI fragment, which we had isolated previously from A. xylanus Ep01 chromosomal DNA (20), and was cloned into plasmid pUC119 to yield pPU9. Nucleotide sequencing of the fragment revealed an ORF, which is immediately preceded by a typical ribosomal binding site, GGAGG (Shine-Dalgarno sequence). The ORF encodes a protein homologous to the Salmonella AhpC (64.4% [data not shown]). The two ORFs encoding AhpC and NADH oxidase are separated by 17 bp and would be transcribed in the same direction. To investigate the possibility that two ORFs are cotranscribed, we cloned the fragment containing the two ORFs into the expression vector pTTQ18 as follows. The EcoRI-BamHI fragment from pPU9 was partially digested with SspI to release a 2.5-kb fragment. This fragment was cloned into pTTQ18. The resultant plasmid, pAHNO2.5, contained the complete AhpC and NADH oxidase gene downstream from the tac promoter. The plasmid was transformed into E. coli JM109, which gave consistent IPTG-induced expression of AhpC and NADH oxidase, confirmed by SDS-polyacrylamide gel electrophoresis, which revealed 21- and 56-kDa bands after IPTG induction (data not shown). The above result indicates that both proteins are synthesized from a single transcript.
Purification of recombinant and native AhpC.
The purification of recombinant AhpC is based on a method presented in a previous report (25). The average purification yield from 1 liter of culture broth of E. coli(pAHNO2.5) was 7 mg. SDS-polyacrylamide gel electrophoresis analysis of recombinant AhpC revealed a molecular weight of 21,000 (data not shown), which is consistent with the calculated value of 20,774. However, under nonreducing conditions, the observed molecular weight was 40,000, corresponding to the molecular size of a dimer (data not shown). This dimerization was also observed in Salmonella AhpC (29) and yeast thiol-specific antioxidant (6), which showed 64.4 and 39.3% homology, respectively, to Amphibacillus AhpC. AhpC was also partially purified from aerobically growing cells of A. xylanus Ep01 by the same purification method as that used for recombinant AhpC. The N-terminal amino acid sequence of AhpC from Amphibacillus was determined to be SLIGTEVQPFRA, which is identical to that of recombinant AhpC and indicates that the initiating methionine of AhpC is removed to give mature AhpC, starting with the second amino acid, Ser. Removal of the initiating methionine was also observed in Salmonella AhpC (29).
Effect of free FAD or AhpC on oxygen consumption and hydrogen peroxide reductase activity.
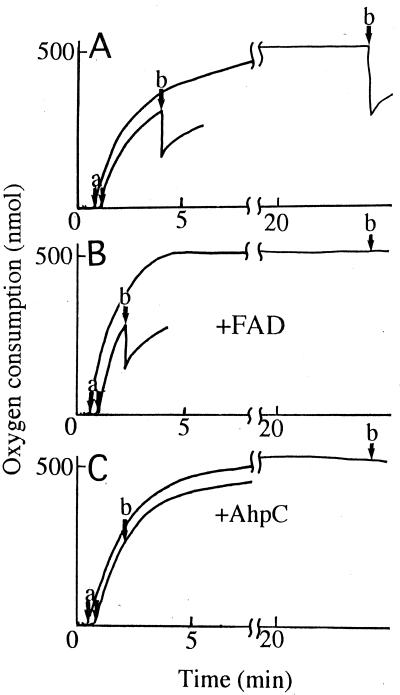
Since the NADH oxidase consumes oxygen to produce hydrogen peroxide, the addition of catalase at the end of the reaction results in the release of oxygen equivalent to half of that consumed (20). As shown in Fig. 1A, such an effect is independent of whether catalase is added during or at the end of the reaction. In the presence of free FAD, the rate of oxygen consumption was enhanced considerably at all experimentally obtainable concentrations of oxygen (23). When catalase was added after complete consumption of oxygen, there was no release of oxygen; however, when it was added during the course of the reaction, there was partial release of oxygen (Fig. 1B). In contrast, when the auxiliary protein, AhpC, which binds with the flavoprotein to constitute a highly reactive alkyl hydroperoxide reductase was added, catalase had no effect regardless of the time of its addition (Fig. 1C). The effect of FAD is due to the ability of NADH oxidase to function as a flavin reductase and to the ability of reduced flavin to reduce hydrogen peroxide nonenzymatically, as shown in subsequent sections.
FIG. 1.
Oxygen consumption during the catalysis of A. xylanus NADH oxidase in the presence of FAD or AhpC. The reaction mixture (2.5 ml) contained 50 mM sodium phosphate buffer (pH 7.0), 0.5 mM EDTA, 0.02% bovine serum albumin, 2% ammonium sulfate, and 600 μM NADH, with 50 μM FAD (B), 30 μM AhpC (C), or neither (A). The reaction was started at 25°C by the addition of 52.8 μg (A), 5.3 μg (B), or 105.6 μg (C) of NADH oxidase, as indicated by arrows a. During oxygen consumption, 60 μg of catalase was added, as indicated by arrows b.
Flavin reductase activity of NADH oxidase.
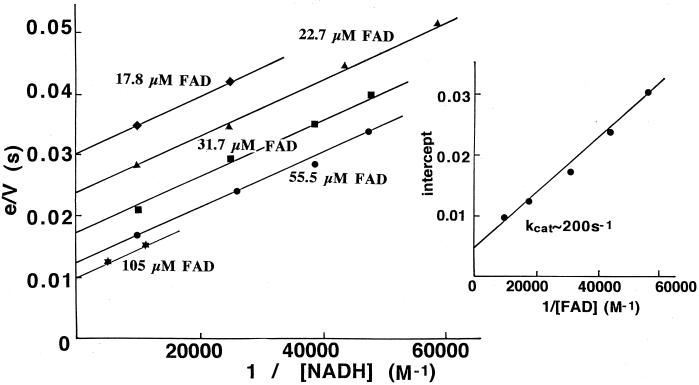
The NADH oxidase was found to be very active as a FAD reductase under anaerobic conditions. The steady-state kinetics of this reaction were determined by measuring the initial rates of FAD reduction at various concentrations of NADH and with a catalytic concentration of enzyme (in the range from 0.4 to 1.0 μM), utilizing the ability of the stopped-flow spectrophotometer to operate under anaerobic conditions. The results of such a study are shown in Fig. 2. From a secondary plot of the primary intercepts against the reciprocal FAD concentration (Fig. 2 inset), kcat is estimated to be ∼200 s−1, with Km values of ∼100 μM, for both NADH and FAD. The value of kcat is the same as that for reduction of the enzyme-bound flavin by NADH (22) and implies that the reaction of the reduced enzyme with free FAD is approximately second order:
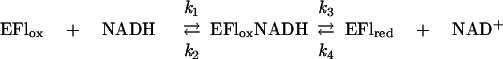 |
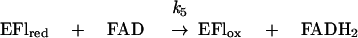 |
For such a kinetic mechanism, kcat = k3, Km (NADH) = (k2 + k3)/k1, and Km (FAD) = k3/k5. In previous studies (22), k3 has been determined to be 200 s−1 and Kd for NADH binding has been determined to be 1.2 × 10−4 M. The value for Km (FAD) of ∼100 μM would require a second-order rate constant (k5) of ∼2 × 106 M−1 s−1 for reoxidation of the reduced enzyme by FAD. Reaction traces of FAD reduction in the presence of high concentrations of NADH in fact show a single exponential decay, implying a direct relationship between catalytic turnover and residual FAD concentration, and yield a second-order rate constant of 1.6 × 106 M−1 s−1. We made use of the different spectral characteristics of 6-OH-FAD (λmax = 424 nm) and 8-dimethylamino-FAD (λmax = 505 nm) compared to that of NADH oxidase (λmax = 448 nm) to measure directly their reaction rate constants with reduced NADH oxidase. Second-order rate constants at pH 7.0 and 25°C of 1.9 × 106 and 1.7 × 106 M−1 s−1 were obtained for the reactions with 6-OH-FAD and 8-dimethylamino-FAD, respectively (results not shown).
FIG. 2.
Steady-state kinetics of NADH-FAD reductase activity. Initial rates of reduction of FAD were monitored at 450 nm on mixing 1.0 μM NADH oxidase with the concentrations of NADH and FAD shown in the figure in a stopped-flow spectrophotometer under anaerobic conditions (the concentrations given are those after mixing). The enzyme was made anaerobic and stored under argon in a tonometer, and the NADH-FAD mixtures were made anaerobic by bubbling for 15 min with argon before introduction into the stopped-flow apparatus. The reaction conditions were 0.05 M sodium phosphate (pH 7.0), 0.5 mM EDTA, and 25°C.
Flavin reductase activity of C337S/C340S NADH oxidase.
The mutant enzyme also catalyzed rapid NADH-FAD reductase activity under anaerobic conditions. Although a complete steady-state study like that of Fig. 4 was not carried out, a Vmax value of 42 s−1 and an apparent Km (NADH) of 25 μM were obtained in the presence of 25 μM FAD. These values are similar to those obtained with wild-type enzyme under similar conditions (see the results with 22.6 μM FAD in Fig. 2). Thus, although reducing equivalents from FADH2 are passed rapidly to the C337-C340 disulfide of the native enzyme (22), it is clear that the flavin moiety alone is required for the FAD reductase activity.
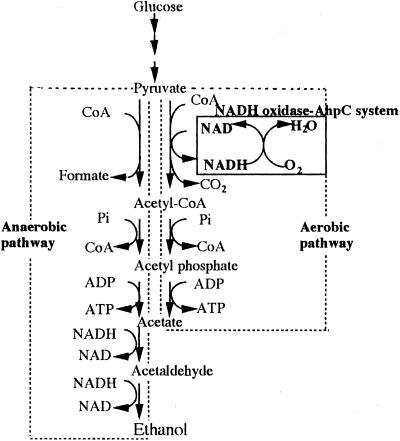
FIG. 4.
Proposed pathway for pyruvate metabolism in A. xylanus. The NADH oxidase-AhpC system is enclosed by the solid rectangle. CoA, coenzyme A.
Reaction of FADH2 with hydrogen peroxide.
From the results presented in Fig. 1B, it is clear that in the presence of FAD, hydrogen peroxide accumulates as an intermediate and is only slowly removed in a subsequent step. This is probably due to the slow reduction of H2O2 by FADH2 in a nonenzymatic reaction. We determined by stopped-flow spectrophotometry that under our standard experimental conditions, i.e., pH 7.0 and 25°C, H2O2 reoxidized FADH2 anaerobically with a rate constant of 21 M−1 s−1 (data not shown). This value is consistent with the slow reoxidation of reduced flavin by H2O2 reported by Dixon (9).
Stoichiometry of NADH consumption in the presence of FAD and oxygen.
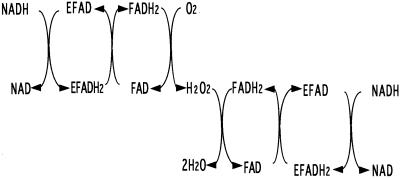
Under anaerobic conditions the catalytic NADH-FAD reductase reaction resulted in the consumption of NADH in stoichiometric amounts with respect to the amount of FAD reduced. However, in the presence of excess NADH, limiting concentrations of oxygen, and catalytic amounts of FAD, approximately 2 equivalents of NADH were consumed for reduction of 1 equivalent of oxygen. Thus, in a solution equilibrated with 5% O2 (O2 concentration, 61 μM) containing 10 μM FAD, 310 μM NADH, and 1.2 μM NADH oxidase, a total of 145 μM NADH was consumed (half in the first 15 s and the remainder over a period of 10 min) as estimated by the reduction in absorbance at 340 nm. After accounting for the 24 μM NADH required for the reduction of FAD and the enzyme, it can be calculated that almost exactly 2 equivalents of NADH are consumed for each equivalent of oxygen initially present. It is of course long established that the reaction of reduced flavin with O2 gives rise to a stoichiometric amount of H2O2 and that the latter can be further reduced to H2O by reduced flavin (9). Therefore, the NADH-peroxide reductase reaction catalyzed by the NADH oxidase, with the fast phase represented by the top section and the slower phase by the lower section, can be formulated as in Fig. 3.
FIG. 3.
Proposed reaction mechanism of the NADH oxidase in the presence of FAD and oxygen.
Alkyl hydroperoxide reductase activity in the presence of AhpC from A. xylanus.
The ability of NADH oxidase to catalyze the reduction of hydrogen peroxide or alkyl hydroperoxide in the presence of Amphibacillus AhpC was tested using a stopped-flow assay system that allowed continuous monitoring of NADH oxidation in the presence of peroxide substrates such as hydrogen peroxide or cumene hydroperoxide. The activity was found to be markedly dependent on the ionic strength, similar to the result previously found with Salmonella AhpC (21). The data described below were obtained in the presence of optimal concentrations of ammonium sulfate (300 mM). NADH oxidase is able to reduce both hydrogen peroxide and cumene hydroperoxide to give similar turnover numbers in the presence of Amphibacillus AhpC to those found previously with Salmonella AhpC. The Vmax values for hydrogen peroxide and cumene hydroperoxide were both approximately 150 s−1, extrapolated to saturating Amphibacillus AhpC concentration (results not shown). These Vmax values are almost the same as the values in the presence of Salmonella AhpC (21). The Km values for Amphibacillus AhpC in the reaction with hydrogen peroxide or cumene hydroperoxide were 8.9 and 10.0 μM, respectively and were similar in magnitude to those for Salmonella AhpC. On the other hand, the Km values for hydrogen peroxide, cumene hydroperoxide, and NADH were too low to allow an accurate determination of the values by the assay system used; the rates were the same with hydrogen peroxide or cumene hydroperoxide in the range from 0.1 to 1 mM and with NADH in the range from 37.5 to 150 μM.
Activity of SOD and the other peroxidases in Cell-Free Extracts of Amphibacillus.
Strong SOD activity was found in cell extracts of Amphibacillus. The activity was induced markedly under aerobic conditions, i.e., 45.8 U/mg of protein under aerobic conditions and 4.50 U/mg of protein under anaerobic conditions. Hydrogen peroxide formed by this reaction must be removed in the living cells of Amphibacillus. The possibility was considered that some peroxidase might be induced in aerobic growing cells, because the bacterium lacks catalase. Several kinds of peroxidases have been purified and characterized from bacteria or mammalian sources (1, 4, 15, 16, 28, 34, 37). Peroxidase activities were tested as previously described (1, 4, 15, 16, 23, 28, 30, 34, 37), but no peroxidase activity except NADH:hydrogen peroxide reductase depending on free FAD or AhpC was detected in cell free extracts of Amphibacillus. These two peroxidase activities of purified NADH oxidase have been reported in previous papers (21, 23). Proposals for their physiological role in aerobic metabolism of Amphibacillus will be considered in Discussion.
Induction of NADH oxidase and AhpC by oxygen.
Immunoblot analysis revealed that cell extracts reacted with antibodies against NADH oxidase from A. xylanus (results not shown). The NADH oxidase-like protein was induced markedly under aerobic conditions but not under anaerobic conditions. Immunoblot analysis using antibodies against AhpC from A. xylanus also show that the AhpC-like protein was induced significantly under aerobic conditions, indicating that both proteins are actually involved in aerobic metabolism of Amphibacillus (see Discussion for further detail).
DISCUSSION
Although A. xylanus can grow well in aerobic media, no catalase or known peroxidase activities could be observed in the cells. In a previous study, we found peroxidase activity in the presence of free FAD and showed that 13 μM free FAD is actually contained in the cells of A. xylanus (25). The peroxidase activity results from reduction of peroxide by FADH2 formed from the flavin reductase activity of the enzyme. The peroxidase activity exhibited in the presence of FAD is, however, unlikely to be of much physiological significance, since FADH2 is required for the reduction of peroxide but this reaction is much slower than the competing reaction of FADH2 with oxygen to produce H2O2. In contrast, the peroxidase activity in the presence of AhpC is not affected by oxygen. A partial ORF homologous to the Salmonella AhpC structural gene was previously found upstream of the Amphibacillus NADH oxidase structural gene (5). In fact, AhpC exists in the cells of A. xylanus, and both AhpC and NADH oxidase are induced markedly under aerobic conditions. Thus, it seems clear that these proteins are involved in removing peroxides formed in aerobic metabolism.
High SOD activity was also observed in aerobically growing cells of A. xylanus. Hydrogen peroxide is formed by the SOD reaction, as well as by other oxidase reactions. The scavenging activity for hydrogen peroxide produced by such reactions should be high to permit cells to survive under aerobic conditions. In the presence of saturating concentrations of AhpC, the Vmax values for reduction of hydrogen peroxide and alkyl hydroperoxide are similar to the rate constant for the reduction of the enzyme-bound FAD by NADH in the NADH oxidase, suggesting that these values may be limited by the reduction rate of the flavoprotein component. The Km values for hydrogen peroxide and cumene hydroperoxide are too low to be determined by the analytical method employed. The yeast thiol-specific antioxidant (6) which is 39.3% homologous to Amphibacillus AhpC was renamed thioredoxin peroxidase because it reduced hydrogen peroxide and alkyl hydroperoxides with thioredoxin, thioredoxin reductase, and NADPH (7). These activities, however, are low in comparison to that of the NADH oxidase-AhpC system of Amphibacillus. The high turnover numbers and low Km values of the Amphibacillus enzyme system are unusual for known peroxide-reducing enzymes and provide a rationale for the vigorous aerobic growth of Amphibacillus, despite its lack of both catalase and conventional peroxidases.
In our early physiological study of A. xylanus, large amounts of hydrogen peroxide were presumed to be produced in aerobically growing cells, because the NADH oxidase must regenerate NAD from NADH formed in the aerobic pathway of A. xylanus, which lacks a conventional respiratory chain (see Fig. 4). Hydrogen peroxide must be formed as an intermediate but must be removed by the NADH oxidase-AhpC system as fast as it is formed. Based on a metabolic study of A. xylanus, we proposed the presence of anaerobic and aerobic pathways producing similar amounts of ATP (Fig. 4) (21). While NADH formed from NAD+ in the glycolytic pathway is reoxidized by the functioning of NAD-linked aldehyde dehydrogenase and NAD-linked alcohol dehydrogenase in anaerobic metabolism, NADH produced both from the glycolytic pathway and by pyruvate metabolism of the aerobic pathway (19) is oxidized to NAD during the reduction of oxygen to water by the NADH oxidase-AhpC system (Fig. 4), which provides metabolic balance in the aerobic pathway.
Previously, we found that the Salmonella alkyl hydroperoxide reductase system containing AhpC catalyzes the reduction of hydrogen peroxide with a high turnover number (21). However, its NADH oxidase activity is very low compared to that of Amphibacillus NADH oxidase, indicating that the Salmonella enzyme system mainly catalyzes the reduction of hydrogen peroxide. Because Salmonella has a respiratory chain, providing an effective oxidizing system for NADH, the Salmonella enzyme probably functions mainly in the reduction of peroxides rather than in the regeneration of NAD. Sporolactobacillus inulinus, lacking a respiratory system, catalase, and peroxidases, grows well under both anaerobic and aerobic conditions, as does A. xylanus. In the presence of AhpC, Sporolactobacillus NADH oxidase also shows high turnover numbers and low Km values for peroxide (24). Therefore, the NADH oxidase-AhpC system probably plays an important role not only in removing peroxides but also as an NAD-regenerating system in bacteria, which lack both a respiratory chain and conventional hydrogen peroxide-removing enzymes.
ACKNOWLEDGMENTS
We thank Kunio Matsui (Osaka International University for Women) and Sabu Kasai (Osaka City University) for 8-dimethylamino-riboflavin and Sandro Ghisla (University of Konstanz) for 6-hydroxyriboflavin.
This work was supported in part by a grant from the U.S. Public Health Service (GM-11106) to V.M.
REFERENCES
- 1.Alexander N M, Corcoran B J. The reversible dissociation of thyroid iodide peroxidase into apoenzyme and prosthetic group. J Biol Chem. 1962;237:243–248. [PubMed] [Google Scholar]
- 2.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 3.Bevington . Data reduction and error analysis for the physical sciences. New York, N.Y: McGraw Hill, Inc.; 1969. pp. 235–242. [Google Scholar]
- 4.Castelfranco P, Stumpf P K, Contopouleu R. Fat metabolism in higher plants. J Biol Chem. 1955;214:567–587. [PubMed] [Google Scholar]
- 5.Chae H Z, Robison K, Poole L B, Church G, Storz G, Rhee S G. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chae H Z, Kim I, Kim K, Rhee S G. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J Biol Chem. 1993;268:16815–16821. [PubMed] [Google Scholar]
- 7.Chae H Z, Chung S J, Rhee S G. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- 8.Cocco D, Rinaldi A, Savini I, Cooper J M, Bannister V. NADH oxidase from the extreme thermophile Thermus aquaticus YT-1, purification and characterization. Eur J Biochem. 1981;174:267–271. doi: 10.1111/j.1432-1033.1988.tb14093.x. [DOI] [PubMed] [Google Scholar]
- 9.Dixon M. The acceptor specificity of flavins and flavoproteins. Biochim Biophys Acta. 1971;226:259–268. doi: 10.1016/0005-2728(71)90093-4. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi M, Shimada M, Yamamoto Y, Hayashi T, Koga T, Kamio Y. Identification of two distinct NADH oxidase corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J Gen Microbiol. 1993;139:2343–2351. doi: 10.1099/00221287-139-10-2343. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson F S, Morgan R W, Christman M F, Ames B N. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]
- 12.Koike K, Kobayashi T, Ito S, Saitoh M. Purification and characterization of NADH oxidase from a strain of Leuconostoc mesenteroides. J Biochem. 1985;97:1279–1288. doi: 10.1093/oxfordjournals.jbchem.a135179. [DOI] [PubMed] [Google Scholar]
- 13.Koyama N, Niimura Y, Kozaki M. Bioenergetic properties of a facultatively anaerobic alkalophile. FEMS Microbiol Lett. 1988;49:123–126. [Google Scholar]
- 14.Matsui K, Juri N, Kubo Y, Kasai S. Formation of roseoflavin from guanine through riboflavin. J Biochem. 1979;86:167–175. [PubMed] [Google Scholar]
- 15.Morris D R, Hager L P. Chloroperoxidase. J Biol Chem. 1966;241:1763–1768. [PubMed] [Google Scholar]
- 16.Nakamura W, Hosoda S, Hayashi K. Purification and properties of rat liver glutathione peroxidase. Biochim Biophys Acta. 1974;358:251–261. [Google Scholar]
- 17.Niimura Y, Yanagida F, Uchimura T, Ohara N, Suzuki K, Kozaki M. A new facultative anaerobic xylan-using alkalophile lacking cytochrome, quinone, and catalase. Agric Biol Chem. 1987;51:2271–2275. [Google Scholar]
- 18.Niimura Y, Koh E, Yanagida F, Suzuki K, Komagata K, Kozaki M. Amphibacillus xylanus gen. nov., sp. nov., which lacks cytochrome, quinone, and catalase. Int J Syst Bacteriol. 1990;40:297–301. [Google Scholar]
- 19.Niimura Y, Koh E, Uchimura T, Ohara N, Kozaki M. Aerobic and anaerobic metabolism in a facultative anaerobe, Ep01, lacking cytochrome, quinone and catalase. FEMS Microbiol Lett. 1989;61:79–84. [Google Scholar]
- 20.Niimura Y, Ohnishi K, Yarita Y, Hidaka M, Masaki H, Uchimura T, Suzuki H, Kozaki M, Uozumi T. A flavoprotein functional as NADH oxidase from Amphibacillus xylanus Ep01: purification and characterization of the enzyme and structural analysis of its gene. J Bacteriol. 1993;175:7945–7950. doi: 10.1128/jb.175.24.7945-7950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niimura Y, Poole L B, Massey V. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkylhydroperoxide reductase 22-kDa protein component. J Biol Chem. 1995;270:25645–25650. doi: 10.1074/jbc.270.43.25645. [DOI] [PubMed] [Google Scholar]
- 22.Niimura Y, Massey V. Reaction mechanism of Amphibacillus xylanus NADH oxidase/alkyl hydroperoxide reductase flavoprotein. J Biol Chem. 1996;271:30459–30464. doi: 10.1074/jbc.271.48.30459. [DOI] [PubMed] [Google Scholar]
- 23.Niimura Y, Yokoyama K, Ohnishi K, Massey V. A flavoprotein functional as NADH oxidase from Amphibacillus xylanus scavenges hydrogen peroxide in the presence of free FAD. Biosci Biotechnol Biochem. 1994;58:2310–2311. [Google Scholar]
- 24.Nishiyama Y, Massey V, Anzai Y, Watanabe T, Miyaji T, Uchimura T, Kozaki M, Suzuki S, Niimura Y. Purification and characterization of Sporolactobacillus inulinus NADH oxidase and its physiological role in aerobic metabolism of the bacterium. J Ferment Bioeng. 1997;84:22–27. [Google Scholar]
- 25.Ohnishi K, Niimura Y, Yokoyama K, Hidaka M, Masaki H, Uchimura T, Suzuki H, Uozumi T, Kozaki M, Komagata K, Nishino T. Purification and analysis of a flavoprotein functional as NADH oxidase from Amphibacillus xylanus overexpressed in Escherichia coli. J Biol Chem. 1994;269:31418–31423. [PubMed] [Google Scholar]
- 26.Ohnishi K, Niimura Y, Hidaka M, Masaki H, Suzuki H, Uozumi T, Nishino T. Role of cysteine 337 and cysteine 340 in flavoprotein that functions as NADH oxidase from Amphibacillus xylanus studied by site-directed mutagenesis. J Biol Chem. 1995;270:5812–5817. doi: 10.1074/jbc.270.11.5812. [DOI] [PubMed] [Google Scholar]
- 27.Park H J, Reiser C O A, Kondoruweit S, Erdomann H, Schmidt R, Sprinzl M. Purification and characterization of a NADH oxidase from the thermophile, Thermus thermophilus HB8. Eur J Biochem. 1992;205:881–885. doi: 10.1111/j.1432-1033.1992.tb16853.x. [DOI] [PubMed] [Google Scholar]
- 28.Parsonage D, Miller H, Ross R P, Claiborne A. Purification and analysis of streptococcal NADH peroxidase expressed in Escherichia coli. J Biol Chem. 1993;268:3161–3167. [PubMed] [Google Scholar]
- 29.Poole L B, Ellis H R. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 30.Putter J, Becker R. Peroxidases. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 3th ed. Vol. 3. Weinheim, Germany: Verlag Chemie; 1983. pp. 286–302. [Google Scholar]
- 31.Reinards R, Kubicki J, Ohlenbusch H. Purification and characterization of NADH oxidase from membranes of Acholeplasma laidlawii, a copper-containing iron-sulfur flavoprotein. Eur J Biochem. 1981;120:329–337. doi: 10.1111/j.1432-1033.1981.tb05708.x. [DOI] [PubMed] [Google Scholar]
- 32.Saeki Y, Nozaki M, Matsumoto K. Purification and properties of NADH oxidase from Bacillus megaterium. J Biochem. 1985;98:1433–1440. doi: 10.1093/oxfordjournals.jbchem.a135411. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt H L, Stocklein W, Danzer J, Kirch P, Limbach B. Isolation and properties of an H2O-forming NADH oxidase from Streptococcus faecalis. Eur J Biochem. 1986;156:149–155. doi: 10.1111/j.1432-1033.1986.tb09560.x. [DOI] [PubMed] [Google Scholar]
- 34.Shigeoka S, Nakano Y, Kitaoka S. Metabolism of hydrogen peroxide in Euglena gracilis z by l-ascorbic acid peroxidase. Biochem J. 1980;186:377–380. doi: 10.1042/bj1860377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer R, Fisher J, Walsh C. Preparation, characterization, and chemical properties of the flavin coenzyme analogues 5-deazariboflavin, 5-deazariboflavin 5′-phosphate, and 5-deazariboflavin 5′-diphosphate, 5′→5′-adenosine ester. Biochemistry. 1976;15:1043–1053. doi: 10.1021/bi00650a015. [DOI] [PubMed] [Google Scholar]
- 36.Tartaglia L A, Storz G, Brodsky M H, Lai A, Ames B N. Alkyl hydroperoxide reductase from Salmonella typhimurium. J Biol Chem. 1990;265:10535–10540. [PubMed] [Google Scholar]
- 37.Yonetani T. Cytochrome c peroxidase. Adv Enzymol. 1970;33:309–335. doi: 10.1002/9780470122785.ch6. [DOI] [PubMed] [Google Scholar]