Abstract
Klebsiella pneumoniae (Kp) has emerged as a global life-threatening pathogen owing to its multidrug resistance and hypervirulence phenotype. Several fatal outbreaks of carbapenem-resistant hypervirulent Kp have been reported recently. Hypermucoviscosity (HMV) is a phenotype commonly associated with hypervirulence of Kp, which is usually regulated by rmpA or rmpA2 (regulators of the mucoid phenotype). Here, we found that temperature was important in the HMV phenotype of Kp, and the impact of temperature on HMV was not uniform among strains. We investigated the HMV phenotype at 37 °C and room temperature (20–25 °C) in 170 clinically isolated hypermucoviscous Kp strains in Japan and analysed the association between the HMV phenotype, virulence genes and antimicrobial resistance (AMR) genes. String length distribution at different temperatures was correlated with the genomic population of Kp. The strains carrying rmpA/rmpA2 frequently showed the HMV phenotype at 37 °C, while the strains negative for these genes tended to show the HMV phenotype at room temperature. Hypervirulent Kp clusters carrying rmpA/rmpA2 without extended-spectrum beta-lactamases (ESBL)/carbapenemases produced higher string lengths at 37 °C than at room temperature, and were mostly isolated from the respiratory tract. Other HMV strains showed distinct characteristics of not carrying rmpA/rmpA2 but were positive for ESBL/carbapenemases, with a higher string length at room temperature than at 37 °C, and were frequently isolated from bloodstream infections. In total, 21 (13.5 %) HMV isolates carried ESBL and carbapenemases, among which five isolates were carbapenem-resistant hypervirulent Kp with a pLVPK-like plasmid (an epidemic virulence plasmid) and a pKPI-6-like plasmid (an epidemic bla IMP-6-bearing plasmid in Japan), suggesting the convergence of worldwide hypervirulence and epidemic AMR in Japan.
Keywords: Klebsiella pneumoniae, hypermucoviscous, hypervirulent, multidrug resistance, string test
Data Summary
The authors confirm that all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
Hypermucoviscosity is considered a phenotype usually associated with hypervirulence of the pathogen Klebsiella pneumoniae ; however, the effect of temperature on hypermucoviscosity has not been studied. We investigated the hypermucoviscosity phenotype at both 37 °C and room temperature (20–25 °C) and measured the length of the strings. We discovered temperature-dependent differences in string length patterns, with the hypervirulent clones showing higher hypermucoviscosity at 37 °C, whereas the other clones, including the isolates with antimicrobial resistance, showed higher hypermucoviscosity at room temperature. These findings will facilitate the discovery of unknown factor(s) responsible for hypermucoviscosity at room temperature and may explain the different virulence tactics of K. pneumoniae infection.
Introduction
Klebsiella pneumoniae (Kp) is currently regarded as a significant threat to global health because of the emergence of hypervirulent (hv) clones causing severe community-acquired infections and multidrug-resistant (MDR) clones related to hospital outbreaks [1]. Kp infections can be community- or hospital-acquired, leading to serious diseases such as pneumonia, primary pyogenic liver abscess or distinctive invasive syndrome. The virulence and resistance determinants are distributed in distinct subpopulations of Kp [1]; however, the emergence of Kp with both carbapenem-resistant and hv phenotypes was recently reported in China [2], South and South-East Asian countries [3], and various other regions globally [4]. The geographical focus of this convergence is likely to occur in Asia because of the common existence of both hv and MDR clones. As Japan is a part of Asia, hv/MDR Kp might have been disseminated into the main continent and evolved. In West Japan, carbapenemase-producing Kp has emerged in the past decade; these strains carry extended-spectrum β-lactamases (ESBL) (bla CTX-M-2) and carbapenemase (bla IMP-6) and are resistant to all β-lactam antibiotics, except imipenem [5]. Nevertheless, no studies have provided a complete molecular epidemiological investigation of the Kp population in Japan so far, leaving a gap in knowledge about the specific regional population and diversity.
A variety of Kp virulence factors have been identified, including capsular polysaccharide, colibactin, ferric ion uptake, salmochelin (iro) and aerobactin (iuc) [6]. hvKp strains display an increased ability to acquire iron than classical Kp (cKp) strains due to the synthesis of iron-acquisition factors, such as aerobactin or salmochelin, that ‘steal’ the iron from the host [7]. Another factor reported to contribute to the virulence of Kp is the hypermucoviscosity (HMV) phenotype [6–8]. HMV has traditionally been attributed to overexpression of the capsule, which assists these bacteria in colonizing the mucosa and protects them from phagocytosis and human defensin-mediated bactericidal activity [6]. The HMV phenotype is sometimes associated with the hypervirulence of Kp [8], although not all Kp with the HMV phenotype are hvKp [9]. The magA gene (mucoviscosity-associated gene A) was first identified to code for a factor responsible for HMV [10], but was later found to be specific to the capsular serotype K1 [11]. Nevertheless, strains other than the K1 serotype (magA-negative) have also been reported to have HMV and hv phenotypes [12], suggesting that another factor may be associated with the HMV phenotype besides magA. Recent studies have suggested that unknown factors other than capsule production also play a role in the HMV phenotype. The KpnO porin, an outer membrane protein, was found to contribute to capsular polysaccharide production in the hvKp NTUH-K2044 [13]. However, it remains unclear whether there is any difference in the distribution of this factor between hvKp and cKp. The HMV phenotype is reportedly enhanced by expression of the plasmid-borne loci rmpADC [14] (where rmpA is the regulator of the mucoid phenotype A) [7, 15–17] or rmpA2 [7, 9, 17, 18], which are considered some of the major factors involved in the HMV phenotype [6, 7, 19]. Nevertheless, the relationship between the rmpA genes and the HMV phenotype remains unclear, as some HMV-positive isolates do not harbour these genes [9, 20]. In this context, the regulatory mechanism of HMV in Kp is not fully understood.
From the viewpoint of clinical practice, an effective diagnostic tool to predict hvKp and its related characteristics could provide valuable early warnings about the hypervirulence and potential metastatic infections; therefore, comprehensive knowledge of the genomic population, virulence determinants and resistance determinants may significantly contribute to the infection control strategy. Furthermore, elaborating our understanding of the regulatory mechanisms of HMV and virulence of Kp may advance the development and implementation of new chemotherapies targeting these factors, which would improve the prognosis of serious infections. From our preliminary data, we found that temperature was a key factor affecting the HMV of Kp. Here, we present a genomic epidemiological study of hypermucoviscous Kp that examines the genomic characteristics (sequence types, capsular types, rmpA/rmpA2, virulence genes and resistance genes) of hypermucoviscous Kp in correlation with the temperature-dependent HMV phenotype.
Methods
Bacterial isolates
A total of 236 Kp isolates, obtained from patient specimens, such as blood, respiratory tract (RT), urine, bile, pus or puncture fluid from different hospitals in the Kansai (southern-centre area of Japan) and Chugoku (middle-west area of Japan) regions between 2006 and 2017, were used for HMV evaluation by string tests. Strains were collected after completion of routine microbiological diagnostics. This study was approved by the ethical committees of the National Institute of Infectious Diseases Committee of Ethics (approval number 972).
Modified string test
The isolates were cultured on agar containing 5 % sheep blood (Becton, Dickinson) and incubated at 37 °C and room temperature (20–25 °C). Each blood agar plate was divided into four parts, and a full loop of each bacterial strain was streaked evenly in each quarter with a 2 mm inoculating loop. After 24, 48 and 72 h of incubation, a string test was performed with a cotton swab. A 5-mm-diameter cotton swab was used to collect all the colonies of a strain within the area and was stretched upward. The string test was deemed positive when a viscous string of ≥5 mm was generated. The string test at each time point was repeated five times, and the mean string length was recorded. The maximum string length at three time-points of each strain was used for data analysis. The strains showing negative string tests at all growth conditions were excluded, and 170 Kp isolates with a positive string test in at least one condition were included in subsequent investigations (Fig. 1).
Fig. 1.
Flow chart of sample selection and data analysis processes.
Based on the string length at different temperatures, the isolates were separated into three classes: string length at room temperature was lower than that at 37 °C (Class I), string lengths at room temperature were equal to those at 37 °C (Class II) and string length at room temperature was higher than that at 37 °C (Class III). Class I was further divided into subgroups based on the pattern of string length: group A (positive string test at 37 °C but negative string test at room temperature) and group B (positive string test at both temperatures but higher string length at 37 °C). Similarly, Class III was subdivided into group D (positive string test at both temperatures but higher string length at room temperature) and group E (positive string test at room temperature but negative string test at 37 °C). Class II included only one group, namely group C (Fig. 1).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the broth microdilution method. The following drugs were tested: piperacillin (PIPC), cefazolin (CAZ), cefepime (CFPM), imipenem (IPM), meropenem (MEPM), doripenem (DRPM), aztreonam (AZT), ampicillin/sulbactam (ABPC/SBT), gentamycin (GM), tobramycin (TOB), amikacin (AMK), minocycline (MINO), levofloxacin (LVFX), ciprofloxacin (CPFX), sulfamethoxazole (ST), fosfomycin (FOM) and florfenicol (CP). The results of the minimum inhibitory concentrations (MICs) were interpreted based on the Clinical and Laboratory Standards Institute (CLSI) M100-S28 guidelines [21].
DNA extraction, whole-genome sequencing and genomic analysis
All Kp isolates were cultured overnight in Luria-Bertani (LB) broth at 37 °C, followed by total DNA extraction using the phenol–chloroform method. Libraries were constructed using Nextera DNA kits (Illumina,) following the manufacturer’s instructions, and whole-genome sequencing (WGS) was performed using Illumina MiSeq, generating 150 bp paired-end reads. The samples in this study achieved a sequencing depth of coverage ranging from 30× to 110×. De novo assemblies were generated using Shovill v1.0.9 and subsequently annotated using the PATRIC RAST-tk-enabled Genome Annotation Service. The whole-genome sequences of our isolates were analysed using Kleborate v2.1.0 [22] with the Kaptive option [23] for their sequence types, K (capsule) serotype prediction, virulence loci and antimicrobial resistance (AMR) genes. The multi-locus sequence type (MLST) alleles (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) and sequence type (ST) profiles that had not been previously described were submitted to the curator of the official Kp BIGSdb-Pasteur database (http://bigsdb.pasteur.fr/klebsiella/) to assign new designations. The virulence and resistance scores were automatically calculated using Kleborate based on the types of virulence and AMR genes carried by each isolate (https://github.com/katholt/Kleborate/wiki/Scores-and-counts). From the results of Kleborate, 14 isolates were identified as K. quasipneumoniae or K. variicola and hence were excluded from this study. The remaining 156 isolates were confirmed as K. pneumoniae and were used for further analysis (Fig. 1).
Whole-genome SNP analysis was performed with CSIPhylogeny 1.4 [24] from the Center for Genomic Epidemiology with default settings [minimum depth at SNP positions: 10, relative depth at SNP positions: 10, minimum distance between SNPs (prune): 10, minimum SNP quality: 30, minimum read mapping quality: 25, and minimum Z-score: 1.96]. A phylogenetic tree was created and was annotated with the Interactive Tree of Life (iTOL) [25]. Related nodes within the phylogenetic tree were clustered using RAMI [26]. With the threshold of 0.1, RAMI produced 48 clusters, among which eight predominant clusters were found, and their key features were compared.
Isolates carrying ESBL and/or carbapenemase genes were selected for long-read sequencing. Total genomic DNA was extracted using the Genomic DNA buffer set (Qiagen) and Qiagen Genomic-tip 20 G−1, according to the manufacturer’s instructions. The DNA library was prepared using the SQK-RBK004 Rapid Barcoding Kit, loaded onto a FLO-MIN106D R9.4.1 flow cell, and sequenced with MinION (Oxford Nanopore Technologies) for 72 h. Hybrid assembly of Illumina short reads and MinION long reads was performed using the hybrid assembler Unicycler v0.4.8 [27]. The complete sequences of virulence plasmids were selected and compared with the available virulence plasmids pLVPK (accession number AY378100), a well-characterized virulence plasmid in Kp, using blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi), Blast Ring Image Generator (BRIG) v0.95 [28]. Additionally, the virulence plasmids from our ST36 Kp were compared with WCHKP13F2 (MF943217), an ST36Kp-derived virulence plasmid reported from China. The AMR plasmids were also extracted, and complete plasmid comparisons were performed. The bla IMP-6-carrying plasmids were compared with pKPI-6 (AB616660), a bla IMP-6-carrying epidemic plasmid in Japan. The plasmids encoding bla KPC-2 were compared with some publicly available bla KPC-2-harbouring plasmids such as pKPC-HvKP4 (MF437312), p675920-1 (MF133495), pHN7A8 (JN232517) and pKPC-LK30 (KC405622). The bla CTX-M-15-carrying plasmids were compared with two available plasmids encoding bla CTX-M-15, pAMA1416 (MG462728) and pCTX15_DHQP14000954 (CP016925).
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics v28.0.1.0. Comparisons between either Class I and Class III isolates, group A and group B isolates, or RT and blood isolates were performed using the chi-square test and Fisher’s exact test (for key features) and Mann–Whitney test for mean string length. Mean string lengths at 37 °C and room temperature between the rmpA/rmpA2 positive and negative groups were analysed using the Mann–Whitney test and Wilcoxon signed-rank test.
Accession number(s)
The whole-genome sequence FASTQ files were deposited in DDBJ under BioProject nos. PRJDB12075, DRX309353–DRX309536 and DRX317549–DRX317564.
Results
Distribution of multi-locus STs and predicted capsular serotypes (K) within the HMV Kp population
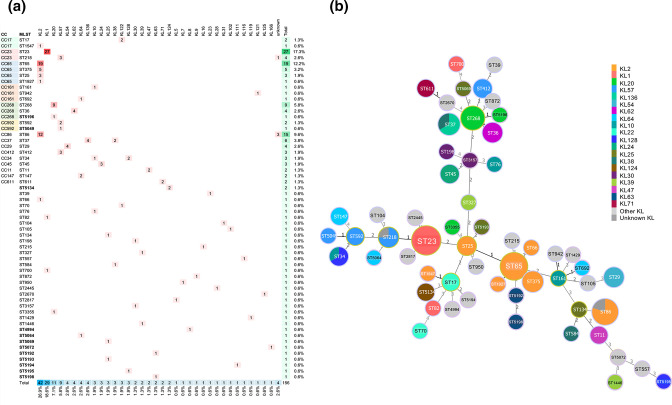
Genotypic analysis using WGS on 156 HMV Kp strains showed that HMV Kp was highly diverse, comprising 58 STs and belonging to 35 known K-loci and four unknown K-loci (Fig. 2). Twelve (20.69%) of the 58 STs had not been previously identified (ST4994, ST5049, ST5064, ST5069, ST5072, ST5134, ST5192, ST5193, ST5194, ST5195, ST5196 and ST5198). The most prevalent STs were ST23 (n=27, 17.3%), ST65 (n=19, 12.2%), ST86 (n=15, 9.6%), ST268 (n=9, 5.8%), ST37 (n=6, 3.8%) and ST375 (n=5, 3.2%). The most prevalent K-loci, which accounted for >50 % of the isolates, were KL2 (n=42, 26.9%), KL1 (n=29, 18.6%), KL20 (n=11, 7.1%) and KL57 (n=9, 5.8%). While ST23, ST65 and ST268 were each associated with a single K-locus (KL1, KL2 and KL20, respectively), some other STs were associated with multiple K-loci; for example, ST37 included four KL136 and two KL38 (Fig. 2). Furthermore, 67.2 % (39/58) and 42.9 % (15/35) of the STs and known K-loci, respectively, were represented by a single isolate.
Fig. 2.
Sequence type (ST), clonal complex (CC) and capsular locus (KL) diversity among 156 hypermucoviscous K. pneumoniae isolates. (a) The number of genomes representing each KL by ST as well as the prevalence of each ST and KL are demonstrated. STs in bold indicate the newly identified STs in our study. Four strains having KL with low or no confidence as assessed by Kleborate and Kaptive are shown as ‘unknown KL’. Among these unidentified KL isolates, three belong to ST86 and only had 14/18 expected genes for KL2, and one belongs to ST218 and only carried 12/18 expected genes for KL57. (b) Minimum spanning tree showing the correlation between ST and KL present in our study.
AMR determinants and phenotypes
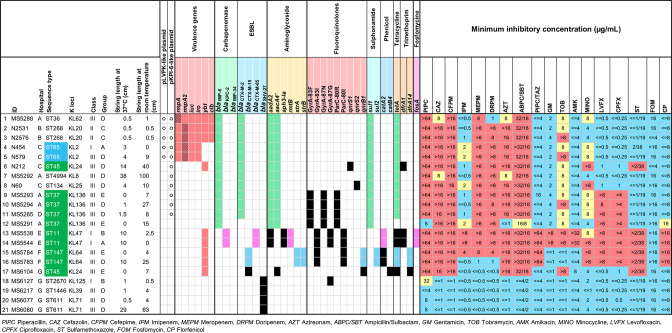
In this collection, we detected 21 isolates (13.5%) that carried ESBL and/or carbapenemase genes. Among them, 13 isolates carried both ESBL and carbapenemase genes, seven isolates carried only ESBL, and two isolates carried only carbapenemases. The ESBL genes included bla CTX-M-2 (n=11, 52.4%), bla SHV-27 (n=4, 19.0%), bla CTX-M-15 (n=3, 14.3%) and bla CTX-M-65 (n=2, 9.5%), and the carbapenemase genes included bla IMP-6 (n=12, 57.1%) and bla KPC-2 (n=2, 9.5%) (Fig. 3).
Fig. 3.
Key features of AMR-HMV K. pneumoniae isolates. Characteristics of 21 HMV-AMR isolates that carried ESBL/carbapenemase genes are demonstrated. The isolates from hypervirulence-background sequence types (STs) and AMR-background STs are highlighted in blue and green in the sequence type column, respectively. The presence of pLVPK-like and pKPI-6-like plasmids is marked with circles. The presence of intact or truncated rmpA and rmpA2 genes is displayed as plain red colour or striped red colour, respectively. For the AMR genes, the same colours, except black, indicate that genes belonged to the same plasmid: green, pKPI-6-like plasmid; pink, bla KPC-2-carrying plasmid; blue, bla CTX-M-15-carrying plasmid; black indicates that genes belonged to the other plasmids or chromosome. Minimum inhibitory concentrations (µg ml–1) are interpreted as susceptible (blue), intermediate (yellow) or resistant (red).
Half of the ESBL- and/or carbapenemase gene-carrying isolates (n=11/21) carried both bla CTX-M-2 and bla IMP-6, along with aacA4, aadA2, sul1 and tetA, all of which showed resistance to cephalosporins, meropenem and doripenem, as well as tobramycin and minocycline. Furthermore, four of these isolates (MS5293, MS5294, MS5265 and MS5291) also carried mutations associated with fluoroquinolone resistance (GyrA-83F, GyrA-87N and ParC-80R), resulting in resistance to levofloxacin and ciprofloxacin.
Two isolates (MS5538 and MS5544) possessed bla KPC-2, bla CTX-M-65 and bla TEM-1B, together with aadA2, aph3-Ia, rmtB (aminoglycoside resistance), sul2 (sulfonamide resistance) and catA2 (phenicol resistance). These also displayed extended resistance to all tested cephalosporins, carbapenems, aminoglycosides, sulfamethoxazole and florfenicol. Additionally, these two isolates carried chromosomal mutations (GyrA-83I, GyrA-87G and ParC-80I) consistent with the observed fluoroquinolone resistance phenotype.
Genotypic convergence of AMR and HMV
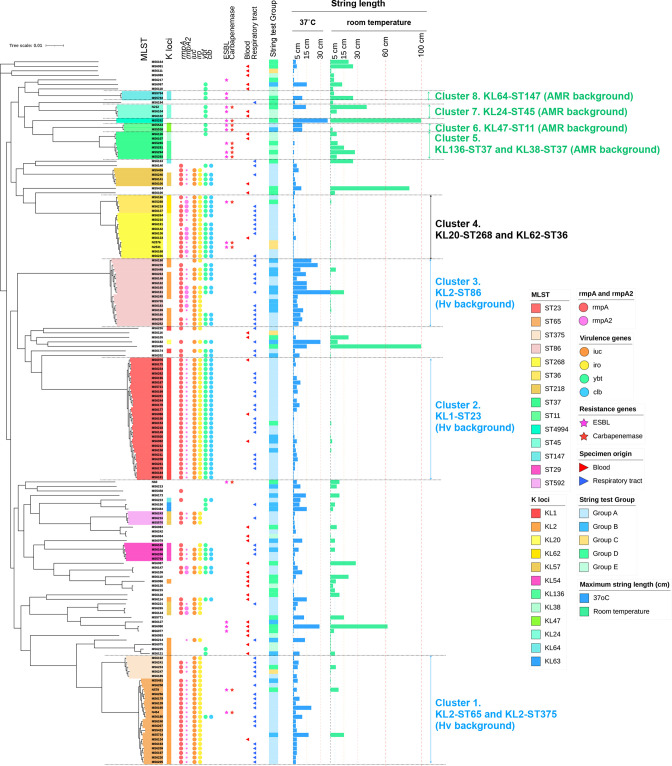
Phylogenetic analysis (Fig. 4) revealed dominant clusters corresponding to the dominant STs described above and represented well-known hvKp and cKp clones: hv-KL2-ST65 and -KL2-ST375 (cluster 1), -KL1-ST23 (cluster 2) and -KL2-ST86 (cluster 3) possessed rmpA/rmpA2 and other virulence determinants (ybt, clb, iuc and iro), but only a few carried ESBL/carbapenemases. Additionally, all KL2-ST65 and KL1-ST23 isolates carried an intact rmpA and a truncated rmpA2, while the other clusters did not show a consensus pattern. In contrast, the other clusters, including KL136-ST37 and KL38-ST3, KL47-ST11, KL24-ST45, and KL64-ST147 (clusters 5, 6, 7 and 8, respectively), mostly lacked rmpA/rmpA2 and all other acquired virulence loci except ybt, but had a high prevalence of ESBL and/or carbapenemases (10/13 strains, 76.9%). Another cluster comprising KL20-ST268 and KL62-ST36 (cluster 4) displayed characteristics similar to those of cluster 2 (KL1-ST23), with a high prevalence of rmpA/rmpA2 and other virulence determinants. These findings were generally consistent with the traditional view of Kp, for which acquired virulence and acquired resistance genes are usually found in distinct subsets of the population. However, we also identified several so-called ‘convergent’ isolates harbouring both acquired virulence and acquired AMR genes.
Fig. 4.
Phylogenetic analysis of 156 HMV K. pneumoniae isolates. Isolates are annotated with the datasets of sequence types (ST), capsular locus (KL), virulence genes, resistance genes, specimen origins, string test group, and maximal string length at 37 °C and room temperature, from left to right. Different STs are highlighted in different colours, and the most prominent clusters are marked as follows: cluster 1, KL2-ST65 and KL2-ST375; cluster 2, KL1-ST23; cluster 3, KL2-ST86; cluster 4, KL20-ST268 and KL62-ST36; cluster 5, KL136-ST37 and KL38-ST37; cluster 6, KL47-ST11; cluster 7, KL24-ST45; and cluster 8, KL64-ST147. For the rmpA and rmpA2 genes, the large circle indicates an intact gene, while the small circle indicates a truncated gene.
The 21 AMR-HMV isolates belonged to 14 different STs and 13 corresponding K-loci (Fig. 3). These STs included AMR-background STs [4] such as ST37 (n=4, 19.0%), ST11 (n=2, 9.5%), ST45 (n=2, 9.5%) and ST147 (n=2, 9.5%) (coloured in green), the hypervirulent-background STs [1, 4] such as ST65 (n=2, 9.5%) (coloured in blue), and some other miscellaneous STs such as ST268, ST36, ST134 and ST611.
Among 21 AMR-HMV isolates, five were positive for the rmpA (N2531, N2576, N454 and N579) or rmpA2 (MS5288) genes and some other virulence genes (MS5288, N2531 and N2576: yersiniabactin, colibactin, aerobactin and salmochelin; N454 and N579: aerobactin and salmochelin) (Fig. 3). The complete plasmid sequences of these five strains were obtained and used for plasmid comparison with BRIG. The results demonstrated that these isolates carried a plasmid homologous to the virulence plasmid pLVPK with ≥82 % coverage and ≥99.52 % sequence identity (Fig. 5a, Table S1, available in the online version of this article). The remaining 16 AMR-HMV isolates were negative for both rmpA and rmpA2 genes (Fig. 3).
Fig. 5.
Plasmid comparisons of (a) rmpA/rmpA2-carrying plasmids, (b) bla IMP-6-carrying plasmids, (c) bla KPC-2 , bla CTX-M-65 and bla TEM-1B-carrying plasmids, and (d) bla CTX-M-15-carrying plasmids. (a) Structural comparison of the plasmid pLVPK, WCHKP13F2-pVir, and the rmpA/rmpA2-harbouring plasmids from MS5288, N2531, N2576, N454 and N579 strains using complete plasmid sequences. pLVPK was used as the reference and is shown as the innermost ring. The outermost circle displays the locations of rmpA, rmpA2 and other virulence-coding genes (iro, iuc and iut) in pLVPK. (b) Structural comparison of the plasmid pKPI-6 and the bla IMP-6-carrying plasmids from MS5288, N2531, N2576, N454, N579, MS5292, N60, MS5293, MS5294, MS5265 and MS5291 strains using complete plasmid sequences. pKPI-6 was used as the reference and is shown as the innermost ring. The outermost circle displays the locations of AMR genes (bla IMP-6 integron, bla CTX-M-2, tetA and tetR) in pKPI-6. (c) Comparison of bla KPC-2 , bla CTX-M-65 and bla TEM-1B-carrying complete plasmids from MS5538 with four publicly available plasmids. The plasmid from MS5538 was used as the reference and is shown as the innermost ring. (d) Comparison of bla CTX-M-15-carrying complete plasmids from MS5783 and MS5784 strains with two publicly available plasmids. The plasmid from MS5783 was used as the reference and is shown as the innermost ring.
The complete plasmid nucleotide sequence comparison by BRIG confirmed that 11 isolates possessed pKPI-6-like plasmids with ≥96 % coverage and ≥99.81 % sequence identity to pKPI-6, which possessed a bla IMP-6-carrying integron and bla CTX-M-2 (Fig. 5b, Table S1). The plasmid pKPI-6 is an IncN plasmid that has caused an epidemic in West Japan due to its dissemination within the family Enterobacteriaceae [29].
Strikingly, 5/11 isolates carrying pKPI-6-like plasmids also carried the pLVPK-like virulence plasmid (Fig. 3), suggesting the convergence of hypervirulence and antimicrobial resistance in these clones. Among these, one ST36 isolate (MS5288) carried a plasmid with very high homology to the virulence plasmid from WCHKP13F2, an ST36 hypervirulent-carbapenem-resistant Kp reported from China [30] (100 % coverage, 99.96 % identity) (Fig. 5a). Strain WCHKP13F2 was reported to have an intact rmpA and truncated rmpA2, whereas our isolate MS5288 carried an intact rmpA2 and a truncated rmpA due to the insertion of a stop codon. In terms of AMR elements, WCHKP13F2 carried a bla KPC-2-harbouring plasmid, whereas MS5288 carried a plasmid co-expressing bla CTX-M-2 and bla IMP-6.
Two isolates carried plasmids with the coexistence of bla KPC-2, bla CTX-M-65, and bla TEM-1B (MS5538 and MS5544). The plasmid derived from MS5538 showed homology with the publicly available pKPC-HvKP4 (96 % coverage, 99.94 % identity) and p675920-1 (96 % coverage, 99.97 % identity) (Fig. 5c). Plasmid p675920-1 was reported to be a hybrid of pHN7A8 from Escherichia coli (carrying bla CTX-M-65 and bla TEM-1B) and pKPC-KL30 (carrying bla KPC-2) [31].
Two isolates (MS5783 and MS5784) carried bla CTX-M-15 and bla TEM-1B on the same plasmid, with 100 % homology between the two isolates (Fig. 5d). The MDR region, which harbours bla CTX-M-15 (β-lactam resistance), bla TEM-1B (β-lactam resistance), sul2 (sulphonamide resistance), aph(6)-Id (aminoglycoside resistance), aph(3’)-Ib (aminoglycoside resistance), qnrB1 (fluoroquinolone resistance), tetA (tetracycline resistance) and dfrA14 (trimethoprim resistance), was partially homologous to that of pAMA1416 (44 % coverage, 100 % identity) and pCTX15_DHQP14000954 (44 % coverage, 99.99 % identity) (Fig. 5d).
String level and temperatures
We re-ordered the isolates according to string length at 37 °C or room temperature and combined this with the other key features, such as specimen origins, virulence factors, ESBL and carbapenemase genes, ST and K-loci (Fig. 6).
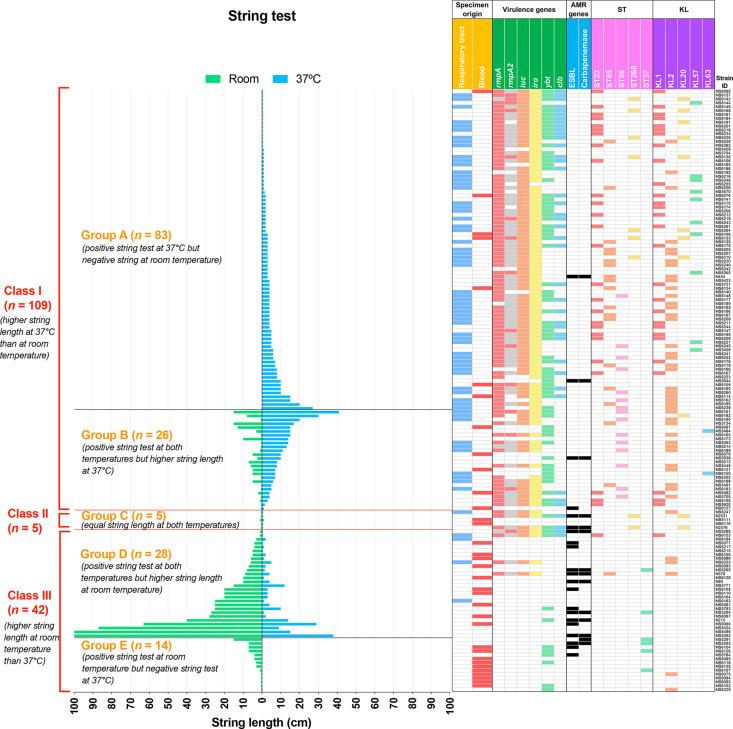
Fig. 6.
Correlation among temperature, string length and other characteristics. String lengths of each isolate at room temperature (green bars) and 37 °C (blue bars) are presented and separated into three classes: Class I, higher string length at 37 °C than at room temperature; Class II, equal string length at both 37 °C and room temperature; and Class III, higher string length at room temperature than at 37 °C. Temperature-dependent string patterns within Class I and Class III are further divided into subgroups: group A, positive string test at 37 °C but negative string test at room temperature; group B, positive string test at both temperatures but higher string length at 37 °C; group D, positive string test at both temperatures but higher string length at room temperature; and group E, positive string test at room temperature but negative string test at 37 °C. Class II comprises only one group, namely group C. The corresponding characteristics of each isolate, including specimen origin, virulence genes, AMR genes, ST and KL, are displayed in the right panel. For the rmpA and rmpA2 columns, red colour indicates an intact gene, and grey colour indicates a truncated gene.
According to string test categorization (see Methods), among 156 string test-positive isolates, 69.9 % (n=109) belonged to Class I, 3.2 % (n=5) belonged to Class II and 26.9 % (n=42) belonged to Class III. More specifically, 53.2 % (n=83) of isolates showed positive string tests only at 37 °C (negative string test at room temperature) (group A), 16.7 % (n=26) of isolates showed positive string tests at both temperatures but higher string length at 37 °C than at room temperature (group B), 3.2 % (n=5) of isolates produced equal string lengths at both temperatures (group C), 17.9 % (n=28) of isolates showed positive string tests at both temperatures but higher string length at room temperature than at 37 °C (group D), and 9.0 % (n=14) of isolates showed positive string tests only at room temperature (negative string tests at 37 °C) (group E).
The mean string length of Class I at 37 °C was significantly higher than that of Class III (5.67±0.63 vs 4.04±1.20 cm, P<0.001), while the mean string length of Class III at room temperature was significantly higher than that of Class I (17.32±3.91 vs 0.10±0.27 cm, P<0.001, Table 1). Within Class I, group B produced a significantly higher mean string length at 37 °C than group A (11.21±1.74 vs 3.93±0.48, P<0.001, Table 2).
Table 1.
Comparison of key features between K . pneumoniae Class I and Class III
|
Total (n=151) |
Class I (n=109) |
Class III (n=42) |
OR (95 % CI) |
p-value |
||
|---|---|---|---|---|---|---|
|
Sequence type | ||||||
|
ST23 |
27 (17.9 %) |
26 (23.9 %) |
1 (2.4 %) |
12.843 (1.683–97.990) |
0.002 |
** |
|
ST65 |
19 (12.6 %) |
18 (16.5 %) |
1 (2.4 %) |
8.110 (1.047–62.817) |
0.019 |
* |
|
ST86 |
14 (9.3 %) |
14 (12.8 %) |
0 (0.0 %) |
n/a |
0.011 |
* |
|
ST268 |
6 (4.0 %) |
6 (5.5 %) |
0 (0.0 %) |
n/a |
0.187 |
|
|
ST37 |
6 (4.0 %) |
0 (0.0 %) |
6 (14.3 %) |
n/a |
3.52×10−4 |
*** |
|
K loci | ||||||
|
KL1 |
29 (19.2 %) |
28 (25.7 %) |
1 (2.4 %) |
14.173 (1.862–107.883) |
0.001122 |
*** |
|
KL2 |
39 (25.8 %) |
35 (32.1 %) |
4 (9.5 %) |
4.493 (1.487–13.579) |
0.004 |
** |
|
KL20 |
9 (6.0 %) |
9 (8.3 %) |
0 (0.0 %) |
n/a |
0.063 |
|
|
KL57 |
10 (6.6 %) |
10 (9.2 %) |
0 (0.0 %) |
n/a |
0.062 |
|
|
Regulator of mucoid phenotype | ||||||
|
intact rmpA (+) |
95 (62.9 %) |
92 (84.4 %) |
3 (7.1 %) |
70.353 (19.497–253.856) |
1.285×10−18 |
*** |
|
intact rmpA2 (+) |
15 (9.9 %) |
14 (12.8 %) |
1 (2.4 %) |
6.042 (0.769–47.480) |
0.069 |
|
|
intact rmpA and/or rmpA2 (+) |
97 (64.2 %) |
93 (85.3 %) |
4 (9.5 %) |
55.219 (17.331–175.936) |
3.104×10−18 |
*** |
|
Virulence determinants | ||||||
|
Yersiniabactin (ybt) |
77 (51.0 %) |
67 (61.5 %) |
10 (23.8 %) |
5.105 (2.275–11.453) |
0.000034 |
*** |
|
Colibactin (clb) |
39 (25.8 %) |
37 (33.9 %) |
2 (4.8 %) |
10.278 (2.353–44.899) |
0.000241 |
*** |
|
Aerobactin (iuc) |
95 (62.9 %) |
91 (83.5 %) |
4 (9.5 %) |
48.028 (15.243–151.324) |
3.425×10−17 |
*** |
|
Salmochelin (iro) |
101 (66.9 %) |
97 (89.0 %) |
4 (9.5 %) |
76.792 (23.312–252.959) |
1.437×10−20 |
*** |
|
Virulence score 0 |
37 (24.5 %) |
7 (6.4 %) |
30 (71.4 %) |
0.027 (0.010–0.076) |
8.645×10−17 |
*** |
|
Virulence score 1 |
19 (12.6 %) |
11 (10.1 %) |
8 (19.0 %) |
0.477 (0.177–1.285) |
0.137 |
|
|
Virulence score 2 |
0 (0.0 %) |
0 (0.0 %) |
0 (0.0 %) |
n/a |
n/a |
|
|
Virulence score 3 |
37 (24.5 %) |
35 (32.1 %) |
2 (4.8 %) |
9.459 (2.162–41.387) |
4.63×10−4 |
*** |
|
Virulence score 4 |
19 (12.6 %) |
19 (17.4 %) |
0 (0.0 %) |
n/a |
0.002 |
** |
|
Virulence score 5 |
39 (25.8 %) |
37 (33.9 %) |
2 (4.8 %) |
10.278 (2.353–44.899) |
2.41×10−4 |
*** |
|
Antimicrobial resistance | ||||||
|
ESBL |
18 (11.9 %) |
4 (3.7 %) |
14 (33.3 %) |
0.076 (0.023–0.250) |
4.639×10−7 |
*** |
|
Carbapenemase |
12 (7.9 %) |
3 (2.8 %) |
9 (21.4 %) |
0.104 (0.027–0.406) |
1.43×10−4 |
*** |
|
EBSL and Carbapenemase |
11 (7.3 %) |
3 (2.8 %) |
8 (19.0 %) |
0.120 (0.030–0.479) |
5.55×10−4 |
** |
|
Temperature and string test | ||||||
|
Mean string length at 37 °C (cm) |
5.67 ± 0.63 |
4.04 ± 1.20 |
1.29×10−4 |
*** |
||
|
Mean string length at room temperature (cm) |
0.10 ± 0.27 |
17.32 ± 3.91 |
<0.0001 |
*** |
||
Virulence score: 0, no acquired virulence loci detected; 1, only yersiniabactin (ybt); 2, only colibactin (clb) or colibactin and yersiniabactin; 3, only aerobactin (iuc) and/or salmochelin (iro) (without yersiniabactin or colibactin); 4, aerobactin and/or salmochelin with yersiniabactin (without colibactin); and 5, yersiniabactin, colibactin, aerobactin, and/or salmochelin.
Resistance score: 0, no ESBL and no carbapenemase, irrespective of colistin resistance; 1, only ESBL, but no carbapenemase, irrespective of colistin resistance; 2, carbapenemase without colistin resistance; and 3, carbapenemase with colistin resistance.*p < 0.05, **p < 0.01, ***p < 0.001.
Table 2.
Comparison of key features between K. pneumoniae Class I group A and Class I group B
|
Total (n=109) |
Group A (n=83) |
Group B (n=26) |
OR (95 % CI) |
p-value |
||
|---|---|---|---|---|---|---|
|
Sequence type | ||||||
|
ST23 |
26 (23.9 %) |
23 (27.7 %) |
3 (11.5 %) |
2.939 (0.804–10.736) |
0.091 |
|
|
ST65 |
18 (16.5 %) |
16 (19.3 %) |
2 (7.7 %) |
2.866 (0.613–13.396) |
0.165 |
|
|
ST86 |
14 (12.8 %) |
6 (7.2 %) |
8 (30.8 %) |
0.175 (0.054–0.568) |
3.738×10−3 |
** |
|
ST268 |
6 (5.5 %) |
6 (7.2 %) |
0 (0.0 %) |
n/a |
0.332 |
|
|
K loci | ||||||
|
KL1 |
28 (25.7 %) |
25 (30.1 %) |
3 (11.5 %) |
3.305 (0.908–12.020) |
0.058 |
|
|
KL2 |
35 (32.1 %) |
27 (32.5 %) |
8 (30.8 %) |
1.085 (0.419–2.808) |
0.867 |
|
|
KL20 |
9 (8.3 %) |
8 (9.6 %) |
1 (3.8 %) |
2.667 (0.318–22.385) |
0.683 |
|
|
Regulator of mucoid phenotype | ||||||
|
intact rmpA (+) |
92 (84.4 %) |
77 (92.8 %) |
15 (57.7 %) |
9.411 (3.015–29.373) |
9.9×10−5 |
*** |
|
intact rmpA2 (+) |
14 (12.8 %) |
11 (13.3 %) |
3 (11.1 %) |
1.171 (0.301–4.564) |
1.000 |
|
|
intact rmpA and/or rmpA2 (+) |
93 (85.3 %) |
78 (94.0 %) |
15 (57.7 %) |
11.440 (3.470–37.711) |
4.1×10−5 |
*** |
|
Virulence prediction | ||||||
|
Yersiniabactin (ybt) |
67 (61.5 %) |
52 (62.7 %) |
15 (57.7 %) |
1.230 (0.502–3.014) |
0.650 |
|
|
Colibactin (clb) |
37 (33.9 %) |
34 (41.0 %) |
3 (11.5 %) |
5.320 (1.479–19.137) |
0.006 |
** |
|
Aerobactin (iuc) |
91 (83.5 %) |
75 (90.4 %) |
16 (61.5 %) |
5.859 (2.000–17.168) |
5.52×10−4 |
*** |
|
Salmochelin (iro) |
97 (89.0 %) |
80 (96.4 %) |
17 (65.4 %) |
14.118 (3.488–57.686) |
9.8×10−5 |
*** |
|
Temperature and string test | ||||||
|
Mean string length at 37 °C (cm) |
3.93 ± 0.48 |
11.21 ± 1.74 |
2.898×10−7 |
*** |
||
*p< 0.05, **p< 0.01, ***p< 0.001.
HMV phenotype and rmpA/rmpA2 genes
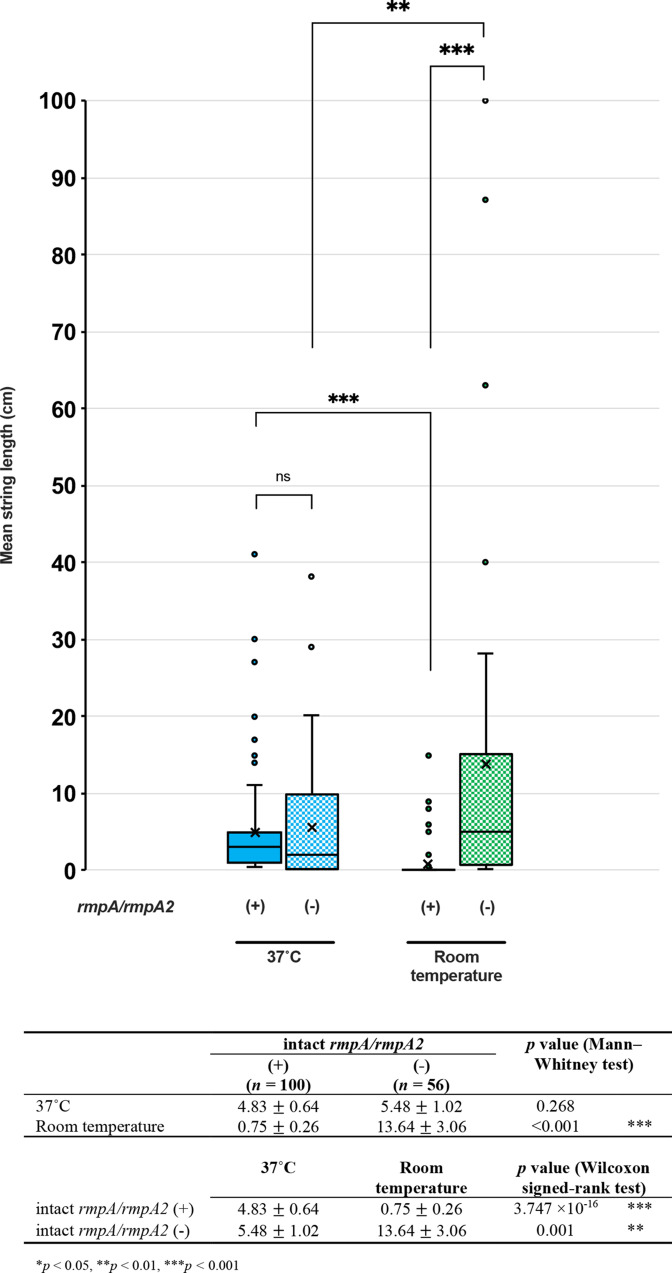
We detected 101 isolates carrying the rmpA gene, among which three isolates (3.0%) possessed truncated rmpA genes (Fig. 6). Conversely, 88 isolates carried rmpA2 genes, 73 (83.0 %) of which were truncated rmpA2 genes. Thirteen strains (8.3%) carried both intact rmpA and intact rmpA2. Altogether, 100 isolates harboured an intact rmpA and/or rmpA2 [henceforth referred to as the rmp (+) group], and 56 isolates carried a truncated or were negative or for both rmpA and rmpA2 [henceforth referred to as the rmp (-) group]. A comparison of the mean string length between the rmpA-positive and rmpA-negative groups revealed no significant difference in string length at 37 °C, but the string length at room temperature in the rmp (-) group was significantly higher than that of the rmp (+) group (Mann–Whitney test, P<0.001) (Fig. 7). Within the rmp (+) group, the mean string length at 37 °C was significantly higher than that at room temperature (Wilcoxon signed-rank test, P<0.001), whereas, within the rmp (-) group, the mean string length at room temperature was significantly higher than that at 37 °C (Wilcoxon signed-rank test, P<0.001).
Fig. 7.
The presence of rmpA/rmpA2 genes and mean string length at 37 °C and room temperature. The isolates are divided into rmpA/rmpA2-positive and -negative groups, and the mean string lengths (cm) of each group at 37 °C and room temperature are demonstrated. Comparison of the mean string length between two groups at the same temperature was analysed with a Mann–Whitney test, and comparison of the mean string length between two temperatures of the same group was analysed with a Wilcoxon signed-rank test. ns, Not significant; ***P<0.001.
Genomic comparisons of Class I and Class III isolates
Class I isolates were commonly distributed in specific STs, such as ST23 (n=26, 23.9%), ST65 (n=18, 16.5%), ST86 (n=14, 12.8%) and ST268 (n=6, 5.5%) (Fig. 6). Conversely, Class III isolates were highly diverse, with the most common STs being ST37 (n=6, 14.3%), ST45 (n=3, 7.1%), and certain sporadic STs such as ST147, ST17, ST34 and ST611. ST23, ST65 and ST86 were significantly more prevalent among Class I isolates, while ST37 was more so among Class III isolates (Table 1). Within Class I, ST86 was significantly more common in group B than in group A (P=3.738×10−3, Table 2).
Given the known associations between the STs and K-loci, particularly among the STs that were overrepresented in the Class I group (described above), it is unsurprising that KL2 (n=35, 32.1%), KL1 (n=28, 25.7%), KL57 (n=10, 9.2%) and KL20 (n=9, 8.3%) were most frequently found in Class I, whereas Class III isolates were associated with the more diverse K-loci. Similarly, isolates carrying rmp (+), which were overrepresented among the well-known hv-clones (ST23, ST65 and ST86), were significantly more prevalent among Class I isolates than Class III isolates (85.3 % vs. 9.5 %, P=3.104×10−18, Table 1, Fig. 6). This was also the case for the key acquired virulence determinants coding for yersiniabactin (ybt), colibactin (clb), aerobactin (iuc) and salmochelin (iro) (P<0.001, Table 1, Fig. 6). A virulence score of 0 (VS0) was significantly associated with Class III (P<0.001), while VS3, VS4 and VS5 were significantly associated with Class I (Table 1). Conversely, ESBLs and carbapenemases, which were rare among the hv-clones, were both significantly more common among Class III than among Class I isolates (P<0.001, Table 1, Figs 3 and 6).
Within Class I, the rmp (+) groups were significantly more common in group A than in group B (94.0 % vs. 57.7 %, P=4.1×10−5, Table 2), as were the genes for colibactin, aerobactin and salmochelin.
Specimen origins
Among the 156 isolates, 125 with available information were analysed to investigate the characteristics associated with the specimen origins, e.g. the samples from the RT, bloodstream infection (BSI), urinary tract infection, bile or ascitic fluid. The majority of specimens were collected from the RT (n=67, 53.6%) and BSI (n=34, 27.2%), with the rest from urine (n=9, 7.2%), bile (n=5, 4.0%), ascitic fluid (n=3, 2.4%), pus (n=2, 1.6%), drainage (n=2, 1.6%) and other miscellaneous origins.
The isolates derived from the RT and BSI showed clear differences with regard to the STs, K-loci, rmpA/rmpA2 genes, virulence genes, AMR genes and favourable/unfavourable temperature for string formation (Table 3, Fig. 3). KL2-ST65 and KL2-ST86 were significantly associated with the RT isolates (P<0.05, Table 3). Similarly, KL1 and KL2 were significantly associated with the RT isolates (P<0.05, Table 3). KL1-ST23 originated from 22.4 % of the RT isolates but only from 8.8 % of the BSI isolates; however, the difference was not statistically significant (P=0.092, Table 3). On the other hand, isolates from BSI were distributed in a diverse set of STs, including ST37 (n=2, 5.9%, Table 3) and some other uncommon STs (data not shown). Remarkably, isolates from the RT showed higher virulence than isolates from BSI; VS3 was significantly more prevalent among isolates from the RT (P<0.001), while VS0 was significantly more common among isolates from BSI (P<0.001, Table 3). In contrast, ESBL was significantly more common in the BSI isolates (11.8%, P=0.011). Regarding the string length under the two temperatures, Class I was significantly associated with the RT isolates (92.5 % vs 35.3 %, P<0.001), whereas Class III was significantly associated with the BSI isolates (58.8 % vs 6.0 %, P<0.001, Table 3 and Fig. 6). More precisely, group A was significantly associated with the RT origin (P<0.001), and groups D and E were significantly more common in the BSI isolates (P<0.01, Table 3). Similarly, 86.6 % of isolates from the RT possessed the rmpA gene, whereas only 23.5 % of isolates from BSI carried the rmpA gene (P<0.001, Table 3). Nonetheless, the key driver associated with the specimen types is unascertainable because of the strong associations among the ST, K-locus, rmpA/rmpA2, virulence genes and AMR genes.
Table 3.
Comparison of key features of hypermucoviscous K. pneumoniae derived from the respiratory tract and blood specimens
|
Total (n=101) |
Respiratory tract (n=67) |
Blood (n=34) |
OR (95 % CI) |
p-value |
||
|---|---|---|---|---|---|---|
|
Sequence type |
||||||
|
ST23 |
18 (17.8 %) |
15 (22.4 %) |
3 (8.8 %) |
2.981 (0.799–11.124) |
0.092 |
|
|
ST65 |
14 (13.9 %) |
13 (19.4 %) |
1 (2.9 %) |
7.944 (0.993–63.562) |
0.031 |
* |
|
ST86 |
10 (9.9 %) |
10 (14.9 %) |
0 (0.0 %) |
n/a |
0.015 |
* |
|
ST268 |
5 (5.0 %) |
4 (6.0 %) |
1 (2.9 %) |
2.095 (0.225–19.513) |
0.661 |
|
|
ST37 |
2 (2.0 %) |
0 (0.0 %) |
2 (5.9 %) |
3.094 (2.326–4.114) |
0.111 |
|
|
K loci |
||||||
|
KL1 |
20 (19.8 %) |
17 (25.4 %) |
3 (8.8 %) |
3.513 (0.951–12.977) |
0.049 |
* |
|
KL2 |
31 (30.7 %) |
26 (38.8 %) |
5 (14.7 %) |
3.678 (1.263–10.709) |
0.013 |
* |
|
KL20 |
8 (7.9 %) |
7 (10.4 %) |
1 (2.9 %) |
3.850 (0.454–32.655) |
0.261 |
|
|
Regulator of mucoid phenotype |
||||||
|
intact rmpA (+) |
66 (65.3 %) |
58 (86.6 %) |
8 (23.5 %) |
20.944 (7.265–60.379) |
3.151×10−10 |
*** |
|
intact rmpA2 (+) |
8 (7.9 %) |
6 (9.0 %) |
2 (5.9 %) |
1.574 (0.300–8.248) |
0.714 |
|
|
Virulence prediction |
||||||
|
Yersiniabactin (ybt) |
52 (51.5 %) |
39 (58.2 %) |
13 (38.2 %) |
2.250 (0.966–5.238) |
0.058 |
|
|
Colibactin (clb) |
27 (26.7 %) |
22 (32.8 %) |
5 (14.7 %) |
2.836 (0.965–8.328) |
0.052 |
|
|
Aerobactin (iuc) |
67 (66.3 %) |
59 (88.1 %) |
8 (23.5 %) |
23.969 (8.114–70.803) |
8.861×10−11 |
*** |
|
Salmochelin (iro) |
71 (70.3 %) |
63 (94.0 %) |
8 (23.5 %) |
51.188 (14.172–184.885) |
2.350×10−13 |
*** |
|
Virulence score 0 |
23 (22.8 %) |
3 (4.5 %) |
20 (58.8 %) |
0.033 (0.009–0.126) |
7.531×10−10 |
*** |
|
Virulence score 1 |
11 (10.9 %) |
5 (7.5 %) |
6 (17.6 %) |
0.376 (0.106–1.337) |
0.175 |
|
|
Virulence score 2 |
0 (0.0 %) |
0 (0.0 %) |
0 (0.0 %) |
n/a |
n/a |
|
|
Virulence score 3 |
26 (25.7 %) |
25 (37.3 %) |
1 (2.9 %) |
19.643 (2.528–152.602) |
1.89×10−4 |
*** |
|
Virulence score 4 |
14 (13.9 %) |
12 (17.9 %) |
2 (5.9 %) |
3.491 (0.734–16.597) |
0.132 |
|
|
Virulence score 5 |
27 (26.7 %) |
22 (32.8 %) |
5 (14.7 %) |
2.836 (0.965–8.328) |
0.052 |
|
|
Antimicrobial resistance |
||||||
|
ESBL |
4 (4.0 %) |
0 (0.0 %) |
4 (11.8 %) |
n/a |
0.011 |
* |
|
Carbapenemase |
0 (0.0 %) |
0 (0.0 %) |
0 (0.0 %) |
n/a |
n/a |
|
|
Temperature and string test |
||||||
|
Class I |
74 (73.3 %) |
62 (92.5 %) |
12 (35.3 %) |
22.733 (7.190–71.874) |
8.110×10−10 |
*** |
|
Group A |
59 (58.4 %) |
52 (77.6 %) |
7 (20.6 %) |
13.371 (4.868–36.729) |
3.914×10−8 |
*** |
|
Group B |
15 (14.9 %) |
10 (14.9 %) |
5 (14.7 %) |
1.018 (0.318–3.255) |
0.977 |
|
|
Class III |
24 (23.8 %) |
4 (6.0 %) |
20 (58.8 %) |
0.044 (0.013–0.151) |
3.693×10−9 |
*** |
|
Group D |
14 (13.9 %) |
4 (6.0 %) |
10 (29.4 %) |
0.152 (0.044–0.533) |
0.004 |
** |
|
Group E |
10 (9.9 %) |
0 (0.0 %) |
10 (29.4 %) |
n/a |
7×10−6 |
*** |
|
Class II (Group C) |
3 (3.0 %) |
1 (1.5 %) |
2 (5.9 %) |
0.242 (0.021–2.774) |
0.261 |
|
|
Mean string length at 37 °C (cm) |
5.68 ± 0.91 |
3.63 ± 1.03 |
0.006 |
** |
||
|
Mean string length at room temperature (cm) |
1.08 ± 0.46 |
7.12 ± 2.12 |
1.995×10−8 |
** |
||
Discussion
This study represents the first investigation of string length distribution across different temperatures (37 °C and room temperature) and its correlation with the genomic population of Kp. We detected clear differences in the multi-locus STs, capsular serotype, string level, string test temperature, rmpA, virulence score, AMR factors and specimen origins by combining phenotypic and genotypic analysis of 156 HMV Kp isolates collected in Japan (shown in Fig. 4). The predominant clusters, cluster 1 (KL2-ST65), cluster 2 (KL1-ST23), cluster 3 (KL2-ST86) and cluster 4 (KL20-ST268 and KL62-ST36), shared similar characteristics, such as positive rmpA, iuc and iro, and the general absence of ESBL/carbapenemase (with some exceptions). These observations were consistent with the other findings among the hvKp population in Asia and Japan, especially in the case of KL1-ST23, KL2-ST65 and KL2-ST86, which have frequently been associated with severe clinical complications [3, 9, 32–34]. Among these, KL1-ST23 is well known for severe community-acquired diseases such as pyogenic liver abscesses and distinct invasive syndromes in Southeast Asia, especially in Taiwan, Hong Kong, Singapore, Korea and Vietnam [32, 35]. We found that the isolates were mostly derived from the RT and produced higher string lengths at 37 °C than at room temperature (Class I isolates). In contrast to the stereotypical hv-clones, a proportion of the isolates, especially clusters 5, 6, 7 and 8 (ST37, ST11, ST45 and ST147), displayed distinct characteristics such as the absence of rmpA, iuc and iro and the presence of ESBL and/or carbapenemases. Strikingly, these isolates were frequently derived from BSI and produced higher string lengths at room temperature than at 37 °C (Class III isolates).
When investigating the molecular mechanism of HMV, we observed an interesting correlation between mean string length and the presence of rmpA (Figs. 6 and 7). The strains carrying rmpA produced short strings at 37 °C, while they were nearly negative at room temperature. In contrast, the strains lacking rmpA could produce a certain amount of HMV at 37 °C, similar to the rmpA (+) isolates, but produced significantly higher HMV at room temperature. RmpA may be responsible for HMV mainly at 37 °C, which can produce relatively short strings, while a novel factor may be responsible for the HMV phenoype at room temperature, which can produce considerably high string lengths. However, the strong associations among the ST, K-locus, rmpA, virulence genes and AMR genes make it difficult to specify the single genetic factor that drives the association with different HMV patterns and specimen origins. Nonetheless, we hypothesize that Class III isolates are more likely to stick to the instruments because of their high HMV at room temperature, which may enhance their propagation and eventually result in nosocomial outbreaks of AMR Kp in hospital settings. Meanwhile, Class I isolates are likely to stick to the alveolar epithelial cells and pulmonary endothelial cells because of their HMV production at 37 °C, providing them with resistance against excretion from the human body.
To the best of our knowledge, no study has investigated the temperature-dependent HMV phenotype or mechanism of temperature-dependent capsule expression of Kp. The HMV phenotype is important for virulence inside the human body (37 °C), but its significance at room temperature, which may facilitate their long endurance and propagation on the surfaces and in the hospital environment (e.g. on catheters, medical equipment, water sinks, tables and chairs) remains unknown. From our observations, in most strains that lacked rmpA and produced high HMV at room temperature, string length could either reach its peak after 1 or 2 days or be maintained at a high level over 3 days (unpublished data), suggesting their high survival capacity and endurance under unfavourable conditions outside the human body. Furthermore, approximately two-thirds of these clusters were able to produce HMV at 37 °C (group D), implying their ability to invade the human immunity defence similar to that of the rmp-positive clone. There were some rare phenomena in previous reports that liver abscesses are not significantly associated with HMV or that metastasis infection was developed from ‘classical Kp’ (non-hv-Kp) [9]. Previous studies evaluated string tests by stretching a single [2, 14] or multiple colonies [7] using an inoculation loop [2, 7, 14] or a toothpick [36]. These methods provide a qualitative measurement of HMV with ‘positive’ or ‘negative’ results; however, comparative evaluation between strains with ‘positive’ string tests is difficult because of the short string length generated by only one or a few colonies. Therefore, to make the string test quantifiable, we used a cotton swab to collect an area of bacterial colonies (one-quarter of a standard agar plate) and stretched upwards so that longer strings could be generated to make the string length measurement easier. Additionally, based on our findings, we suggest a definition of HMV that is not only limited to the result of the string test at 37 °C but also at room temperature, and the novel factor responsible for HMV in the absence of rmpA, if discovered, should be considered as a biomarker for defining a strain as hvKp or cKp. A different study is being conducted to discover novel factors responsible for the HMV phenotype at room temperature within these clusters.
Carbapenem antibiotics have been the last resort against MDR Kp; however, carbapenemase-producing Kp has been continuously reported over the past decade. Our study detected 13.5 % (21/156 isolates) of the HMV Kp carrying ESBL/carbapenemase genes, among which 23.8 % (5/21 strains) displayed the convergence of hypervirulence and carbapenem resistance by the acquisition of pLVPK-like and pKPI-6 plasmids. These five isolates belonged to three sequence types and three capsular serotypes [ST36-KL62 (one isolate), ST65-KL2 (two isolates) and ST268-KL20 (two isolates)] (Figs 2 and 3). The pLVPK plasmid has been broadly disseminated worldwide [37], while pKPI-6 is an epidemic AMR plasmid in Japan [29]. In Japan, in addition to the pKPI-6-like plasmid-carrying ST23-hv-Kp, which was reported in 2019 [38], our study revealed that the acquisition of this AMR plasmid also occurred in ST65-hv-Kp, posing a critical threat to the extended emergence of carbapenem resistance in hvKp clones. While most carbapenem-resistant hvKp strains in China and some other countries harboured bla KPC-2 [37, 39], all five carbapenem-resistant hvKp in this study carried bla IMP-6 and bla CTX-M-2 in pKPI-6-like plasmids, suggesting a distinct epidemiological feature of Japanese clones compared with the clones from other countries. In addition, we detected six classical Kp carrying pKPI-6 like plasmids (N212, MS5292, N60, MS5293, MS5294 and MS5265) and two ST11 isolates carrying bla KPC-2 (MS5538 and MS5544), although none of them carried a pLVPK-like plasmid or virulence genes such as rmpA/rmpA2, aerobactin-, and salmochelin-encoding genes (Fig. 3).
A limitation of our study is the inability to access patient clinical data, hindering the interpretation of the association between infection with different clonal Kp and clinical outcomes. Additionally, the isolates in our study were mainly collected from West Japan, which may not precisely reflect the HMV Kp population in Japan. Future surveillance will be carried out to investigate the characteristics of HMV Kp from all over Japan, as well as to compare the geographical epidemiology among different areas. Another limitation is that the results of the modified string test may not be entirely compatible with those of the conventional string test. In this study, we performed the string test at two different temperatures and three different time points, making it complex and time-consuming, and unsuitable for screening in clinical laboratories. Nevertheless, we standardized the string test protocol to make it reliable for evaluating the temperature-dependent HMV phenotype, which may contribute to the future exploration of the HMV regulatory mechanism under different conditions.
In summary, our analysis of HMV Kp showed that the low HMV level is usually related to rmpA/rmpA2, while the high HMV level is not. Strains carrying rmpA/rmpA2 are likely to express HMV at 37 °C, whereas those negative for these genes are likely to express HMV at room temperature. On the one hand, some clusters shared similar characteristics, such as positivity for rmpA, mostly without ESBLs/carbapenemases, mostly derived from the RT, and produced higher string lengths at 37 °C than at room temperature. Some clusters, however, showed distinct characteristics such as testing negative for rmpA, positive for ESBL and/or carbapenemases, frequently derived from BSI, and produced a higher string length at room temperature than at 37 °C. Our findings may facilitate the future discovery of unknown factor(s) responsible for HMV at room temperature and may explain different virulence tactics of Kp infection.
Supplementary Data
Funding information
This study was supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japanese Agency for Medical Research and Development (AMED) under grant numbers #20fk0108132j0001 and #21fk0108604j0001, and JSPS KAKENHI Grant Numbers JP21K16947 and JP18K07112.
Acknowledgements
We thank M. Nakazawa, S. Aoki, S. Wakai and E. Anzai (AMR-RC, NIID) for susceptibility testing, T. Ueshimo (Hyogo Prefectural Amagasaki Hospital), H. Nishio (Clinical Laboratory, Shiga Medical Center for Adults, Moriyama, Shiga, Japan), K. Yamasaki (Department of Clinical Laboratory, Wakayama Rosai Hospital, Wakayama, Japan), Y. Wada (Clinical Laboratory, Hyogo Medical University Hospital, Nishinomiya, Hyogo, Japan), S. Kashiyama (Clinical Laboratory, Hiroshima University Hospital, Hiroshima City, Hiroshima, Japan), Y. Koba (Clinical Laboratory, Hiroshima University Hospital, Hiroshima City, Hiroshima, Japan), K. Tadera (National Hospital Organization Kure Medical Center and Chugoku Cancer Center), K. Kimura (Hiroshima City Hiroshima Citizens Hospital), K. Yokota (Hiroshima City Hiroshima Citizens Hospital), M. Shiba (Hiroshima Red Cross Hospital and Atomic-bomb Survivors Hospital) and T. Harino (Clinical Laboratory, Hiroshima City Asa Hospital, Asakita-ku, Hiroshima City, Hiroshima, Japan) for supplying the Klebsiella isolates, and Y. Sugawara and N. Sakamoto (National Institute of Infectious Diseases) for genome submission to DDBJ.
Author contributions
Conceptualization: M.S., S.Ka., H.O. Data curation: M.N-T.L., S.K., K.L.W. Formal analysis: M.N-T.L., S.K., K.L.W. Funding acquisition: M.S., S.K., M.N-T.L. Investigation: M.N-T.L., S.K., L.Y., J.H., M.S., K.Y. Methodology: M.S., S.K., M.N-T.L. Project administration: M.S., S.K., M.N-T.L. Resources: T.T., K.S., S.T., T.O., K.K., T.M., H.O., K.L.W., K.E.H. Software: K.L.W., K.E.H. Supervision: M.S. Validation: M.N-T.L., S.K., M.S., K.L.W. Visualization: M.N-T.L. Writing – original draft: M.N-T.L. Writing – review and editing: M.N-T.L., S.K., K.L.W., M.S.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was approved by the ethical committees of the National Institute of Infectious Diseases Committee of Ethics (approval number 972).
Footnotes
Abbreviations: AMR, antimicrobial resistance; BSI, bloodstream infection; cKp, classical Kp; CLSI, Clinical and Laboratory Standards Institute; ESBL, extended-spectrum beta-lactamase; HMV, hypermucoviscosity; hv, hypervirulent; KL, K-loci; Kp, Klebsiella pneumoniae; LB, Luria-Bertani; MDR, multi-drug resistance; MIC, minimum inhibitory concentration; MLST, multi-locus sequence typing; RT, respiratory tract; ST, sequence type; VS, Virulence Score; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. A supplementary table is available with the online version of this article.
References
- 1.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–81. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu D, Dong N, Zheng Z, Lin D, Huang M, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in A Chinese hospital: A molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 3.Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 2020;12:11. doi: 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae . Nat Rev Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 5.Shigemoto N, Kuwahara R, Kayama S, Shimizu W, Onodera M, et al. Emergence in Japan of an imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae carrying blaIMP-6. Diagn Microbiol Infect Dis. 2012;72:109–112. doi: 10.1016/j.diagmicrobio.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae . Clin Microbiol Rev. 2019;32 doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai T. Hypermucoviscosity: an extremely sticky phenotype of Klebsiella pneumoniae associated with emerging destructive tissue abscess syndrome. Clin Infect Dis. 2006;42:1359–1361. doi: 10.1086/503429. [DOI] [PubMed] [Google Scholar]
- 9.Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8:1111–1123. doi: 10.1080/21505594.2017.1317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang Y-P, Fang C-T, Lai S-Y, Chang S-C, Wang J-T. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2006;193:645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 12.Liu YM, Li BB, Zhang YY, Zhang W, Shen H, et al. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother. 2014;58:5379–5385. doi: 10.1128/AAC.02523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan VB, Venkataramaiah M, Mondal A, Vaidyanathan V, Govil T, et al. Functional characterization of a novel outer membrane porin KpnO, regulated by PhoBR two-component system in Klebsiella pneumoniae NTUH-K2044. PLoS One. 2012;7:e41505. doi: 10.1371/journal.pone.0041505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio. 2019;10:1–16. doi: 10.1128/mBio.00089-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nassif X, Honoré N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol Microbiol. 1989;3:1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 16.Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42:1351–1358. doi: 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 17.Russo TA, Olson R, Fang CT, Stoesser N, Miller M, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae . J Clin Microbiol. 2018;56:e00776-18. doi: 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wacharotayankun R, Arakawa Y, Ohta M, Tanaka K, Akashi T, et al. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect Immun. 1993;61:3164–3174. doi: 10.1128/iai.61.8.3164-3174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker KA, Miller VL. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae . Curr Opin Microbiol. 2020;54:95–102. doi: 10.1016/j.mib.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcántar-Curiel MD, Girón JA. Klebsiella pneumoniae and the pyogenic liver abscess: implications and association of the presence of rpmA genes and expression of hypermucoviscosity. Virulence. 2015;6:407–409. doi: 10.1080/21505594.2015.1030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute (CLSI) M100Ed30 | Performance Standards for Antimicrobial Susceptibility Testing. 2018. [Google Scholar]
- 22.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:1–16. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pommier T, Canbäck B, Lundberg P, Hagström A, Tunlid A. RAMI: A tool for identification and characterization of phylogenetic clusters in microbial communities. Bioinformatics. 2009;25:736–742. doi: 10.1093/bioinformatics/btp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:1–10. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kayama S, Shigemoto N, Kuwahara R, Oshima K, Hirakawa H, et al. Complete nucleotide sequence of the IncN plasmid encoding IMP-6 and CTX-M-2 from emerging carbapenem-resistant Enterobacteriaceae in Japan. Antimicrob Agents Chemother. 2015;59:1356–1359. doi: 10.1128/AAC.04759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother. 2018;62:e02644-17. doi: 10.1128/AAC.02644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi L, Feng J, Zhan Z, Zhao Y, Zhou H, et al. Comparative analysis of blakpc-2-and rmtb-carrying incfii-family pkpc-LK30/phn7a8 hybrid plasmids from Klebsiella pneumoniae CG258 strains disseminated among multiple chinese hospitals. Infect Drug Resist. 2018;11:1783–1793. doi: 10.2147/IDR.S171953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siu LK, Fung C-P, Chang F-Y, Lee N, Yeh K-M, et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol. 2011;49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moradigaravand D, Martin V, Peacock SJ, Parkhill J. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. mBio. 2017;8:e01976-16. doi: 10.1128/mBio.01976-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harada S, Aoki K, Yamamoto S, Ishii Y, Sekiya N, et al. Clinical and molecular characteristics of Klebsiella pneumoniae isolates causing bloodstream infections in Japan: occurrence of hypervirulent infections in health care. J Clin Microbiol. 2019;57:e01206-19. doi: 10.1128/JCM.01206-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: A new invasive syndrome. Lancet Infect Dis. 2012;12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 36.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, et al. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192:3144–3158. doi: 10.1128/JB.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Dong N, Chan EWC, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae . Trends Microbiol. 2021;29:65–83. doi: 10.1016/j.tim.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Harada S, Aoki K, Ishii Y, Ohno Y, Nakamura A, et al. Emergence of IMP-producing hypervirulent Klebsiella pneumoniae carrying a pLVPK-like virulence plasmid. Int J Antimicrob Agents. 2019;53:873–875. doi: 10.1016/j.ijantimicag.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Lee C-R, Lee JH, Park KS, Jeon JH, Kim YB, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:11. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.