Abstract
Moraxella catarrhalis is a common cause of respiratory tract infection, particularly otitis media in children, whilst it is also associated with the onset of exacerbation in chronic obstructive pulmonary disease in adults. Despite the need for an efficacious vaccine against M. catarrhalis , no candidates have progressed to clinical trial. This study, therefore, aimed to characterize the diversity of M. catarrhalis isolated from the upper respiratory tract of healthy children and adults, to gain a better understanding of the epidemiology of M. catarrhalis and the distribution of genes associated with virulence factors, to aid vaccine efforts. Isolates were sequenced and the presence of target genes reported. Contrary to prevailing data, this study found that lipooligosaccharide (LOS) B serotypes are not exclusively associated with 16S type 1. In addition, a particularly low prevalence of LOS B and high prevalence of LOS C serotypes was observed. M. catarrhalis isolates showed low prevalence of antimicrobial resistance and a high gene prevalence for a number of the target genes investigated: ompB2 (also known as copB), ompCD, ompE, ompG1a, ompG1b, mid (also known as hag), mcaP, m35, tbpA, lbpA, tbpB, lbpB, msp22, msp75 and msp78, afeA, pilA, pilQ, pilT, mod, oppA, sbp2, mcmA and mclS.
Keywords: AMR, carriage, epidemiology, Moraxella catarrhalis, virulence
Data Summary
All genomes have been deposited in GenBank under BioProject ID PRJEB39742: accession numbers SAMEA7160394, SAMEA7160393, SAMEA7160392, SAMEA7160391, SAMEA7160390, SAMEA7160389, SAMEA7160388, SAMEA7160387, SAMEA7160386, SAMEA7160385, SAMEA7160384, SAMEA7160383, SAMEA7160382, SAMEA7160381, SAMEA7160380, SAMEA7160379, SAMEA7160378 and SAMEA7160377 (https://www.ncbi.nlm.nih.gov/biosample?Db=biosample&DbFrom=bioproject&Cmd=Link&LinkName=bioproject_biosample&LinkReadableName=BioSample&ordinalpos=1&IdsFromResult=655879).
Impact Statement.
This paper gives new insight into the diversity and epidemiology of Moraxella catarrhalis , an increasingly important opportunistic human pathogen. These data help to clarify the distribution of 16S type, lipooligosaccharide (LOS) type and multilocus sequence type in community settings, and provide novel insight into the prevalence of antibiotic resistance and virulence factors in isolates circulating in the general healthy population. Contrary to prevailing data, this study found that LOS B serotypes are not exclusively associated with 16S type 1. In addition, a low prevalence of LOS B and a high prevalence of LOS C serotypes was observed. M. catarrhalis isolates showed a low prevalence of antimicrobial resistance and a high gene prevalence for a number of the target genes investigated. This is, to our knowledge, the first study to focus on carriage isolates, especially using strains isolates from people of all ages, and should now be followed by a similar analysis on a larger set of community isolates.
Introduction
Moraxella catarrhalis is a Gram-negative, non-encapsulated diplococci and opportunistic human pathogen [1]. A common commensal of the upper respiratory tract [2], M. catarrhalis was once considered non-pathogenic. However, in the 1970s and 1980s, the pathogenic potential of M. catarrhalis was demonstrated through isolation from cases of disease [3–9]; it is now recognized as one of the most common causes of respiratory tract infection (RTI) [10, 11]. As the third largest bacterial cause of otitis media (OM) [12], M. catarrhalis causes 709 million cases of acute OM (AOM) globally each year; 51 % of which are in those ages four and under [13]. AOM is considered one of the most prevalent childhood conditions, with approximately 80 % of all children suffering at least one episode of AOM by 3 years of age [14, 15]. M. catarrhalis is also the second most common cause of exacerbation in chronic obstructive pulmonary disease (COPD) [15, 16], which is the third largest cause of global morbidity, responsible for 3 million deaths in 2016 [17].
M. catarrhalis has two distinct lineages that evolved independently, whilst divergent strains with lower homology have also been identified. Lineage one is complement resistant and adheres to epithelial cells; thus, it is known as the seroresistant subpopulation [18, 19]. Lineage two is less pathogenic, adheres less efficiently to the epithelium and is commonly complement sensitive; thus, it is known as the serosensitive subpopulation [18, 20, 21]. The seroresistant lineage comprises of 16S type 1 isolates, whereas 16S type 2 and 3 isolates fall into the serosensitive lineage [19, 21]. Whilst disease burden is greater from strains belonging to the seroresistant lineage, all 16S types can cause disease [19, 22]. Despite their separate evolution, distinct core genomes and differing genome size (~1.89 Mb for the seroresistant lineage, ~1.93 Mb for the serosensitive), both lineages show regular horizontal gene transfer [19]. M. catarrhalis is commonly typed according to the expression of highly conserved lipooligosaccharide (LOS) surface antigens, which forms the basis for the classification of M. catarrhalis into serotypes A, B or C [23, 24].
In recent years, the need for an efficacious vaccine against M. catarrhalis has been highlighted, yet no vaccine candidates have progressed to clinical trial [25, 26]. To help identify suitable vaccine targets, a better understanding of the epidemiology of M. catarrhalis and the distribution and diversity of virulence factors is required. Current data regarding the prevalence of M. catarrhalis and the distribution and expression of virulence genes across carriage and disease isolates remains inconclusive [18, 21, 22, 27]. For example, the gene for ubiquitous surface protein A1 (UspA1) has been reported present in 87–98 % of 16S type 1 isolates and 23–36 % 16S type 2 and 3 isolates [18, 21] in some research, whilst it is reported to be present in 100 % of 16S type 1 and >89 % of 16S type 2 and 3 isolates in other studies [22, 27]. However, the expression of uspA1 appears similar in carriage and disease with expression of uspA1 in 95 % of M. catarrhalis isolated in child carriage versus 97 % expression in M. catarrhalis isolated from child RTI [22].
Furthermore, whilst previous studies suggest 16S type 1 is most commonly associated with disease [18], it is unknown whether pathogenesis is implicitly associated with a particular type, what the roles of additional subpopulations or strains of M. catarrhalis are in disease epidemiology, or indeed what can be considered as the gene repertoire for virulence [28]. For example, most M. catarrhalis isolates, from 16S types 1, 2 and 3 (both seroresistant and serosensitive isolates), contain conserved genes for the majority of known virulence factors, suggesting perhaps all M. catarrhalis strains/16S types have equal pathogenic potential [19]. For instance, Uspa2 is vital for serum resistance, yet the gene encoding it is equally present in serum-resistant and serum-sensitive strains [21]. Similarly, the gene for M. catarrhalis immunoglobulin D binding outer membrane protein (MID) (also known as haemagglutinin/Hag) [29] is present in at least 90 % of child RTI isolates, 91 % of adult RTI isolates and 80 % of child carriage isolates, indicating no clear link between gene presence and carriage or disease [22, 27, 29]. This highlights the importance of looking at the genotypes and phenotypes of both disease and carriage isolates. As virulence is based on multiple factors, the balance between the harmless carriage of M. catarrhalis and the development of disease may be determined by the combination of the virulence genes present, differences in expression of these genes and environmental factors [19]. Understanding the prevalence and distribution of numerous virulence factors and their importance in carriage and disease and, thus, their potential use in vaccine development is vital.
The aim of this study was to characterize a collection of M. catarrhalis isolated from people of all ages, to give new insight into the diversity and epidemiology of this important human pathogen. As the first study, to our knowledge, to focus on carriage isolates, especially using strains isolated from people of all ages, it provides important data to improve our knowledge of the diversity of M. catarrhalis and to update the literature.
Methods
Sample collection and bacterial identification
A large population-based cross-sectional carriage study, the ‘Analysis of the microbial community of the upper respiratory tract to support the development of effective vaccine policy’ study (Bupa SMART study; REC reference 11/SC/0518), was undertaken as described previously [30, 31]. Briefly, swabbing was undertaken over two time-points: May to August 2012 and February to April 2013. Study participants were identified from general practice lists in the Wessex Primary Care Research Network and were randomized into one of two study arms. For one arm, each participant took a self-taken nasal swab and for the other arm a nasopharyngeal (NP) swab was taken by a trained healthcare professional [30, 31]. Prior to culture, each swab was immersed and vortexed in skim milk, tryptone, glucose and glycerine (STGG) storage media. For each, 10 µl was plated onto Columbia blood agar with horse blood (Oxoid) and Columbia blood agar with chocolated horse blood (Oxoid). Plates were incubated for 24–48 h at 37 °C in 5 % CO2. M. catarrhalis were initially identified by standard morphology: non-haemolytic colonies that appear grey or white on blood agar or pinkish brown on chocolated agar, opaque, flat, smooth, dry, stay as complete colonies when pushed across agar and are 1–3 mm in diameter after 24 h of incubation [32]. Isolates were then verified as M. catarrhalis by being confirmed as Gram-negative, and oxidase, tributyrin and DNase positive. Tests were done using oxidase strips (Oxoid), tributyrin strips (Sigma- Aldrich) and DNase/methyl green plates (VWR; EOLAPP0560) as per manufacturers' instructions. Pure growth was frozen at −80 °C in STGG for future analysis.
Isolates
A subset (n=24) of M. catarrhalis were selected for whole-genome analysis. Equal numbers (n=6) of M. catarrhalis were drawn from participants in the following age ranges; 0–4 years, 5–16 years, 17–59 years and 60+ years. M. catarrhalis isolated from NP swabs were preferentially chosen, as the nasopharynx is the recommended sampling site for these bacteria [33]. Where this was not possible, M. catarrhalis isolated from nasal swabs were used.
Whole-genome sequencing
DNA extraction was performed for each isolate using an overnight culture of a single colony pick, grown on Columbia blood agar with horse blood (Oxoid). Here, the Qiagen mini prep kit (Qiagen) was used in accordance with the manufacturer's instructions. DNA quantification was done using a Qubit fluorometer (Invitrogen). DNA extracts were then diluted to 0.2 ng µl−1 in distilled water. Library preparation was done using a Nextera XT kit (Illumina). Sequencing was done in house using an Illumina MiSeq to generate 2×250 bp V2 paired-end read data.
Bioinformatics
FastQC v. 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess read metrics. Reads were trimmed to remove adapters using Trimmomatic v. 0.32 [34]. Multilocus sequence type (MLST) was obtained by submitting the Fastq files to EnteroBase (https://enterobase.warwick.ac.uk/species/index/mcatarrhalis); accessed February 2017. Genome assembly was undertaken using SPAdes v. 3.1 [35] and the quality of assembly was checked using Quast v. 4.2 [36]. LOS serotyping was done with in silico PCR using Ipcress (with a primer mismatch tolerance of three) on assembled genomes, using published primer sequences [37]. Expected product lengths for serotypes A, B and C were 1.9, 3.3 and 4.3 kbp, respectively. 16S typing, a common method for the classification of M. catarrhalis based on the homology of 16S rRNA sequence, was similarly undertaken using Ipcress with published primers [21]. Resulting sequences were mapped against the 16S sequences of known types [21] using srst2 v. 0.1.3 [38]. ParSNP v. 1.2 [39] was used to align and construct a core-genome phylogeny, using BBH18 as a reference genome. The resultant phylogeny was visualized using iTOL version 4.2.3 [40].
The M. catarrhalis pangenome was defined and analysed using Anvi’o v7 with reference strains NCTC 11020 and BBH18 included for comparison [41]. Identification of ORFs and annotation with Clusters of Orthologous Genes (COGs) function was done within Anvi’o. Amino acid sequence comparisons were done using blastp using the --use-ncbi-blast flag with additional initial parameters including a minbit score of 0.5 and an Markov Clustering (MCL) inflation of 10 for clustering of genes into gene clusters. Functional enrichment analysis was done using the ‘anvi-get-enriched-functions-per-pan-group’ using the COGs as the function annotation source. This contrasts the prevalence of gene clusters and associated functional annotation, rather than genes, between user-defined groups of isolates using a generalized linear model with the logit linkage. This outputs both an enrichment score and P value for each function. Benjamini–Hochberg false discovery rate (FDR) corrected P values (<0.05) were used to determine enriched functions between phylogenetic clades. Finally, average nucleotide identity (ANI) was computed using ‘anvi-compute-genome-similarity.
Antibiotic resistance
srst2 v. 0.1.3 [38] using the ARGannot.r1.fasta database for acquired resistance genes and ResFinder v. 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/; accessed May 2021 with an 80 % ID threshold) [42] were used to detect the presence of antibiotic-resistance genes. Additionally, reads were mapped, using srst2 v. 0.1.3 [38], to the bla BRO-1 (GenBank accession no. Z54180.1) and bla BRO-2 (GenBank accession no. Z54181.1) gene sequences, which can confer resistance to β-lactam antibiotics as they code for the production of β-lactamase. Consensus sequences from srst2 were aligned in Clustal Omega [43] for manual confirmation of gene presence, the sequences obtained were verified as bla BRO-1 or bla BRO-2 using blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Antibiotic resistance was also tested phenotypically. Here, 10 µl of each isolate (a suspension of cells in liquid STGG) was plated onto Columbia blood agar with horse blood (Oxoid). Plates were incubated for 24 h at 37 °C in 5 % CO2. Pure colonies were added to 1 ml saline to get an inoculum of 0.5 McFarland. A sterile swab was used to spread this inoculum over Mueller Hinton F plates [Mueller-Hinton agar, 5 % defibrinated horse blood and 20 mg β-NAD l−1 (Oxoid)]. Antibiotic discs, four per plate, were added and plates were incubated at 35 °C in 5 % CO2 for 18 h (±2 h). Each M. catarrhalis isolate was tested with amoxicillin/clavulanic acid, 2/1 μg; cefotaxime, 5 μg; ceftriaxone, 30 μg; erythromycin, 15 μg; tetracycline, 30 μg; chloramphenicol, 30 μg; ciprofloxacin, 5 μg; and meropenem, 10 μg (all from Oxoid). EUCAST (European Committee on Antimicrobial Susceptibility Testing) breakpoints were used to assess susceptibility and resistance. β-Lactamase production was confirmed using β-lactamase identification sticks (Oxoid), as per the manufacturer's instructions [44].
Virulence factors
Isolates were tested for the presence of genes for UspA1, UspA2 and UspA2H, outer membrane proteins (OMP) B2 (also known as C̱atarrhalis o̱uter membrane p̱rotein Ḇ/CopB), CD, E, G1a and G1b, MID/Hag, M. catarrhalis adherence protein (McaP), outer membrane porin M35, Moraxella haemagglutinin-like proteins (Mha) B1, B2 and C; transferrin binding proteins (Tbp) A and B, lactoferrin binding proteins (Lbp) A and B, Moraxella surface proteins (Msp) 22, 75 and 78, chelated iron ABC transporter substrate binding protein AfeA, M. catarrhalis metallopeptidase-like adhesin (McmA), M. catarrhalis cardiolipin synthase (MclS), M. catarrhalis type IV pilin (PilA), M. catarrhalis type IV pilus biogenesis secretin (PilQ), M. catarrhalis type IV pilus retraction ATPase (PilT), M. catarrhalis type III restriction-modification system methyltransferase (ModM), oligopeptide permease protein A (OppA) and substrate binding protein 2 (SBP2). This was done by read mapping of raw sequence reads (.fastq.gz) using BBMap v36.59 [45] and BWA v0.7.17 [46] against the following NCBI, EMBL or ENA references: AF113610.1, EU430059.1, U61725.1, AF113610.1 and AF352398.1 (uspA1), AY730666.1, AF352399.1, AF410950.1, AF352399.1, AF181073.1 and AF113609.1 (uspA2), AF181075.1, DQ811779.1, AF410951.1, AF181074.1 and AF181075.1 (uspA2H), AY726667.1, AY726666.1 and AY726664.1 (copB), AY493741.1 (ompCD), L31788.1 and CP002005.1 (ompE), AY275816.1 and AY275809.1 (ompG1a), AY462077.1, AY462072.1 and AY462066.1 (ompG1b), AY862881.1, AY077638.2 and AY077637.1 (mid/hag), EF075933.1, EF075940.1 and EF075937.1 (mcaP), AY905613.1 (m35), EF362385.1 and EF362386.1 (mhaB1), EF362388.1 and EF362389.1 (mhaB2), EF362396.1 and EF362391.1 (mhaC), AF039315.1 (tbpAB), AAC34275.1 (tbpA), AF039314.1 and AF039311.1 (tbpB), AF043131.1 (lbpAB), AAC31366.1 (lbpA), AF043133.1 (lbpB), ACA52193.1 (msp22), EU339314.1 (msp75), EU339313.1 (msp78), EMBL ADG62123.1 (afeA), EF017300.1 (mcmA), KC692996.1 (mclS), AY647185.1 and HQ442298.1 and ADG60664.1 (pilA), AY647186.1 (pilQ), AY647187.1 (pilT), AY049056.1 and CP018059.1 (modM), ADG61563.1 (oppA) and STY81838.1 (sbp2). SAMtools v0.1.19 [47] was used to view, sort and index .bam files. Coverage statistics were generated with pileup. Cut-offs for sequence coverage were specific for each gene, based on the level of variation expected for each based on prior publication as detailed in Table 1. Therefore, genes known to be less conserved were permitted a lower sequence coverage as an indication of gene presence.
Table 1.
Gene identity and cut-off points for read mapping analysis
|
Gene |
Identity (%) |
Reference |
Cut-off used (%) |
|---|---|---|---|
|
hag |
56.6–85 |
[24, 28] |
55 |
|
uspA1 |
Modular |
[28] |
– |
|
uspA2 |
Modular |
[28] |
– |
|
uspa2H |
Modular |
[28] |
– |
|
modM |
70 |
[28] |
70 |
|
mhaB1 |
68.8 |
[28] |
68 |
|
mhaB2 |
98 |
[28] |
90 |
|
mhaC |
Not published |
– |
70 |
|
tbpA |
98 |
[28] |
90 |
|
tbpB |
51 |
[28] |
50 |
|
lbpA |
99 |
[28] |
90 |
|
lbpB |
77 |
[28] |
75 |
|
pilA |
>78 |
[28, 82] |
70 |
|
pilQ |
Not published |
– |
70 |
|
pilT |
Not published |
– |
70 |
|
ompG1a |
90 |
[28] |
90 |
|
ompG1b |
92 |
[28] |
90 |
|
mcmA |
Not published |
[28] |
70 |
|
sbp2 |
99.8 |
[28] |
90 |
|
copB |
76.1 |
[28] |
70 |
|
ompCD |
97.1 |
[28] |
95 |
|
ompE |
96.6–100 |
[28] |
97 |
|
mcaP |
98–100 |
[28] |
98 |
|
m35 |
92.8–99.4 |
[28] |
90 |
|
msp22 |
99 |
[28] |
90 |
|
msp75 |
97 |
[28] |
90 |
|
msp78 |
99 |
[28] |
90 |
|
afeA |
87–100 |
[28] |
85 |
|
mclS |
99 |
[83] |
90 |
|
oppA |
98.7 |
[28] |
90 |
Whilst the current literature suggests mhaB2, tbpA, lpbA, SBP2, ompE, ompCD, mcaP, msp78, mclS and oppA all have conservation identities of >95 %, a 90 % coverage cut-off was used to ensure genes showing slight variation were not excluded. A 90 % coverage means the reads overlap 90 % the length of the gene supporting the presence of the gene.
Reference strains
Reference strains NCTC 11020 and BBH18 were included for comparison in figures and as controls to assess the accuracy of bioinformatic analysis. However, the data presented within the text of the results are solely for the M. catarrhalis isolated as part of the SMART study; thus, data are representative of carriage isolates only. Whilst BB818 was included as a reference as a known serum-resistant strain, no data on serum sensitivity was available for NCTC 11020. NCTC 11020 has previously been sequenced and this data published, so was included as an additional reference to be used as a quality control for in-house sequencing.
Results and Discussion
Carriage of M. catarrhalis
From the 314 NP swabs obtained during the Bupa SMART study, 14 M . catarrhalis were isolated representing an overall carriage prevalence of 4.5 %. Of the 314 participants who had NP swabs taken, 56 were 0–4 years, 24 were aged 5–16, 59 were aged 17–59 and 175 were aged 60 or over. The NP carriage prevalence of M. catarrhalis for each age group was 10.7 % (6/56) for those aged 0–4, 4.2 % (1/24) for those aged 5–16, 3.4 % (2/59) for those aged 17–59 and 2.9 % (5/175) for those aged 60 years and over. Of the 2103 nasal swabs received, 96 M . catarrhalis were isolated: an overall carriage prevalence of 4.6 %. Of the 2103 participants who took nasal swabs, 497 were 0–4 years, 248 were aged 5–16, 614 were aged 17–59 and 708 were aged 60 or over. No age was provided by 36 participants. The nasal carriage prevalence of M. catarrhalis for each age group was 10.1 % (50/497) for those aged 0–4, 6.9 % (17/248) for those aged 5–16, 1.5 % (9/614) for those aged 17–59 and 2.4 % (17/708) for those aged 60 years and over. All NP swabs were taken between May and August 2012 (the first time-point). Nasal swabs were taken over both time-points; 1260 nasal swabs were taken between May and August 2012 and 843 were taken between February and April 2013 [30, 31].
Of the 24 pre-selected M. catarrhalis isolates, 18 were successfully sequenced; the number of contigs ranged from 27 to 141 with a mean of 58.9, whilst the N50 ranged from 31 207 to 214 693 with a mean of 108 622.8. Metadata for these isolates, including information about the participants from whom they were obtained, can be seen in Table 2.
Table 2.
Isolate metadata
|
Participant data |
||||||
|---|---|---|---|---|---|---|
|
Isolate no. |
Swab type |
Age (years) |
Antibiotic use* |
Vaccination up to date |
Date of culture (dd.mm.yy) |
Co-carriage† |
|
57 |
NP |
2 |
No |
Yes |
07.06.12 |
None |
|
626 |
NP |
1 |
No |
Yes |
29.06.12 |
|
|
628 |
NP |
3 |
No |
Yes |
29.06.12 |
None |
|
1077 |
NP |
4 |
No |
Yes |
16.07.12 |
None |
|
1080 |
NP |
2 |
Yes (flucloxacillin) |
Yes |
16.07.12 |
None |
|
1227 |
NP |
1 |
No |
Yes |
24.07.12 |
|
|
1343 |
Nasal |
6 |
No |
Yes |
30.07.12 |
|
|
1592 |
Nasal |
5 |
No |
Yes |
19.02.13 |
|
|
1648 |
Nasal |
6 |
No |
Yes |
20.02.13 |
None |
|
1833 |
Nasal |
7 |
No |
Yes |
27.02.13 |
None |
|
18 |
NP |
26 |
No |
Yes |
02.07.12 |
None |
|
608 |
Nasal |
34 |
No |
Yes |
29.06.12 |
None |
|
19 |
Nasal |
41 |
No |
No |
05.03.13 |
None |
|
20 |
Nasal |
43 |
No |
Yes |
16.03.13 |
None |
|
687 |
NP |
85 |
No |
Yes |
03.07.12 |
None |
|
10 309 |
NP |
75 |
No |
No |
27.07.12 |
None |
|
1470 |
NP |
81 |
No |
Yes |
07.08.12 |
None |
|
37 |
Nasal |
81 |
No |
Yes |
24.05.12 |
None |
*Indicates use in the month prior to swabbing.
†H. influenzae, Haemophilus influenzae; S. aureus, Staphylococcus aureus; S. pneumoniae, Streptococcus pneumoniae.
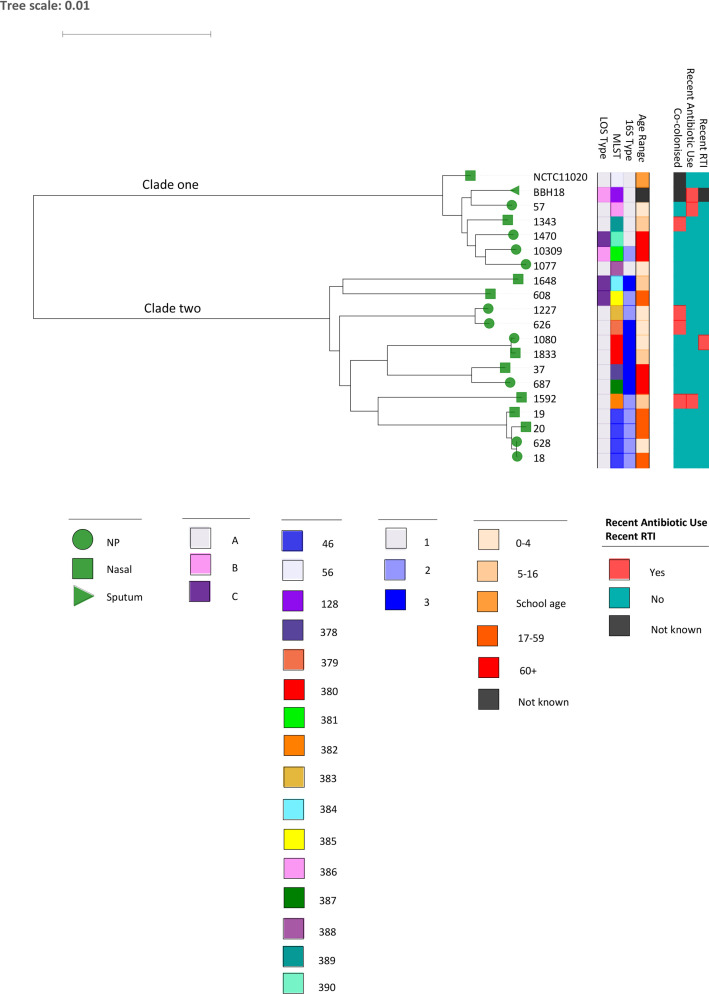
All 18 isolates were assigned a sequence type (ST) (Table 3); four isolates were ST46, all of which were LOS type A and 16S type 2. Two isolates were identified as ST380, both of which were LOS type A and 16S type 3. The remaining isolates were singleton STs. No clear correlation between ST and metadata was observed. Similarly, no clear correlation can be seen between LOS or 16S type and any of the metadata, as shown by the core-genome phylogeny in Fig. 1. As the subset of M. catarrhalis was chosen to ensure we had isolates obtained from participants in the following age ranges, 0–4 years, 5–16 years, 17–59 years and 60+ years, no associations can be made between carriage and prior antibiotic use, respiratory infection and vaccine status. All M. catarrhalis isolates from the study and all related metadata would be needed for such associations to be made.
Table 3.
Virulence and typing results
|
Isolate |
BBH18 |
NCTC 11020 |
57 |
1343 |
1470 |
10 309 |
1077 |
1648 |
608 |
1227 |
626 |
1080 |
1833 |
37 |
687 |
1592 |
19 |
20 |
628 |
18 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
copB |
|||||||||||||||||||||
|
ompCD |
|||||||||||||||||||||
|
ompE |
|||||||||||||||||||||
|
ompG1a |
|||||||||||||||||||||
|
ompG1b |
|||||||||||||||||||||
|
mid/hag |
|||||||||||||||||||||
|
mcaP |
|||||||||||||||||||||
|
m35 |
|||||||||||||||||||||
|
mhaB1 |
|||||||||||||||||||||
|
mhaB2 |
|||||||||||||||||||||
|
mhaC |
|||||||||||||||||||||
|
tbpA |
|||||||||||||||||||||
|
tbpB |
|||||||||||||||||||||
|
lbpA |
|||||||||||||||||||||
|
lbpB |
|||||||||||||||||||||
|
msp22 |
|||||||||||||||||||||
|
msp75 |
|||||||||||||||||||||
|
msp78 |
|||||||||||||||||||||
|
afeA |
|||||||||||||||||||||
|
mcmA |
|||||||||||||||||||||
|
mclS |
|||||||||||||||||||||
|
pilA |
|||||||||||||||||||||
|
pilQ |
|||||||||||||||||||||
|
pilT |
|||||||||||||||||||||
|
modM |
|||||||||||||||||||||
|
oppA |
|||||||||||||||||||||
|
sbp2 |
|||||||||||||||||||||
|
LOS type |
B |
A |
A |
A |
C |
B |
A |
C |
C |
A |
A |
A |
A |
A |
A |
A |
A |
A |
A |
A |
|
|
MLST |
128 |
56 |
386 |
389 |
390 |
381 |
388 |
384 |
385 |
383 |
379 |
380 |
380 |
378 |
387 |
382 |
46 |
46 |
46 |
46 |
|
|
16S type |
1 |
1 |
1 |
1 |
1 |
2 |
1 |
3 |
2 |
2 |
3 |
3 |
3 |
3 |
3 |
2 |
2 |
2 |
2 |
2 |
|
|
β-Lactamase (bla BRO) |
2 |
1 |
1 |
2 |
1 |
1 |
2 |
2 |
1 |
1 |
1 |
1 |
2 |
1 |
2 |
1 |
1 |
1 |
1 |
||
|
Age (years) |
School age |
2 |
6 |
81 |
75 |
4 |
6 |
34 |
1 |
1 |
2 |
7 |
81 |
85 |
5 |
41 |
43 |
3 |
26 |
||
|
Clade |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
|
|
Gene presence |
LOS type |
MLST |
16S type |
β-Lactamase gene |
Age range (years) |
Core-genome phylogeny clade |
|||||||||||||||
|
Present |
A |
46 |
1 |
bla BRO-1 |
0–4 |
1 |
|||||||||||||||
|
Absent |
B |
56 |
2 |
bla BRO-2 |
5–16 |
2 |
|||||||||||||||
|
C |
128 |
3 |
17–59 |
||||||||||||||||||
|
378 |
60+ |
||||||||||||||||||||
|
379 |
|||||||||||||||||||||
|
380 |
|||||||||||||||||||||
|
381 |
|||||||||||||||||||||
|
382 |
|||||||||||||||||||||
|
383 |
|||||||||||||||||||||
|
384 |
|||||||||||||||||||||
|
385 |
|||||||||||||||||||||
|
386 |
|||||||||||||||||||||
|
387 |
|||||||||||||||||||||
|
388 |
|||||||||||||||||||||
|
389 |
|||||||||||||||||||||
|
390 |
|||||||||||||||||||||
Fig. 1.
Core-genome phylogeny.
Distribution of LOS types
In silico analysis of the glycosyltransferase (lgt) genes showed that 77.8 % (14/18) of isolates were LOS type A, 5.6 % (1/18) were LOS type B and 16.7 % (3/18) were LOS type C. Overall, 90 % (9/10) of M. catarrhalis isolated from children were LOS A, with the remaining 10 % (1/10) being LOS C, whilst 62.5 % (5/8) of M. catarrhalis isolated from adults were LOS A, 12.5 % (1/8) were LOS B and 25 % (2/8) were LOS C. When distribution was considered by age range, all M. catarrhalis isolated from 0 to 4 year olds (6/6) were identified as LOS type A. Of the isolates from the 5–16 age group, 75 % (3/4) were LOS type A, with the remainder being LOS type C. This matches the data from 17 to 59 year olds for whom LOS type A represented 75 % (3/4) of the isolates with the remaining 25 % (1/4) being LOS type C. Of the four isolates from the 60+ age group, 50 % (2/4) were LOS type A, with 25 % (1/4) being type B and 25 % (1/4) type C.
Prior publications suggest that the majority of clinical isolates express LOS type A (61–70 %) while few express LOS type C (2–7 %) [22, 37, 48, 49]. The data here agree with regards to the majority prevalence of LOS type A, regardless of the age of the carrier. However, in contrast, our data show a much lower prevalence of LOS type B (5.6 % here versus 19–30 %) and a much higher prevalence of LOS type C (16.7 % here versus 2–7 %) [22, 37, 48, 49]. There are numerous reasons why the results in this dataset may differ to that previously published, firstly the low number of isolates in our dataset. Alternatively, isolates in numerous prior publications are now at least 16 years old [22, 37, 48]; therefore, previously published LOS prevalences may not be reflective of current epidemiology. Conversely, dissimilarities could be due to geographical differences in the distribution on LOS types, or differences in carriage versus clinical isolates.
Research using clinical isolates from the USA, showed higher proportions of LOS type A in M. catarrhalis isolates from adults versus isolates from children (81 % in adults versus 64 % in children) and a lower proportion of LOS type B in adults versus children (15 % in adults versus 34 % in children) [37]. However, our carriage data show an opposite trend, which is comparable to clinical data obtained globally, which observed that 81 % of M. catarrhalis from children were LOS type A whilst the percentage was lower (63%) in adults. This was reversed for LOS B where 13 % of isolates from children were LOS type B whilst the percentage was higher (28%) for isolates from adults, again fitting the trend (but not the values) seen in our data [22]. Consequently, there may be little difference in distribution of LOS in carriage and disease, as suggested by Mitov et al. [49]. This highlights the potential use of carriage data to provide insight into disease, certainly for LOS distributions. Furthermore, the majority of clinical isolates may be LOS type A, not because strains of this type are more pathogenic, but simply because they are more prevalent.
Distribution of 16S types
In total, 22.2 % (4/18) of isolates were 16S type 1, 44.4 % (8/18) were 16S type 2 and 33.3 % (6/18) were 16S type 3. It has previously been reported that 92 % of clinical isolates are 16S type 1, 4 % type 2 and 4% type 3 [22]. The proportion of 16S type 1 seen here is substantially lower (22.2%) with a higher prevalence of type 2 and 3 isolates. These differences could be a result of our data being based on carriage isolates.
Overall, 30 % (3/10) of isolates from children were 16S type 1, 30 % (3/10) were 16S type 2 and 40 % (4/10) were 16S type 3, whilst 12.5 % (1/8) of isolates from adults were 16S type 1, 62.5 % (5/8) were 16S type 2 and 25 % (2/8) were 16S type 3. When distribution of 16S type is considered by set age range, an equal proportion of 16S types was found in young children (0–4 years old); 33.3 % (2/6) for each. In comparison, 25 % (1/4) of the M. catarrhalis from older children aged 5–16 were identified as 16S type 1, 25 % (1/4) as type 2 and 50 % (2/4) as type 3. All isolates (n=4) from adults aged 17–59 were 16S type 2. For the 60+ age range, 25 % (1/4) of isolates were identified as 16S type 1, 25 % (1/4) as type 2 and 50 % (2/4) as type 3.
LOS versus 16S type
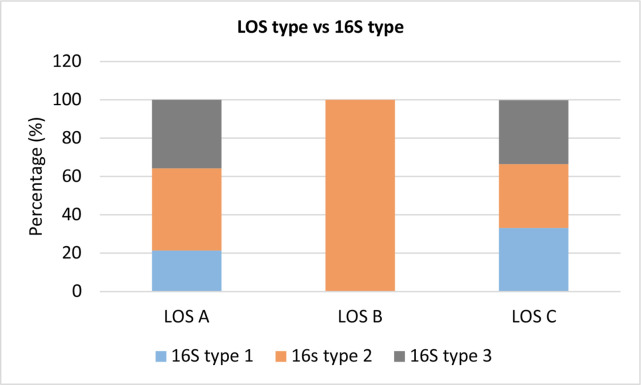
From the 14 isolates identified as LOS type A, 21.4 % (3/14) were found to be 16S type 1, 42.9 % (6/14) were identified as 16S type 2 and 35.7 % (5/14) 16S type 3. The LOS type B isolate was found to be 16S type 2. From the 3 LOS type C isolates, 33.3 % (1/3) were identified as 16S type 1, 33.3 % (1/3) were identified as 16S type 2 and 33.3 % (1/3) identified as 16S type 3. This is visualized in Fig. 2.
Fig. 2.
A graph illustrating the distribution of 16S types found for each LOS type.
Previous research found 91% of LOS type A M. catarrhalis were 16S type 1, 4 % were 16S type 2 and 5 % were 16S type 3 [22]. Here, however, a much lower proportion (21.4%) of the LOS type A isolates were also identified as 16S type 1 (3/14), with 42.9 % (6/14) being 16S type 2 and 35.7 % (5/14) identified as 16S type 3.
It was believed that LOS B was exclusively associated with 16S type 1, as previously 100 % of isolates were identified as such [22]. However, the LOS type B isolate from this study was identified as 16S type 2. This study has, therefore, importantly highlighted that LOS B serotypes may not exclusively be associated with 16S type 1, although further phenotypic analysis is required for confirmation of LOS expression.
Previous data show M. catarrhalis LOS type C isolates to have an even split, with one half reported as 16S type 1 and the other 16S type 2 [22]. Our data, however, suggest an even split across all 16S types with 33.3 % (1/3) being attributed to all three types.
Pangenome
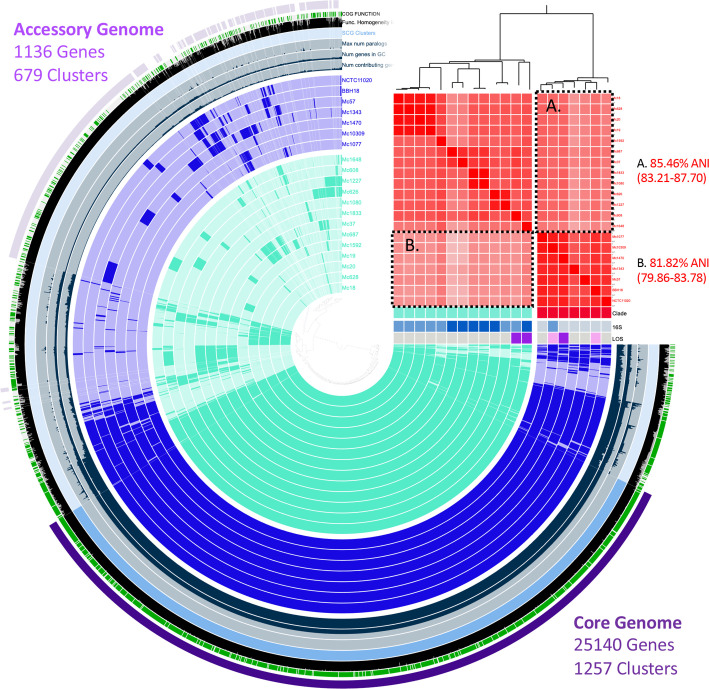
The M. catarrhalis pangenome is shown in Fig. 3. A total of 36 535 genes were used in the analysis, resulting in 2776 gene clusters. The core genome (genes occurring in all strains) was 68.8 %, comprising 25 140 genes in 1257 clusters. Genes that occurred in less <15 % of isolates (n=3) equated to 1136 genes in 679 clusters.
Fig. 3.
Pangenome of M. catarrhalis . Red-red clade, 93.91 % (91.49–96.32); blue to blue clade, 89.15 % (84.88–93.41); red to blue (box A in the figure); and blue to red (box B) are shown in the figure.
The functional enrichment analysis highlights the presence of multiple virulence genes that differentiate clade one and two (Table 4). There were 10 genes that were present in all seven clade one M. catarrhalis isolates, but were absent from all 13 clade two M. catarrhalis isolates. Genes only present in clade one seem to be involved in adaptation and the formation of resistance mechanisms. rayt encodes a rep element-mobilizing transposase that catalyses cleavage and recombination of bacterial interspersed mosaic elements (BIMEs), which may be important in the creation of BIME variability and amplification. BIMEs also act as binding sites for several proteins and are thought to be involved in organization of the chromosome and transcription termination [50]. Moreover, many of the genes in this clade are involved in the biosynthesis of antibiotics. For instance, the alkaline phosphatase (AlkP) family catalyse the hydrolysis of polyphosphates and phosphonates into antibacterial agents. Enzymes in this family are known to be involved in the biosynthesis of antibiotics such as streptomycin, bialaphos and mitomycin [51, 52]. Furthermore, PhoX family phosphatases catalyse the hydrolysis of phosphomonoester to a phosphate [53], whilst membrane-associated phospholipid phosphatases have a similar function [54]. As the name suggests, the ABC-type phosphate transport system and its ATPase component transports phosphate into the cell and hydrolyse it [55]. Restriction endonuclease Mrr cleaves DNA leaving double-stranded fragments with terminal 5'-phosphates. Mrr appears to be a final component of a bacterial SOS response to DNA damage. It is also an essential mechanism in generating genetic variability that facilitates adaptation and the development of resistance [56–58].
Table 4.
Functional enrichment analysis
Only significant data are reported in this table (those with a P value <0.05). All had P values of 0.00, corrected P values of 0.03 and an enrichment score of 3.49.
|
Clade |
COGs function |
Portion occurrence in group |
Portion occurrence outside of group |
Occurrence in group |
Occurrence outside of group |
Gene clusters IDs |
Core in group |
Core |
|---|---|---|---|---|---|---|---|---|
|
C2 |
ABC-type bacteriocin/lantibiotic exporters, contain an N-terminal double-glycine peptidase domain |
1 |
0.14 |
13 |
1 |
GC_00001559, GC_00002662 |
TRUE |
FALSE |
|
C2 |
Uncharacterized membrane protein YjiH, contains nucleoside recognition GATE domain |
1 |
0 |
13 |
0 |
GC_00001582 |
TRUE |
FALSE |
|
C2 |
Predicted transposase YdaD |
1 |
0 |
13 |
0 |
GC_00001531, GC_00001547 |
TRUE |
FALSE |
|
C2 |
DNA-binding transcriptional regulator, XRE-family HTH domain |
1 |
0.14 |
13 |
1 |
GC_00001561, GC_00002218, GC_00002755 |
TRUE |
FALSE |
|
C2 |
Uncharacterized membrane protein YczE |
1 |
0.14 |
13 |
1 |
GC_00001541 |
TRUE |
FALSE |
|
C2 |
Protein involved in initiation of plasmid replication |
1 |
0.14 |
13 |
1 |
GC_00001513, GC_00001543, GC_00001550, GC_00001716, GC_00002180, GC_00002518, GC_00002635, GC_00002672 |
TRUE |
FALSE |
|
C2 |
Cys-tRNA(Pro) deacylase, prolyl-tRNA editing enzyme YbaK/EbsC |
1 |
0 |
13 |
0 |
GC_00001567 |
TRUE |
FALSE |
|
C2 |
Phage-related protein, tail component |
0.92 |
0.14 |
12 |
1 |
GC_00000083, GC_00001528, GC_00001843, GC_00002183, GC_00002230, GC_00002339 |
FALSE |
FALSE |
|
C2 |
Predicted epimerase YddE/YHI9, PhzF superfamily |
1 |
0 |
13 |
0 |
GC_00001569 |
TRUE |
FALSE |
|
C2 |
Cell division protein DamX, binds to the septal ring, contains C-terminal SPOR domain |
1 |
0 |
13 |
0 |
GC_00001573 |
TRUE |
FALSE |
|
C2 |
NADP-dependent 3-hydroxy acid dehydrogenase YdfG |
1 |
0 |
13 |
0 |
GC_00001588 |
TRUE |
FALSE |
|
C2 |
DNA-binding transcriptional regulator, XRE family |
1 |
0 |
13 |
0 |
GC_00001511 |
TRUE |
FALSE |
|
C2 |
Uncharacterized conserved protein, phosphatidylethanolamine-binding protein (PEBP) family |
1 |
0 |
13 |
0 |
GC_00001584 |
TRUE |
FALSE |
|
C1 |
REP element-mobilizing transposase RayT |
1 |
0 |
7 |
0 |
GC_00001768, GC_00002736 |
TRUE |
FALSE |
|
C1 |
ABC-type phosphate transport system, permease component |
1 |
0 |
7 |
0 |
GC_00001724, GC_00001726 |
TRUE |
FALSE |
|
C1 |
Membrane-associated phospholipid phosphatase |
1 |
0 |
7 |
0 |
GC_00001777 |
TRUE |
FALSE |
|
C1 |
Predicted pyrophosphatase or phosphodiesterase, AlkP superfamily |
1 |
0 |
7 |
0 |
GC_00001732 |
TRUE |
FALSE |
|
C1 |
Secreted phosphatase, PhoX family |
1 |
0 |
7 |
0 |
GC_00001783 |
TRUE |
FALSE |
|
C1 |
Predicted endonuclease, GIY-YIG superfamily |
1 |
0 |
7 |
0 |
GC_00001627 |
TRUE |
FALSE |
|
C1 |
ABC-type spermidine/putrescine transport system, permease component II |
1 |
0 |
7 |
0 |
GC_00001725 |
TRUE |
FALSE |
|
C1 |
Restriction endonuclease Mrr |
1 |
0 |
7 |
0 |
GC_00001772 |
TRUE |
FALSE |
|
C1 |
ABC-type phosphate transport system, ATPase component |
1 |
0 |
7 |
0 |
GC_00001727 |
TRUE |
FALSE |
|
C1 |
DNA modification methylase |
1 |
0.08 |
7 |
1 |
GC_00001685 |
TRUE |
FALSE |
|
C1 |
Dienelactone hydrolase |
1 |
0 |
7 |
0 |
GC_00001721 |
TRUE |
FALSE |
Colour highlights the genes present in all isolates in one clade but completely absent in the other clade. Two separate colours were used to differentiate those in clade one (red) and those in clade two.(blue).
There were eight genes that were present in all 13 clade two M. catarrhalis , but were absent in all 7 clade one M. catarrhalis . Genes only present in clade two are involved in stress responses and replication. For instance, ydaD is induced as a response to different stress conditions such as heat shock, oxidative stress, glucose limitation and oxygen limitation [59]. Furthermore, YdfG produces a dehydrogenase/reductase that catalyses oxidation–reduction reactions, this counteracts oxidative stress [60]. The DNA-binding transcriptional regulators, the XRE family, have been shown to play a role in oxidative and high temperature stress tolerance [61]. yjiH encodes a nucleoside recognition pore and gate family inner membrane transporter and YbaK functions in trans to edit the amino acid from incorrectly charged Cys-tRNA(Pro) via a Cys-tRNA(Pro) deacylase activity [62, 63]. YddE/YHI9 epimerases (part of the PhzF family) are involved in the production of phenazine derivative antibiotic compounds [64], whilst DamX protein plays a role in DNA adenine methylation, carbohydrate metabolism and tRNA charging [65].
Antimicrobial-resistance (AMR) profiles
No AMR genes were detected using srst2 with the associated ARGannot. ARGannot uses an extensive list of antibiotic-resistance genes (n=1689) collected from published data and online resources with the nucleotide and protein sequences taken from the NCBI GenBank database including bla genes [66]. However, ARGannot is not specific for M. catarrhalis so may not include bla BRO, which comparison with other β-lactamases suggests is unique [67]. Again, whilst not specific for M. catarrhalis , ResFinder identifies the presence of an extensive list of acquired AMR genes and is continuously updated as new resistance genes are identified. β-Lactam-resistance genes were found in all 18 isolates (100%), with these genes having predicted phenotypes for resistance against ampicillin, penicillin, piperacillin and amoxicillin [68]. The production of β-lactamase is known as a leading source of resistance for M. catarrhalis . This enzyme digests β-lactam antibiotics rendering them ineffective, conferring resistance to antibiotics such as penicillins and cephamycins. M. catarrhalis produces two distinct β-lactamases: BRO-1 and BRO-2 [69]. A total of 72 % (13/18) of isolates were positive for the β-lactamase-encoding gene (bla) BRO-1 and 28 % (5/18) for bla BRO-2. No isolates had both BRO-1 and BRO-2 genes.
Additionally, srst2 was used to map the bla gene in all isolates again to confirm gene presence and BRO-1 or BRO-2 status. Gene mapping supported the findings from ResFinder, showing all 18 isolates were bla positive. The 100 % prevalence of β-lactamase resistance genes is comparable to the 98 % seen in other publications [49], and in agreement with the widespread nature of β-lactam resistance in M. catarrhalis . We observed a lower proportion of bla BRO-1 isolates than previously reported: 72 % (13/18) in this dataset versus the 91 % previously seen [49]. It remains to be seen whether this difference in BRO-1 and BRO-2 prevalence is a reflection of differences between carriage and disease. However, it potentially enforces the importance of β-lactamase resistance as a virulence factor giving M. catarrhalis a potential clinical edge for causing disease; particularly as bla BRO-1 is the stronger of the two, producing more β-lactamase than BRO-2 counterparts [67, 70, 71]. What is interesting is that prior data has suggested that BRO-2 isolates are associated with the 16S type 1 lineage [28]; however, this was not seen here (Table 3). To confirm the expression of β-lactamase, isolates were tested with β-lactamase identification sticks. All 18 isolates tested positive for the production of β-lactamase. Fortunately, β-lactamase production does not automatically mean resistance, β-lactamase does not render all β-lactam antibiotics ineffective; second-, third- and fourth-generation cephalosporins are still effective [72].
When tested phenotypically, all isolates were sensitive to amoxicillin/clavulanic acid, cefotaxime, ceftriaxone, erythromycin, tetracycline, chloramphenicol, ciprofloxacin and meropenem. These antibiotics were used to test resistance to cephalosporins, macrolide, tetracycline, chloramphenicol, fluoroquinolones and carbapenem antibiotics, whilst amoxicillin/clavulanic acid is a combination antibiotic containing potassium clavulanate, a β-lactamase inhibitor; thus, is commonly used to treat β-lactam-resistant bacteria. Although cefotaxime and ceftriaxone are β-lactam antibiotics, they are third-generation cephalosporins, which are known to still be effective for M. catarrhalis , so data are in keeping with current literature.
Distribution of virulence factors
Table 3 highlights the presence and absence of virulence genes tested in all isolates, including reference strains BBH18 and NCTC 11020. copB, ompCD, ompE, ompG1a, ompG1b, mcaP, m35, tbpA, lbpA, tbpB, lbpB, msp75, msp78, afeA, pilA, pilQ, pilT, oppA and sbp2 were present in 100 % of the carriage isolates, which is comparable to other studies, most of which focus on clinical isolates [21, 22, 28, 73, 74]. In total, 88.9 % (16/18) of isolates contained mid/hag, this is in agreement with prior publications that report mid/hag to be present in 80 % of child carriage isolates [27] and in up to 100 % of clinical strains [29]. The two isolates ‘missing’ did have signs of the gene; however, they were below the coverage cut-off. This either means the gene is only partially present or could mean that the gene is present but is altered. Less is published on the prevalence of mcmA and mclS; however, these were also present in all isolates here.
Due to the overlap of gene sequence between uspA1 and uspA2H, and uspA2 and uspA2H, it was not possible to confirm the presence or absence of uspA genes using read mapping (Table 5). To try to clarify gene presence, coverage was visually inspected in Tablet v. 1.19.09.93 [75] and in silico PCR using published primers was attempted; however, results remain unclear.
Table 5.
uspA read mapping results (percentage of gene coverage)
|
Accession numbers |
Gene |
Percentage of gene coverage |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
57 |
1343 |
1470 |
10 309 |
1077 |
1648 |
608 |
1227 |
626 |
1080 |
1833 |
37 |
687 |
1592 |
19 |
20 |
628 |
18 |
||
|
uspA1 |
42.9 |
46.6 |
72.0 |
17.6 |
70.5 |
49.2 |
69.9 |
0.0 |
0.0 |
63.5 |
10.6 |
26.8 |
15.4 |
29.5 |
41.9 |
25.1 |
22.6 |
48.9 |
|
|
61.8 |
43.6 |
93.5 |
62.8 |
81.6 |
59.0 |
48.6 |
10.9 |
14.6 |
56.2 |
52.4 |
63.7 |
65.8 |
54.4 |
58.9 |
63.5 |
62.0 |
61.3 |
||
|
96.4 |
79.0 |
73.3 |
76.7 |
69.3 |
42.8 |
53.0 |
9.5 |
7.6 |
52.2 |
48.8 |
49.1 |
54.7 |
42.3 |
44.9 |
45.4 |
45.9 |
41.1 |
||
|
55.2 |
66.7 |
78.0 |
41.7 |
82.8 |
50.0 |
46.2 |
8.0 |
10.3 |
55.9 |
33.4 |
51.7 |
62.0 |
54.2 |
48.1 |
51.1 |
54.1 |
50.8 |
||
|
uspA2 |
62.6 |
67.1 |
58.1 |
48.2 |
79.9 |
39.7 |
24.1 |
31.2 |
47.5 |
39.7 |
43.0 |
36.0 |
59.9 |
35.6 |
31.0 |
30.4 |
35.2 |
35.2 |
|
|
24.9 |
43.2 |
30.8 |
43.5 |
53.6 |
34.4 |
57.9 |
38.1 |
31.4 |
25.9 |
31.8 |
39.6 |
35.6 |
33.4 |
32.4 |
33.1 |
34.8 |
32.5 |
||
|
32.5 |
33.8 |
38.7 |
15.0 |
39.5 |
41.2 |
43.1 |
34.0 |
33.5 |
37.5 |
30.8 |
42.3 |
40.4 |
34.6 |
25.9 |
28.0 |
38.9 |
38.7 |
||
|
99.0 |
47.9 |
29.3 |
36.6 |
98.4 |
50.3 |
37.9 |
30.0 |
33.3 |
33.8 |
41.9 |
36.2 |
33.6 |
47.0 |
28.8 |
28.3 |
32.8 |
31.2 |
||
|
40.7 |
86.5 |
34.0 |
46.9 |
55.6 |
49.1 |
52.2 |
38.2 |
39.9 |
37.8 |
34.7 |
49.4 |
40.7 |
41.9 |
35.6 |
36.9 |
42.6 |
40.0 |
||
|
uspA2H |
88.9 |
75.1 |
100.0 |
100.0 |
83.7 |
65.0 |
73.1 |
24.6 |
23.7 |
65.5 |
66.3 |
63.1 |
63.0 |
80.8 |
64.6 |
68.5 |
75.0 |
73.4 |
|
|
63.8 |
55.4 |
71.3 |
73.8 |
77.9 |
54.9 |
58.4 |
19.1 |
26.9 |
77.5 |
73.2 |
63.4 |
24.9 |
31.4 |
33.3 |
38.6 |
47.6 |
31.7 |
||
|
32.1 |
38.7 |
54.2 |
73.4 |
36.8 |
35.0 |
27.1 |
10.4 |
18.0 |
23.2 |
19.6 |
21.9 |
24.4 |
25.4 |
22.3 |
25.8 |
23.5 |
24.7 |
||
|
29.4 |
44.9 |
78.6 |
54.4 |
39.1 |
31.9 |
23.9 |
13.7 |
13.9 |
20.4 |
26.7 |
22.8 |
24.3 |
36.6 |
29.0 |
31.9 |
29.6 |
33.5 |
||
|
51.4 |
45.5 |
68.7 |
75.8 |
47.1 |
32.1 |
30.0 |
22.6 |
21.4 |
25.8 |
26.1 |
27.5 |
24.1 |
44.6 |
37.9 |
33.6 |
41.1 |
40.8 |
||
The M. catarrhalis two-partner secretion (TPS) system comprises MhaC (transporter), MhaB1 (exoprotein) and MhaB2 (exoprotein) [28, 76, 77]. Here, mhaB1 was present in 44.4 % (8/18) of isolates, mhaB2 was present in none of the isolates and mhaC was present in 55.6 % (10/18) of isolates. Other studies have shown both MhaB1 and MhaB2 expression in M. catarrhalis strain O35E; it is, therefore, interesting that mhaB2 was not found in any of our isolates [76]. Our data further contrast previous research, which showed 100 % presence of mhaC, mhaB1 and mhaB2 in isolates, although these were of clinical origin [28, 77]. Overall, mhaB1 and mhaC were present in 30 % (3/10) and 50 % (5/10) of isolates from children and 62.5 % (5/8) and 62.5 % (5/8) of isolates from adults, respectively. When distribution is considered by age range, mhaB1 and mhaC were present in 33.3 % (2/6) and 50 % (3/6) of isolates from 0 to 4 year olds, 25 % (1/4) and 50 % (2/4) of isolates from 5 to 16 year olds, 75 % (3/4) and 75 % (3/4) of isolates from 17 to 59 year olds, and 50 % (2/4) and 50 % (2/4) of isolates from 60+ year.
Both tbpA and tbpB were present in all isolates, which is comparable to other studies [28, 78]. Again, in agreement with prior research, lbpA and lbpB were present in 100 % of isolates [28, 79]. msp22 was present in 94.4 % (17/18) of isolates, which is lower than the 100 % found in other studies [80]; however, the isolate classed as negative for msp22 did have a coverage of 82 %, which was below the cut-off. When visually inspected in Tablet v. 1.19.09.93 [75] in the isolates ‘missing’ msp22, the gene appeared partially present; however, it showed a lack of coverage in the final ~80 bp. Overall, 90 % (9/10) of isolates from children and 100 % (8/8) of isolates from adults possessed msp22. When distribution is considered by age range 83.3 % (5/6) of isolates from 0 to 4 year olds, 100 % (4/4) of isolates from 5 to 16, 100 % (4/4) of isolates from 17 to 59 and 100 % (4/4) of isolates from 60+ year olds possessed msp22.
modM was present in 61.1 % (11/18) of isolates, much lower than previous reports suggesting modM to be present in 100 % of the carriage and clinical strains tested [81]. Overall, 70 % (7/10) of isolates from children and 62.5 % (5/8) of isolates from adults possessed modM here. When distribution is considered by age range, 50 % (3/6) of isolates from 0 to 4 year olds, 75 % (3/4) of isolates from 5 to 16, 25 % (1/4) of isolates from 17 to 59 and 100 % (4/4) of isolates from 60+ year olds possessed modM. It is unclear why the data from this study should be different to prior publication [81].
From the data presented here, there appears to be no association between LOS type, MLST and 16S type and the presence or absence of any of the virulence factors; however, the limited number of isolates analysed impedes the conclusions that can be drawn. Further research is needed with a greater number of samples. An important caveat that should be made is that just because homology exists between a reference gene and an isolate’s genome, it does not mean that the gene exists in an ORF in that genome. The results here highlight potential gene presence but not evidence of intact ORFs.
Conclusion
This is the only study, to our knowledge, to focus on the epidemiology of and distribution of virulence factors in M. catarrhalis carriage isolates from all ages. As carriage is considered a prerequisite for disease [31], it is important to better understand the epidemiology of M. catarrhalis in the community. Especially as no particular type, subpopulation nor strain has been implicitly associated with disease or virulence. The key limitation of this study is the low number of isolates analysed, which restricts how accurately the data reflects the epidemiology and distribution of virulence factors for M. catarrhalis . Therefore, this paper should be followed by further analysis on a larger set of community isolates. This study does, however, provide a novel and timely insight into the types of M. catarrhalis carried, the AMR profiles of such isolates and the distribution of virulence factors, which can be used to aid our understanding of the disease potential of community isolates and to inform vaccine development. Further to this, a comparison of carriage and clinical isolates would be beneficial and likely to provide additional data to facilitate a better understanding of the differences between carriage and disease and the identification of markers of pathogenic strains of M. catarrhalis .
Funding information
D.E.M. and K.L.O. were supported by the Solent NHS Trust in the form of NIHR Research Capability Funding. D.W.C. is supported by funding from NIHR Southampton Biomedical Research Centre. The study and collection of the M. catarrhalis samples was made possible via investigator-initiated research grants from the Bupa Foundation to S.C.C.
Acknowledgements
We would like to acknowledge Neil Churchill and Keith Manship, the Patient and Public Involvement (PPI) representatives involved in the design of the ‘Analysis of the microbial community of the upper respiratory tract to support the development of effective vaccine policy’ study, REC reference 11/SC/0518, from which the isolates in this study were obtained. We would also like to acknowledge the NIHR Southampton Clinical Research Facility and CRN Wessex for their support for sample collection and study oversight.
Author contributions
D.E.M., conceptualization, data curation, formal analysis, investigation, methodology, software, project administration, visualization, writing – original draft, and writing – review and editing. K.L.O., formal analysis, methodology, software, and writing – review and editing. D.W.C., conceptualization, formal analysis, methodology, software, supervision, visualization, and writing – review and editing. S.C.C., conceptualization, funding acquisition, methodology, project administration, resources, supervision, and writing – review and editing. SMART group, funding acquisition, methodology and writing – review.
Conflicts of interest
S.C.C. acts as principal investigator for clinical trials and other studies conducted on behalf of University Hospital Southampton NHS Foundation Trust/University of Southampton that are sponsored by vaccine manufacturers, but receives no personal payments from them. S.C.C. has participated in advisory boards for vaccine manufacturers but received no personal payments for this work. S.C.C. has received financial assistance from vaccine manufacturers to attend conferences. All grants and honoraria are paid into accounts within the respective NHS Trusts or Universities, or to independent charities. All other authors have no conflicts of interest.
Ethical statement
UK NHS Research Ethics 11/SC/0518. Informed consent was gained in accordance with good clinical practice and was followed for all participants, with parents/guardians providing consent for those aged 16 years and younger. Following consent and the completion of a questionnaire, the samples were taken from participants.
Footnotes
Abbreviations: AMR, antimicrobial resistance; AOM, acute otitis media; BIME, bacterial interspersed mosaic element; COGs, Clusters of Orthologous Genes; LOS, lipooligosaccharide; MLST, multilocus sequence type; NIHR, National Institute for Health and Care Research; NP, nasopharyngeal; OM, otitis media; RTI, respiratory tract infection; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files.
References
- 1.Aebi C. Moraxella catarrhalis – pathogen or commensal? Adv Exp Med Biol. 2011;697:107–116. doi: 10.1007/978-1-4419-7185-2_9. [DOI] [PubMed] [Google Scholar]
- 2.Frosch PKW. Die Mikrokokken. 1896. [Google Scholar]
- 3.Catlin BW. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin Microbiol Rev. 1990;3:293–320. doi: 10.1128/CMR.3.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feder HM, Garibaldi RA. The significance of nongonococcal, nonmeningococcal Neisseria isolates from blood cultures. Rev Infect Dis. 1984;6:181–188. doi: 10.1093/clinids/6.2.181. [DOI] [PubMed] [Google Scholar]
- 5.McLeod DT, Ahmad F, Power JT, Calder MA, Seaton A. Bronchopulmonary infection due to Branhamella catarrhalis . Br Med J (Clin Res Ed) 1983;287:1446–1447. doi: 10.1136/bmj.287.6403.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson MA, Drew WL, Roberts M. Branhamella (Neisseria) catarrhalis – a lower respiratory tract pathogen? J Clin Microbiol. 1981;13:1066–1069. doi: 10.1128/jcm.13.6.1066-1069.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onofrio JM, Shulkin AN, Heidbrink PJ, Toews GB, Pierce AK. Pulmonary clearance and phagocytic cell response to normal pharyngeal flora. Am Rev Respir Dis. 1981;123:222–225. doi: 10.1164/arrd.1981.123.2.222. [DOI] [PubMed] [Google Scholar]
- 8.Hager H, Verghese A, Alvarez S, Berk SL. Branhamella catarrhalis respiratory infections. Rev Infect Dis. 1987;9:1140–1149. doi: 10.1093/clinids/9.6.1140. [DOI] [PubMed] [Google Scholar]
- 9.McNeely DJ, Kitchens CS, Kluge RM. Fatal Neisseria (Branhamella) catarrhalis pneumonia in an immunodeficient host. Am Rev Respir Dis. 1976;114:399–402. doi: 10.1164/arrd.1976.114.2.399. [DOI] [PubMed] [Google Scholar]
- 10.Bosch AATM, Biesbroek G, Trzcinski K, Sanders EAM, Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9:e1003057. doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy TF, Bakaletz LO, Smeesters PR. Microbial interactions in the respiratory tract. Pediatr Infect Dis J. 2009;28:S121–S126. doi: 10.1097/INF.0b013e3181b6d7ec. [DOI] [PubMed] [Google Scholar]
- 12.Ngo CC, Massa HM, Thornton RB, Cripps AW. Predominant bacteria detected from the middle ear fluid of children experiencing otitis media: a systematic review. PLoS One. 2016;11:e0150949. doi: 10.1371/journal.pone.0150949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monasta L, Ronfani L, Marchetti F, Montico M, Vecchi Brumatti L, et al. Burden of disease caused by otitis media: systematic review and global estimates. PLoS One. 2012;7:e36226. doi: 10.1371/journal.pone.0036226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faden H, Duffy L, Boeve M. Otitis media: back to basics. Pediatr Infect Dis J. 1998;17:1105–1112. doi: 10.1097/00006454-199812000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Murphy TF, Parameswaran GI. Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis. 2009;49:124–131. doi: 10.1086/599375. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson TMA, Aris E, Bourne S, Clarke SC, Peeters M, et al. A prospective, observational cohort study of the seasonal dynamics of airway pathogens in the aetiology of exacerbations in COPD. Thorax. 2017;72:919–927. doi: 10.1136/thoraxjnl-2016-209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization The Top 10 Causes of Death 2018 ( http://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death) Geneva: World Health Organization; 2018. [Google Scholar]
- 18.Wirth T, Morelli G, Kusecek B, van Belkum A, van der Schee C, et al. The rise and spread of a new pathogen: seroresistant Moraxella catarrhalis . Genome Res. 2007;17:1647–1656. doi: 10.1101/gr.6122607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl JP, de Vries SPW, Ahmed A, Powell E, Schultz MP, et al. Comparative genomic analyses of the Moraxella catarrhalis serosensitive and seroresistant lineages demonstrate their independent evolution. Genome Biol Evol. 2016;8:955–974. doi: 10.1093/gbe/evw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaller A, Troller R, Molina D, Gallati S, Aebi C, et al. Rapid typing of Moraxella catarrhalis subpopulations based on outer membrane proteins using mass spectrometry. Proteomics. 2006;6:172–180. doi: 10.1002/pmic.200500086. [DOI] [PubMed] [Google Scholar]
- 21.Bootsma HJ, van der Heide HG, van de Pas S, Schouls LM, Mooi FR. Analysis of Moraxella catarrhalis by DNA typing: evidence for a distinct subpopulation associated with virulence traits. J Infect Dis. 2000;181:1376–1387. doi: 10.1086/315374. [DOI] [PubMed] [Google Scholar]
- 22.Verhaegh SJC, Streefland A, Dewnarain JK, Farrell DJ, van Belkum A, et al. Age-related genotypic and phenotypic differences in Moraxella catarrhalis isolates from children and adults presenting with respiratory disease in 2001–2002. Microbiology. 2008;154:1178–1184. doi: 10.1099/mic.0.2007/015057-0. [DOI] [PubMed] [Google Scholar]
- 23.Fomsgaard JS, Fomsgaard A, Høiby N, Bruun B, Galanos C. Comparative immunochemistry of lipopolysaccharides from Branhamella catarrhalis strains. Infect Immun. 1991;59:3346–3349. doi: 10.1128/iai.59.9.3346-3349.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Vries SPW, Bootsma HJ, Hays JP, Hermans PWM. Molecular aspects of Moraxella catarrhalis pathogenesis. Microbiol Mol Biol Rev. 2009;73:389–406. doi: 10.1128/MMBR.00007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez AC, Murphy TF. A Moraxella catarrhalis vaccine to protect against otitis media and exacerbations of COPD: an update on current progress and challenges. Hum Vaccin Immunother. 2017;13:2322–2331. doi: 10.1080/21645515.2017.1356951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez AC, Murphy TF. Potential impact of a Moraxella catarrhalis vaccine in COPD. Vaccine. 2019;37:5551–5558. doi: 10.1016/j.vaccine.2016.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhaegh SJC, Snippe ML, Levy F, Verbrugh HA, Jaddoe VWV, et al. Colonization of healthy children by Moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with Haemophilus influenzae . Microbiology. 2011;157:169–178. doi: 10.1099/mic.0.042929-0. [DOI] [PubMed] [Google Scholar]
- 28.Blakeway LV, Tan A, Peak IRA, Seib KL. Virulence determinants of Moraxella catarrhalis: distribution and considerations for vaccine development. Microbiology. 2017;163:1371–1384. doi: 10.1099/mic.0.000523. [DOI] [PubMed] [Google Scholar]
- 29.Möllenkvist A, Nordström T, Halldén C, Christensen JJ, Forsgren A, et al. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J Bacteriol. 2003;185:2285–2295. doi: 10.1128/JB.185.7.2285-2295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coughtrie AL, Whittaker RN, Begum N, Anderson R, Tuck A, et al. Evaluation of swabbing methods for estimating the prevalence of bacterial carriage in the upper respiratory tract: a cross sectional study. BMJ Open. 2014;4:e005341. doi: 10.1136/bmjopen-2014-005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coughtrie AL, Morris DE, Anderson R, Begum N, Cleary DW, et al. Ecology and diversity in upper respiratory tract microbial population structures from a cross-sectional community swabbing study. J Med Microbiol. 2018;67:1096–1108. doi: 10.1099/jmm.0.000773. [DOI] [PubMed] [Google Scholar]
- 32.Public Health England Identification of Moraxella species and Morphologically Similar Organisms. London: Standards Unit, Microbiology Services, PHE; 2015. [Google Scholar]
- 33.van den Bergh MR, Bogaert D, Dun L, Vons J, Chu MLJN, et al. Alternative sampling methods for detecting bacterial pathogens in children with upper respiratory tract infections. J Clin Microbiol. 2012;50:4134–4137. doi: 10.1128/JCM.02376-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, et al. In: Research in Computational Molecular Biology. Deng M, Jiang R, Sun F, Zhang X, editors. Berlin, Heidelberg: Springer; 2013. Assembling genomes and mini-metagenomes from highly chimeric reads. [Google Scholar]
- 36.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards KJ, Schwingel JM, Datta AK, Campagnari AA. Multiplex PCR assay that identifies the major lipooligosaccharide serotype expressed by Moraxella catarrhalis clinical isolates. J Clin Microbiol. 2005;43:6139–6143. doi: 10.1128/JCM.43.12.6139-6143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, et al. Anvio: an advanced analysis and visualization platform foromics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oxoid Beta-lactamase identification sticks. 2018. http://www.oxoid.com/UK/blue/prod_detail/prod_detail.asp?pr=BR0066&c=UK&lang=EN
- 45.Bushnell B. BBMap. https://sourceforge.net/projects/bbmap/
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaneechoutte M, Verschraegen G, Claeys G, Van Den Abeele AM. Serological typing of Branhamella catarrhalis strains on the basis of lipopolysaccharide antigens. J Clin Microbiol. 1990;28:182–187. doi: 10.1128/jcm.28.2.182-187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitov IG, Gergova RT, Ouzounova-Raykova VV. Distribution of genes encoding virulence factors ompB2, ompCD, ompE, β-lactamase and serotype in pathogenic and colonizing strains of Moraxella catarrhalis . Arch Med Res. 2010;41:530–535. doi: 10.1016/j.arcmed.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Nunvar J, Huckova T, Licha I. Identification and characterization of repetitive extragenic palindromes (REP)-associated tyrosine transposases: implications for REP evolution and dynamics in bacterial genomes. BMC Genomics. 2010;11:44. doi: 10.1186/1471-2164-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galperin MY, Jedrzejas MJ. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins. 2001;45:318–324. doi: 10.1002/prot.1152. [DOI] [PubMed] [Google Scholar]
- 52.Harsági N, Keglevich G. The hydrolysis of phosphinates and phosphonates: a review. Molecules. 2021;26:2840. doi: 10.3390/molecules26102840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaheer R, Morton R, Proudfoot M, Yakunin A, Finan TM. Genetic and biochemical properties of an alkaline phosphatase PhoX family protein found in many bacteria. Environ Microbiol. 2009;11:1572–1587. doi: 10.1111/j.1462-2920.2009.01885.x. [DOI] [PubMed] [Google Scholar]
- 54.Icho T, Raetz CR. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol. 1983;153:722–730. doi: 10.1128/jb.153.2.722-730.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 56.Williams RJ. Restriction endonucleases: classification, properties, and applications. Mol Biotechnol. 2003;23:225–243. doi: 10.1385/mb:23:3:225. [DOI] [PubMed] [Google Scholar]
- 57.Waite-Rees PA, Keating CJ, Moran LS, Slatko BE, Hornstra LJ, et al. Characterization and expression of the Escherichia coli Mrr restriction system. J Bacteriol. 1991;173:5207–5219. doi: 10.1128/jb.173.16.5207-5219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aertsen A, Michiels CW. Mrr instigates the SOS response after high pressure stress in Escherichia coli . Mol Microbiol. 2005;58:1381–1391. doi: 10.1111/j.1365-2958.2005.04903.x. [DOI] [PubMed] [Google Scholar]
- 59.Briolat V, Reysset G. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J Bacteriol. 2002;184:2333–2343. doi: 10.1128/JB.184.9.2333-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujisawa H, Nagata S, Misono H. Characterization of short-chain dehydrogenase/reductase homologues of Escherichia coli (YdfG) and Saccharomyces cerevisiae (YMR226C) Biochim Biophys Acta. 2003;1645:89–94. doi: 10.1016/s1570-9639(02)00533-2. [DOI] [PubMed] [Google Scholar]
- 61.Hu Y, Hu Q, Wei R, Li R, Zhao D, et al. The XRE family transcriptional regulator SrtR in Streptococcus suis is involved in oxidant tolerance and virulence. Front Cell Infect Microbiol. 2018;8:452. doi: 10.3389/fcimb.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruan B, Söll D. The bacterial YbaK protein is a Cys-tRNAPro and Cys-tRNA Cys deacylase. J Biol Chem. 2005;280:25887–25891. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 63.Kumar S, Das M, Hadad CM, Musier-Forsyth K. Aminoacyl-tRNA substrate and enzyme backbone atoms contribute to translational quality control by YbaK. J Phys Chem B. 2013;117:4521–4527. doi: 10.1021/jp308628y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liger D, Quevillon-Cheruel S, Sorel I, Bremang M, Blondeau K, et al. Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily. Proteins. 2005;60:778–786. doi: 10.1002/prot.20548. [DOI] [PubMed] [Google Scholar]
- 65.López-Garrido J, Casadesús J. The DamX protein of Escherichia coli and Salmonella enterica . Gut Microbes. 2010;1:285–288. doi: 10.4161/gmic.1.4.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bootsma HJ, van Dijk H, Verhoef J, Fleer A, Mooi FR. Molecular characterization of the BRO beta-lactamase of Moraxella (Branhamella) catarrhalis . Antimicrob Agents Chemother. 1996;40:966–972. doi: 10.1128/AAC.40.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGregor K, Chang BJ, Mee BJ, Riley TV. Moraxella catarrhalis: clinical significance, antimicrobial susceptibility and BRO beta-lactamases. Eur J Clin Microbiol Infect Dis. 1998;17:219–234. doi: 10.1007/BF01699978. [DOI] [PubMed] [Google Scholar]
- 70.Ejlertsen T, Skov R. The beta-lactamases of Moraxella (Branhamella) catarrhalis isolated from Danish children. APMIS. 1996;104:557–562. doi: 10.1111/j.1699-0463.1996.tb04911.x. [DOI] [PubMed] [Google Scholar]
- 71.Wallace RJ Jr, Steingrube VA, Nash DR, Hollis DG, Flanagan C, et al. BRO beta-lactamases of Branhamella catarrhalis and Moraxella subgenus Moraxella, including evidence for chromosomal beta-lactamase transfer by conjugation in B. catarrhalis, M. nonliquefaciens, and M. lacunata . Antimicrob Agents Chemother. 1989;33:1845–1854. doi: 10.1128/AAC.33.11.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi W, Wen D, Chen C, Yuan L, Gao W, et al. β-Lactamase production and antibiotic susceptibility pattern of Moraxella catarrhalis isolates collected from two county hospitals in China. BMC Microbiol. 2018;18:77. doi: 10.1186/s12866-018-1217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Easton DM, Smith A, Gallego SG, Foxwell AR, Cripps AW, et al. Characterization of a novel porin protein from Moraxella catarrhalis and identification of an immunodominant surface loop. J Bacteriol. 2005;187:6528–6535. doi: 10.1128/JB.187.18.6528-6535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luke NR, Howlett AJ, Shao J, Campagnari AA. Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation. Infect Immun. 2004;72:6262–6270. doi: 10.1128/IAI.72.11.6262-6270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, et al. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 76.Balder R, Hassel J, Lipski S, Lafontaine ER. Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect Immun. 2007;75:2765–2775. doi: 10.1128/IAI.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plamondon P, Luke NR, Campagnari AA. Identification of a novel two-partner secretion locus in Moraxella catarrhalis . Infect Immun. 2007;75:2929–2936. doi: 10.1128/IAI.00396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Myers LE, Yang YP, Du RP, Wang Q, Harkness RE, et al. The transferrin binding protein B of Moraxella catarrhalis elicits bactericidal antibodies and is a potential vaccine antigen. Infect Immun. 1998;66:4183–4192. doi: 10.1128/IAI.66.9.4183-4192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du RP, Wang Q, Yang YP, Schryvers AB, Chong P, et al. Cloning and expression of the Moraxella catarrhalis lactoferrin receptor genes. Infect Immun. 1998;66:3656–3665. doi: 10.1128/IAI.66.8.3656-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruckdeschel EA, Kirkham C, Lesse AJ, Hu Z, Murphy TF. Mining the Moraxella catarrhalis genome: identification of potential vaccine antigens expressed during human infection. Infect Immun. 2008;76:1599–1607. doi: 10.1128/IAI.01253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blakeway LV, Power PM, Jen FE-C, Worboys SR, Boitano M, et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J. 2014;28:5197–5207. doi: 10.1096/fj.14-256578. [DOI] [PubMed] [Google Scholar]
- 82.Luke-Marshall NR, Sauberan SL, Campagnari AA. Comparative analyses of the Moraxella catarrhalis type-IV pilus structural subunit PilA. Gene. 2011;477:19–23. doi: 10.1016/j.gene.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 83.Buskirk SW, Lafontaine ER. Moraxella catarrhalis expresses a cardiolipin synthase that impacts adherence to human epithelial cells. J Bacteriol. 2014;196:107–120. doi: 10.1128/JB.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]