Abstract
Native American individuals in the Southwestern USA experience a higher burden of invasive Staphylococcus aureus disease than the general population. However, little is known about S. aureus carriage in these communities. A cross-sectional study was conducted to determine the carriage prevalence, risk factors and genomic epidemiology of S. aureus among Native American children (<5 years, n=121) and adults (≥18 years, n=167) in the Southwestern USA. Short- and long-read sequencing data were generated using Illumina and Oxford Nanopore Technology platforms to produce high-quality hybrid assemblies, and antibiotic-resistance, virulence and pangenome analyses were performed. S. aureus carriage prevalence was 20.7 % among children, 30.2 % among adults 18–64 years and 16.7 % among adults ≥65 years. Risk factors among adults included recent surgery, prior S. aureus infection among household members, and recent use of gyms or locker rooms by household members. No risk factors were identified among children. The bacterial population structure was dominated by clonal complex 1 (CC1) (21.1 %), CC5 (22.2 %) and CC8 (22.2 %). Isolates from children and adults were intermixed throughout the phylogeny. While the S. aureus population was diverse, the carriage prevalence was comparable to that in the general USA population. Genomic and risk-factor data suggest household, community and healthcare transmission are important components of the local epidemiology.
Keywords: carriage, genome epidemiology, Native American, phylogeny, Staphylococcus aureus
Data Summary
The Staphylococcus aureus genome sequences are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive. The accession numbers are included in File S1 (available with the online version of this article). Participant data collected from Native American individuals is governed by the participating Tribal Nations. Data can be made available upon request (contact lhammitt@jhu.edu or csutcli1@jhu.edu), if consistent with the Institutional Review Board-approved protocol and if the disclosure is approved by the participating Tribes.
Impact Statement.
Staphylococcus aureus , a bacteria carried by an estimated 30 % of the population, can cause a variety of infections, which can be difficult to treat due to antibiotic resistance (e.g. meticillin-resistant S. aureus ). Native American individuals experience a higher burden of S. aureus disease than the general population. Little is known about the proportion of Native American children and adults colonized by S. aureus , the types of S. aureus found in these communities, or the individual and household factors associated with carriage. Here, we address these gaps and find that carriage prevalence was 20.7 % among children <5 years, 30.2 % among adults 18–64 years and 16.7 % among adults ≥65 years of age. We also identify several risk factors associated with carriage among adults but none among children. Last, we define the types of S. aureus found within the community and describe the relevance of those types based on recent findings. Overall, this study lays the foundation for future investigations of the relatedness between S. aureus types causing carriage and disease. In addition, this study provides contemporary community carriage data, which are extremely limited in the USA, emphasizing the need to update national data.
Introduction
Staphylococcus aureus is a human commensal and pathogen that is asymptomatically carried in the nose by 20–30 % of the general USA population [1, 2]. In addition, an estimated 1.5–3 % of individuals carry difficult-to-treat meticillin-resistant S. aureus (MRSA). Each year, S. aureus causes millions of infections in the USA, and at least 72 000 cases of severe invasive disease and 10 000 deaths are attributed to MRSA [3–5]. Carriage is the greatest risk factor for infection [6, 7], and individuals are most often infected by the strain they carry [2]. In the USA, a community-associated MRSA (CA-MRSA) clone, USA300, is known to be a common, hypervirulent strain type that spreads easily and persists for prolonged periods in households [8].
The population structure of S. aureus is diverse, comprised of ‘clones’ defined by the genetic relatedness of seven housekeeping genes used to assign multilocus sequence types (STs). While dominant clones of epidemic MRSA are widely recognized, meticillin resistance has emerged among diverse lineages due to the horizontal transfer of the staphylococcal cassette chromosome mec (SCCmec) element that confers resistance. The difficulty in treating invasive disease caused by antibiotic-resistant S. aureus strains has led to a narrow focus of epidemiological and population genomic studies on MRSA, leaving an important gap in our understanding of meticillin-susceptible S. aureus (MSSA), as the population dynamics of this pathogen involve interplay among all lineages, and not just MRSA strains. In addition, MSSA is associated with a considerable burden of invasive disease, especially among indigenous communities in the USA and globally [9–11].
Indigenous populations have historically been burdened by high rates of infectious disease [12]. Indeed, outbreaks of CA-MRSA were first described among these communities [13, 14]. In the USA, recent data have shown that rates of invasive S. aureus disease among Native American individuals are up to seven times the national average, likely due to a higher prevalence of risk factors [10–12, 14, 15]. This trend is observed globally, extending to indigenous populations in Canada [16, 17] and Northern Australia [18, 19]. Yet, data on S. aureus remain limited among Native American populations. To compound this deficiency, S. aureus epidemiology in the USA has changed significantly since the last carriage surveys among Native American individuals were conducted 20 years ago [20]. In particular, incidence of S. aureus skin and soft tissue infections has increased, and CA-MRSA genotypes have infiltrated the healthcare setting [8, 21–23]. To improve understanding of S. aureus invasive disease rates and inform disease prevention strategies, we investigated the contribution of individual, household and pathogen factors associated with S. aureus carriage. The objectives of the study were to estimate the S. aureus carriage prevalence, distribution of strain types, prevalence of antibiotic resistance and S. aureus population structure among Native American individuals on the Navajo Nation and White Mountain Apache (N/WMA) Tribal lands.
Methods
Study population and sample collection
A cross-sectional study of S. aureus carriage was conducted between March and September of 2017 among Native American children and adults living on Tribal lands in the Southwestern USA who were co-enrolled in a pneumococcal carriage study [24]. A convenience sample of participants was recruited at Indian Health Service (IHS) facilities and Tribal Health Organizations during well-child or routine visits and at local community events. Participants were eligible if they were Native American, <5 or ≥18 years of age, did not have a congenital anomaly of the nasopharynx, had not previously been enrolled in the study, and did not have a household member currently or previously enrolled in the study. An equal number of participants were targeted for recruitment each month in five age groups (0–1, 2–4, 18–39, 40–64 and ≥65 years old). After enrolment, a risk factor questionnaire was administered, focusing on demographics, household characteristics, livestock contact, day-care attendance, hygiene practices and participation in high-risk activities for S. aureus transmission (e.g. sports, sweat lodges). Staff collected anterior nares (AN) and nasopharyngeal (NP) swabs from children <5 years old, and AN, NP and oropharyngeal (OP) swabs from adults ≥18 years old. Swabs were inoculated into transport medium and stored at −80 °C. Healthcare exposures and other clinical risk factors were obtained through medical record review. Study data were collected and managed using REDCap electronic data capture tools hosted at the Johns Hopkins School of Public Health (USA) [25, 26].
S. aureus isolation and antibiotic-susceptibility testing
Swabs were shipped to the University of Central Florida (USA) for testing. BBL CHROMagar Staph aureus and MRSA selective media plates (Becton Dickinson) were used according to the manufacturer’s instructions to isolate S. aureus and MRSA, respectively, and isolation was performed for each AN, OP and NP sample independently. Up to three colonies were picked from positive plates and confirmed using catalase and BBL Staphyloslide latex agglutination test (Becton Dickinson). One isolate per positive participant was selected for susceptibility testing, bacterial genomic DNA (gDNA) extraction and whole-genome sequencing (WGS). Isolate selection was performed in the following order: AN, OP, NP. In instances where we detected dual colonization with MRSA and MSSA, an isolate of each was taken forward. Phenotypic susceptibility to seven antibiotics was assessed using disc diffusion after plating isolates on Mueller Hinton agar (MHA). Antibiotic-susceptibility breakpoints were determined based on the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing (27th edition) [27].
Bacterial gDNA isolation and WGS
gDNA extraction, Illumina sequencing, Oxford Nanopore Technology (ONT) MinION sequencing, hybrid genome assembly, genome annotation and multilocus sequence typing (MLST) were performed as previously described [28]. Briefly, gDNA was quantified using a Qubit, and quality was assessed with the TapeStation. WGS short-read libraries were constructed using the Illumina Nextera Flex library prep kit, which were sequenced on an Illumina MiSeq using a 600 cycle V3 chemistry flow cell to produce 2×250 paired-end reads with a target idealized coverage of 75×. Long-read libraries were constructed using an ONT ligation sequencing kit (SQK-LSK109) and barcode kits (EXP-NB104 and EXP-NB114), and sequenced on the ONT MinION with MinIT using R9.4.1 flow cells. Base-calling and demultiplexing was carried out using Guppy v0.5.1 with FAST mode. Adapter sequences were trimmed with Porechop v0.2.2, long-reads were filtered with Filtlong v0.2.0 and read quality was investigated with nanostat v1.0 [29]. Hybrid assembly of short- and long-read WGS data was performed using Unicycler v0.4.8 with default options [30]. Pangenome analysis was performed using Roary v.3.12 [31]. A maximum-likelihood core-genome phylogeny was inferred with iq-tree v1.6.8 using the ASC_GTRGAMMA substitution model with 100 bootstrap replicates and population structure was assessed using fastBAPS [32]. Based on the population structure and MLST analyses, dominant lineages were identified. For these lineages, we repeated the pangenome analysis and inferred a core-genome phylogeny as described above. ABRicate v.1.0.1 was used to detect the antibiotic resistance and virulence determinants (https://github.com/tseemann/abricate) [33] using arg-annot [34] and VFDB [35] databases. Finally, for MRSA isolates, SCCmec type was determined by assessing ccr and mec gene complexes from assemblies using the SCCmec finder tool (www.genomicepidemiology.org/). MLST, SCCmec and antibiotic-resistance results were mapped on the core-genome phylogeny using ggTree implemented in Rstudio running R v 3.6.0 [36].
Carriage prevalence and risk-factor analysis
An initial analysis was conducted to evaluate the performance of different body sites in detecting S. aureus colonization. As OP swabs were only collected from adults, the analysis was stratified by age group (<5 years and ≥18 years). The sensitivity of each site (AN, OP or NP) was assessed compared to the ‘gold standard’ of positive at any body site.
To evaluate and compare age-specific prevalence, only body sites sampled for all participants (AN and NP samples) were included, and carriage was defined as isolation of S. aureus from swabs collected from either site. Age-specific proportions and binomial 95 % confidence intervals (CIs) were calculated. To evaluate all other individual and household risk factors, carriage was defined as isolation of S. aureus (MSSA or MRSA) from any body site (AN, OP or NP). As potential risk factors for evaluation differed by age, the analysis was stratified by age group. Log-binomial regression was used to estimate univariable and multivariable prevalence ratios (PRs) and 95 % CIs. As the sample size within age groups was small, a limited multivariable analysis was conducted focusing on statistically significant (P<0.05) covariates from the univariable analysis. Among adults, age remained a driver of carriage prevalence and, therefore, age-adjusted PRs are presented. sas software, version 9.4 of the SAS System for Windows (SAS Institute), was used.
Results
Study population
Two hundred and eighty-eight individuals (121 children and 167 adults) were enrolled, and 288 AN, 287 NP and 167 OP swabs were collected. Most participants were enrolled from the Navajo Nation (89.9 %), were female (66.3 %) and lived in a private residence (99.9 %) with a median household size of 4 (interquartile range: 2, 6), and had indoor piped water (78.1 %). Few participants reported owning pets (29.8 %) or livestock (20.9 %) (Tables 1, S1 and S2).
Table 1.
Individual risk factors for colonization with S. aureus among Native American participants
P values represent the difference in the prevalence of colonization between those with and without the characteristic of interest. The P values are reported from log-binomial regression or from Fisher’s exact test where regression was not possible. For values <0.05, the results are presented in bold. Ref, Reference value.
|
Factor |
Children |
Adults |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
Total population (N=121) [n (%)] |
Positive at any site (N=25) [n (%)] |
Crude PR (95 % CI) |
P value |
Total population (N=167) [n (%)] |
Positive at any site (N=66) [n (%)] |
Crude PR (95 % CI) |
P value |
||
|
Characteristic |
|||||||||
|
Age group |
0–1 years |
58 (47.9) |
12 (20.7) |
Ref |
– |
– |
– |
– |
– |
|
2–4 years |
63 (52.1) |
13 (20.6) |
1.00 (0.50, 2.01) |
0.99 |
– |
– |
– |
– |
|
|
18–39 years |
– |
– |
– |
– |
53 (31.7) |
26 (49.1) |
Ref |
– |
|
|
40–64 years |
– |
– |
– |
– |
54 (32.3) |
25 (46.3) |
0.94 (0.63, 1.40) |
0.78 |
|
|
≥65 years |
– |
– |
– |
– |
60 (35.9) |
15 (25.0) |
0.51 (0.30, 0.85) |
0.01 |
|
|
Sex |
Male |
60 (49.6) |
13 (21.7) |
1.10 (0.55, 2.22) |
0.79 |
37 (22.2) |
14 (37.8) |
0.95 (0.60, 1.50) |
0.81 |
|
Female |
61 (50.4) |
12 (19.7) |
Ref |
– |
130 (77.8) |
52 (40.0) |
Ref |
– |
|
|
Type of living facility |
Residence |
121 (100.0) |
25 (20.7) |
Ref |
– |
164 (98.2) |
66 (40.2) |
Ref |
– |
|
Currently homeless |
0 |
0 |
– |
– |
3 (1.8) |
0 |
– |
0.28 |
|
|
Times likely to bathe or shower in a typical week |
1–2 |
16 (13.2) |
1 (6.3) |
Ref |
– |
29 (17.8) |
12 (41.4) |
Ref |
– |
|
3–4 |
42 (34.7) |
8 (19.1) |
3.05 (0.41, 22.46) |
0.27 |
46 (28.2) |
18 (38.1) |
0.95 (0.54, 1.66) |
0.85 |
|
|
5–6 |
21 (17.4) |
5 (23.8) |
3.81 (0.49, 29.48) |
0.20 |
23 (14.1) |
7 (30.4) |
0.74 (0.35, 1.56) |
0.42 |
|
|
7+ |
42 (34.7) |
11 (26.2) |
4.19 (0.59, 29.88) |
0.15 |
65 (39.9) |
28 (43.1) |
1.04 (0.62, 1.74) |
0.88 |
|
|
Current tobacco smoker |
No |
– |
– |
– |
– |
148 (88.6) |
54 (36.5) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
19 (11.4) |
12 (63.2) |
1.73 (1.16, 2.59) |
0.008 |
|
|
Current chewing tobacco user |
No |
– |
– |
– |
– |
154 (92.2) |
61 (39.6) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
13 (7.8) |
5 (38.5) |
0.97 (0.48, 1.98) |
0.94 |
|
|
Self-reported history of skin infections or complications (e.g. boils, spider bites, eczema) |
No |
94 (77.7) |
19 (20.2) |
Ref |
– |
136 (81.4) |
52 (38.2) |
Ref |
– |
|
Yes |
27 (22.3) |
6 (22.2) |
1.10 (0.49, 2.48) |
0.82 |
31 (18.6) |
14 (45.2) |
1.18 (0.76, 1.84) |
0.46 |
|
|
Documented S. aureus infection in the past 5 years |
No |
118 (98.3) |
24 (20.3) |
Ref |
– |
162 (97.0) |
64 (39.5) |
Ref |
– |
|
Yes |
2 (1.7) |
1 (50.0) |
2.46 (0.69, 10.28) |
0.22 |
5 (3.0) |
2 (40.0) |
1.01 (0.34, 3.01) |
0.98 |
|
|
Antibiotic use in the past 14 days |
No |
101 (88.6) |
24 (23.8) |
Ref |
– |
144 (89.4) |
61 (42.4) |
Ref |
– |
|
Yes |
13 (11.4) |
0 |
– |
0.07 |
17 (10.6) |
2 (11.8) |
0.28 (0.07, 1.04) |
0.06 |
|
|
Any underlying health condition |
No |
104 (86.7) |
20 (19.2) |
Ref |
61 (36.5) |
24 (39.3) |
Ref |
– |
|
|
Yes |
16 (13.3) |
5 (31.3) |
1.63 (0.71, 3.71) |
0.25 |
106 (63.5) |
42 (39.6) |
1.01 (0.68, 1.49) |
0.97 |
|
|
Diabetes |
No |
120 (100.0) |
25 (20.8) |
Ref |
– |
114 (68.3) |
47 (41.2) |
Ref |
– |
|
Yes |
0 |
0 |
– |
– |
53 (31.7) |
19 (35.9) |
0.87 (0.57, 1.33) |
0.52 |
|
|
Obesity |
No |
118 (98.3) |
24 (20.3) |
Ref |
– |
93 (55.7) |
33 (35.5) |
Ref |
– |
|
Yes |
2 (1.7) |
1 (50.0) |
2.46 (0.59, 10.28) |
0.22 |
74 (44.3) |
33 (44.6) |
1.26 (0.86, 1.83) |
0.23 |
|
|
Behaviours in the past 6 months |
|||||||||
|
Hospitalization |
No |
106 (87.6) |
24 (22.6) |
Ref |
– |
145 (86.8) |
56 (38.6) |
Ref |
– |
|
Yes |
15 (12.4) |
1 (6.7) |
0.29 (0.04, 2.02) |
0.21 |
22 (13.2) |
10 (45.5) |
1.18 (0.71, 1.94) |
0.52 |
|
|
Surgery |
No |
119 (98.4) |
25 (21.0) |
Ref |
– |
148 (88.6) |
55 (37.2) |
Ref |
– |
|
Yes |
2 (1.7) |
0 |
– |
1.00 |
19 (11.4) |
11 (57.9) |
1.56 (1.01, 2.41) |
0.05 |
|
|
Long-term care residence |
No |
– |
– |
– |
– |
163 (97.6) |
65 (39.9) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
4 (2.4) |
1 (25.0) |
0.63 (0.11, 3.46) |
0.59 |
|
|
Shared a towel |
No |
98 (81.0) |
22 (22.5) |
Ref |
– |
142 (85.0) |
52 (36.6) |
Ref |
– |
|
Yes |
23 (19.0) |
3 (13.0) |
0.58 (0.19, 1.78) |
0.34 |
25 (15.0) |
14 (56.0) |
1.53 (1.02, 2.30) |
0.04 |
|
|
Shared a bed |
No |
65 (53.7) |
14 (21.5) |
Ref |
– |
108 (64.7) |
39 (36.1) |
Ref |
– |
|
Yes |
56 (46.3) |
11 (19.6) |
0.91 (0.45, 1.84) |
0.80 |
59 (35.3) |
27 (45.8) |
1.27 (0.87, 1.84) |
0.21 |
|
|
Worn clothes more than once without washing |
No |
103 (85.1) |
21 (20.4) |
Ref |
– |
116 (69.5) |
43 (37.1) |
Ref |
– |
|
Yes |
18 (14.9) |
4 (22.2) |
1.09 (0.42, 2.80) |
0.86 |
51 (30.5) |
23 (45.1) |
1.22 (0.83, 1.79) |
0.32 |
|
|
Shared clothes |
No |
118 (97.5) |
25 (21.2) |
Ref |
– |
162 (97.0) |
65 (40.1) |
Ref |
– |
|
Yes |
3 (2.5) |
0 |
– |
1.00 |
5 (3.0) |
1 (20.0) |
0.50 (0.09, 2.89) |
0.44 |
|
|
Gym use |
No |
– |
– |
– |
– |
147 (88.0) |
56 (38.1) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
20 (12.0) |
10 (50.0) |
1.31 (0.81, 2.13) |
0.27 |
|
|
Used a locker room |
No |
– |
– |
– |
– |
159 (95.2) |
61 (38.4) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
8 (4.8) |
5 (62.5) |
1.63 (0.92, 2.89) |
0.09 |
|
|
Went to a sweat lodge |
No |
– |
– |
– |
– |
164 (98.2) |
65 (39.6) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
3 (1.8) |
1 (33.3) |
0.84 (0.17, 4.21) |
0.83 |
|
|
Participated in organized team sports or activities |
No |
– |
– |
– |
– |
153 (92.7) |
60 (39.2) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
12 (7.3) |
5 (41.7) |
1.06 (0.53, 2.14) |
0.86 |
|
|
Received any artistic (body-art) or cosmetic tattoos |
No |
– |
– |
– |
– |
156 (94.6) |
60 (38.5) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
9 (5.4) |
5 (55.6) |
1.44 (0.78, 2.68) |
0.24 |
|
|
Received any body or facial piercings |
No |
– |
– |
– |
– |
160 (97.0) |
62 (38.8) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
5 (3.0) |
3 (60.0) |
1.55 (0.74, 3.25) |
0.25 |
|
|
Used drugs for non-medical purposes |
No |
– |
– |
– |
– |
158 (95.8) |
61 (38.6) |
Ref |
– |
|
Yes |
– |
– |
– |
– |
7 (4.2) |
4 (57.1) |
1.48 (0.76, 2.90) |
0.25 |
|
|
Attended day care |
No |
118 (97.5) |
24 (20.3) |
Ref |
– |
– |
– |
– |
– |
|
Yes |
3 (2.5) |
1 (33.3) |
1.64 (0.32, 8.45) |
0.55 |
– |
– |
– |
– |
|
Prevalence and risk factors for S. aureus carriage
Overall, 31.6 % (n=91) and 3.5 % (n=10) of participants were culture positive at any body site for S. aureus and MRSA, respectively. In one adult participant, we identified co-carriage of a MRSA and a MSSA strain. Among children, the proportion culture positive was similar for AN (14.1 %) and NP (15.7 %) samples (Table 2). Seven per cent (n=9/121) of children were positive at both sites. Among adults, the proportion culture positive was similar for AN (22.8 %) and OP (20.4 %), and lower for NP (16.3 %) samples (Table 2). One quarter (24.6 %; n=41/167) of all adults were positive at only one body site [the majority of which were at the OP site (n=23)], 10.2 % (n=17) were positive at two body sites, and 4.8 % (n=8) were positive at all three body sites. Consequently, the proportion culture positive was higher when samples from all sites were considered (39.2 %) compared to AN (22.8 %) or NP (16.3%) samples alone, or AN in combination with NP samples (25.3 %). Table S1 shows the sensitivity of sampling each body site alone and in combination.
Table 2.
S. aureus carriage among Native American participants, by body site and age group
|
Specimen type(s) sampled |
No. of samples |
Any S. aureus /MRSA isolated from sampled site(s) [N (%)] |
S. aureus /MRSA isolated only from this site [N (%)] |
|---|---|---|---|
|
Children (0–4 years) |
|||
|
AN |
121 |
17 (14.1)/1 (0.8) |
6 (5.0)/0 |
|
NP |
121 |
19 (15.7)/2 (1.7) |
8 (5.6)/1 (0.8) |
|
AN and NP |
121 |
25 (20.7)/2 (1.7) |
– |
|
Adults (≥18 years) |
|||
|
AN |
167 |
38 (22.8)/5 (3.0) |
13 (7.8)/3 (1.8) |
|
NP |
166 |
27 (16.3)/2 (1.2) |
5 (3.0)/0 |
|
OP |
167 |
34 (20.4)/4 (2.4) |
23 (13.8)/2 (1.2) |
|
AN and NP |
166 |
42 (25.3)/5 (3.0) |
– |
|
AN and OP |
167 |
61 (36.5)/8 (4.8) |
– |
|
NP and OP |
166 |
53 (31.9)/5 (3.0) |
– |
|
AN, NP and OP |
166 |
65 (39.2)/8 (4.8) |
– |
|
Adults (18–64 years) |
|||
|
AN |
107 |
30 (28.0)/3 (2.8) |
11 (10.3)/2 (1.9) |
|
NP |
106 |
19 (17.9)/1 (0.9) |
3 (2.8)/0 |
|
OP |
107 |
26 (24.3)/2 (1.9) |
18 (16.8)/2 (1.9) |
|
AN and NP |
106 |
32 (30.2)/3 (2.8) |
– |
|
AN and OP |
107 |
48 (44.9)/5 (4.7) |
– |
|
NP and OP |
106 |
40 (37.7)/3 (2.8) |
– |
|
AN, NP and OP |
106 |
50 (47.2)/5 (4.7) |
– |
|
Adults (≥65 years) |
|||
|
AN |
60 |
8 (13.3)/2 (3.3) |
2 (3.3)/1 (1.7) |
|
NP |
60 |
8 (13.3)/1 (1.7) |
2 (3.3)/0 |
|
OP |
60 |
8 (13.3)/2 (3.3) |
5 (8.3)/1 (1.7) |
|
AN and NP |
60 |
10 (16.7)/2 (3.3) |
– |
|
AN and OP |
60 |
13 (21.7)/3 (5.0) |
– |
|
NP and OP |
60 |
13 (21.7)/2 (3.3) |
– |
|
AN, NP and OP |
60 |
15 (25.0)/3 (5.0) |
– |
The age-specific prevalence of S. aureus carriage (based on AN and NP swabs only) was 20.7 % (95 % CI: 13.8, 29.0) among children, 30.2 % (95 % CI: 21.7, 39.9) among adults 18–64 years of age, and 16.7 % (95 % CI: 8.3, 28.5) among adults ≥65 years of age (P=0.09). The age-specific prevalence of MRSA carriage was 1.7 % (95 % CI: 0.2, 5.8) among children, 2.8 % (95 % CI: 0.6, 8.0) among adults 18–64 years of age, and 3.3 % (95 % CI: 0.4, 11.5) among adults ≥65 years of age (P=0.75) (Table 2). There was no significant difference in S. aureus carriage prevalence between Navajo Nation and White Mountain Apache participants.
No significant individual nor household risk factors for S. aureus carriage were identified among children (Tables 1 and S2). Among adults, S. aureus carriage was significantly associated with current use of tobacco products, and history of surgery and sharing towels within the past 6 months in the univariable analysis (Table 1). After adjusting for age, only surgery in the past 6 months remained significantly associated with S. aureus carriage (PR 1.91; 95 % CI 1.23, 2.97) (Table S3). Household factors significantly associated with carriage in the univariable analysis included history of S. aureus infection among household members, and household members sharing a bed, using a gym and using a locker room in the past 6 months (Table S4). After adjusting for age, history of S. aureus infection among household members (PR 1.87; 95 % CI 1.08, 3.22) and household members using a gym (PR 1.80; 95 % CI 1.23, 2.64) or locker room (PR 1.98; 95 % CI 1.29, 3.04) remained significantly associated with S. aureus carriage (Table S4).
Genomic analyses
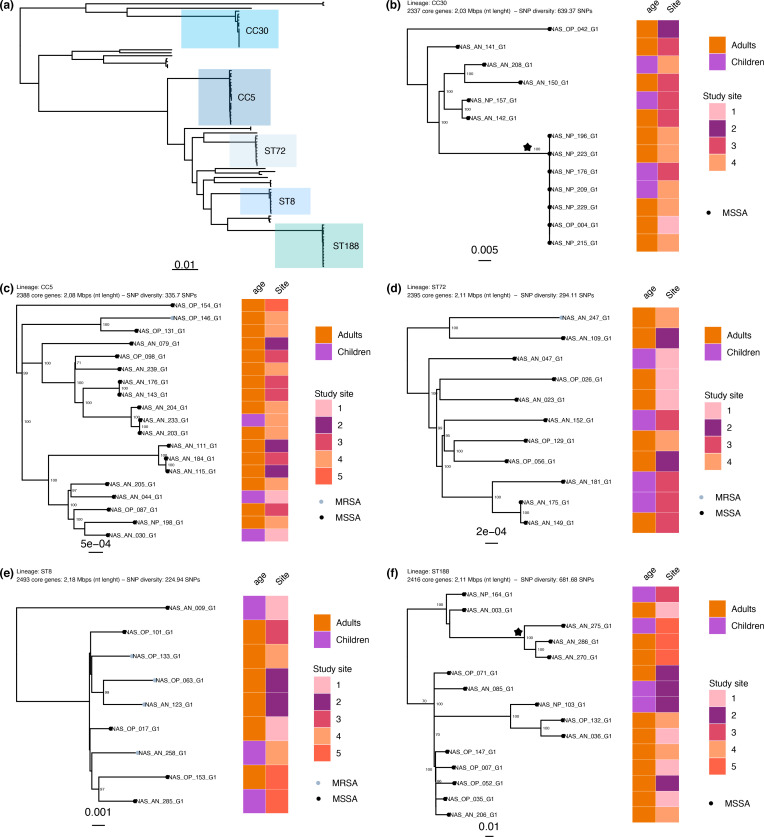
Performing hybrid sequencing and assembly of 92 isolates (82 MSSA and 10 MRSA) collected from 91 participants, 73 genomes were circularized and 17 were assembled into linear unitigs (File S1). Two samples were excluded from phylogenetic analysis due to poor genome assembly statistics. The phylogeny, inferred from a core-genome alignment of 1808 Clusters of Orthologous Groups of proteins (COGs) and 80 116 SNP sites, showed several well-supported clades representing the dominant S. aureus clonal complexes (CCs) (Fig. 1). The S. aureus population structure was dominated by three major lineages belonging to CC1 (21.1 %: 3.3 % ST1, 1.1 % ST9, 16.7 % ST188), CC5 (22.2 %: 20.0 % ST5, 1.1 % ST6, 1.1 % ST6177) and CC8 (22.2%: 10.0 % ST8, 12.2 % ST72) (Table 3). Two novel STs were found and assigned: ST6176 and ST6177. The ST6176 strain clustered outside the CC30 clade, while the ST6177 strain clustered within the CC5 clade and is a single locus variant of ST5 (Fig. 1). Analysis of the co-carriage episode identified the MRSA strain as ST6177 and the MSSA strain as ST9. These two strains were separated by 9013 SNPs, indicating separate acquisition events. Isolates sampled from children and adults were intermixed throughout the phylogeny, suggesting overlapping transmission networks.
Fig. 1.
Maximum-likelihood phylogeny of the S.aureus isolates (n=90) inferred from an alignment of 80 116 SNPs present in the core genome. The tip colours indicate strains with meticillin susceptibility (MSSA; grey) or resistance (MRSA; black). Bootstrap values indicating statistical support of dominant clades in the phylogeny are shown on the branches. Lineages identified through statistical analysis of population structure are highlighted. Clades are annotated with multilocus ST. The heatmap around the tree shows the CC, where identifiable, and age group associated with the isolate. The scale bar indicates genetic distance.
Table 3.
CC distribution of the S. aureus isolates
|
Clonal complex |
Children (<5 years old) [N (%)] |
Adults (≥18 years old) [N (%)] |
Total [N (%)] |
|---|---|---|---|
|
CC1 |
5 (20.8) |
14 (21.2) |
19 (21.1) |
|
CC5 |
3 (12.5) |
17 (25.8) |
20 (22.2) |
|
CC8 |
7 (29.2) |
13 (19.7) |
20 (22.2) |
|
CC15 |
1 (4.1) |
1 (1.5) |
2 (2.2) |
|
CC30 |
4 (16.7) |
9 (13.6) |
13 (14.4) |
|
CC45 |
– |
2 (3) |
2 (2.2) |
|
CC97 |
– |
3 (4.5) |
3 (3.3) |
|
Unknown |
4 (16.7) |
7 (10.6) |
11 (12.2) |
|
Total |
24 |
66 |
90 |
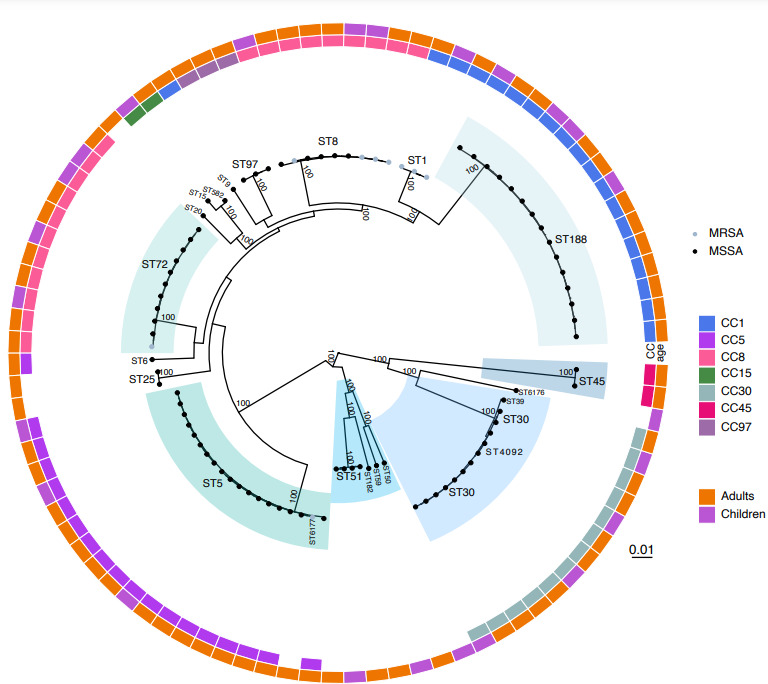
Of the nine MRSA isolates, three (33.3 %) were ST1 (representing 100 % of the ST1 isolates), four (44.4 %) were ST8 (representing 40 % of ST8 isolates), one was ST72 (representing 9.1 % of ST72 isolates) and one was ST6177. Genotypic SCCmec typing identified that six (77.8 %; four ST8 and two ST1) MRSA isolates carried SCCmec IVa (2B), one (11.1 %; 1 ST1) carried SCCmec Vb (5C and 5), and one (11.1 %; one ST6177) carried SCCmec II (2A) (Fig. 2). Interestingly, the ST6177 isolate, which was obtained from the MRSA–MSSA co-carriage episode, was the only MRSA strain from our sample found in CC5. Further, it was only 188 SNPs diverged from the next closest MSSA strain in the CC5 phylogeny, suggesting a recent acquisition of SCCmec (Fig. 3c).
Fig. 2.
Maximum-likelihood phylogeny illustrating population structure, phenotypic antibiotic susceptibility, and genotypic virulence and antibiotic-resistance determinants. Dominant clades identified through analysis of population structure are highlighted with MLST type annotated. Strains identified as MRSA or MSSA are indicated by circular and square tip shapes, respectively, with SCCmec type shown by the tip colour of MRSA strains. The heatmap to the right shows the phenotypic antibiotic (Abx) susceptibility results, genotypic antibiotic determinants and virulence determinants. Phenotypic antibiotic testing is reported for erythromycin (erm), clindamycin (clinda), trimethoprim/sulfamethoxazole (TMP_SMX), ciprofloxacin (cipro), gentamicin (gent), tetracycline (tet) and oxacillin. The presence of genotypic antibiotic-resistance determinants is indicated by light pink. Antibiotic-resistance determinants include: aminoglycoside resistance (AadD, Aph3.III, Sat4A, Spc), β-lactam resistance (BlaZ, Dha1), fosfomycin resistance (FosfB), macrolide resistance (ErmA, ErmC, MphC, MsrA), fluoroquinolone resistance (NorA) and tetracycline resistance (TetM). Virulence factor presence is indicated in purple, and includes: (i) adherence – collagen binding protein (Cna); (ii) toxin – PVL Panton-Valentine leucocidin (LukF-PV), staphylococcal enterotoxin (enterotoxin), exfoliative toxin b (Etb), toxic shock syndrome toxin-1 (TSST-1); (iii) exoenzyme – von Willebrand factor-binding protein (vWbp), staphylocoagulase (Coa); (iv) immune evasion – CHIPS chemotaxis inhibitory protein of staphylococcus (Chp), SCIN staphylococcal complement inhibitor (Scn); - plasminogen activator – staphylokinase (Sak). The scale bar indicates genetic distance.
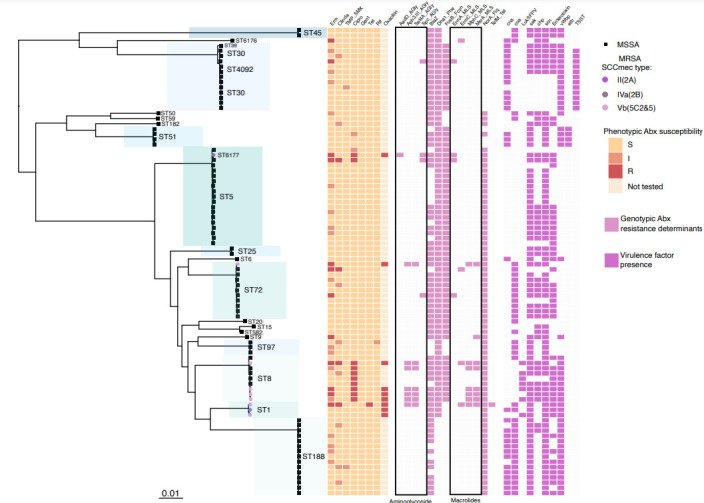
Fig. 3.
Population structure of the S. aureus isolates carried (a) and five dominant lineages (b–f). (a) The maximum-likelihood phylogeny from Fig. 1, with the dominant lineages indicated. (b–f) Lineage (ST or CC), number of core genes, nucleotide length of the core genome and the mean pairwise SNP diversity are shown. The tips of the phylogeny are coloured according to meticillin susceptibility (MSSA; black) or resistance (MRSA; grey). Bootstrap statistical support values are annotated on the branches of the phylogeny and the branch length scales are shown at the bottom of each panel. The heatmap to the right shows the age group and deidentified study site associated with the isolate. The clades annotated with a star have a mean SNP difference of 0 SNPs (CC30) and 306 SNPs (ST188). The scale bar indicates genetic distance.
We assessed phenotypic and genotypic antibiotic resistance as well as genotypic virulence determinants in the context of population structure (Fig. 2). MRSA strains, as expected, demonstrated phenotypic resistance to oxacillin and β-lactam antibiotics. Phenotypic macrolide, clindamycin and tetracycline resistance were sporadic throughout the population, suggesting a limited spread of mobile genetic elements harbouring their respective resistance determinants. Most notably, eight of nine CC8 strains and two of twenty CC5 strains demonstrated phenotypic resistance to ciprofloxacin, and we confirmed the Ser84→Leu substitution in gyrA, which confers fluoroquinolone resistance. Aminoglycoside-resistance determinants were largely confined to MRSA strains belonging to CC8. No resistance to gentamicin, rifampicin and trimethoprim/sulfamethoxazole was found. Among virulence determinants, toxic shock syndrome toxin (TSST-1) was harboured by ST30 strains. ST8 and ST1 were found to sporadically possess lukSF-PV, the gene encoding the bacterial toxin Panton-Valentine leukocidin. Exfoliative toxin b (Etb), which is associated with staphylococcal scalded skin syndrome, was present is all four ST51 strains. Last, staphylococcal enterotoxins were present among 54.8 % of strains interspersed throughout the population.
To further resolve the population structure of carriage isolates, we inferred phylogenies for five dominant lineages: CC5, CC8, CC30, ST72, ST188 (Fig. 3a–f). We found that CC30 and ST188 populations demonstrated the greatest nucleotide diversity, with a mean core-genome pairwise SNP distance of 639 and 681, respectively, as compared to <335 SNPs for all other prevalent lineages (Fig. 3b, f). In general, there was no significant phylogenetic structure in relation to geographical location or age group apart from a subclade on the ST188 lineage that included isolates collected only from White Mountain Apache participants. In addition, the CC30 lineage was comprised of a subclade with little genetic diversity (mean CC30 core-genome SNP distance of 0) as represented by the polytomy of seven isolates collected from adult and paediatric participants. Further examination of the participants’ risk factors, date of sample collection, age group and geographical location did not identify any overt epidemiological links.
Discussion
This study provides contemporary estimates for the carriage prevalence and genomic epidemiology of S. aureus in two tribal communities in the Southwestern USA where S. aureus has been shown to be an important cause of skin and soft tissue infections, as well as severe invasive disease [10]. While direct comparisons between studies are challenging due to differing anatomical sites for sample collection, laboratory techniques and time period, our estimates of carriage prevalence and the major S. aureus lineages are not dissimilar to what has been reported nationally in the USA.
Two decades ago, a study of S. aureus disease among Native Americans in the Midwest found that 74 % of cases were acquired within the community, providing early evidence of the CA-MRSA epidemic in the USA [14, 37, 38]. Since then, few studies have investigated S. aureus epidemiology among indigenous people and the reasons for the high rates of S. aureus disease are not fully understood. A single carriage study among Native American individuals in Washington State found a 27.3 and 1.9% carriage prevalence of MSSA and MRSA, respectively, in 2001 [15]. Unfortunately, USA national data are equally dated with the most recent estimates obtained from the National Health and Nutrition Examination Survey (NHANES) conducted from 2001 to 2004 [1]. NHANES, which used sampling from only the AN, estimated S. aureus and MRSA carriage prevalence among individuals 1–19 years as 34.6 and 1.3 %, respectively, and 27.4 and 1.1 % for individuals 20–59 years. Here, in this study of 2017 data, we found that the carriage prevalence of S. aureus among Native American children and adults in the Southwestern USA was generally consistent with these estimates. However, MRSA carriage among adults 18–64 years of age (4.8 %) approached the upper limits of reported statistics [1], even if data were limited to the AN (3.5 %) to control for sampling variation. Of note, pneumococcal carriage rates in these same communities were ~20 % higher than the general USA population prior to the introduction of the pneumococcal conjugate vaccines [39, 40].
The AN is the primary site of S. aureus carriage, as well as the most common site for screening; however, the skin, throat, gastrointestinal tract and urogenital tract of women have all been identified as carriage sites [41–45]. The inclusion of multiple anatomical sites in S. aureus screening may increase the detection rate [46], especially since up to 25 % of colonized individuals may have exclusive OP carriage [47–50]. Indeed, we found that sampling of the oropharynx and nasopharynx increased the detection sensitivity as we would have missed up to 40 % of carriers using AN sampling alone. Further, to our knowledge, we are the first to compare NP carriage of S. aureus to other anatomical sites, finding 13 individuals (14.3 %) positive only at that anatomical site. More investigation is needed to determine whether these strains are adapted for colonization at that anatomical site or whether their detection was the result of passing the swab through the AN on the way to the nasopharynx.
The significant risk factors for carriage in our study included both individual and household factors among adults [51]. A recent history of surgery was associated with carriage, which may point to a healthcare association for these strains. Previous studies have identified the household setting as a primary reservoir for S. aureus transmission [8]; household size has been associated with higher rates of S. aureus [15] and pneumococcal [39, 40] carriage among Native American populations. While household size was not a risk factor for carriage here, other risk factors, including history of S. aureus infection among household members, and use of gyms and locker rooms by household members, suggest household transmission. Interestingly, while strains from both children and adults were intermixed in the phylogeny, indicating overlapping transmission networks, no significant risk factors were identified among children, possibly due to the small sample size or our focus on adult-specific risk factors. Further, population genomic analysis in the context of participant demographics and risk factors did not reveal any overt epidemiological association with lineage. Together, these findings suggest that individual behaviours, healthcare exposures and the household all play a role in transmission. In consideration of healthcare and household risk factors, one dynamic worth investigating is the migration of strains from the healthcare setting into the household via individuals with recurrent S. aureus infections or recent healthcare exposure. Overall, the findings of similar carriage prevalence and carriage risk factors in these communities to that of the general USA population suggest that carriage prevalence alone does not account for the comparatively high rates of S. aureus invasive disease and underscores the role of social determinants of health as drivers of disparities.
While the population structure of S. aureus is diverse, much of our knowledge is focused on MRSA due to its clinical relevance as an antibiotic-resistant pathogen. Further, at present, there are no comparable population genomic studies of S. aureus carriage in the USA, precluding detailed comparison to the general population. In the USA, the most well recognized clones are the epidemic CA-MRSA strain ST8 (PFGE type USA300) belonging to CC8 and healthcare-associated MRSA strain ST5 (PFGE type USA100) belonging to CC5 [52]. Here, we considered all S. aureus , recognizing the importance of MSSA as a cause of invasive disease as well as its role in species-level ecology and evolution. We found that CC1, CC5 and CC8 were the major lineages found in carriage, with MRSA strains largely belonging to CC1 (ST1) and CC8 (ST8). Interestingly, ST1 strains belonging to CC1 were previously identified as a prevalent MRSA lineage in rural south-western Alaska before being replaced by MRSA strains belonging to ST8 [53]. MSSA strain ST188, also belonging to CC1 and making up 17 % of isolates in this study, has been recognized as a bovine livestock-associated strain in China and a significant cause of community-onset MSSA infections in South-East Asia [54, 55]. In the USA, its distribution and role in community-associated disease is unknown and requires further investigation. Strain ST72, belonging to CC8, was found in 12 % of all isolates and 11 % of MRSA isolates in this study. There is limited data on this strain in the USA, but it has been associated with community-associated infections and contact with livestock [56]. Other prevalent lineages included ST30, a well-known community lineage of MSSA in Europe and South America that is notable for possessing toxic shock syndrome toxin (TSST), which is also harboured by the strains we identified. Last, we identified another virulent lineage carried by four participants – ST51 possessing Etb, which has recently been associated with cases of staphylococcal scalded skin syndrome in the USA [57]. Our previous studies of S. aureus invasive disease in the same communities found that 33–75 % of infections were caused by MRSA and that more than 85 % of individuals had at least one medical condition [10, 11]. The low prevalence of MRSA carriage found in this study, and lack of association between carriage and comorbidities, suggest that other factors are driving disease. Further genomic investigation of isolates from S. aureus infection in the context of risk factors would reveal the relative contribution of each of these lineages to the high burden of S. aureus disease observed among Native American communities.
Our study is not without limitations. First, our participant population was limited to individuals <5 and ≥18 years of age, which likely omits an important component of the S. aureus transmission network. Second, our enrolment was largely healthcare-facility based, which may introduce selection bias, and the use of questionnaires may suffer from recall bias. However, we minimized recall bias by conducting a review of medical records. Lastly, we did not obtain OP swabs from child participants and, therefore, may have underestimated carriage prevalence in this age group considering the increased detection observed with OP swabs from adult participants in this study and from children in other studies [58].
Overall, our study elucidates the carriage prevalence and population structure of S. aureus among Native American communities in the Southwestern USA and provides essential data for developing and assessing interventions targeting the high burden of S. aureus disease. In addition, we assessed sampling multiple anatomical sites for the first time, to our knowledge, showing that inclusion of NP swabs improves detection of S. aureus carriage. We resolve a missing component of the evolutionary history of S. aureus in the USA and establish the foundation for future genomic comparisons of S. aureus strains from carriage and disease.
Supplementary Data
Funding information
This work was supported through funding from the Johns Hopkins Center for American Indian Health. T.A. and E.C. were funded in part by the USA National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) award K22 AI141582 to T.A.
Acknowledgements
The authors wish to express our deep gratitude to the children and family members from the Navajo and White Mountain Apache Tribes who participated in the study. We also thank the Center for American Indian Health staff involved in conducting the study. The study could not have taken place without the guidance from the Institutional Review Boards (IRBs) of the Navajo Nation, the Phoenix Area IHS and the Johns Hopkins Bloomberg School of Public Health.
Author contributions
E.C., formal analysis and writing – original draft preparation. C.G.S., conceptualization, project administration, formal analysis and writing – review and editing. C.T., E.P., N.R., J.C., E.D., investigation and writing – review and editing. L.R.G., R.C.W., project administration and writing – review and editing. L.L.H., conceptualization, funding, supervision and writing – review and editing. T.A., conceptualization, funding, formal analysis and writing – review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Approval was obtained from the Navajo Nation and White Mountain Apache communities and the IRBs of the Johns Hopkins Bloomberg School of Public Health (IRB no. 6771), the Navajo Nation (IRB no. NNR-16.238) and the Phoenix Area IHS (IRB no. PXR 16.02). Written informed consent was obtained from adult participants and parents or guardians of child participants. A Health Insurance Portability and Accountability Act (HIPAA) authorization was obtained to conduct medical chart reviews.
Footnotes
Abbreviations: AN, anterior nares; CA-MRSA, community-associated meticillin-resistant Staphylococcus aureus; CC, clonal complex; CI, confidence interval; gDNA, genomic DNA; IHS, Indian Health Service; IRB, Institutional Review Board; MLST, multilocus sequence typing; MRSA, meticillin-resistant Staphylococcus aureus; MSSA, meticillin-sensitive Staphylococcus aureus; NP, nasopharyngeal; ONT, Oxford Nanopore Technology; OP, oropharyngeal; PR, prevalence ratio; SCCmec, staphylococcal cassette chromosome mec; ST, sequence type; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. A supplementary file and four supplementary tables are available with the online version of this article.
References
- 1.Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J Infect Dis. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 2.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31:e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention Atlanta, GA: CDC; 2014. Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Neisseria meningitidis .www.cdc.gov/abcs/reports-findings/survreports/mrsa14.html [Google Scholar]
- 4.Landrum ML, Neumann C, Cook C, Chukwuma U, Ellis MW, et al. Epidemiology of Staphylococcus aureus blood and skin and soft tissue infections in the US military health system, 2005-2010. JAMA. 2012;308:50–59. doi: 10.1001/jama.2012.7139. [DOI] [PubMed] [Google Scholar]
- 5.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz SA, Epplin EK, Garbutt J, Storch GA, Kaplan SL, et al. Skin infection in children colonized with community-associated methicillin-resistant Staphylococcus aureus . J Infect. 2009;59:394–401. doi: 10.1016/j.jinf.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 8.Alam MT, Read TD, Petit RA, 3rd, Boyle-Vavra S, Miller LG, et al. Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. mBio. 2015;6:e00054. doi: 10.1128/mBio.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanjilal S, Sater MRA, Thayer M, Lagoudas GK, Kim S, et al. Trends in antibiotic susceptibility in Staphylococcus aureus in Boston, Massachusetts, from 2000 to 2014. J Clin Microbiol. 2018;56:e01160-17. doi: 10.1128/JCM.01160-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutcliffe CG, Grant LR, Reid A, Douglass GK, Weatherholtz RC, et al. The burden of Staphylococcus aureus among Native Americans on the Navajo Nation. PLoS One. 2019;14:e0213207. doi: 10.1371/journal.pone.0213207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutcliffe CG, Grant LR, Reid A, Douglass G, Brown LB, et al. High burden of Staphylococcus aureus among Native American individuals on the White Mountain Apache tribal lands. Open Forum Infect Dis. 2020;7:ofaa061. doi: 10.1093/ofid/ofaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler JC, Crengle S, Cheek JE, Leach AJ, Lennon D, et al. Emerging infectious diseases among indigenous peoples. Emerg Infect Dis. 2001;7:554–555. doi: 10.3201/eid0707.017732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landen MG, McCumber BJ, Asam ED, Egeland GM. Outbreak of boils in an Alaskan village: a case-control study. West J Med. 2000;172:235–239. doi: 10.1136/ewjm.172.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groom AV, Wolsey DH, Naimi TS, Smith K, Johnson S, et al. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA. 2001;286:1201–1205. doi: 10.1001/jama.286.10.1201. [DOI] [PubMed] [Google Scholar]
- 15.Leman R, Alvarado-Ramy F, Pocock S, Barg N, Kellum M, et al. Nasal carriage of methicillin-resistant Staphylococcus aureus in an American Indian population. Infect Control Hosp Epidemiol. 2004;25:121–125. doi: 10.1086/502361. [DOI] [PubMed] [Google Scholar]
- 16.Muileboom J, Hamilton M, Parent K, Makahnouk D, Kirlew M, et al. Community-associated methicillin-resistant Staphylococcus aureus in northwest Ontario: a five-year report of incidence and antibiotic resistance. Can J Infect Dis Med Microbiol. 2013;24:169409. doi: 10.1155/2013/169409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abelson WH, Banerji A, Baydala LT, Jetty R, Schroter HM, et al. Community-associated methicillin-resistant Staphylococcus aureus in indigenous communities in Canada. Paediatr Child Health. 2012;17:395–398. doi: 10.1093/pch/17.7.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng JWS, Holt DC, Lilliebridge RA, Stephens AJ, Huygens F, et al. Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in northern Australia. J Clin Microbiol. 2009;47:2295–2300. doi: 10.1128/JCM.00122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong SYC, Varrone L, Chatfield MD, Beaman M, Giffard PM. Progressive increase in community-associated methicillin-resistant Staphylococcus aureus in indigenous populations in northern Australia from 1993 to 2012. Epidemiol Infect. 2015;143:1519–1523. doi: 10.1017/S0950268814002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leman R, Alvarado-Ramy F, Pocock S, Barg N, Kellum M, et al. Nasal carriage of methicillin-resistant Staphylococcus aureus in an American Indian population. Infect Control Hosp Epidemiol. 2004;25:121–125. doi: 10.1086/502361. [DOI] [PubMed] [Google Scholar]
- 21.Nerby JM, Gorwitz R, Lesher L, Juni B, Jawahir S, et al. Risk factors for household transmission of community-associated methicillin-resistant Staphylococcus aureus . Pediatr Infect Dis J. 2011;30:927–932. doi: 10.1097/INF.0b013e31822256c3. [DOI] [PubMed] [Google Scholar]
- 22.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 23.Otter JA, French GL. Community-associated meticillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated infection. J Hosp Infect. 2011;79:189–193. doi: 10.1016/j.jhin.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 24.Grant L, Weatherholtz R, Alexander-Parrish R, Jacobs M, Good C, et al. Melbourne, Australia: 2018. Nasopharyngeal pneumococcal carriage among American Indian children and adults during routine use of the 13-valent pneumococcal conjugate vaccine (PCV13). International Symposium on Pneumococci & Pneumococcal Diseases (ISPPD) [Google Scholar]
- 25.Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, et al. Research electronic data capture (REDCap) – a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, 19th informational supplement, 27th edn. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 28.Cella E, David MZ, Jubair M, Baines SL, Pegues DA, et al. Complete genome sequence of exfoliative toxin-producing Staphylococcus aureus strain MSSA_SSSS_01, obtained from a case of staphylococcal scalded-skin syndrome. Microbiol Resour Announc. 2021;10:e01335-20. doi: 10.1128/MRA.01335-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 2019;47:5539–5549. doi: 10.1093/nar/gkz361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. ABRicate: Mass Screening of Contigs for Antimicrobial Resistance or Virulence Genes. GitHub. 2017 https://github.com/tseemann/abricate
- 34.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis – 10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Core Team R: a Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2021. [Google Scholar]
- 37.Nimmo GR, Bergh H, Nakos J, Whiley D, Marquess J, et al. Replacement of healthcare-associated MRSA by community-associated MRSA in Queensland: confirmation by genotyping. J Infect. 2013;67:439–447. doi: 10.1016/j.jinf.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 38.Pallin DJ, Egan DJ, Pelletier AJ, Espinola JA, Hooper DC, et al. Increased US emergency department visits for skin and soft tissue infections, and changes in antibiotic choices, during the emergence of community-associated methicillin-resistant Staphylococcus aureus . Ann Emerg Med. 2008;51:291–298. doi: 10.1016/j.annemergmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Millar EV, O’Brien KL, Zell ER, Bronsdon MA, Reid R, et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Navajo and White Mountain Apache children before the introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2009;28:711–716. doi: 10.1097/INF.0b013e3181a06303. [DOI] [PubMed] [Google Scholar]
- 40.Scott JR, Millar EV, Lipsitch M, Moulton LH, Weatherholtz R, et al. Impact of more than a decade of pneumococcal conjugate vaccine use on carriage and invasive potential in Native American communities. J Infect Dis. 2012;205:280–288. doi: 10.1093/infdis/jir730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nilsson P, Ripa T. Staphylococcus aureus throat colonization is more frequent than colonization in the anterior nares. J Clin Microbiol. 2006;44:3334–3339. doi: 10.1128/JCM.00880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadimpalli ML, Stewart JR, Pierce E, Pisanic N, Love DC, et al. Face mask use and persistence of livestock-associated Staphylococcus aureus nasal carriage among industrial hog operation workers and household contacts, USA. Environ Health Perspect. 2018;126:127005. doi: 10.1289/EHP3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Acton DS, Plat-Sinnige MJT, van Wamel W, de Groot N, van Belkum A. Intestinal carriage of Staphylococcus aureus: how does its frequency compare with that of nasal carriage and what is its clinical impact? Eur J Clin Microbiol Infect Dis. 2009;28:115–127. doi: 10.1007/s10096-008-0602-7. [DOI] [PubMed] [Google Scholar]
- 44.Young BC, Wu C-H, Gordon NC, Cole K, Price JR, et al. Severe infections emerge from commensal bacteria by adaptive evolution. Elife. 2017;6:e30637. doi: 10.7554/eLife.30637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, et al. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus . Infect Genet Evol. 2009;9:32–47. doi: 10.1016/j.meegid.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Ide L, Lootens J, Thibo P, on behalf of the Infection Control Team of the Jan Palfijn Ziekenhuis Gent The nose is not the only relevant MRSA screening site. Clin Microbiol Infect. 2009;15:1192–1193. doi: 10.1111/j.1469-0691.2009.02954.x. [DOI] [PubMed] [Google Scholar]
- 47.Mertz D, Frei R, Periat N, Zimmerli M, Battegay M, et al. Exclusive Staphylococcus aureus throat carriage: at-risk populations. Arch Intern Med. 2009;169:172–178. doi: 10.1001/archinternmed.2008.536. [DOI] [PubMed] [Google Scholar]
- 48.Mertz D, Frei R, Jaussi B, Tietz A, Stebler C, et al. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus . Clin Infect Dis. 2007;45:475–477. doi: 10.1086/520016. [DOI] [PubMed] [Google Scholar]
- 49.Esposito S, Terranova L, Zampiero A, Ierardi V, Rios WP, et al. Oropharyngeal and nasal Staphylococcus aureus carriage by healthy children. BMC Infect Dis. 2014;14:723. doi: 10.1186/s12879-014-0723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Widmer AF, Mertz D, Frei R. Necessity of screening of both the nose and the throat to detect methicillin-resistant Staphylococcus aureus colonization in patients upon admission to an intensive care unit. J Clin Microbiol. 2008;46:835. doi: 10.1128/JCM.02276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francisco S, Foley D, Antone Nez R, Kinlacheeny J, Yazzie D. Report of the Navajo Behavioral Risk Factor Surveillance Survey, 2013, 2015, 2016. Window Rock, AZ: Navajo Epidemiology Center; 2017. [Google Scholar]
- 52.Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J Antimicrob Chemother. 2009;64:441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 53.David MZ, Rudolph KM, Hennessy TW, Boyle-Vavra S, Daum RS. Molecular epidemiology of methicillin-resistant Staphylococcus aureus, rural southwestern Alaska. Emerg Infect Dis. 2008;14:1693–1699. doi: 10.3201/eid1411.080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Liu Q, Liu Q, Gao Q, Lu H, et al. Phylogenetic analysis and virulence determinant of the host-adapted Staphylococcus aureus lineage ST188 in China. Emerg Microbes Infect. 2018;7:45. doi: 10.1038/s41426-018-0048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen FJ, Siu LKK, Lin JC, Wang CH, Lu PL. Molecular typing and characterization of nasal carriage and community-onset infection methicillin-susceptible Staphylococcus aureus isolates in two Taiwan medical centers. BMC Infect Dis. 2012;12:343. doi: 10.1186/1471-2334-12-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller LG, Eells SJ, Taylor AR, David MZ, Ortiz N, et al. Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin Infect Dis. 2012;54:1523–1535. doi: 10.1093/cid/cis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hultén KG, Kok M, King KE, Lamberth LB, Kaplan SL. Increasing numbers of staphylococcal scalded skin syndrome cases caused by ST121 in Houston, Texas. Pediatr Infect Dis J. 2020;39:30–34. doi: 10.1097/INF.0000000000002499. [DOI] [PubMed] [Google Scholar]
- 58.Williamson DA, Ritchie S, Keren B, Harrington M, Thomas MG, et al. Persistence, discordance and diversity of Staphylococcus aureus nasal and oropharyngeal colonization in school-aged children. Pediatr Infect Dis J. 2016;35:744–748. doi: 10.1097/INF.0000000000001173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.