Abstract
Purpose
Bile acids are steroid synthesized in liver, which are essential for fat emulsification, cholesterol excretion and gut microbial homeostasis. However, the role of bile acids in leukemia progression remains unclear. We aim at exploring the effects and mechanisms of chenodeoxycholic acid (CDCA), a type of bile acids, on acute myeloid leukemia (AML) progression.
Results
Here, we found that CDCA was decreased in feces and plasma of AML patients, positively correlated with the diversity of gut microbiota, and negatively associated with AML prognosis. We further demonstrated that CDCA suppressed AML progression both in vivo and in vitro. Mechanistically, CDCA bound to mitochondria to cause mitochondrial morphology damage containing swelling and reduction of cristae, decreased mitochondrial membrane potential and elevated mitochondrial calcium level, which resulted in the production of excessive reactive oxygen species (ROS). Elevated ROS further activated p38 MAPK signaling pathway, which collaboratively promoted the accumulation of lipid droplets (LDs) through upregulating the expression of the diacylglycerol O-acyltransferase 1 (DGAT1). As the consequence of the abundance of ROS and LDs, lipid peroxidation was enhanced in AML cells. Moreover, we uncovered that CDCA inhibited M2 macrophage polarization and suppressed the proliferation-promoting effects of M2 macrophages on AML cells in co-cultured experiments.
Conclusion
Our findings demonstrate that CDCA suppresses AML progression through synergistically promoting LDs accumulation and lipid peroxidation via ROS/p38 MAPK/DGAT1 pathway caused by mitochondrial dysfunction in leukemia cells and inhibiting M2 macrophage polarization.
Keywords: Acute myeloid leukemia, Chenodeoxycholic acid, Lipid droplets, Mitochondria
1. Introduction
Acute myeloid leukemia (AML) is a highly lethal hematologic malignancy characterized by the accumulation of poorly differentiated myeloid cells in the bone marrow (BM) and blood, as well as other tissues and organs, ultimately leading to dysfunction of the body [1]. Currently, there are a considerable number of researches and clinical trials aiming at improving the treatment outcomes in AML [2], but complete remission (CR) rate is still low. In consequence, it is imperative to explore more effective treatments and more comprehensive mechanisms of AML.
Bile acids, hydroxylated steroids synthesized in hepatocytes, are classified into the primary bile acids (chenodeoxycholic acid (CDCA) and cholic acid (CA)) and the secondary bile acids (deoxycholic acid (DCA) and lithocholic acid (LCA)) [3]. Accumulating evidence obtained from researches pointed to a strong interaction between bile acids and gut microbiota homeostasis. The primary bile acids, such as CDCA synthesized in hepatocytes, are transported into the intestine where they are metabolized by the microbiota into the secondary bile acids [4]. The alteration of microbiome affects the composition of bile acids, which may further cause the dysregulation of the host [5]. Many studies demonstrated that the abnormalities of CDCA played a significant role in the development and progression of many diseases, such as the endometrial cancer, esophageal cancer, and metabolic disease [[6], [7], [8]]. However, the relationship between CDCA and AML has not been fully clarified. Therefore, it is of vital importance to demonstrate the role of CDCA in AML.
Mitochondria, the “powerhouse of the cell”, fuel the metabolic needs for most cancers. It was reported that unique mitochondrial alterations including mitochondrial metabolism and reactive oxygen species (ROS) generation resulted in metabolic vulnerabilities in AML [9]. In addition to mitochondria, lipid droplets (LDs) are another essential organelles involved in lipid metabolism and energy homeostasis. LDs are dynamic and multifunctional organelles, originated from endoplasmic reticulum, which are surrounded by a single monolayer of phospholipids and contain a hydrophobic core consisting of neutral lipid, mainly triacylglycerol and cholesteryl esters [10]. Among cells, there exists association between LDs and mitochondria. Mitochondrial function has been shown to be determinant to lipogenesis and triacylglyceride storage in LDs [11]. In addition, mitochondria could bind to LDs to regulate its function [[12], [13], [14]]. Recently, mounting literatures have documented the significant role of mitochondria and LDs in immunity, metabolic diseases, as well as malignances [[15], [16], [17]]. However, mitochondrial dysregulation and LDs dynamics have not been extensively studied in AML yet. In consequence, it is worth exploring the mechanism and interaction between LDs and mitochondria in AML, which will provide a prospective future.
Mitogen-activated protein kinase (MAPK), belonging to serine-threonine kinases family, plays a significant role in signal transduction. MAPK contains a group of tertiary phosphorylation-dependent kinases—MAP kinase kinase kinase (MKKK), MAP kinase kinase (MKK) and MAP kinase (MAPK), which is classified into ERK, JNK and p38 MAPK pathway. It was documented that MAPK could be activated by ROS, which had an essential effect on many diseases, such as inflammation, carcinoma and lung injury [[18], [19], [20], [21]]. However, whether MAPK is involved in the LDs biogenesis has still not been fully clarified, especially in AML. Therefore, exploring the relationship between MAPK and LDs will provide an interesting viewpoint for this field.
Studies pointed that macrophage was tightly correlated with the development and progression of AML [22,23], and bile acids had an important impact on macrophage [24]. Macrophage can be characterized into M1 and M2 macrophage traditionally. M2 macrophage acts as tumor-promoting function, while M1 macrophage has an anti-tumor effect. There have been numerous studies focusing on macrophage immunity, especially M2 macrophage in tumorigenesis and therapeutics [25]. Emerging evidence indicates that immunomodulation of macrophages against cancer and modulating the polarization of macrophages will be a promising anti-tumor strategy [26,27].
In this study, we aimed to explore the effects of CDCA on AML. We found that the level of CDCA was significantly decreased in feces and plasma of AML patients compared with normal people, and its level was correlated with AML prognosis, which meant that CDCA might be an indicator for AML. Further researches uncovered that CDCA promoted LDs accumulation and lipid peroxidation through ROS/p38 MAPK/DGAT1 pathway, which was initiated by mitochondrial dysfunction. Moreover, we further clarified that CDCA could inhibit M2 macrophages polarization. Taken together, CDCA suppressed AML progression through activating ROS/p38 MAPK/LDs pathway in leukemia cells and inhibiting M2 macrophage polarization.
2. Materials and methods
2.1. Cell culture
Human AML cell lines THP1 and Molm-13 were cultured in RPMI-1640 medium (Gibco, USA) with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin/streptomycin (Gibco, USA) in an incubator at 37 °C and 5% CO2. Bone marrow derived macrophages (BMDMs) were gathered from the femurs of C57BL/6 mice, and cultured in DMEM medium (Gibco, USA) containing 10% FBS and 1% penicillin/streptomycin as well as 100 ng/mL M-CSF (PeproTech, USA). After 7 days, cells were pretreated with 0.12 mmol/L CDCA for 1h before adding 20 ng/mL IL-4 (PeproTech, USA) to induce macrophage polarizing towards M2 subtype. Murine peritoneal derived macrophages were extracted from C57BL/6 mice after being intraperitoneally injected with 3% thioglycollate broth (Solarbio, China) for 3 days, and cultured with DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin for 24 h. Then the supernatant was discarded, and the cells were pretreated with 0.12 mmol/L CDCA for 1h before adding IL-4 as mentioned above.
2.2. Patient specimens
Peripheral blood samples utilized in this study were obtained from 17 newly diagnosed and non-APL AML patients and 21 normal people. Bone marrow samples were collected from 8 newly diagnosed and non-APL AML patients. These specimens were collected at Qilu Hospital of Shandong University. The plasma of peripheral blood was used for quantitative detection of bile acids by targeted MRM analysis. Bone marrow samples were used for apoptosis assay after treatment with different concentrations of CDCA. The informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The study was approved by the Medical Ethical Committee of Qilu Hospital of Shandong University.
2.3. Murine model and treatment
C57BL/6 mice (male, 6∼8-week old) and NOD-Prkdcscid-Il2rgem1IDMO (NPI) mice (male, 6∼8-week old) were purchased from the Beijing Vital River Laboratory Animal Technology company. C1498 cells transfected with GFP (106 cells/host) were transplanted into mice via tail vein injection to establish homograft AML murine model. After 24 days, mice were sacrificed and mononuclear cells of bone marrow, spleen and liver were gathered by erythrocyte lysate. The percentage of GFP + cells among mononuclear cells was detected by flow cytometry. Besides, liver and spleen samples of mice were obtained for weighing and HE as well as Ki67 staining. THP1 cells transfected with GFP were transplanted into NPI immunodeficient mice to establish xenograft AML murine model via tail vein injection. Animal protocols were authorized by the Animal Ethics Committee of Qilu Hospital of Shandong University.
2.4. Cells transfection
C1498 cells or THP1 cells were seeded into 24-well plates at the density of 1 × 105 cells/well. Lentivirul-GFP was constructed by Shanghai Genechem company (Shanghai, China). Then we transfected C1498 cells or THP1 cells with Lentivirul-GFP with multiplicity of infection (MOI) of 100 or 30 respectively. Cells labeled with GFP were selected by puromycin. The transfection fluorescence was detected by fluorescence microscope.
2.5. Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using Trizol (Invitrogen, USA) and then was reverse transcribed into cDNA through the PrimeScript RT reagent Kit Perfect Real Time (Takara Bio, Japan). The quantitative PCR was performed on the LightCycler 480II real-time PCR system (Roche, Switzerland). The primers were purchased from BioSune Company (Shanghai, China) and shown as follows: CD163: forward primer: TTT-GTC-AAC-TTG-AGT–CCC–TTC-AC, reverse primer: TCC-CGC-TAC-ACTT-GTT-TTC-AC. ARG-1: forward primer: TGT–CCC–TAA-TGA-CAG-CTC-CTT, reverse primer: GCA-TCC-ACC-CAA-ATG-ACA-CAT. CD206: former primer: ACG-AGC-AGG-TGC-AGT-TTA-CA, reverse primer: ACA-TCC-CAT-AAG-CCA-CCT-GC. GAPDH (mouse): former primer: AGG-TCG-GTG-TGA-ACG-GAT-TTG, reverse primer: GGG-GTC-GTT-GAT-GGC-AAC-A. GAPDH (human): former primer: GCT-CTC-TGC-TCC-TCC-TGT-TC, reverse primer: GTT-GAC-TCC-GAC-CTT-CAC-CT IP3R1: former primer: GAG-AAT-AAA-GGC-AGT-AAC-GTG-ATG-A, reverse primer: GCT-CTG-CCT-GCT-TCA-CAT-TG. IP3R2: former primer: TCA-GCA-CCT-TGG-GGT-TAG-TG, reverse primer: TGT-GGT-TCC-CTT-GTT-TGG-CT. Gene expression data were analyzed using 2−ΔCT method.
2.6. Cell viability assay
Cell viability was determined by CCK8 assay. Briefly, THP1 and Molm-13 cells were adjusted to 1 × 105 cells/mL and treated with CDCA at the concentration of 0, 0.1, 0.12, 0.15, 0.17 and 0.2 mmol/L for 24h or 48h. The cells were seeded into 96-well plates, and OD value was determined after adding 10 μL CCK8 (Bestbio Company, China) for 4 h in a humidified incubator at 37 °C and 5% CO2. The absorbance was measured at 450 nm.
2.7. The determination of EdU
Cell proliferation was determined by the way of EdU to examine DNA synthesis. EdU assay was detected by flow cytometry using iClick™ EdU Andy Fluor 647 Flow Cytometry Assay Kit (ABP-Biosciences, China).
2.8. Colony formation assay
For colony formation assay, cells were planted into 6-well plate at the density of 5 × 105 cells/well treated with 0, 0.12 and 0.17 mmol/L CDCA for 24 h, and then 10 μL suspension was seeded into solid medium in an incubator at 37 °C and 5% CO2 for 2 weeks. The colony formation was detected by microscope to measure the formation of clusters. Image J was used to analyze the number of clusters.
2.9. Apoptosis assay
Apoptosis assay was detected by flow cytometry using Annexin V/propidium iodide (PI) apoptosis detection kit (BestBio Company, China). Cells were seeded into 6-well plates at the concentration of 5 × 105 cells/well for 24h, and treated with different concentrations of CDCA with or without pretreatment with N-acetylcysteine (NAC) (Selleck Chemicals, USA), SB203580 (MCE, China), Ferrostatin-1 (Selleck Chemicals, USA), or Z-VAD-FMK (Selleck Chemicals, USA) for 1h. Then the cells were collected and suspended with 300 μL binding buffer including 5 μL Annexin-V-FITC and 10 μL PI for 15min at 4 °C. The percentage of apoptotic cells was analyzed using a Gallios flow cytometer (Beckman Coulter, CA, USA).
2.10. BODIPY 493/503 analysis
We first treated THP1 and Molm-13 cells with 0, 0.12 and 0.17 mmol/L CDCA for 24h. Then AML cells were gathered and incubated with 1 μmol/L BODIPY 493/503 (Shanghai Maokang Biotechnology, China) for 30 min at 37 °C. After being washed twice by PBS, the cells were counterstained with DAPI and covered with glass slides for confocal microscope detection. The mean fluorescence intensity was used for analysis. Besides, AML cells were pretreated with A922500 (MCE, China) for 1h before CDCA treatment to examine the level of LDs using flow cytometry.
2.11. The determination of Oil Red O staining
For Oil Red O staining assay, the cells were collected, covered on glass slides and stained using Oil Red O staining Kit (Bestbio Company, China) following a standard protocol. Microscope was used to document the results.
2.12. BODIPY 581/591 C11 assay
AML cells were seeded into 6-well plates at the concentration of 1 × 106 cells/well for 24h and incubated with 5 μmol/L BODIPY 581/591 C11 (Invitrogen, USA) at 37 °C for 15 min. Then we collected cells and observed the ratio of green to red fluorescence using confocal microscope.
2.13. Measurement of GSH level
A total of 5 × 106 cells were added into every well and treated with different concentrations of CDCA. After 24h, the cells were managed following the procedure of GSH detection kit (Solarbio, China) and detected by microplate reader.
2.14. Mitochondrial mass assay
AML cells were incubated with 0.5 μg/mL mitotracker deep red (MDR) staining (Invitrogen, USA) at 37 °C for 30 min. MDR was detected using confocal microscope imaging and was analyzed by mean fluorescence intensity.
2.15. Measurement of mitochondrial calcium level
AML cells were incubated with 1 μmol/L Rhod-2 AM (Abcam, USA), a fluorescent Ca2+ indicator, at 37 °C for 30 min to examine the level of calcium among mitochondria. Rhod-2 AM was detected by confocal microscope imaging and was analyzed by mean fluorescence intensity.
2.16. Mitochondrial membrane potential assay
We determined the mitochondrial membrane potential with TMRE or JC-1 staining. AML cells were incubated with 0.1 mmol/L TMRE (Beyotime, China) at 37 °C for 30 min, and then were detected by flow cytometry to analyze the mitochondrial membrane potential. For JC-1 detection, cells were collected and suspended with JC-1 working solution following the protocol of JC-1 kit (Bestbio, China), and then detected by flow cytometry.
2.17. Measurement of cytoplasm ROS level
AML cells were incubated with 0.1 mmol/L DCFH-DA (Beyotime, China) at 37 °C for 30 min, and then were detected by flow cytometry to examine the cytoplasm ROS production by analyzing mean fluorescence intensity.
2.18. Measurement of mitochondrial superoxide (mitoSOX) level
AML cells were incubated with 0.1 mmol/L mitoSOX (Bestbio company, China) at 37 °C for 30 min, and then were collected and covered on glass slides for confocal microscope detection to analyze the level of mitochondrial ROS.
2.19. FITC-CDCA synthesis
CDCA was labeled with FITC fluorescence by Xianqiyuebio Company (Xian, China), and then added into AML cells to visualize its location in AML cells.
2.20. Bile acids and microbiota diversity measurement
The data of bile acids and microbiota diversity of feces were obtained by using LC-MS method according to the procedure of our previous study [28]. For bile acids detection in peripheral blood serum, extracts were analyzed using MRM technology by Applied Protein Technology company (Shanghai, China).
2.21. Transmission electron microscope (TEM) analysis
THP1 and Molm-13 cells were fixed in 2.5% glutaraldehyde at 4 °C for 4h and then were post-fixed with 1% osmic acid in 0.1 M phosphate buffer (PH7.4) for 2h at room temperature. After dehydration and embedding, 60–80 nm ultrathin slices were cut and stained by uranium-lead double staining. TEM (Hitachi, HT7700) was used to observe the morphology changes of AML cells.
2.22. Western blot analysis
Cells were obtained and lysed with the Total Protein Extraction Kit (Bestbio Company, China). Protein concentration was determined by BCA kit. Equal amount of each sample protein (30 μg) added with loading buffer was loaded onto 7.5%, 10% or 15% SDS-PAGE gels, and then electrotransferred onto 0.2 μm PVDF membranes. After blocking for 1h at room temperature, the PVDF membranes were incubated with antibodies against DGAT1 (ab181180, Abcam, USA), p38 (#8690, CST, USA), p-p38 (#4511, CST, USA), GPX4 (#59735S, CST, USA), ASK1 (A12458, ABclonal, China), Plin1 (A16295, ABclonal, China), β-tubulin (AB0058, Abways technology, China) or GAPDH (AB0038, Abways technology, China) overnight at 4 °C and detected by SageCapture Chemiluminescent imaging system (ChampChemi 610 Plus, China) the next day after conjugating with secondary antibodies.
2.23. Statistical analysis
Data were analyzed by GraphPad and SPSS 15.0 (SPSS, Science, Chicago, IL, USA) and represented as mean ± SEM. The significance of difference was determined by the student's t-test and the Mann-Whitney U test. Difference was meaningful at P < 0.05.
3. Results
3.1. Microbiota-associated CDCA is decreased in feces and plasma of AML patients and its low level predicts poor prognosis
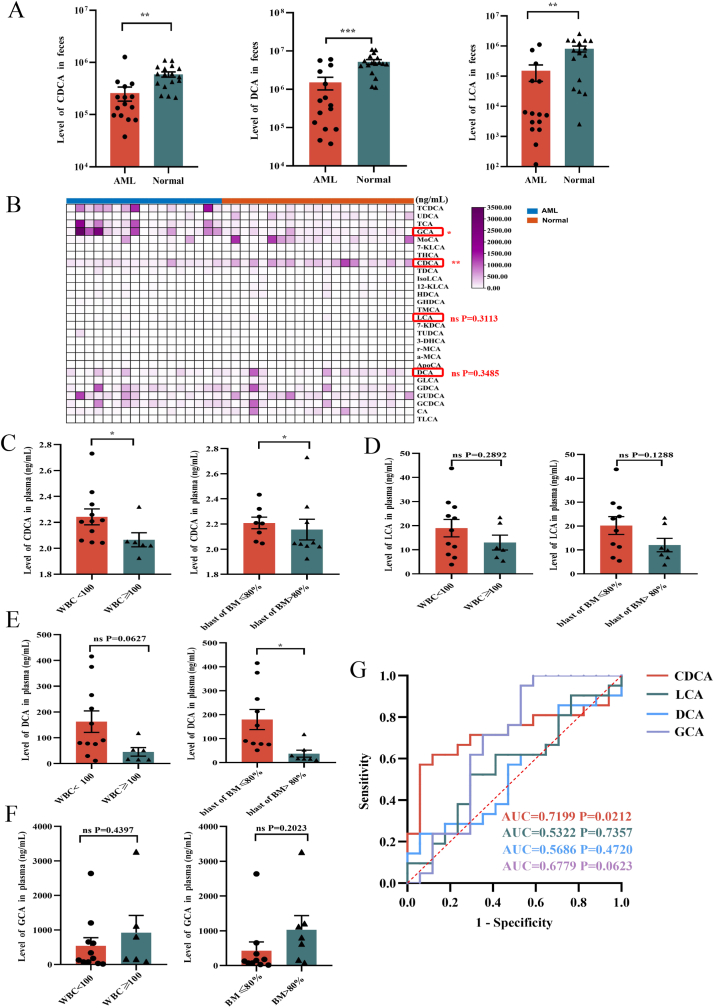
Bile acids have been reported to have essential effects on many diseases, such as metabolic diseases and gastrointestinal diseases [29,30], and little is known about their roles in AML. To investigate whether bile acids play a key role in AML progression, we first utilized the data of our previous LC-MS determination to analyze three typical bile acids, CDCA, DCA and LCA in the stool samples from 15 newly diagnosed and untreated AML patients and 17 normal people. The results demonstrated that the level of CDCA, DCA or LCA was significantly reduced in AML patients compared to normal people (Fig. 1A). Our previous results revealed that the diversity of gut microbiota was significantly decreased in AML patients and was correlated with the change of metabolites [28]. Since bile acids were tightly correlated with microbiota metabolism, we analyzed Shannon index, the alpha diversity index of microbiota, in the feces of participants to clarify the association between microbiota with CDCA, DCA or LCA. Our data showed that there existed a significant positive correlation between Shannon index with CDCA or LCA, while DCA didn't show obvious difference (Table 1), indicating that the metabolism of bile acids including CDCA and LCA in feces was involved in the gut microbiota homeostasis.
Fig. 1.
Microbiota-associated CDCA is decreased in feces and plasma of AML patients and its low level predicts poor prognosis
(A)The levels of chenodeoxycholic acids (CDCA), Lithocholic acids (LCA), and Deoxycholic acids (DCA) in fecal samples of AML patients (n = 15) and normal people (n = 17) were determined by LC-MS. (B) Heatmap of abundance of different bile acids in periphery blood serum from AML patients (n = 17) and normal people (n = 21). (C) The relation between the level of CDCA and the white blood cells in peripheral blood or the blast cells in bone marrow was shown. (D–F) The relation between the level of DCA, LCA or GCA with the white blood cells in peripheral blood or the blast cells in bone marrow was shown. (G) The ROC curves of CDCA, LCA, DCA or GCA were shown. P-values were determined using student's t-test and Wilcoxon rank test. Error bars represent as mean ± SEM. ns: Not significant; *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
Table 1.
Linear correlation analysis.
| Variables |
Correlations |
|||||
|---|---|---|---|---|---|---|
| CDCA |
LCA |
DCA |
||||
| r | p | r | p | r | p | |
| Shannon index | 0.367 | 0.039 | 0.371 | 0.043 | 0.307 | 0.099 |
r: correlation coefficient.
To confirm whether the differences of bile acids in feces could affect their level in blood, we further collected peripheral blood serum from newly diagnosed and untreated AML patients and normal people for quantitative measurements of CDCA, DCA and LCA by targeted MRM analysis. We found that the level of CDCA was significantly decreased in the serum of AML patients compared with controls, while the change of DCA or LCA didn't reach statistical significance. In addition, we observed that there existed an obvious increase of GCA in AML patients (Fig. 1B, Fig. S1A). To further determine the relationship between bile acids and AML, we analyzed the correlation between the levels of bile acids and the clinical characteristics of AML patients. The level of CDCA in AML patients was negatively correlated with the white blood cells in peripheral blood or the blast cells in bone marrow, while there was no significant association between DCA, LCA or GCA levels with AML patients' clinical characteristics (Fig. 1C–F). To further analyze the association of bile acids and prognosis, the achievement of complete remission (CR) after first therapy was employed as the outcome index. The results showed that the CR rate of the patients with favorable and intermediate risk was higher than that of the patients with poor risk (Table 2). Moreover, the level of CDCA was higher in CR patients compared with non-CR patients, while DCA, LCA or GCA exhibited less obvious difference, but all data didn't reach statistical significance. The results suggested that CDCA might have an influence to the achievement of AML patients' CR after treatment (Fig. S1B). To analyze the impacts of bile acids on clinical diagnosis, we made Receiver Operating Characteristic Curve (ROC curve) of CDCA, LCA, DCA, as well as GCA for prediction. We observed that CDCA had much higher accuracy and sensitivity for diagnosis compared with DCA, LCA or GCA (Fig. 1G). Consequently, these data demonstrate that CDCA plays an important role in the progression of AML and might act as a biomarker for AML prognosis.
Table 2.
CR rate analysis of patients.
| Risk stratification | CR | Total | CR/total |
|---|---|---|---|
| Favorite and Intermediate | 6 | 13 | 0.46 |
| Poor/adverse | 1 | 4 | 0.25 |
3.2. CDCA inhibits the proliferation and promotes the apoptosis of AML cells
To investigate the biological effects of CDCA on AML cells, we used CCK8 assay to determine the effects of different concentrations of CDCA (0, 0.1, 0.12, 0.15, 0.17, 0.2 mmol/L) on the viability of AML cells, THP1 and Molm-13 cells, after 24h or 48h. We found that CDCA significantly reduced the frequency of viability of THP1 or Molm-13 cells compared to control groups (Fig. 2A). According to the results of CCK8 assay, we used the intermediate concentration of 0.12 and 0.17 mmol/L for the following study. To further clarify the proliferation-inhibiting effects of CDCA, we employed EdU cell proliferation assay to measure DNA synthesis. The results showed that CDCA obviously inhibited the percentage of AML cells in S phase (Fig. 2B). Moreover, we used the colony formation assay to detect the effect of CDCA on the colony formation ability of AML cells, and the results showed that clone number was significantly decreased in AML cells treated with CDCA compared with controls (Fig. 2C). All these results demonstrate that CDCA inhibits AML cells proliferation. Next, we used flow cytometry with Annexin V/PI staining to determine the apoptotic changes after CDCA treatment. Our results showed that CDCA increased the frequency of apoptosis of AML cells compared with controls (Fig. 2D). Moreover, we further employed apoptosis inhibitor Z-VAD-FMK to validate the role of CDCA on apoptosis. The data revealed that Z-VAD-FMK reduced the frequency of death cells caused by CDCA (Fig. S1C).
Fig. 2.
CDCA inhibits the proliferation and promotes the apoptosis of AML cells
(A) CCK8 analysis of cell viability of THP1 (n = 4) and Molm-13 (n = 4) treated with CDCA at different concentrations of 0, 0.10, 0.12, 0.15, 0.17, 0.20 mmol/L at 0, 24, 48h was shown. (B) EdU analysis of cell proliferation of THP1 (n = 3) and Molm-13 (n = 6) at concentrations of 0, 0.12, 0.17 mmol/L CDCA for 24h was shown. And the corresponding histograms were shown on the right. (C) The colony formation assays of THP1 (n = 3) and Molm-13 (n = 3) at concentrations of 0, 0.12, 0.17 mmol/L CDCA for 24h were shown. And the corresponding histograms were shown on the right. (D) The apoptosis analysis of THP1 (n = 6) and Molm-13 (n = 3) at concentrations of 0, 0.12, 0.17 mmol/L CDCA for 24h was shown. And the statistical histograms were shown on the right. (E) The apoptosis assay of primary leukemia cells after being treated with 0, 0.12, 0.17 mmol/L CDCA for 24h was shown (n = 8). And the corresponding histograms were shown on the right. P-values were determined using student's t-test or Wilcoxon rank test. Error bars represent mean ± SEM. ns: Not significant; *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
Furthermore, to validate the generalizability of our findings, we separated the primary leukemia cells of bone marrow from eight newly diagnosed and untreated AML patients and treated the cells with CDCA at the same manner. Our results showed that the apoptotic frequency of the primary AML cells was significantly increased after treatment with CDCA (Fig. 2E). Taken together, all these results demonstrate that CDCA inhibits the proliferation and promotes the apoptosis of AML cells lines and primary AML cells in vitro, which suggests that CDCA might be a promising therapeutic agent for AML.
3.3. CDCA suppresses AML progression in homograft and xenograft mouse models
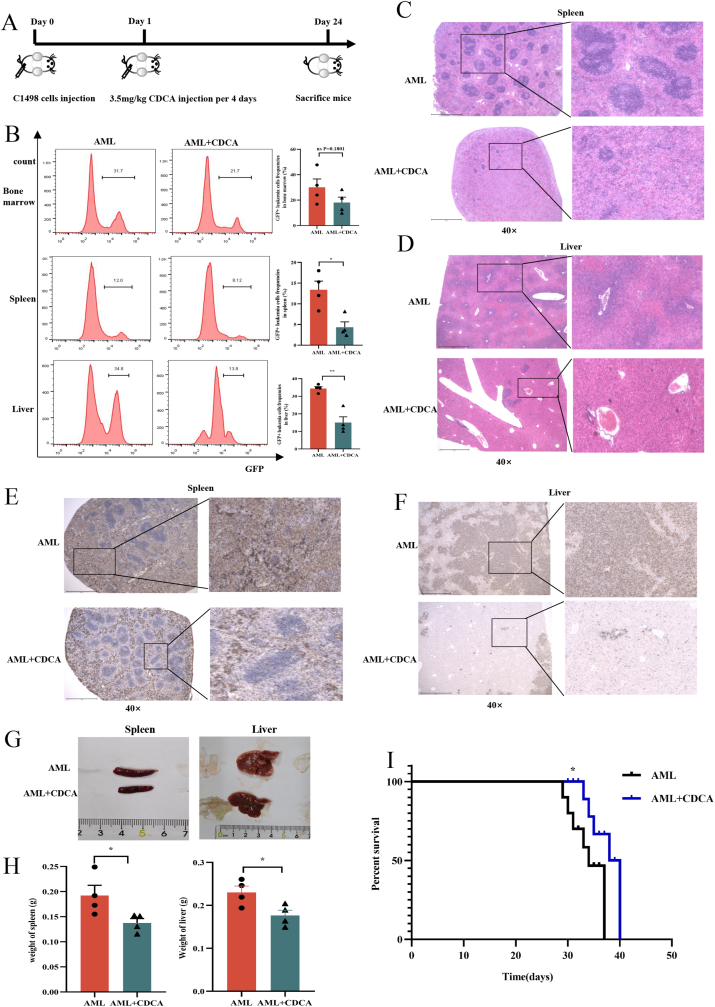
To further validate the clinical significance of CDCA in AML, we designed homograft and xenograft AML mouse models to examine its effects. First, we injected C57BL/6 mice with murine leukemia cells C1498. GFP to establish homograft AML mouse model, and then applied CDCA on them via tail vein injection every 4 days to evaluate its effect on AML progression (Fig. 3A). After sacrificing the mice, the results showed that the frequency of GFP+ leukemia cells in bone marrow, spleen and liver in CDCA-treated AML mice was significantly lower than that in control AML mice (Fig. 3B). Furthermore, we used haematoxylin and eosin (HE) staining and Ki67 immunohistochemical staining methods to evaluate the infiltration and proliferation of leukemia cells in spleen or liver. As expected, the results showed that the infiltration and proliferation of leukemia cells were attenuated in CDCA-treated AML mice (Fig. 3C–F). Consistently, the decreased size and weight of the spleen or liver in CDCA-treated AML mice compared with controls were also verified (Fig. 3G and H).
Fig. 3.
CDCA suppresses AML progression in homograft and xenograft mouse models
(A) Schematic diagram of the mouse experimental processes. (B) The frequency of leukemia cells (GFP + leukemia cells) in bone marrow, spleen and liver from AML-control mice (n = 4) and AML-CDCA mice (n = 4) was shown. And the corresponding histograms were shown on the right. (C–F) Representative images of HE histopathology and Ki67 immunohistochemical staining sections of spleen and liver from AML-CDCA group and AML-control group (n = 4) were shown. All microscopic analysis was performed with original magnification × 40. Scale bar = 1000 μm. (G–H) The photographs and weights of spleens and livers from AML-control mice (n = 4) and AML-CDCA mice (n = 4) were shown. (I) The Kaplan-Meier survival curve of AML mice was shown (n = 6). P-values were determined using student's t-test and Wilcoxon rank test. Error bars represent mean ± SEM. ns: Not significant; *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
Moreover, we established xenograft AML mouse model through transplanting THP1. GFP cells into NPI immunodeficient mice via tail vein injection. Then we administered CDCA on the AML mice to determine its role on the survival. Our results showed that AML mice with CDCA treatment had a longer overall survival compared with control AML mice [median 33 (range 29–37) days vs median 36.5 (range 33–40) days (P = 0.0309)] (Fig. 3I). Taken together, all these results demonstrate that CDCA exhibits suppressive effects on AML progression in vivo.
3.4. CDCA binds to mitochondria and induces mitochondrial dysfunction in AML cells
Since mitochondria are essential for the cell proliferation and apoptosis [31], we next examined whether CDCA could induce the mitochondrial changes of AML cells. We first used FITC-labeled CDCA and Mitotracker Deep Red (MDR) to visualize the localization of CDCA and mitochondria in AML cells. Our results showed that CDCA could directly bind to the mitochondria (Fig. 4A).
Fig. 4.
CDCA binds to mitochondria and induces mitochondrial dysfunction in AML cells
(A) THP1 or Molm-13 cell were treated with 0.17 mmol/L FITC-labeled CDCA. After 24h, AML cells were collected and incubated with MDR and were detected by confocal microscope to visualize the location of CDCA and mitochondria. The representative images were shown. Scale bar = 5 μm (B) Representative images of mitochondria in THP1 and Molm-13 detected by TEM were shown. Scale bar = 1 μm or 0.2 μm. (C) Representative images of mitochondrial mass of THP1 and Molm-13 using MDR detected by confocal microscope were shown (n = 3). And the corresponding histograms were shown on the right. Scale bar = 10 μm. (D) Mitochondrial membrane potential of THP1 and Molm-13 using JC-1 measured by flow cytometry was shown (n = 3). And the corresponding histograms were shown on the right. (E) Rhod-2 AM kit was used to examine calcium among mitochondria in THP1 (n = 3) and Molm-13 (n = 4) cells. Representative images detected by confocal microscope were shown. And the corresponding histograms were shown on the right. Scale bar = 10 μm. P-values were determined using student's t-test and Wilcoxon rank test. Error bars represent mean ± SEM. ns: Not significant; *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
We next determined the morphology and function of mitochondria in AML cells after being treated with CDCA. The results showed that CDCA induced the obviously morphological abnormalities of mitochondria including mitochondrial swelling and the decrease of cristae in AML cells (Fig. 4B). To investigate whether CDCA was involved in mitochondrial dynamics and activities, we first examined mitochondrial mass with MDR staining. The fluorescence density of MDR was significantly decreased after CDCA treatment, suggesting that CDCA caused the reduction of mitochondrial mass (Fig. 4C). Next, we determined the mitochondrial membrane potential with JC-1 or TMRE staining. The results from both staining tests demonstrated that the mitochondrial membrane potential was significantly decreased in CDCA-treated AML cells compared with control groups (Fig. 4D, Fig. S1D).
As reported, the relationship between mitochondrial integrity and calcium level was deeply intertwined [32]. To clarify the subsequent calcium variation caused by mitochondrial membrane damages, we detected the level of calcium among mitochondria by using Rhod-2 AM assay. Our results exhibited that the level of calcium among mitochondria was significantly increased after CDCA treatment in AML cells (Fig. 4E). Mechanistically, we further detected the mRNA expression of calcium related receptors, Inositol 1,4,5-trisphosphate receptor1 (IP3R1) and IP3R2. As predicted, the expression levels of IP3R1 and IP3R2 were significantly increased after CDCA treatment (Fig. S1E). All these results demonstrate that CDCA binds to mitochondria and induces mitochondrial morphological and functional abnormality.
3.5. CDCA-induced mitochondrial dysfunction causes excessive ROS production, which further activates p38 MAPK in AML cells
It was reported that mitochondrial integrity was vital for cellular energy production and cell survival, and mitochondrial dysfunction would promote ROS production [33]. Therefore, we first analyzed the levels of ROS in cytoplasm of AML cells using DCFH-DA kit by flow cytometry. Our results showed that the fluorescence density was significantly increased in CDCA-treated AML cells compared with controls (Fig. 5A), indicating that CDCA promoted ROS production in AML cells. Furthermore, we used mitoSOX indicator to analyze the ROS level among mitochondria of AML cells. The results showed that the level of mitoSOX was obviously elevated after being treated with CDCA, which was consistent with the change of ROS in cytoplasm (Fig. 5B). All these results suggest that CDCA induces higher ROS levels both in cytoplasm and mitochondria of AML cells.
Fig. 5.
CDCA-induced mitochondrial dysfunction causes excessive ROS production, which further activates p38 MAPK in AML cells
(A) DCFH-DA was used to examine ROS in cytoplasm of THP1 (n = 4) and Molm-13 (n = 3) cells after being treated with 0, 0.12, 0.17 mmol/L CDCA for 24h. Representative images detected by flow cytometry were shown. And the corresponding histograms were shown below the images. (B) MitoSOX assay was employed to determine ROS in mitochondria of THP1 and Molm-13 cells after being treated with 0, 0.12, 0.17 mmol/L CDCA for 24h. Representative images detected by confocal microscope were shown (n = 3). And the corresponding histograms were shown below the images. Scale bar = 10 μm. (C) Western blot analysis of p38 and p-p38 in THP1 and Molm-13 cells after being treated with 0, 0.12, 0.17 mmol/L CDCA for 24h was shown. (D) Western blot analysis of p38 and p-p38 in THP1 and Molm-13 cells after being treated with 0, 0.12, 0.17 mmol/L CDCA for 24h with or without NAC pretreatment was shown. (E) Apoptosis assays of THP1 (n = 6) and Molm-13 (n = 4) cells after being treated with 0, 0.12, 0.17 mmol/L CDCA for 24h with or without NAC pretreatment was shown. And the corresponding histograms were shown on the right. (F) Apoptosis assays of THP1 (n = 3) and Molm-13 (n = 3) cells after being treated with 0, 0.12, 0.17 mmol/L CDCA for 24h with or without p38 inhibitor pretreatment was shown. And the corresponding histograms were shown on the right. P-values were determined using student's t-test and Wilcoxon rank test. Error bars represent mean ± SEM. ns: Not significant; *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
As ROS accumulation and mitochondrial damage have been reported to activate MAPK pathway [34], we next determined the expression change of a typical molecule p38 MAPK. Western blot results showed that the level of phosphorylated p38 (p-p38) was significantly elevated after being treated with CDCA in AML cells compared with control group, while the total p38 protein expression had no apparent changes (Fig. 5C). As reported, apoptosis signal-regulating kinase (ASK) 1 regulated the activation of p38 MAPK [35], we detected the ASK1 protein expression by Western blot. We found that the level of ASK1 was increased after CDCA treatment, which further verified the activation of p38 MAPK (Fig. S1F). To clarify the correlation between ROS and p38 MAPK, we next pretreated NAC, an ROS clearance agent, to the CDCA-treated AML cells to examine the change of phosphorylation of p38. Our data exhibited that the level of p-p38 was remarkably decreased after NAC pretreatment, while the expression of total p38 didn't show significant difference, which indicated that ROS activated p38 MAPK pathway in AML cells (Fig. 5D).
Since ROS and p38 MAPK played an important role in leukemia-inhibition caused by CDCA, we next used NAC or P38 inhibitor (SB203580) to clear ROS or inhibit p38 activation respectively. The results showed that when adding NAC or p38 inhibitor, the apoptotic rate induced by CDCA was decreased compared with control groups, which meant that inhibition of ROS or p38 reversed the cytotoxicity of CDCA on AML cells (Fig. 5E and F).
3.6. CDCA causes LDs accumulation and lipid peroxidation through ROS/p38 MAPK/DGAT1 pathway in AML cells
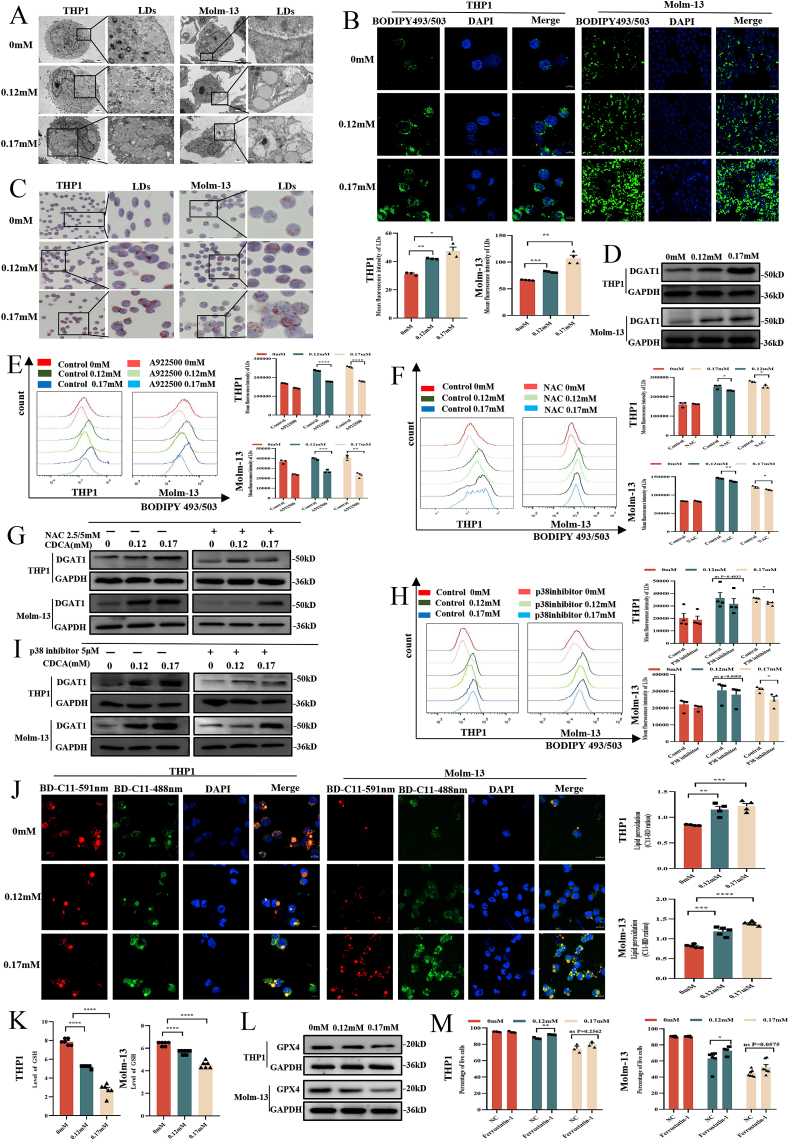
Increasing evidence has shown that CDCA caused lipid abnormalities in tumor progression [36], and lipid droplets abundance has been closely related to ROS accumulation and mitochondria [37]. Therefore, we further detected the effects of CDCA on LDs in AML cells. The transmission electron microscope (TEM) showed a significant accumulation of LDs with round, homogenous and low-density structures in AML cells treated with CDCA compared with control groups (Fig. 6A). Meanwhile, cells stained with BODIPY 493/503 or Oil Red O staining further confirmed the results that CDCA caused LDs accumulation in AML cells (Fig. 6B and C). Since it was reported that Perilipin1 (Plin1) was a kind of LDs surface protein [38], we next detected Plin1 protein expression in AML cells. Our results showed that the level of Plin1 was increased after CDCA treatment compared with control groups (Fig. S1G). These results suggest that CDCA promotes LDs accumulation in AML cells.
Fig. 6.
CDCA causes LDs accumulation and lipid peroxidation through ROS/p38 MAPK/DGAT1 pathway in AML cells
(A) Representative images of LDs in THP1 and Molm-13 detected by transmission electron microscope (TEM) were shown. The scale bar was 1 μm. (B) Representative images of LDs of THP1 (n = 3) and Molm-13 (n = 4) using BODIPY 493/503 measured by confocal microscope were shown. And the corresponding histograms were shown below the images. Scale bar = 10 μm. (C) Representative images of THP1 and Molm-13 using Oil Red O staining were shown. (D) Western blot analysis of DGAT1 protein after CDCA treatment was shown. (E) The representative images of LDs in THP1 and Molm-13 cells after 0, 0.12, 0.17 mmol/L CDCA treatment for 24h with or without A922500 pretreatment were shown (n = 3). And the corresponding histograms were shown on the right. (F) The representative images of LDs in THP1 and Molm-13 cells after being treated with 0, 0.12, 0.17 mmolm/L CDCA with or without NAC pretreatment were shown (n = 3). And the corresponding histograms were shown on the right. (G) Western blot analysis of DGAT1 in THP1 and Molm-13 cells after 0, 0.12, 0.17 mmol/L CDCA treatment for 24h with or without NAC pretreatment was shown. (H) The representative images of LDs in THP1 and Molm-13 cells after 0, 0.12, 0.17 mmol/L CDCA treatment for 24h with or without p38 inhibitor pretreatment were shown (n = 4). And the corresponding histograms were shown on the right. (I) Western blot analysis of DGAT1 in THP1 and Molm-13 cells after 0, 0.12, 0.17 mmol/L CDCA treatment for 24h with or without p38 inhibitor pretreatment was shown. (J) Representative images of THP1 (n = 4) and Molm-13 (n = 5) using BODIPY 581/591 C11(BD-C11) staining were shown. Red (591 nm) and green (488 nm) fluorescence indicate total and peroxidized lipids respectively. Quantitation of lipid peroxidation (the ratio of green to red) was exhibited on the right. Scale bar = 10 μm. (K) Quantitation of GSH levels are shown after CDCA treatment (n = 6). (L) Western blot analysis of GPX4 protein after CDCA treatment was shown. (M) The apoptosis analysis of the corresponding statistical histograms of THP1 (n = 3) and Molm-13 (n = 7) after ferroptosis inhibitor ferrostatin-1 treatment at concentrations of 0, 0.12, 0.17 mmol/L CDCA for 24h were shown. P-values were determined using student's t-test and Wilcoxon rank test. Error bars represent mean ± SEM. ns: Not significant; *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
It has been reported that the DGAT1 played a significant role in LDs synthesis [14]. To explore the mechanism of LDs accumulation caused by CDCA, we first examined DGAT1 protein expression with Western blot in AML cells treated with different doses of CDCA. Our results showed that DGAT1 protein expression was increased after CDCA treatment (Fig. 6D). We further used DGAT1 inhibitor A922500 to clarify the influence of DGAT1 on LDs biogenesis in leukemia cells. Our data revealed that CDCA-induced LDs accumulation was alleviated after pretreatment with DGAT1 inhibitor, which further verified the promoting effect of DGAT1 on LDs accumulation in AML cells (Fig. 6E).
Since ROS was vital for LDs biogenesis, we first employed NAC to CDCA-treated AML cells to identify the role of ROS in LDs accumulation. We observed that LDs accumulation induced by CDCA in AML cells was shown to be significantly decreased after eliminating ROS by NAC (Fig. 6F). Moreover, the level of DGAT1 expression was evidently down-regulated after NAC treatment (Fig. 6G). As shown above that p38 MAPK was activated by ROS, we further verified the effect of p38 MAPK on LDs accumulation. Our results showed that p38 MAPK inhibitor SB203580 reduced LDs accumulation and downregulated DGAT1 expression as well (Fig. 6H and I). Taken together, the accumulation of LDs in AML cells is the result of the activation of ROS/p38 MAPK/DGAT1 pathway.
Since the excessiveness of ROS and LDs could lead to the lipid peroxidation [39], we next explored whether accumulated lipids would be peroxidated. We used BODIPY 581/591 C11 by confocal microscope to visualize peroxidation of lipids. Following peroxidation, the fluorescence emission shifts from red to green [40]. The results showed that there existed an abnormal abundance of peroxidized lipids in THP1 and Molm-13 cells treated with CDCA compared with control groups (Fig. 6J). Moreover, as GSH and GPX4 was reported to be the regulators of the lipid peroxidation [41], we detected the GSH and GPX4 levels in AML cells. We observed that the levels of GSH and GPX4 were significantly decreased after CDCA treatment (Fig. 6K and L). To further testify whether ferroptosis played a role in lipid peroxidation caused by CDCA treatment, we next employed ferroptosis inhibitor ferrostatin-1. Our results revealed that the frequency of cell death caused by CDCA reduced after ferrostatin-1 pretreatment (Fig. 6M, Fig. S1H). Collectively, the increased level of peroxidated lipid and the reduced level of GSH or GPX4, as well as ferrostatin-1, together demonstrate the involvement of ferroptosis in lipid peroxidation caused by CDCA treatment.
3.7. CDCA inhibits AML progression by alleviating M2 macrophage polarization
Bile acids have also been reported to play an essential role in macrophages polarization [24]. Considering the leukemia-inhibiting effects caused by CDCA as shown above, we determined the role of CDCA in M2 macrophage polarization. We first isolated murine bone marrow derived macrophages (BMDMs), and stimulated BMDMs with M-CSF for seven days. Then, we pretreated the cells with 0.12 mmol/L CDCA for 1h, and stimulated cells with IL-4 to induce cells polarizing towards M2 macrophage. We determined the macrophage phenotype using RT-qPCR and flow cytometry. Our results showed that the mRNA expression levels of M2 macrophage signature genes, CD206, CD163 and ARG-1, were significantly down-regulated in macrophages treated with CDCA compared with controls (Fig. 7A). Meanwhile, flow cytometry results revealed that the frequency of CD206+ cells was decreased in CDCA-treated macrophages compared with control groups (Fig. 7C). Moreover, in murine peritoneal derived macrophages, we also found that CDCA exhibited M2-inhibiting effects (Fig. 7B). To further investigate the effects of M2 macrophage polarization on AML proliferation, M2 macrophages were induced with or without 0.12 mmol/L CDCA pretreatment, and then after 24h, M2 macrophages were replaced with fresh media and co-cultured with C1498 cells for another 24h. The cell viability of C1498 cells was detected by CCK8 assay. We found that CDCA could inhibit the AML-promoting effects of M2 macrophages (Fig. 7D). Therefore, CDCA inhibits M2 macrophage polarization, which might play synergistic effects on alleviating AML progression
Fig. 7.
CDCA inhibits AML progression by alleviating M2 macrophage polarization
(A) The mRNA expression results of CD206, CD163 and ARG-1 in M2 macrophages (n = 3) derived from BMDMs with or without CDCA (0.12 mmol/L) pretreatment were shown. (B) The mRNA expression results of CD206, CD163 and ARG-1 in M2 macrophages derived from murine peritoneal derived macrophages with or without CDCA (0.12 mmol/L) pretreatment were shown (n = 3). (C) The percentage of M2 macrophages with or without CDCA (0.12 mmol/L) pretreatment detected by flow cytometry were shown. (D) M2 macrophages were induced with or without CDCA(0.12 mmol/L) pretreatment. After that, C1498 cells were co-cultured with M2 macrophages. CCK8 assays were shown (n = 7). P-values were determined using student's t-test and Wilcoxon rank test. Error bars represent mean ± SEM. ns: Not significant; *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
4. Discussion
AML is a group of highly heterogeneous malignancies, and the early manifestations are non-specific and vary from patients. Although the discoveries and applications of new therapies for AML are increasing, the five-year survival is still low [1]. In consequence, it is urgent to explore new methods for diagnosis and treatments. In our study, we found that the level of CDCA was lower in the peripheral blood serum and feces of AML patients compared with normal people. Moreover, the level of CDCA was negatively correlated with AML prognosis. Remarkably, there also existed a significant association between CDCA and gut microbiota, which was consistent with our previous results [28] and might further indicate the role of microbiota playing in AML progression. Besides, when reaching a certain concentration, CDCA would exhibited cytotoxic effects on AML cells. In general, CDCA may be a biomarker and a therapeutic strategy for AML.
It has been shown that unique mitochondrial alterations including mitochondrial metabolism and ROS generation resulted in metabolic vulnerabilities in AML [42], which was well verified in our research. In our study, we found that CDCA could bind to mitochondria and cause mitochondrial damage, including morphological abnormalities, decreased mitochondrial mass and mitochondrial membrane potential, as well as increased ROS accumulation, which indicated that administrating CDCA to target mitochondria might be a new treatment. Besides, LDs, a type of unique organelles correlated with mitochondria, are crucial for lipid metabolism and energy homeostasis, and are associated with many diseases, such as neurodegenerative diseases, metabolic diseases and tumors [43,44]. Study has reported that LDs could be regulated by kaempferol to prevent dopaminergic neuronal degeneration in Parkinson's disease [40]. Furthermore, increasing LDs abundance through CDCP1 knockdown could inhibit triple-negative breast cancer migration. However, little is known about the effects of LDs on AML. In our study, we demonstrated that ROS production could promote LDs accumulation and ROS clearance agent NAC reserved this effect, which was consistent with what has been described in previous studies in other diseases [39], and provided an interesting viewpoint for AML. Most importantly, the presence of elevated ROS and LDs promoted lipid peroxidation, which was proven by the abundance of peroxidated lipids and the reduction of GSH or GPX4. Overall, mitochondria and LDs, two significant organelles, exert tremendous effects on organism and diseases and provide a bright perspective in AML researches.
Since the interrelation between LDs and mitochondria is complicated, we aimed to explore the specific mechanism between mitochondria and LDs. It was reported that p38 MAPK activation was a classical signal transduction pathway in carcinoma [45], and in our study, we found that excessive ROS could activate p38 MAPK in AML. Further researches manifested that p38 MAPK could act on LDs accumulation through upregulating DGAT1 which was essential to LDs synthesis. Taken together, we identified the role of ROS/p38 MAPK/LDs axis in AML cells, which would be a supplement for AML researches and treatments. Collectively, we have demonstrated that CDCA could delay AML progression in vivo and in vitro. And due to the possible effect of pharmacokinetics, CDCA exhibited less suppressive effects on AML progression in AML mice compared with cell cultures experiments. As we know, the mechanisms and pathogenesis of AML are intricate and unclear, and the occurrence and progression of AML are the result of dysfunction of multisystem [46], including hematopoietic dysfunction, gene mutation, as well as immunological abnormality, among which immune system has a vital impact. Previous studies have revealed that bile acids played a crucial role in macrophage immunology and its polarization in many other diseases [24]. Here, we showed that CDCA could inhibit M2 macrophage polarization, which was the novel discovery clarified in AML and clearly supported our assumption that CDCA acted on AML progression through various channels.
5. Conclusions
In conclusion, we find that CDCA is decreased in feces and plasma of AML patients and correlated with the diversity of gut microbiota and AML prognosis. In mechanism, CDCA binds to mitochondria and causes mitochondrial abnormality, which further induces ROS elevation. The enhanced ROS promotes the activation of p38 MAPK, which collaboratively promotes the LDs accumulation through upregulating DGAT1 expression. Consequently, the existence of increased ROS and LDs promotes lipid peroxidation in AML cells. Moreover, CDCA modulates M2 macrophages polarization. Taken together, our study might be useful for the prognosis and treatments of AML (Fig. 8).
Fig. 8.
Chenodeoxycholic acid suppresses AML progression through promoting lipid peroxidation via ROS/p38 MAPK/DGAT1 pathway and inhibiting M2 macrophage polarization
Chenodeoxycholic acid (CDCA) is decreased in feces and plasma of AML and is correlated with gut microbiota. In mechanism, CDCA promotes LDs accumulation via ROS/p38 MAPK/DGAT1 pathway caused by mitochondrial dysfunction in leukemia cells, and the enrichment of excessive ROS and LDs promotes lipid peroxidation. Moreover, CDCA inhibits M2 macrophage polarization, which alleviates the AML-promoting effects of M2 macrophage.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers: No. 81873439, No. 82173135).
Authorship contributions
Professor Daoxin Ma designed and funded the research. Professor Fabao Liu helped design and funded the research. Professor Chunyan Ji helped design the research. Jinting Liu performed the research and wrote the manuscript. Yihong Wei and Can Can assisted with the research. Ruiqing Wang and Xinyu Yang analyzed the data. Chaoyang Gu edited the paper.
Declaration of competing interest
The authors declare no competing financial interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102452.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Data availability
No data was used for the research described in the article.
References
- 1.CDCP1 drives triple-negative breast cancer metastasis through reduction of lipid-droplet abundance and stimulation of fatty acid oxidation. www.pnas.org/cgi/doi/10.1073/pnas.1703791114 [DOI] [PMC free article] [PubMed]
- 2.Short N.J., Konopleva M., Kadia T.M., Borthakur G., Ravandi F., DiNardo C.D., et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 2020;10(4):506–525. doi: 10.1158/2159-8290.CD-19-1011. [DOI] [PubMed] [Google Scholar]
- 3.Chiang J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013;3(3):1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K., et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metabol. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Cai J., Sun L., Gonzalez F.J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. 2022;30(3):289–300. doi: 10.1016/j.chom.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soma T., Kaganoi J., Kawabe A., Kondo K., Tsunoda S., Imamura M., et al. Chenodeoxycholic acid stimulates the progression of human esophageal cancer cells: a possible mechanism of angiogenesis in patients with esophageal cancer. Int. J. Cancer. 2006;119(4):771–782. doi: 10.1002/ijc.21917. [DOI] [PubMed] [Google Scholar]
- 7.Casaburi I., Avena P., Lanzino M., Sisci D., Giordano F., Maris P., et al. Chenodeoxycholic acid through a TGR5-dependent CREB signaling activation enhances cyclin D1 expression and promotes human endometrial cancer cell proliferation. Cell Cycle. 2012;11(14):2699–2710. doi: 10.4161/cc.21029. [DOI] [PubMed] [Google Scholar]
- 8.Broeders E.P., Nascimento E.B., Havekes B., Brans B., Roumans K.H., Tailleux A., et al. The bile acid chenodeoxycholic acid increases human Brown adipose tissue activity. Cell Metabol. 2015;22(3):418–426. doi: 10.1016/j.cmet.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 9.de Beauchamp L., Himonas E., Helgason G.V. Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia. Leukemia. 2022;36(1):1–12. doi: 10.1038/s41375-021-01416-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019;20(3):137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veliova M., Petcherski A., Liesa M., Shirihai O.S. The biology of lipid droplet-bound mitochondria. Semin. Cell Dev. Biol. 2020;108:55–64. doi: 10.1016/j.semcdb.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benador I.Y., Veliova M., Mahdaviani K., Petcherski A., Wikstrom J.D., Assali E.A., et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metabol. 2018;27(4):869–885 e6. doi: 10.1016/j.cmet.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benador I.Y., Veliova M., Liesa M., Shirihai O.S. Mitochondria bound to lipid droplets: where mitochondrial dynamics regulate lipid storage and utilization. Cell Metabol. 2019;29(4):827–835. doi: 10.1016/j.cmet.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen T.B., Louie S.M., Daniele J.R., Tran Q., Dillin A., Zoncu R., et al. DGAT1-Dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell. 2017;42(1):9–21 e5. doi: 10.1016/j.devcel.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch M., Sanchez-Alvarez M., Fajardo A., Kapetanovic R., Steiner B., Dutra F., et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science. 2020;370(6514) doi: 10.1126/science.aay8085. [DOI] [PubMed] [Google Scholar]
- 16.Mejhert N., Kuruvilla L., Gabriel K.R., Elliott S.D., Guie M.A., Wang H., et al. Partitioning of MLX-family transcription factors to lipid droplets regulates metabolic gene expression. Mol Cell. 2020;77(6):1251–12564 e9. doi: 10.1016/j.molcel.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills E.L., Kelly B., O'Neill L.A.J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 2017;18(5):488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 18.Leilei Gong, Lei Yueyue, Liu Yixiao, Tan Fanggen, Li Shuangshuang. Vaccarin prevents ox-LDL-induced HUVEC EndMT, inflammation and apoptosis by suppressing ROS p38 MAPK signaling. Am. J. Transl. Res. 2019;11(4):2140–2154. [PMC free article] [PubMed] [Google Scholar]
- 19.Jalmi S.K., Sinha A.K. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Front. Plant Sci. 2015;6:769. doi: 10.3389/fpls.2015.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao Y., Cui L., Ye J., Yang G., Lu G., Fang X., et al. Dioscin facilitates ROS-induced apoptosis via the p38-MAPK/HSP27-mediated pathways in lung squamous cell carcinoma. Int. J. Biol. Sci. 2020;16(15):2883–2894. doi: 10.7150/ijbs.45710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z., Nian Q., Chen G., Cui S., Han Y., Zhang J. Klotho alleviates lung injury caused by paraquat via suppressing ROS/P38 MAPK-regulated inflammatory responses and apoptosis. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/1854206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi S.K., Nechiporuk T., Bottomly D., Piehowski P.D., Reisz J.A., Pittsenbarger J., et al. The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer Cell. 2021;39(7):999–1014 e8. doi: 10.1016/j.ccell.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalton W.B., Ghiaur G., Resar L.M. Taking the STING out of acute myeloid leukemia through macrophage-mediated phagocytosis. J. Clin. Invest. 2022;132(5) doi: 10.1172/JCI157434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Gong Z., Zhang X., Zhu F., Liu Y., Jin C., et al. Gut microbial bile acid metabolite skews macrophage polarization and contributes to high-fat diet-induced colonic inflammation. Gut Microb. 2020;12(1):1–20. doi: 10.1080/19490976.2020.1819155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehla K., Singh P.K. Metabolic regulation of macrophage polarization in cancer. Trends Cancer. 2019;5(12):822–834. doi: 10.1016/j.trecan.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills C.D., Lenz L.L., Harris R.A. A breakthrough: macrophage-directed cancer immunotherapy. Cancer Res. 2016;76(3):513–516. doi: 10.1158/0008-5472.CAN-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao L., Zhao S.K., Wen C., Tian R., Lin L., Cai B., et al. Activating macrophage-mediated cancer immunotherapy by genetically edited nanoparticles. Adv. Mater. 2020;32(47) doi: 10.1002/adma.202004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R., Yang X., Liu J., Zhong F., Zhang C., Chen Y., et al. Gut microbiota regulates acute myeloid leukaemia via alteration of intestinal barrier function mediated by butyrate. Nat. Commun. 2022;13(1):2522. doi: 10.1038/s41467-022-30240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 2008;7(8):678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 30.Jia W., Xie G., Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018;15(2):111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dijk S.N., Protasoni M., Elpidorou M., Kroon A.M., Taanman J.W. Mitochondria as target to inhibit proliferation and induce apoptosis of cancer cells: the effects of doxycycline and gemcitabine. Sci. Rep. 2020;10(1):4363. doi: 10.1038/s41598-020-61381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bravo-Sagua Roberto, Parra Valentina, López-Crisosto Camila, Díaz Paula, Quest Andrew F.G., Bravo-Sagua Roberto. Calcium Transport and signaling in mitochondria. Compr. Physiol. 2017;7:623–634. doi: 10.1002/cphy.c160013. [DOI] [PubMed] [Google Scholar]
- 33.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X., Fu M., Bi R., Zheng X., Fu B., Tian S., et al. Cadmium induced BEAS-2B cells apoptosis and mitochondria damage via MAPK signaling pathway. Chemosphere. 2021;263 doi: 10.1016/j.chemosphere.2020.128346. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X.S., Lu Y., Li W., Tao T., Peng L., Wang W.H., et al. Astaxanthin ameliorates oxidative stress and neuronal apoptosis via SIRT1/NRF2/Prx2/ASK1/p38 after traumatic brain injury in mice. Br. J. Pharmacol. 2021;178(5):1114–1132. doi: 10.1111/bph.15346. [DOI] [PubMed] [Google Scholar]
- 36.Kumari A., Pal Pathak D., Asthana S. Bile acids mediated potential functional interaction between FXR and FATP5 in the regulation of Lipid Metabolism. Int. J. Biol. Sci. 2020;16(13):2308–2322. doi: 10.7150/ijbs.44774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalyanaraman B., Cheng G., Hardy M., Ouari O., Lopez M., Joseph J., et al. A review of the basics of mitochondrial bioenergetics, metabolism, and related signaling pathways in cancer cells: therapeutic targeting of tumor mitochondria with lipophilic cationic compounds. Redox Biol. 2018;14:316–327. doi: 10.1016/j.redox.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S., Raza S.H.A., Zhao C., Cheng G., Zan L. Overexpression of PLIN1 promotes lipid metabolism in bovine adipocytes. Animals (Basel) 2020;10(11) doi: 10.3390/ani10111944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L., Zhang K., Sandoval H., Yamamoto S., Jaiswal M., Sanz E., et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160(1–2):177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X., Zhao S., Song H., Xu T., Fang Q., Hu G., et al. Kaempferol alleviates LD-mitochondrial damage by promoting autophagy: implications in Parkinson's disease. Redox Biol. 2021;41 doi: 10.1016/j.redox.2021.101911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ursini F., Maiorino M. Lipid peroxidation and ferroptosis: the role of GSH and GPx4. Free Radic. Biol. Med. 2020;152:175–185. doi: 10.1016/j.freeradbiomed.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 42.Panina S.B., Pei J., Kirienko N.V. Mitochondrial metabolism as a target for acute myeloid leukemia treatment. Cancer Metabol. 2021;9(1):17. doi: 10.1186/s40170-021-00253-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cruz A.L.S., Barreto E.A., Fazolini N.P.B., Viola J.P.B., Bozza P.T. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020;11(2):105. doi: 10.1038/s41419-020-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng X., Geng F., Pan M., Wu X., Zhong Y., Wang C., et al. Targeting DGAT1 ameliorates glioblastoma by increasing fat catabolism and oxidative stress. Cell Metabol. 2020;32(2):229–242 e8. doi: 10.1016/j.cmet.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner E.F., Nebreda A.R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 2009;9(8):537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 46.Khwaja A., Bjorkholm M., Gale R.E., Levine R.L., Jordan C.T., Ehninger G., et al. Acute myeloid leukaemia. Nat. Rev. Dis. Prim. 2016;2 doi: 10.1038/nrdp.2016.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.