Graphical abstract

Keywords: Alignment algorithms, Comparative methylomes, Mammals, Whole genome bisulfite sequencing
Abstract
Whole genome bisulfite sequencing (WGBS) is an essential technique for methylome studies. Although a series of tools have been developed to overcome the mapping challenges caused by bisulfite treatment, the latest available tools have not been evaluated on the performance of reads mapping as well as on biological insights in multiple mammals. Herein, based on the real and simulated WGBS data of 14.77 billion reads, we undertook 936 mappings to benchmark and evaluate 14 wildly utilized alignment algorithms from reads mapping to biological interpretation in humans, cattle and pigs: Bwa-meth, BSBolt, BSMAP, Walt, Abismal, Batmeth2, Hisat_3n, Hisat_3n_repeat, Bismark-bwt2-e2e, Bismark-his2, BSSeeker2-bwt, BSSeeker2-soap2, BSSeeker2-bwt2-e2e and BSSeeker2-bwt2-local. Specifically, Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt exhibited higher uniquely mapped reads, mapped precision, recall and F1 score than other nine alignment algorithms, and the influences of distinct alignment algorithms on the methylomes varied considerably at the numbers and methylation levels of CpG sites, the calling of differentially methylated CpGs (DMCs) and regions (DMRs). Moreover, we reported that BSMAP showed the highest accuracy at the detection of CpG coordinates and methylation levels, the calling of DMCs, DMRs, DMR-related genes and signaling pathways. These results suggested that careful selection of algorithms to profile the genome-wide DNA methylation is required, and our works provided investigators with useful information on the choice of alignment algorithms to effectively improve the DNA methylation detection accuracy in mammals.
1. Introduction
As the most widely studied epigenetic mechanism [1], DNA methylation is a basic modification that adds a methyl group to the fifth carbon of cytosine to form 5-methylcytosine [2], but does not change the DNA sequence itself [3]. A series of studies have demonstrated that DNA methylation regulates transcription activity [4], [5], X chromosome inactivation [5], imprinting [5], [6], and chromosome stability [6]. Currently, based on the next-generation sequencing, although several methods such as methylated DNA immunoprecipitation sequencing (MeDIP-Seq) [7], [8], methyl-CpG binding domain sequencing (MBD-Seq) [8], [9], methylation capture sequencing (MethylCap-Seq) [8], [10], reduced representation bisulfite sequencing (RRBS) [11], bisulfite amplicon sequencing [12] and SeqCap Epi CpGiant [8], [13], are developed to cover the preselected genomic regions of interest, the whole genome bisulfite sequencing (WGBS) has been developed to investigate DNA methylation landscape at single-base resolution, which is considered as the gold standard technology [14].
More recently, WGBS is at the forefront of epigenetic analysis and popularly utilized to investigate the genome-wide DNA methylation dynamics of mammalian developments [15], [16] as well as the epigenetic marks of diseases [17]. But the bisulfite treatment converts unmethylated cytosine as thymine, reduces the complexity of reads, and thus causes a mapping challenge to aligners in WGBS [18]. To deal with this challenge, many specific mapping tools have been developed based on three strategies, namely wild-card, three-letter and two-letter [19], [20]. Wild-card strategy allows C or T to match with C of the reference genome [19]; three-letter strategy converts both C on the reads and reference genome into T, namely holding A, G and T (three-letter alphabet) in reads and reference genome [19]; two-letter strategy simultaneously converts purines (As and Gs) to one letter and pyrimidines (Cs and Ts) to another letter during the mapping [20]. Furthermore, using wild-card, three-letter, or two-letter strategy combining with in-house aligner [21], [22], [23] or popular aligners, e.g., Bowtie [24], Bowtie2 [25], BWA [26], HISAT2 [27], SOAP [28], SOAP2 [29], and BatAlign [30], a number of mappers are developed to overcome the mapping challenges caused by bisulfite treatment, e.g., Bwa-meth [31], Bismark [32], BSSeeker2 [33], Walt [23], Batmeth2 [34], BSMAP [35], Abismal [20], BSBolt [36], and Hisat-3n [37]. Previous studies have recommended that the mappers wrapped with different aligners shows distinct mapping precision [38], [39], [40], uniquely mapped reads [39], recall [38] and calculation efficiency [41], as well as the diverse sensibility of read depth [38] in WGBS.
Recently, in humans, Tran et al. compare the uniquely mapped reads and runtime of five alignment algorithms (BSMAP, Bismark, BSSeeker, BRAT-BW, and BiSS), as well as the sensitive to read length and sequencing error [42]; Tsuji et al. evaluate the mapping sensitivity, mapping error, runtime, and memory consumption of five alignment algorithms (Bismark, BSMAP, BRAT-BW, GSNAP and LAST) [41]; Govindarajan et al. mainly focus on the genomic coverage of five bisulfite mappers (Bismark, BSMAP, Pash, BatMeth, and BSSeeker) [21]. Although these studies have assessed several alignment algorithms on reads mapping in one mammalian genome, they have not evaluated and compared them comprehensively in multiple mammals as well as on biological insight into methylomes, such as the calling of methylation levels, differentially methylated CpGs (DMCs), differentially methylated regions (DMRs) as well as the signaling pathways.
Herein, 14 alignment algorithms, which were wildly used to profile the genome-wide DNA methylation in mammalian investigations, were collected in this study. These 14 algorithms were well representative in the alignment algorithms of WGBS. We aimed to comprehensively compare and evaluate the runtime, memory consumption, uniquely mapped reads, unsatisfactory aligned reads, mapped precision, recall as well as F1 score of 14 alignment algorithms in humans, cattle and pigs. Moreover, we explored the accuracies of alignment algorithms on the detection of CpG coordinates and methylation levels, as well as the calling of DMCs, DMRs, DMR-related genes and signaling pathways. These works provide investigators with useful information on the choice of alignment algorithms in mammals.
2. Materials and methods
2.1. Generations of simulated data
In this study, the reference genomes of humans, cattle and pigs were downloaded from UCSC (https://genome.ucsc.edu/), and they varied in genome size, assembly qualities and gene numbers (Table 1). Based on the reference genome of the three mammals, the simulated WGBS data were generated by using Sherman (https://www.bioinformatics.babraham.ac.uk/projects/sherman/), which was a simulator of WGBS data developed by Babraham [39], [40], [42], with the specific parameter (Table S1).
Table 1.
Assembly statistics for Human, Cattle and Pig reference genome.
| Species | Genome Size (Mb) | Scaffolds | Scaffold N50 (Mb) | Gene numbers | Genome version |
|---|---|---|---|---|---|
| Human | 3,209.29 | 473 | 67.79 | 64,252 | hg38 |
| Cattle | 2,715.85 | 2,211 | 103.31 | 27,607 | bosTau9 |
| Pig | 2,502.91 | 706 | 88.23 | 31,908 | susScr11 |
In order to benchmark the runtime, memory consumption, uniquely mapped reads, mapped precision, recall and F1 score, we generated 45 simulated data samples (5 sequencing error rates × 3 mammals × 3 replicates) by using Sherman, and named it as Simulated Dataset A (Table S1). The 5 sequencing error rates were 0, 0.25 %, 0.5 %, 0.75 % and 1.00 %; the 3 mammals were human, cattle and pig; 3 replicates were generated for each sequencing error rate of every mammal. In total, 45 simulated data samples with 630 mapping actions were used to benchmark 14 alignment algorithms (Table S2). After considering the overwork and computation efficiency of 630 mappings, two million reads were generated for each of 45 simulated data samples. The Simulated dataset A had a total of 90 million reads.
To further investigate the mapping performance and the influence of the repetitive sequence and CGIs on mapping efficiency for alignment algorithms, the Simulated Dataset B (Table S1) was generated, which contained 18 data samples (2 sequencing error rates × 3 mammals × 3 replicates), and each sample harbored ∼93.80 million reads. The 2 sequencing error rates were 0 and 1.00 %. The Simulated Dataset B had a total of 1.64 billion reads. In this study, the Simulated Dataset A and B were not used to benchmark the biological insights on methylomes, including the calling of CpG coordinates, DMCs, DMRs, DMR-related genes and signaling pathways.
2.2. Descriptions of Real WGBS data
The real WGBS data of humans [43], cattle [44] and pigs [45] were downloaded from Sequence Read Archive of National Center for Biotechnology Information (NCBI) (Table S3). The Real Dataset A (Table S4) was sampled and extracted from Table S3, and contained 9 data samples (3 mammals × 3 replicates). Each sample had two million reads. The Real Dataset A had a total of 18 million reads, and was used to evaluate the runtime, memory consumption and uniquely mapped reads.
The Real Dataset B (Table S3) contained 18 data samples (3 mammals × 2 groups × 3 replicates). The 2 groups were group1 and group 2, and each group had 3 biological replicates. The data of human came from brains of schizophrenia patients and normal people [43]; the data of cattle came from 6 bull longissimus dorsi muscle [44]; the data of pig came from the skeletal muscle of Landrace pigs [45]. The sequencing depth of the samples in Real Dataset B was > 30. The Real Dataset B had a total of 13.1 billion reads, and was used to explore the impacts of alignment algorithms on downstream analysis of biological interpretations, e.g., CpG coordinates, DMCs, DMRs, DMR-genes and signaling pathways.
2.3. Quality control, mapping, and downstream analysis
In the analysis of real WGBS data, FastQC program (https://www.bio informatics.babraham.ac.uk/p rojects/fastqc/) was used to control the quality of reads, and Fastp [46] was used to trim adaptor sequences and filter low-quality bases/reads with the default parameters. For the mapping of reads, it was undertaken by 14 alignment algorithms, including Bwa-meth, BSMAP, Walt, Batmeht2, Bismark-bwt2-e2e, Bismark-his2, BSSeeker2-bwt, BSSeeker2-soap2, BSSeeker2-bwt2-e2e, BSSeeker2-bwt2-local, Abismal, BSBolt, Hisat_3n, and Hisat_3n_repeat (Table S2). In this study, the mapper was developed based on one certain mapping strategy, including Bwa-meth, BSMAP, Bismark, Walt, BSSeeker2, Batmeth2, Abismal, BSBolt, and Hisat-3n. The aligner was the core alignment of mapper, e.g., Bowtie, Bowtie2, BWA, SOAP, SOAP2, HISAT2, BatAlign, and in-house aligners. The alignment algorithms defined as the mapper warped up with one kind of aligner (Table S2).
In the Simulated Dataset A and Real Dataset A, the reads were mapped to reference genome using 14 alignment algorithms to record and investigate the runtime and memory consumption (resident set size) with one thread. The uniquely mapped reads, mapped precision, recall and F1 score of 14 alignment algorithms were calculated in the Simulated Dataset A, and the uniquely mapped reads were also calculated in the Real Dataset A. Since Bwa-meth, BSMAP, Bismark-bwt2-e2e, Walt, and BSBolt exhibited outstanding in uniquely mapped reads and F1 score, the subsequent analysis in Simulated Dataset B and Real Dataset B were focused on these five alignment algorithms. The Simulated Dataset B was used to count the number of the unsatisfactory aligned reads to further investigate the mapping performance and the influence of the repetitive sequence and CGIs on mapping efficiency.
After mapping, the bam files of the Real Dataset B were sorted and indexed by Samtools [47]. To further evaluate the applications of alignment algorithms in the downstream analysis of WGBS data, the CpG sites, DMCs, DMRs, DMR-related genes and signaling pathways were detected based on these bam files. The CpGs with at least 10 coverage were retained for further analysis, and the methylation level was calculated by using methyldackel (https://github.com/dpryan79/Methyl Dackel). The R package “DSS” [2], [48] with a Wald test and the condition of P ≤ 0.05 (corrected by the false discovery rate) was used to identify the DMCs and DMRs. The DMR-related genes were defined as genes whose coding sequence overlapped with the DMRs by at least one base. The Kyoto Encyclopedia of Gene and Genomes (KEGG) enrichment analysis on DMR-related genes was performed on the R package “clusterProfiler” [49] (P ≤ 0.01). In this study, all alignment algorithms used the default parameters recommended by the original developers, and the workflows were summarized in Fig. 1.
Fig. 1.
The analysis protocol for benchmarking of 14 alignment algorithms in mammals.
2.4. Calculation of uniquely mapped reads, mapped precision, recall and F1 score
In the mapping results, the reads were divided into uniquely mapped reads, multiple mapped reads and unmapped reads. The uniquely mapped reads were defined as the reads that were only mapped to one location of reference genome, and its proportion was equal to the number of uniquely mapped reads divided by the number of all reads. The multiple mapped reads were described as the reads that were mapped to multiple locations of reference genome. The unmapped reads were defined as the reads that could not be mapped to reference genome. For simulated reads generated from Sherman, the original positions of simulated reads were recorded by Sherman (https://www.bioinformatics.babraham.ac.uk/\projects/sherman/), and the predicted positions were recorded by the alignment algorithms. Since the simulated reads did not account for insertions and deletions, we considered only the first base of the reads at its genome position when compared the original position and the predicted position. Then the uniquely mapped reads were further divided into the correct and incorrect uniquely mapped reads by comparing the original position and the predicted position. The incorrect uniquely mapped reads, multiple mapped reads and unmapped reads have merged as the unsatisfactory aligned reads.
The mapped precision, recall and F1 score of alignment algorithms were calculated by the following formula [39]:
where i is a genomic fragment mapped by reads, N is the total number of genomic fragments, TPi is the number of true positive for the reads mapped to the genomic fragment, FPi is the number of false positive for the reads mapped to the genomic fragment, and FNi is the number of false negative reads mapped to the genomic fragment. Specially, all the multiple mapped reads were counted into true negative reads mapped to the genomic fragment.
2.5. Statistics analysis
By calculating the accuracy of five algorithms on CpG sites (Fig. S1a), BSBolt exhibited the lowest accuracy (Fig. 5c). Therefore, the subsequent analysis of CpG coordinates, DMCs, DMRs, DMR-related genes, signaling pathways were focused on Bwa-meth, BSMAP, Bismark-bwt2-e2e, and Walt. In terms of these four algorithms, we defined the results (i.e. CpG coordinates, DMCs, DMRs, DMR-related genes, and signaling pathways) detected by at least three alignment algorithms as accurate results (Fig. S1b). For example, among the CpG sites detected by Walt in humans, A CpG sites (13962703) were co-called by Bwa-meth, BSMAP, Bismark-bwt2-e2e; B CpG sites (14697507) were co-called by Bwa-meth and BSMAP; C CpG sites (14606328) were co-called by BSMAP and Bismark-bwt2-e2e; D CpG sites (14295028) were co-called Bwa-meth, and Bismark-bwt2-e2e. Then the number of accurate CpG sites in Walt was calculated by: the number of accurate CpG sites in Walt = (B + C + D)-2 * A = (14697507 + 14697507 + 14295028) – 2 * 13962703 = 15764636. It was worth noting that the CpG sites co-called by multiple algorithms refereed to those CpG sites, which were detected by these algorithms and the methylation differences of which were <5.00 % in these algorithms.
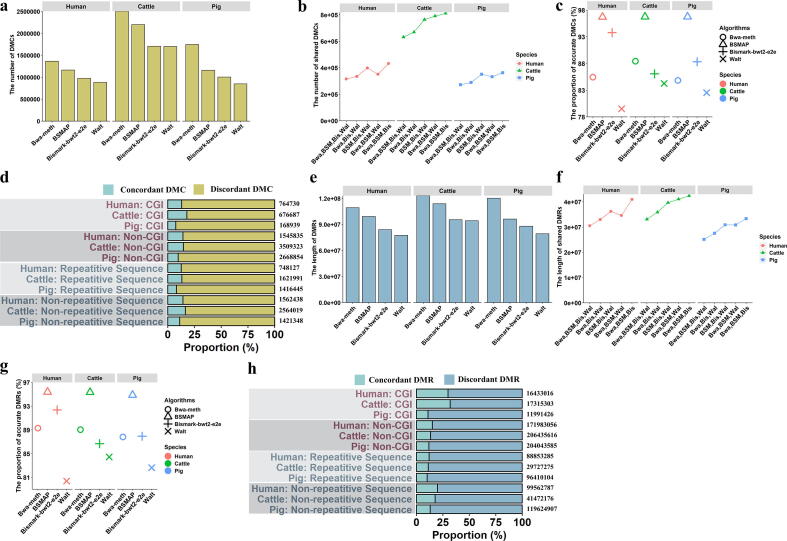
Fig. 5.
The influences of Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt on CpG coordinates and methylation levels in the Real Dataset B. (a) The total number of CpG sites detected by Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt, respectively. (b) The number of shared CpG sites consistently detected by five or four alignment algorithms. (c) The proportion of accurate CpG sites detected by Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt. (d) The number of shared CpG sites consistently detected by four or three alignment algorithms. (e) The proportion of accurate CpG sites detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (f) The proportion of concordant and discordant CpG sites. (g) The proportion of concordant and discordant CpG sites in CGI, non-CGI, repetitive sequence and non-repetitive sequence. (h) The proportion of concordant and discordant CpG sits in the methylation level of high, intermediate and low. Bwa: Bwa-meth; BSM: BSMAP; Bis: Bismark-bwt2-e2e; Wal: Walt: BSB: BSBolt.
Moreover, based on the studies of Sun X et al. [38], we defined the concordant CpG sites as the CpGs which were consistently detected by all four alignment algorithms (Bwa-meth, BSMAP, Bismark-bwt2-e2e, and Walt) and the methylation differences of which were <5.00 %. The remaining CpG sites detected by the four alignment algorithms were regarded as the discordant CpG sites. The DMCs were defined as the concordant DMCs if they were consistently detected by the four alignment algorithms, and the remaining DMCs were termed as the discordant DMCs. The DMRs identified by the four alignment algorithms, which were overlapped genomic regions, were defined as the concordant DMRs, and the remaining DMRs were regarded as the discordant DMRs.
The significant differences of runtime, memory consumption, the number of uniquely mapped reads, mapped precision, recall, the number of unsatisfactory aligned reads and the number of CpG sites were tested by a Student’s t-test with the function of “t.test”. the Pearson’s correlation coefficient was tested using the function of “cor.test”; the enrichment was tested by a two-tail Fisher’s exact test with the function of “fisher.test”, and these functions were utilized from the R “stats package” (https://www.rdocumentation.org/packages/stats/). In order to investigate the influences of the repetitive sequence and CGIs on the unsatisfactory aligned reads, CpG sites, DMCs and DMRs, the locations of repetitive sequences and CGIs was downloaded from UCSC (Table S5). CGIs were defined as the regions >200 bp in length, with a C and G percentage > 0.5, and a ratio of the observed CpG/expected CpG > 0.6. The distributions of unsatisfactory aligned reads, CpG sites, DMCs and DMRs on the repetitive sequence and CGIs were described using BedTools [50]. The visualizations of DNA methylation profiles in genes were showed using the UCSC Genome Browser.
3. Results
3.1. Data sets and analysis protocol
To comprehensively evaluate these 14 alignment algorithms, we used simulated and real data from humans, cattle and pigs, including four data sets: Simulated Dataset A, Simulated Dataset B, Real Dataset A, and Real Dataset B (see Materials and methods). The Simulated Dataset A, which was against at 0, 0.25 %, 0.50 %, 0.75 % and 1.00 % sequencing errors, consisted of 90 million reads with 45 samples in total (Table S1); the Simulated Dataset B, which was against at 0 and 1.00 % sequencing errors, consisted of 1.64 billion reads with 18 samples in total (Table S1); the Real Dataset A was composed of 18 million reads with 9 samples (Table S4), and the Real Dataset B included 18 samples with 13.1 billion reads from humans, cattle, and pigs (Table S3). Among them, the Simulated Dataset A and Real Dataset A was used to evaluate the basic performance of the alignment algorithms, such as the runtime, memory consumption, uniquely mapped reads, mapped precision, recall and F1 score, while the Simulated Dataset B and Real Dataset B were used to further investigate the biological interpretation (i.e. CpG coordinates, DMC, DMR, DMR-related genes, and signaling pathways).
The analysis protocol of this study was shown in Fig. 1. The information of these alignment algorithms was listed in Table S2, and each step of the protocol would be illustrated in the following results.
3.2. Runtime and memory usage cost of alignment algorithms
To benchmark the computational efficiencies of these 14 alignment algorithms, the runtime and memory consumption were calculated using the Simulated Dataset A (Table S1) and Real Dataset A (Table S4). We found that Walt was the fastest, and BSSeeker2-bwt2-local was the slowest algorithms at 0, 0.25 %, 0.50 %, 0.75 % and 1.00 % sequencing errors (Fig. 2a). The runtimes of Bwa-meth, Hisat_3n, Hisat_3n_repeat, BSBolt, BSMAP and Bismark-his2 were positively correlated to the sequencing error rates of 0, 0.25 %, 0.50 %, 0.75 %, and 1.00 % in humans, cattle and pigs (Pearson’s correlation coefficients 0.7, P 0.05) (Fig. 2b and Fig. S2). As shown in Fig. 2c, the average runtime of BSSeeker2-bwt2-e2e, Batmeth2, BSSeeker2-bwt2-local, and Abismal were 5.38 (simulated data) and 3.36 (real data) folds longer than the average runtime of Walt, BSMAP, Bwa-meth, Bismark-his2, BSSeeker2-soap2, BSSeeker2-bwt, Bismark-bwt2-e2e, BSBolt, Hisat_3n, and Hisat_3n_repeat (Student’s t-test, P 2.67e-12).
Fig. 2.
Runtimes and memory consumptions of 14 alignment algorithms based on the Simulated Dataset A and Real Dataset A. (a) Runtimes (left) and memory consumptions (right) of 14 alignment algorithms with five sequencing error rates in Simulated Dataset A. (b) Pearson’s correlation coefficients between the sequencing error rate in Simulated Dataset A and the performance (e.g., runtime, memory consumption, uniquely mapped reads, mapped precision and recall) of 14 alignment algorithms. (c) Average runtimes of 14 alignment algorithms in Simulated Dataset A (left) and Real Dataset A (right). (d) Average memory consumptions of 14 alignment algorithms in Simulated Dataset A (left) and Real Dataset A (right).
Moreover, the runtime of certain alignment algorithm did not significantly change among the humans, cattle and pigs, except for Batmeth2, BSSeeker2-bwt2-local, and Abismal (Fig. 2c). Averaging the results of simulated and real data together (Fig. 2c), BSSeeker2-soap2 was 2.72, 5.78 and 8.95 folds faster than BSSeeker2-bwt, BSSeeker2-bwt2-e2e, and BSSeeker2-bwt2-local (Student’s t-test, P < 3.7e-09), respectively; Bismark-his2 was 2.27 folds faster than Bismark-bwt2-e2e (Student’s t-test, P < 9.5e-07); Bismark-bwt2-e2e was 2.04 folds faster than BSSeeker2-bwt2-e2e (Student’s t-test, P < 2.3e-09), suggesting that the mappers showed diverse runtime by using distinct aligners, as well as that an aligner exhibited dissimilar runtime by wrapping up with distinct mappers.
Besides, at 0, 0.25 %, 0.50 %, 0.75 % and 1.00 % sequencing errors, Walt took up the most memory consumption, while BSSeeker2-bwt2-local took up the lowest, which was contrary to the runtime of Walt and BSSeeker2-bwt2-local (Fig. 2a). The memory consumption of Bwa-meth and BSBolt was highly positively correlated with the sequencing error rates of 0, 0.25 %, 0.50 %, 0.75 % and 1.00 % in humans, cattle and pigs (Pearson’s correlation coefficients , P 0.05, Fig. 2b and Fig. S3). BSSeeker2-soap2, BSMAP, Bwa-meth, Walt, Batmeth2, Hisat_3n, Hisat_3n_repeat, and BSBolt consumed 3.29 (simulated data) and 3.30 (real data) folds more memory than BSSeeker2-bwt, Bismark-bwt2-e2e, BSSeeker2-bwt2-e2e, BSSeeker2-bwt2-local, Bismark-his2, and Abismal in humans, cattle and pigs (Student’s t-test, P 2.2e-16, Fig. 2d). As shown in Fig. 2d, the memory consumption of these alignment algorithms was decreasing along with the genomic sizes of humans, cattle, and pigs. Otherwise, averaging the results of simulated and real data together (Fig. 2d), BSSeeker2-soap2 respectively consumed 2.19, 2.02 and 2.02 folds more memory than BSSeeker2-bwt, BSSeeker2-bwt2-e2e and BSSeeker2-bwt2-local (Student’s t-test, P ); Bismark-his2 consumed 1.33 folds more memory than Bismark-bwt2-e2e (Student’s t-test, P ). These results indicated that the aligner soap2 and his2 consumed more memories than bwt, bwt2-e2e and bwt2-local.
3.3. Uniquely mapped reads, mapped precision, recall and F1 score of alignment algorithms
To investigate mapping performances of the alignment algorithms, the uniquely mapped reads, mapped precision, recall and F1 score were counted using the Simulated Dataset A (Table S1). We found that Bwa-meth exhibited the most, but BSSeeker2-bwt2-e2e expressed the least uniquely mapped reads in three genomes at 0, 0.25 %, 0.50 %, 0.75 % and 1.00 % sequencing errors (Fig. 3a). The sequencing error rates were highly negative correlation with the rates of uniquely mapped reads at Hisat_3n_repeat, Hisat_3n, Walt, BSSeeker2-soap2, BSSeeker2-bwt2-local, Batmeth2 and BSSeeker2-bwt2-e2e (Pearson correlation coefficients , P 0.05, Fig. 2b and Fig. S4). As showed in Fig. 3b, the proportions of uniquely mapped reads of Bwa-meth, BSBolt, BSMAP, Hisat_3n, Hisat_3n_repeat, Abismal, Bismark-bwt2-e2e and Walt dominated 22.68 % higher than others (Student’s t-test, P ). Interestingly, apart from Batmeth2, Hisat_3n_repeat, Bwa-meth, and BSBolt, these alignment algorithms exhibited a higher proportion of uniquely mapped reads in cattle and pigs, comparing to humans (Fig. 3b). It was readable to point out that the four algorithms of BSSeeker2 displayed the similar uniquely mapped reads, but Bismark-bwt2-e2e exhibited 11.00 % more uniquely mapped reads than Bismark-his2 (Fig. 3b). However, the appearances were not observed in real data (Fig. 3c).
Fig. 3.
Uniquely mapped reads, mapped precision, recall and F1 score benchmark of 14 alignment algorithms based on the Simulated Dataset A and Real Dataset A. (a) Uniquely mapped reads with five sequencing error rates in Simulated Dataset A. The average proportions of uniquely mapped reads of these alignment algorithms in Simulated Dataset A (b) and Real Dataset A (c). (d) The average mapped precision and recall for these alignment algorithms in Simulated Dataset A. (e) The average F1 score of these alignment algorithms in Simulated Dataset A. (f) The average rates of uniquely mapped reads and F1 score in Simulated Dataset A.
Moreover, Walt showed the highest mapped precision and recall, while BSSeeker2-bwt2-local expressed the lowest mapped precision and recall (Fig. 3d and Fig. S5a, b). The mapped precision and recall of these alignment algorithms showed a highly and negative correlation with sequencing error rates (Pearson correlation coefficients , P 0.05), except for BSSeeker2-bwt (Fig. 2b and Fig. S6-7). Furthermore, we found that the average F1 score of most alignment algorithms were >90 %, apart from BSSeeker2-bwt2-local and Batmeth2 in three genomes (Fig. 3e). For F1 score, BSSeeker2-bwt2-local showed at least 21.64 % lower than BSSeeker2-soap2, BSSeeker2-bwt and BSSeeker2-bwt2-e2e (Fig. 3e). Interestingly, Batmeth2 and BSSeeker2-bwt2-local performed worse at F1 score in simulated data (Fig. 3e), but exhibited the excellent performance on uniquely mapped reads in real data (Fig. 3c). Averaging the results of uniquely mapped reads and F1 score (Fig. 3b and e) in three genomes, we found that the proportion of uniquely mapped reads of Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e, and Walt were >90 %, as well as the F1 score > 90 % (Fig. 3f). Therefore, for clarity and simplicity, we focused subsequent analyses on Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt to further investigate their performance on biological interpretation of WGBS data.
Fig. 7.
DNA methylation profile of genes revealed by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (a) SOX9 gene was the highest differentially expressed genes between control and schizophrenia in humans (43). (b) MPZ gene was the hub gene related to Wnt signaling pathway of embryonic processes in cattle (44). (c) IGF2BP3 was the key gene that regulated the development of skeletal muscle in pigs (45).
3.4. Influence of the repetitive sequences and CGIs on the mapping performance
Since Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e, and Walt exhibited outstanding in uniquely mapped reads and F1 score, we used the Simulated Dataset B (Table S1) to further investigate the mapping performance of these five algorithms. We found that Bwa-meth exhibited the least, and Walt displayed the most unsatisfactory aligned reads (Fig. 4a). Since the sequencing error rate increased from 0 to 1.00 %, the unsatisfactory aligned reads of BSMAP, BSBolt, Bwa-meth, Bismark-bwt2-e2e and Walt increased by 0.053 %, 1.95 %, 0.144 %, 1.321 % and 5.651 %, respectively (Fig. 4a). With the sequencing error rate 0 and 1.00 %, it was also found that the unsatisfactory aligned reads of Bwa-meth were only composed of incorrect uniquely mapped reads (Fig. 4a). Furthermore, humans harbored more unsatisfactory aligned reads in the five alignment algorithms than cattle and pigs (P , Fig. 4a). Interestingly, we found that 9.2 %–53 % unsatisfactory aligned reads were the same reads in these five algorithms (Fig. S8). This results reflected different mapping performances of these five alignment algorithms.
Fig. 4.
The influence of repetitive sequence and CGI on mapping performance of five alignment algorithms in the Simulated Dataset B. (a) The proportion and class of unsatisfactory aligned reads for Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt. (b) The proportion of unsatisfactory aligned reads in repetitive sequence and non-repetitive sequence for these five alignment algorithms. (c) The proportion of unsatisfactory aligned reads in CGI and non-CGI for these five alignment algorithms.
To investigate the influence of genomic features on unsatisfactory aligned reads, we explored the representations of the unsatisfactory aligned reads on the repetitive sequences and CGIs. More unsatisfactory aligned reads were found in repetitive sequence than in non-repetitive sequence (Student’s t-test, P 2.68e-05, Fig. 4b), and more unsatisfactory aligned reads were found in CGI regions than non-CGI regions (Student’s t-test, P 9.59e-05, Fig. 4c). For Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt, the enrichments of unsatisfactory aligned reads at repetitive sequence were 1.43, 1.33, 1.41, 1.29, and 1.38 in humans (Fisher’s exact test, P 2.2e-16); 4.06, 4.29, 4.37, 2.82, and 3.03 in cattle (Fisher’s exact test, P 2.2e-16); 1.70, 1.69, 1.59, 1.50, and 1.55 in pigs (Fisher’s exact test, P 2.2e-16) (Fig. 4b), compared with non-repetitive sequence. Also, compared with non-CGI (Fig. 4c), the enrichments of unsatisfactory aligned reads at CGI were 1.67, 1.76, 1.64, 1.67, and 1.76 in humans (Fisher’s exact test, P 2.2e-16); 3.18, 2.70, 2.23, 2.56, and 2.00 in cattle (Fisher’s exact test, P 2.2e-16); 1.99, 2.06,1.66, 1.65, and 1.69 in pigs (Fisher’s exact test, P 2.2e-16). These observations indicated that the repetitive sequences and CGI regions were like to facilitate the unsatisfactory aligned reads.
3.5. Influences of alignment algorithms on CpG coordinates and methylation levels
The methylomic profiles of Real Dataset B (Table S3) were evaluated by Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt at the CpG sites covered with 10 reads. We found BSBolt called the most CpG sites in humans (28551216), cattle (36661360) and pigs (31884976); Walt called the least CpG sties in humans (17326301), cattle (20956144) and pigs (18884561) (Fig. 5a and Fig. S9a). In humans (Fig. 5b), these five alignment algorithms consistently detected 8182827 CpG sites, but the CpG sites consistently detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e, and Walt increased by 170.63 % (13962703) after removing BSBolt. This result suggested that BSBolt seems to be responsible for the lack of concordant CpG sites in human, and the similar phenomenon was observed in cattle and pigs (Fig. 5b). Moreover, as shown in Fig. 5c and Fig. S9b, BSMAP exhibited the highest accuracy at the detection of CpG coordinates (human: 15411076, 99.37 %; cattle: 14252815, 98.99 %; pig: 14272746, 99.03 %), while BSBolt obtained the lowest accuracy (human: 9728719, 62.73 %; cattle: 6143665, 42.67 %; pig: 6316216, 43.82 %). Since BSBolt showed a huge difference in CpG sites with the other four algorithms, the following discussion would focus on the Bwa-meth, BSMAP, Bismark-bwt2-e2e, and Walt.
In terms of Bwa-meth, BSMAP, Bismark-bwt2-e2e, and Walt, we found that Walt likely contributed to the discordance of these four alignment algorithms, since the most CpG sites were consistently detected by the other three algorithms after removing Walt (Fig. 5d and Fig. S10a). BSMAP showed higher accuracy than the other three algorithms (Fig. 5e and Fig. S10b). In addition, 26586835, 29736425 and 28674654 CpG sites were respectively detected in humans, cattle and pigs by the four alignment algorithms, of which approximately 51.65 %, 42.02 % and 44.64 % were detected as concordant CpG sites (Fig. 5f). Furthermore, CGIs and repetitive sequences likely retained more discordant CpG sites than non-CGIs and non-repetitive sequences in humans, cattle and pigs (Student’s t-test, P < 6.38e-03, Fig. 5g). In humans, cattle, and pigs, the concordant CpGs were likely to over representation at the low (<1/3) and high methylation (>1/3), but the discordant CpGs were likely to under representation at the intermediate methylation (1/3 ∼ 2/3) (Student’s t-test, P < 0.02, Fig. 5h).
3.6. Influences of alignment algorithms on DMCs and DMRs
The changes and dynamics of methylomes at the points of DMCs and DMRs were evaluated among Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt based on the CpG sites covered with ≥10 reads in Real Dataset B (Table S3). In these four algorithms, Bwa-meth called the most DMCs (human: 1366812; cattle: 2506659; pig: 1747539) and Walt called the least DMCs (human: 884427; cattle: 1702832; pig: 850291) (Fig. 6a and Fig. S11a). For the DMCs co-recognized by four algorithms, 316048, 631985, and 27238 DMCs respectively recognized in humans, cattle and pigs. In terms of the DMCs consistently recognized by three algorithms, Bwa-meth, BSMAP and Bismark-bwt2-e2e consistently recognized the most DMCs (human: 432724; cattle: 811088; pig: 363085), and Bwa-meth, Bismark-bwt2-e2e and Walt consistently recognized the least DMCs (human: 334856; cattle: 669034; pig: 289236). After the remove of Walt, the most DMCs consistently recognized by three algorithms were obtained, indicating that Walt appeared to be responsible for the lack of concordance of DMCs (Fig. 6b). At the calling of DMCs, BSMAP showed the highest accuracy in humans (551145, 96.70 %), cattle (1101408, 96.74 %) and pigs (502505, 96.75 %), while Walt showed the lowest accuracy in humans (453277, 79.53 %), cattle (959354, 84.27 %) and pigs (428656, 82.54 %) (Fig. 6c and Fig. S11b). Moreover, the enrichments of concordant DMCs were 0.92, 0.87, and 0.83 in repetitive sequence of humans, cattle and pigs, respectively (Fig. 6d, Fisher’s exact test, P 2.2e-16), showing that the concordant DMCs were likely to under representation at repetitive sequence. Nonetheless, the concordant DMCs has no clear preference for CGIs (Fig. 6d).
Fig. 6.
Influence of Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt on dynamics of methylomes in the Real Dataset B. (a) The number of DMCs detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (b) The number of shared DMCs consistently detected by multiple alignment algorithms. (c) The proportions of accurate DMCs detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (d) The proportions of concordant and discordant DMCs in CGI, non-CGI, repetitive sequence and non-repetitive sequence. (e) The length of DMRs detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (f) The length of DMRs consistently detected by multiple alignment algorithms. (g) The proportions of accurate DMRs detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (d) The proportions of concordant and discordant DMRs in CGI, non-CGI, repetitive sequence and non-repetitive sequence. Bwa: Bwa-meth; BSM: BSMAP; Bis: Bismark-bwt2-e2e; Wal: Walt.
Among Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt, the longest DMRs were called from the mapping results of Bwa-meth (human: 109239519 bp, cattle: 123175355 bp, pig: 120202580 bp), and the shortest DMRs were called from the mapping results of Walt (human: 77387750 bp, cattle: 94239118 bp, pig: 79180616 bp) (Fig. 6e, Fig. S11c). Moreover, Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt consistently identified 30530480 bp, 33100297 bp and 25135110 bp DMRs in humans, cattle and pigs, respectively (Fig. 6f). In the DMRs co-identified by three algorithms, we found that the DMRs co-identified by Bwa-meth, BSMAP and Bismark-bwt2-e2e was the longest (human: 40957306 bp; cattle: 42379448 bp; pig: 33330430 bp) after the remove of Walt, suggesting that the lack of concordance could be attributed to Walt (Fig. 6f). As shown in Fig. 6g and Fig. S11d, BSMAP exhibited the highest accuracy at the calling of DMRs in humans (50708843 bp, 95.39 %), cattle (56868993 bp, 95.34 %) and pigs (953420 bp, 94.86 %), but Walt showed the lowest accuracy in humans (42734834 bp, 80.39 %), cattle (50368187 bp, 84.44 %) and pigs (39014214 bp, 82.64 %).
The enrichments of these concordant DMRs were 0.74, 0.74, and 0.86 in repetitive sequence of humans, cattle and pigs, respectively (Fig. 6h, Fisher’s exact test, P 2.2e-16), showing that the concordant DMRs were also likely to under representation at repetitive sequence, and the concordant DMRs has no clear preference for CGIs (Fig. 6h). In order to better describe the influences of the four alignment algorithms on the genome-wide DNA methylation profiles, we visually observed the methylation profiles of SOX9 in humans [43], MPZ in cattle [44] and IGF2BP3 in pigs [45] (Fig. 7a-c), and these genes were reported to make sense in the original papers of real WGBS data. The intuitive look showed that the alignment algorithms generated different methylation profiles, in terms of the numbers and methylation level of CpG sites, the number of DMCs, and the length of DMRs. These results suggested that the selection of alignment algorithms had a dramatic effect on methylomes.
3.7. Influence of alignment algorithms on the biological interpretation
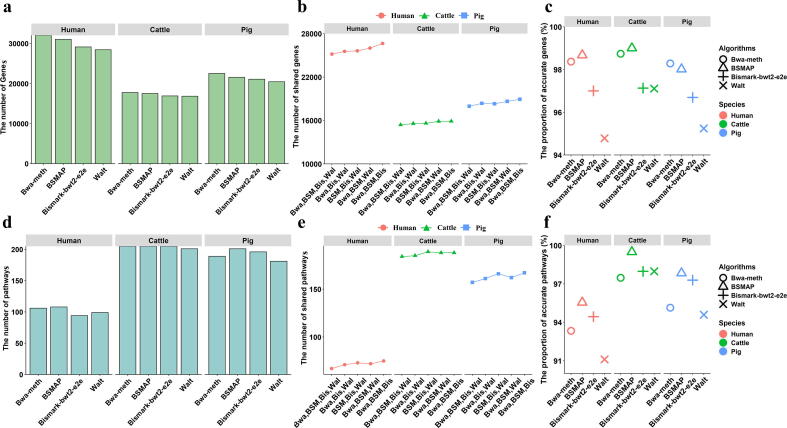
The above results indicated these four alignment algorithms had a dramatic effect on methylomes. To further explore the influence of algorithms on the biological interpretations of WGBS data, the DMRs-related genes were extracted based on Real Dataset B (Table S3), and the biological functions released by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt were interpreted and determined by KEGG enrichment analysis. Consistent with expectations for four algorithms, the most DMR-related genes were detected by Bwa-meth in humans (32055), cattle (17762) and pigs (22510), while the least DMR-related genes were detected by Walt in humans (28446), cattle (16841) and pigs (20444) (Fig. 8a, Fig. S12a). Furthermore, Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt consistently identified 25,161 DMR-related genes in humans (25161), cattle (15409) and pigs (17967) (Fig. 8b). In the DMR-related genes consistently identified by three algorithms, Bwa-meth, BSMAP and Bismark-bwt2-e2e consistently identified the most DMR-related genes after the remove of Walt in humans (26638), cattle (15893) and pigs (18935) (Fig. 8b), showing that the lack of concordance could be attributed to Walt in terms of the calling of DMR-related genes. For the accuracy at the calling of DMR-related genes, BSMAP was the highest in humans (27948, 98.68 %) and cattle (16584, 99.00 %), and Bwa-meth was the highest in pigs (20011, 98.28 %), but Walt was the lowest in humans (26846, 94.78 %), cattle (16267, 97.11 %) and pigs (19392, 95.24 %) (Fig. 8c and Fig. S12b).
Fig. 8.
Influence of Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt on biological interpretation in the Real Dataset B. (a) The number of DMR-related genes detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (b) The number of shared genes consistently detected by multiple alignment algorithms. (c) The proportions of accurate DMR-related genes detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (d) The number of signaling pathways detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. (e) The number of shared pathways consistently detected by multiple alignment algorithms. (f) The proportions of accurate pathways detected by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt. Bwa: Bwa-meth; BSM: BSMAP; Bis: Bismark-bwt2-e2e; Wal: Walt.
In the analysis of KEGG based on the DMR-related genes, 140, 223 and 223 pathways were determined in humans, cattle and pigs, respectively (Fig. 8d and Fig. S12c), of which approximately 67, 184 and 157 were consistently detected by these four alignment algorithms (Fig. 8e). For the consistent pathways determined by three algorithms, Bwa-meth, Bismark-bwt2-e2e and Walt consistently detected the most pathways in humans (75) and pigs (167) after the remove of Walt; BSMAP, Bismark-bwt2-e2e and Walt consistently detected the most pathways in cattle (189) after the remove of Bwa-meth (Fig. 8e). The results suggested that for the analysis of KEGG, Walt seems to be responsible for the lack of concordant in four algorithms. Moreover, BSMAP showed the highest accuracy at the calling of signaling pathways (human: 86, 95.56 %; cattle: 197, 99.49 %; pig: 181, 97.84 %), but Walt exhibited the least accuracy at the calling of signaling pathways (human: 82, 91.11 %; cattle: 194, 97.98 %; pig: 175, 94.59 %) (Fig. 8f and Fig. S12d).
The top 30 pathways with the highest enrichments of each of four alignment algorithms were further explored and discussed. In humans, 15 (50.00 %) signaling pathways (such as Focal adhesion, Axon guidance and Thermogenesis) were consistently interpreted by all four algorithms, but the number of signaling pathways consistently detected by two of four algorithms ranges from 18 (60 %) to 22 (73.33 %) (Fig. S13a, Table S6-7). In cattle, 16 (53.33 %) signaling pathways (such as MAPK signaling pathway, Hepatocellular carcinoma and Axon guidance) were interpreted by all four algorithms, but the number of signaling pathways consistently detected by two of four algorithms ranges from 21 (70 %) to 24 (80.00 %) (Fig. S13b, Table S6-7). In pigs, 15 (50.00 %) signaling pathways (such as Endocytosis, Focal adhesion and Axon guidance) were interpreted by all four algorithms, but the number of signaling pathways consistently detected by two of four algorithms ranges from 19 (63.33 %) to 23 (76.67 %) (Fig. S13c, Table S6-7).
4. Discussion
It is well recognized that DNA methylation plays critical roles in mammalian development [51] and diseases [52]. Compared with other methods to profile the methylomes, WGBS has been accepted as the gold standard at single base resolution [53]. Currently, WGBS is at the forefront of epigenetic analysis and popularly utilized to investigate the genome-wide DNA methylation of mammalian developments [54] and epigenetic marks of diseases [55]. A great number of mappers and aligners have been developed and exploited to handle the mapping challenge caused by bisulfite [18]. It is considered that the mapping efficiencies and performances impress the precision of DNA methylation calculation as well as the calling of candidate DMCs, DMRs, DMR-related genes and signaling pathways. Therefore, it is essential to benchmark the mapping efficiencies of the mappers warped up distinct aligners to provide investigators with useful information. In this study, the runtime, memory consumption, uniquely mapped reads, mapped precision, recall, F1 score, unsatisfactory aligned reads as well as the accuracy at the biological interpretations were compared and evaluated among 14 alignment algorithms in humans, cattle and pigs (Fig. 1). As far as we were concerned, this study provided the most comprehensive information for the selection of alignment algorithms for WGBS data in mammals.
As showed in Fig. 2a and c, Walt and BSMAP were the fastest, and BSSeeker2-bwt2-local was the slowest, which was contrary to the memory consumption. In plants, Grehl et al. [39] and Nunn et al. [40] find that BSMAP is the fastest but consumes the most memory, compared to Bismark-bwt2-e2e, Bwa-meth and BSSeeker2-bwt. These results are in line with our study. It is generally accepted that the shorter runtime of the alignment algorithms, the more memory it consumes. In addition, although the thread of each algorithm was set to one, we found that Bismark-bwt2-e2e, Bismark-his2, BSSeeker2-bwt2-e2e, BSSeeker2-bwt2-local, BSSeeker2-bwt, and BSSeeker2-soap2 natively used 3, 3, 2, 2, 2, and 2 threads, respectively, while other eight algorithms natively used one thread. However, Walt and BSMAP are still the fastest. Previous studies have suggested that the sequencing errors cause the challenge for aligners [56]. In this study, we found that the runtime of Bwa-meth, Hisat_3n, Hisat_3n_repeat, BSBolt, BSMAP and Bismark-his2 was positively correlated with the sequencing error rates, and the memory consumption of Bwa-meth and BSBolt is highly correlated with sequencing error rates (Fig. 2b, Figs. S2-4 and S6-7). The possible explanation for this observation is that the increase of sequencing errors on reads causes aligners to find more candidate positions on the reference genome, and thus cost more runtime and memory consumption. We found that the memory consumption of 14 algorithms were accord with the genome size of humans, cattle and pigs (Fig. 2d), which was in line with the finding that the memory consumption of Bismark-bwt2-e2e, BSMAP and Bwa-meth increased with the increasing size of genomes [39].
In this study, at the alignment model of bwt2-e2e, we found that Bismark was faster than BSSeeker2 with the equal memory consumption (Fig. 2a), and the mapped precision, recall, F1 score and uniquely mapped reads of Bismark are significantly higher than that of BSSeeker2 (Fig. 3b, d and e). Similarly, Grehl et al. find that the uniquely mapped reads of Bismark is higher than BSSeeker2 in five plants, at the alignment model of bwt2-e2e [39]. In terms of Bismark, although bwt2-e2e is slower than his2 (Fig. 2a and c), the uniquely mapped reads of bwt2-e2e increased by 23.94 % than his2 (Fig. 3a and b), with the similar mapped precision, recall and F1 score (Fig. 3d and e, Fig. S5). Also, Keel et al. find that bwt2 has more correctly mapped reads than his2 in cattle and pigs [57]. In terms of BSSeeker2, bwt2-local was the slowest (Fig. 2c) and showed the lowest mapped precision, recall and F1 score (Fig. 3d and e); soap2 was the fast (Fig. 2c) and exhibited the highest uniquely mapped reads (Fig. 3b), but bwt2-e2e displayed the highest mapped precision, recall and F1 score (Fig. 3d and e). In RRBS data, Sun et al. [38] also find that the mapped precision of BSSeeker2-bwt2-e2e is higher than BSSeeker2-bwt and BSSeeker2-bwt2-local. These results demonstrated that the mapping performance of Bismark was better than BSSeeker2, and the aligner bwt2-e2e was better than his2, soap2, bwt and bwt2-local in term of mapped precision.
Moreover, we found that the mapped precision, recall, F1 score and uniquely mapped reads of Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e, and Walt were >90 % in humans, cattle and pigs (Fig. 3d-f), which were more outstanding than other nine alignment algorithms. Previous studies find that the mapped precision and uniquely mapped reads of Bwa-meth, BSMAP, and Bismark-bwt2-e2e exceeds 90 % in four crop plants [39] and three non-modul plants [40]. In humans, Chen et al. [23] find that the mapped precision of Walt, BSMAP and Bismark-bwt2-e2e was > 93 %, and Farrell et al. [36] find that BSBolt aligned the majority of simulated reads with high accuracy (>99 %). These results support that Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt are likely to be more favorable for the methylomes of mammals, with the observations that these five alignment algorithms exhibit outstanding in uniquely mapped reads, mapped precision, recall and F1 score. Compared with simulated data, 14 alignment algorithms contained less uniquely mapped reads in real data (Fig. 3b and c), the reason of which might be that the real data was more complicated and affected by more factors, such as sequencing quality and structural variation of samples, while the simulated data was simple and only affected by sequencing error rate. Although Batmeth2, BSSeeker2-bwt2-local, Hisat_3n, and Hisat_3n_repeat were excellent at uniquely mapped reads in real data, they only had ∼80 % uniquely mapped reads and exhibited lower mapped precision, recall and F1score than other ten alignment algorithms in the simulated data.
Although Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt were excellent at uniquely mapped reads, mapped precision, recall and F1 score (Fig. 3b and d-f), the unsatisfactory aligned reads of these five algorithms were markedly different (Fig. 4a). Bwa-meth was only composed of incorrect uniquely mapped reads, BSBolt mainly included incorrect uniquely mapped reads and unmapped reads, and Walt mainly consisted of unmapped reads, while BSMAP and Bismark-bwt2-e2e mainly consisted of multiple mapped reads (Fig. 4a). We also found that the unsatisfactory aligned reads were significantly over representation at repetitive sequence (Fig. 4b) and CGI regions (Fig. 4c) in mammals. The enrichment of these five algorithms on repetitive sequence and CGI regions were obviously different in mammalian species (Fig. 4b and c). Since CGI is rich in CG, the C base on the CGI will be converted to T base in bisulfite conversion, which severely reduces the complexity of the sequence and ultimately affects the reads mapping on CGI. The repetitive sequence also made it difficult to map reads accurately and uniquely to the reference genome. Previous studies found that the mapped precision was lower in the repeat-rich regions [39], and the repetitive sequences led to a significant reduction in the number of uniquely mapped reads [58], which were consistent with our findings. These results suggested that the repetitive sequences and CGIs might make a difference to the unsatisfactory aligned reads as well as mapping performance.
The discussions for CpG sites, DMCs, DMRs, DMR-related genes, and pathways were focused on Bwa-meth, BSMAP, Bismark-bwt2-e2e, and Walt, because BSBolt exhibited a huge difference in CpG sites with these four algorithms (Fig. 5b-c and Fig. S9b). We found that the distinct alignment algorithms made a significant influence on the methylomes of mammals, since most of CpG sites (Fig. 5d, Fig. S10a), DMCs (Fig. 6b, Fig. S11a) and DMRs (Fig. 6f, Fig. S11c) called by Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt were discordant. Bwa-meth called the most, and Walt called the least CpG sites (Fig. 5a), DMCs (Fig. 6a), DMRs (Fig. 6e) and genes (Fig. 8a). Furthermore, we observed that repetitive sequences retained more discordant CpG sites, discordant DMCs and discordant DMRs than non-repetitive sequence in humans, cattle and pigs (Fig. 5g, Fig 6d, Fig 6h). This observation may be due to the influence of repetitive sequence on the mapping of the unsatisfactory aligned reads (Fig. 4b). Meanwhile, it was found that more discordant CpG sites were distributed in intermediate methylation than low and high methylation (Fig. 5h). This result was in accord with one recent study that the least concordant calling was exhibited on the CpG sites with intermediate methylation for seven alignment algorithms [38]. The rigorous statistical approaches should be developed based on the information of CpG sites, such as coverage, mapping quality of reads and the number of samples, to correct the CpG sites with intermediate methylation. Although most of DMR-related genes (Fig. 8b and Fig. S12a) were detected by all Bwa-meth, BSMAP, Bismark-bwt2-e2e and Walt, the number of signaling pathways consistently detected by two algorithms ranges from 18 (60.00 %) to 24 (80.00 %) in the top 30 pathway with the highest enrichments (Fig. S13a-c, Table S6-7). These findings indicated that the alignment algorithms had a dramatic effect on the dynamics of methylomes, and right after impressed the interpretations of biological functions.
Furthermore, the comparative methylomes are recently popular to investigate the development patterns among multiple species. For example, Zachary et al. present the comparative methylomes of human and mouse in early development, and confirm that the paternal genome demethylation is a general attribute of early mammalian development [59]. Lvanoca et al. revealed species differences in DNA methylation reprogramming by comparing the methylomes of humans, cattle and pigs [60]. In this study, compared with humans, 14 alignment algorithms exhibited higher uniquely mapped reads, mapped precision, recall and F1 score in cattle and pigs (Fig. 3b, d and e). The underlying cause may be related to the complex feature of genomes, such as the number of repetitive sequences, the length of CGIs, and the content of CG. In summary, these results showed that the performance of the alignment algorithms in multiple mammals should be considered in the study of comparative methylomes.
In the Simulated Dataset A, we found that Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt exhibited higher uniquely mapped reads, mapped precision, recall and F1 score than other nine alignment algorithms (Fig. 3b, d, e and f). In terms of Real Dataset B, we found that comparison to Bwa-meth, BSBolt, Bismark-bwt2-e2e and Walt, BSMAP exhibited the highest accuracy at the detection of CpG coordinates and methylation levels (Fig. 5c and Fig. S10b), the calling of DMCs (Fig. 6c and Fig. S11b), DMRs (Fig. 6g and Fig. S11d), DMR-related genes (Fig. 8c and Fig. S12b) and signaling pathways (Fig. 8f and Fig. S12d). Collectively, BSMAP was recommended to undertake the analysis of methylome, especially for the comparative methylomes of multiple mammalian species, not only because it was excellent at uniquely mapped reads, mapped precision, recall and F1 score, but also because it captured more accurate methylation information than Bwa-meth, BSBolt, Bismark-bwt2-e2e and Walt.
5. Conclusions
Based on the real and simulated WGBS data of 14.77 billion reads, we undertook 936 mappings to benchmark and evaluate 14 popularly utilized alignment algorithms in methylomic studies of humans, cattle and pigs, in terms of runtime, memory consumption, uniquely mapped reads, unsatisfactory aligned reads, mapped precision, recall, and F1 score, as well as the accuracies of biological interpretation at the detection of CpG coordinates and methylation levels, the calling of DMCs, DMRs, DMR-related genes and signaling pathways.
It was documented that Bwa-meth, BSBolt, BSMAP, Bismark-bwt2-e2e and Walt exhibited higher uniquely mapped reads, mapped precision, recall and F1 score than other nine alignment algorithms in simulated WGBS data. Comparison to Bwa-meth, BSBolt, Bismark-bwt2-e2e and Walt, BSMAP showed the highest accuracy at the detection of CpG coordinates and methylation levels, the calling of DMCs, DMRs, DMR-related genes and signaling pathways in real WGBS data. These results can provide investigators with useful information on the choice of alignment algorithms, and help to improve the accuracy of mammalian DNA methylation detection.
CRediT authorship contribution statement
Wentao Gong: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Xiangchun Pan: Data curation, Formal analysis, Methodology, Writing – review & editing. Dantong Xu: Data curation, Formal analysis, Methodology. Guanyu Ji: Conceptualization, Supervision, Resources. Yifei Wang: Data curation, Formal analysis, Methodology. Yuhan Tian: Data curation, Formal analysis, Methodology. Jiali Cai: Data curation, Formal analysis, Methodology. Jiaqi Li: Funding acquisition, Supervision, Resources. Zhe Zhang: Funding acquisition, Supervision, Resources, Methodology, Writing – review & editing. Xiaolong Yuan: Conceptualization, Funding acquisition, Resources, Supervision, Methodology, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China [32022078 to Z.Z., 31902131 to X.Y., 32072694 to J.L.], the earmarked fund for China Agriculture Research System [CARS-35 to J.L.], the Natural Science Foundation of Guangdong Province [2019A1515010676 to X.Y., 2021A1515012396 to X.Y.], the Science and Technology Program of Guangzhou [202002030071 to X.Y.], and the China Postdoctoral Science Foundation [2020M672556 to X.Y.].
Footnotes
All scripts used for the benchmarking of alignment algorithms are available online at github (https://github.com/Wentao-Gong/BenchWGBSanimal). The detailed results for Real Dataset B were provided in figshare with DOI (10.6084/m9.figshare.20342715), including the positions, methylation levels, and coverages of CpG sites, as well as the positions of DMCs and DMRs. Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.051.
Contributor Information
Zhe Zhang, Email: zhezhang@scau.edu.cn.
Xiaolong Yuan, Email: yxl@scau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zafon C., Gil J., Perez-Gonzalez B., Jorda M. DNA methylation in thyroid cancer. Endocr Relat Cancer. 2019;26(7):R415–R439. doi: 10.1530/ERC-19-0093. [DOI] [PubMed] [Google Scholar]
- 2.Feng H., Conneely K.N., Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42(8):e69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan R., Gu C., You D., Huang Z., Qian J., Yang Q., et al. Decoding dynamic epigenetic landscapes in human oocytes using single-cell multi-omics sequencing. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Law J.A., Jacobsen S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantone I., Fisher A.G. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol. 2013;20(3):282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 7.Rauluseviciute I., Drablos F., Rye M.B. DNA methylation data by sequencing: experimental approaches and recommendations for tools and pipelines for data analysis. Clin Epigenetics. 2019;11(1):193. doi: 10.1186/s13148-019-0795-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barros-Silva D., Marques C.J., Henrique R., Jeronimo C. Profiling DNA methylation based on next-generation sequencing approaches: new insights and clinical applications. Genes (Basel) 2018;9(9) doi: 10.3390/genes9090429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serre D., Lee B.H., Ting A.H. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinkman A.B., Simmer F., Ma K., Kaan A., Zhu J., Stunnenberg H.G. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52(3):232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee A., Rodger E.J., Stockwell P.A., Le Mee G., Morison I.M. Generating multiple base-resolution DNA methylomes using reduced representation bisulfite sequencing. Methods Mol Biol. 2017;1537:279–298. doi: 10.1007/978-1-4939-6685-1_16. [DOI] [PubMed] [Google Scholar]
- 12.Chatterton Z., Mendelev N., Chen S., Carr W., Kamimori G.H., Ge Y., et al. Bisulfite amplicon sequencing can detect glia and neuron cell-free DNA in blood plasma. Front Mol Neurosci. 2021;14 doi: 10.3389/fnmol.2021.672614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Z., Cunningham J., Slager S., Kocher J.P. Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis. Epigenomics. 2015;7(5):813–828. doi: 10.2217/epi.15.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouil Q., Keniry A. Latest techniques to study DNA methylation. Essays Biochem. 2019;63(6):639–648. doi: 10.1042/EBC20190027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Y., Fu J., Wu W., Wu J. Comparative profiles of DNA methylation and differential gene expression in osteocytic areas from aged and young mice. Cell Biochem Funct. 2020;38(6):721–732. doi: 10.1002/cbf.3539. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y., Liu S., Hu Y., Fang L., Gao Y., Xia H., et al. Comparative whole genome DNA methylation profiling across cattle tissues reveals global and tissue-specific methylation patterns. BMC Biol. 2020;18(1):85. doi: 10.1186/s12915-020-00793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta A., Dobersch S., Romero-Olmedo A.J., Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34(2):229–241. doi: 10.1007/s10555-015-9563-3. [DOI] [PubMed] [Google Scholar]
- 18.Laird P.W. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 19.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13(10):705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 20.de Sena B.G., Smith A.D. Fast and memory-efficient mapping of short bisulfite sequencing reads using a two-letter alphabet. NAR Genom Bioinform. 2021;3(4):lqab115. doi: 10.1093/nargab/lqab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunde-Ramamoorthy G., Coarfa C., Laritsky E., Kessler N.J., Harris R.A., Xu M., et al. Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing. Nucleic Acids Res. 2014;42(6):e43. doi: 10.1093/nar/gkt1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cristian Coarfa, FY, 2, Christopher A Miller, Zuozhou Chen, R Alan Harris, Aleksandar Milosavljevic. Pash 3.0: A versatile software package for read mapping and integrative analysis of genomic and epigenomic variation using massively parallel DNA sequencing. BMC Bioinformatics; 2010. [DOI] [PMC free article] [PubMed]
- 23.Chen H., Smith A.D., Chen T. WALT: fast and accurate read mapping for bisulfite sequencing. Bioinformatics. 2016;32(22):3507–3509. doi: 10.1093/bioinformatics/btw490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv J., Cui W., Liu H., He H., Xiu Y., Guo J., et al. Identification and characterization of long non-coding RNAs related to mouse embryonic brain development from available transcriptomic data. PLoS ONE. 2013;8(8):e71152. doi: 10.1371/journal.pone.0071152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R., Li Y., Kristiansen K., Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 29.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 30.Lim J.Q., Tennakoon C., Guan P., Sung W.K. BatAlign: an incremental method for accurate alignment of sequencing reads. Nucleic Acids Res. 2015;43(16):e107. doi: 10.1093/nar/gkv533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen B.S., Eyring K., De S., Yang I.V., Schwartz D.A. Fast and accurate alignment of long bisulfite-seq reads. Arxiv. 2014:1–2. [Google Scholar]
- 32.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo W., Fiziev P., Yan W., Cokus S., Sun X., Zhang M.Q., et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14(1):774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Q., Lim J.Q., Sung W.K., Li G. An integrated package for bisulfite DNA methylation data analysis with Indel-sensitive mapping. BMC Bioinf. 2019;20(1):47. doi: 10.1186/s12859-018-2593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xi Y., Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinf. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrell C., Thompson M., Tosevska A., Oyetunde A., Pellegrini M. BiSulfite Bolt; A bisulfite sequencing analysis platform. GigaScience. 2021;10(5) doi: 10.1093/gigascience/giab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Park C., Bennett C., Thornton M., Kim D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021 doi: 10.1101/gr.275193.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun X., Han Y., Zhou L., Chen E., Lu B., Liu Y., et al. A comprehensive evaluation of alignment software for reduced representation bisulfite sequencing data. Bioinformatics. 2018;34(16):2715–2723. doi: 10.1093/bioinformatics/bty174. [DOI] [PubMed] [Google Scholar]
- 39.Grehl C., Wagner M., Lemnian I., Glaser B., Grosse I. Performance of Mapping Approaches for Whole-Genome Bisulfite Sequencing Data in Crop Plants. Front Plant Sci. 2020;11:176. doi: 10.3389/fpls.2020.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunn A., Otto C., Stadler P.F., Langenberger D. Comprehensive benchmarking of software for mapping whole genome bisulfite data: from read alignment to DNA methylation analysis. Brief Bioinform. 2021 doi: 10.1093/bib/bbab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuji J., Weng Z. Evaluation of preprocessing, mapping and postprocessing algorithms for analyzing whole genome bisulfite sequencing data. Brief Bioinform. 2016;17(6):938–952. doi: 10.1093/bib/bbv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran H., Porter J., Sun M.A., Xie H., Zhang L. Objective and comprehensive evaluation of bisulfite short read mapping tools. Adv Bioinformatics. 2014;2014 doi: 10.1155/2014/472045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendizabal I., Berto S., Usui N., Toriumi K., Chatterjee P., Douglas C., et al. Cell type-specific epigenetic links to schizophrenia risk in the brain. Genome Biol. 2019;20(1):135. doi: 10.1186/s13059-019-1747-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L., Amorin R., Moriel P., DiLorenzo N., Lancaster P.A., Penagaricano F. Differential network analysis of bovine muscle reveals changes in gene coexpression patterns in response to changes in maternal nutrition. BMC Genomics. 2020;21(1):684. doi: 10.1186/s12864-020-07068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y., Fan X., Yan J., Chen M., Zhu M., Tang Y., et al. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 2021;49(3):1313–1329. doi: 10.1093/nar/gkaa1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park Y., Wu H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics. 2016;32(10):1446–1453. doi: 10.1093/bioinformatics/btw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anvar Z., Chakchouk I., Demond H., Sharif M., Kelsey G. Van den Veyver IB. DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting. Genes (Basel) 2021;12(8) doi: 10.3390/genes12081214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch A., Joosten S.C., Feng Z., de Ruijter T.C., Draht M.X., Melotte V., et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15(7):459–466. doi: 10.1038/s41571-018-0004-4. [DOI] [PubMed] [Google Scholar]
- 53.Miura F., Shibata Y., Miura M., Sangatsuda Y., Hisano O., Araki H., et al. Highly efficient single-stranded DNA ligation technique improves low-input whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2019;47(15):e85. doi: 10.1093/nar/gkz435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 55.Liu B., Liu Y., Pan X., Li M., Yang S., Li S.C. DNA Methylation Markers for Pan-Cancer Prediction by Deep Learning. Genes (Basel) 2019;10(10) doi: 10.3390/genes10100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng X., Wang J., Zhang Z., Xiao Q., Li M., Pan Y. Re-alignment of the unmapped reads with base quality score. BMC Bioinf. 2015;16(Suppl 5):S8. doi: 10.1186/1471-2105-16-S5-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keel B.N., Snelling W.M. Comparison of burrows-wheeler transform-based mapping algorithms used in high-throughput whole-genome sequencing: application to illumina data for livestock genomes. Front Genet. 2018;9:35. doi: 10.3389/fgene.2018.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatterjee A., Stockwell P.A., Rodger E.J., Morison I.M. Comparison of alignment software for genome-wide bisulphite sequence data. Nucleic Acids Res. 2012;40(10):e79. doi: 10.1093/nar/gks150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith Z.D., Chan M.M., Humm K.C., Karnik R., Mekhoubad S., Regev A., et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511(7511):611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivanova E., Canovas S., Garcia-Martinez S., Romar R., Lopes J.S., Rizos D., et al. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin Epigenetics. 2020;12(1):64. doi: 10.1186/s13148-020-00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.