Abstract
CHAMP1 is a gene associated with intellectual disability, which was originally identified as being involved in the maintenance of kinetochore–microtubule attachment. To explore the neuronal defects caused by CHAMP1 deficiency, we established mice that lack CHAMP1. Mice that are homozygous knockout for CHAMP1 were slightly smaller than wild-type mice and died soon after birth on pure C57BL/6J background. Although gross anatomical defects were not found in CHAMP1−/− mouse brains, mitotic cells were increased in the cerebral cortex. Neuronal differentiation was delayed in CHAMP1−/− neural stem cells in vitro, which was also suggested in vivo by CHAMP1 knockdown. In a behavioural test battery, adult CHAMP1 heterozygous knockout mice showed mild memory defects, altered social interaction, and depression-like behaviours. In transcriptomic analysis, genes related to neurotransmitter transport and neurodevelopmental disorder were downregulated in embryonic CHAMP1−/− brains. These results suggest that CHAMP1 plays a role in neuronal development, and CHAMP1-deficient mice resemble some aspects of individuals with CHAMP1 mutations.
Keywords: CHAMP1, mouse model, neuronal development, intellectual disability, behavioural tests
Nagai et al. report the first characterization of a knockout mouse of CHAMP1, a gene mutated in individuals with intellectual disability. CHAMP1 deficiency delayed neuronal development and CHAMP1 heterozygous knockout mice exhibited mild memory defects, altered social interaction, and depression-like behaviours, resembling some aspects of the human phenotype.
Graphical Abstract
Graphical abstract.
Introduction
Intellectual disability (ID) is a common developmental disorder that affects 2–3% of the general population.1,2 As a clinical entity, ID is included in neurodevelopmental disorders (NDDs) together with autism spectrum disorder (ASD) and attention deficit hyperactivity disorder (ADHD). In the Diagnostic and Statistical Manual of Mental Disorders, 5th Edition (DSM-5), ID is defined as NDD that begins in childhood and is characterized by intellectual difficulties as well as difficulties in conceptual, social and practical areas of living.3 Both environmental stress factors, such as poor nutrition and infection, and mutations in genes cause ID.4 Through large-scale studies, a large number of mutated genes implicated in ID have been identified in nearly half of the cases, although mutations of each gene occur in a small subset of cases.5,6 How such a diverse spectrum of genes cause ID is largely unknown, and effective therapeutic interventions are lacking.
CHAMP1 is one of the genes mutated in individuals with ID.7–9 A recent report showed that disorders associated with CHAMP1 mutations also include ASD and ADHD phenotypes.10 We originally reported that this gene product is involved in the maintenance of kinetochore–microtubule attachment on the spindle during mitosis and designated the molecule as CAMP (chromosome alignment-maintaining phosphoprotein),11 registered as CHAMP1 in Human Gene Nomenclature Database. CHAMP1 (CAMP) is conserved in vertebrates, and human CHAMP1 is an 812 aa protein composed of a large intrinsically disordered region with zinc-finger domains at each end (Fig. 1A). The middle region contains several repeat motifs and it is phosphorylated at many serine residues, especially in mitosis, by CDK1 (cyclin-dependent kinase 1).11 CHAMP1 localizes to the nucleus in interphase and to chromosomes and the spindle during mitosis.11 In mice, CHAMP1 expression is detected in brain, thymus, testis and ovary tissues.8 In individuals with ID, most of the mutations identified in CHAMP1 are either nonsense or frameshift de novo mutations that result in the formation of truncated proteins lacking the C-terminal region to varying degrees (Fig. 1A).7–10,12–15 We reported that these C-terminally truncated CHAMP1 mutant proteins cannot properly localize to the chromatin in interphase nor to chromosomes in mitosis.8 Some individuals with CHAMP1 mutations show other symptoms such as microcephaly, facial dysmorphism and convulsions.7–10 It has been reported that microdeletion of the 13q34 locus that includes CHAMP1 also causes a similar phenotype,13,16,17 which is sometimes inherited. Intriguingly, the gene encoding pogo transposable element-derived protein with zinc-finger domain (POGZ), an interacting partner of CHAMP1, is also mutated in individuals with ID, as well as in individuals with ASD,18–21 causing a variety of phenotype known as White-Sutton syndrome.22 The POGZ mutations found in ID are either nonsense or frameshift de novo mutations similar to CHAMP1 mutations. We found that CHAMP1 and POGZ interact with their C-terminal regions,8 suggesting that the interaction between CHAMP1 and POGZ is lost in mutants of both proteins, which may be related to the aetiology of ID.
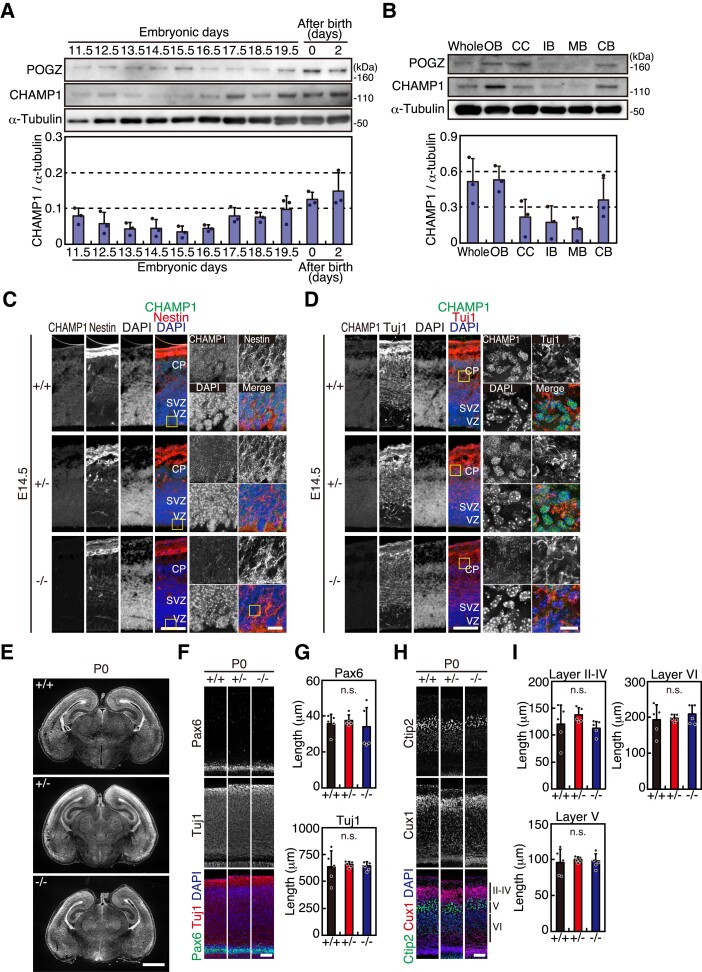
Figure 1.
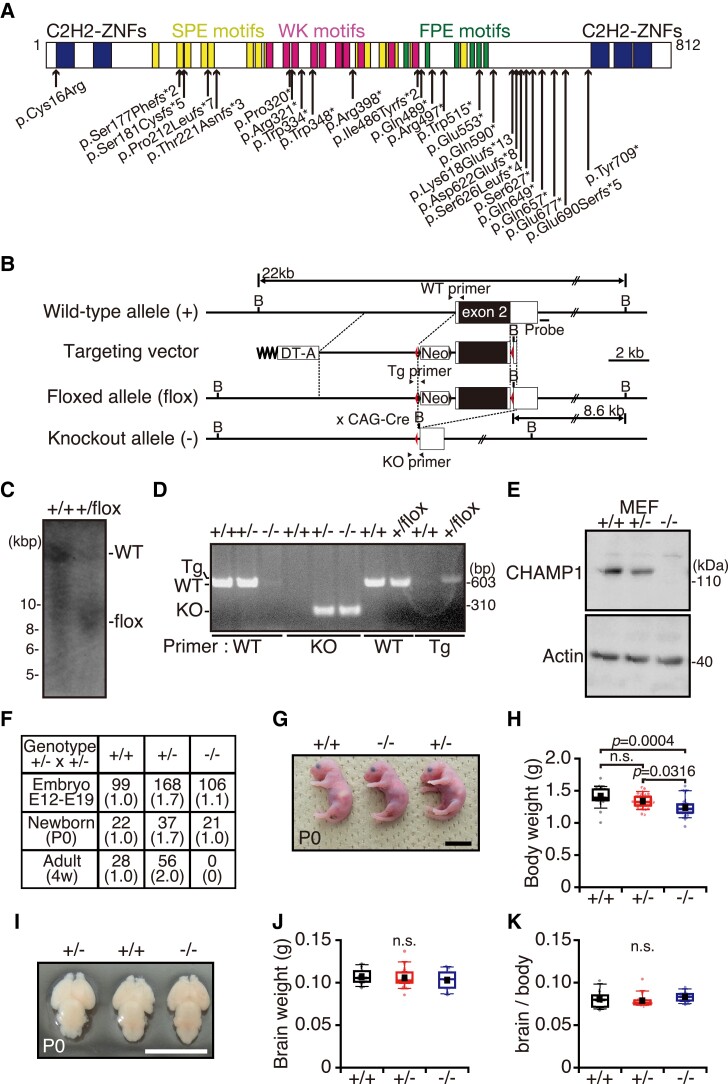
CHAMP1 −/− mice show a newborn lethal defect. (A) CHAMP1 mutations found in individuals with ID. A schematic structure of CHAMP1 and its ID-associated de novo mutations are shown. fs, frameshift; *, premature stop codon. (B) A schematic of construction of the CHAMP1-knockout allele. Positions of the probes for Southern blotting and PCR primers are indicated. Red triangle shows positions of the lox-P sites. Neo, neomycin-resistance gene; DT-A, diphtheria toxin A gene; B, BamHI site. (C) Southern blot analysis of genomic DNA prepared from CHAMP1+/+ and CHAMP1+/flox mice with the probe shown in (B). (D) Genotyping PCR analysis of CHAMP1+/+, CHAMP1+/flox, CHAMP1+/−, and CHAMP1−/− mice using the primer sets shown in (B). (E) IB analysis of lysates prepared from MEFs isolated from CHAMP1+/+, CHAMP1+/−, and CHAMP1−/− mice using antibodies as indicated. (F) Number and ratio of CHAMP1+/+, CHAMP1+/−, and CHAMP1−/− mice from crosses of C57BL/6J CHAMP1+/− mice. (G) Representative picture of newborn CHAMP1+/+ (left), CHAMP1+/− (right), and CHAMP1−/− (middle) mice. Scale bar: 1 cm. (H) Comparison of the body weight of newborn mice (CHAMP1+/+; n = 19, CHAMP1+/−; n = 36, CHAMP1−/−; n = 19). (I) Representative pictures of newborn CHAMP1+/+ (middle), CHAMP1+/− (left), and CHAMP1−/− (right) mouse brains. Scale bar: 1 cm. (J, K) Absolute brain weight (J) and ratios of brain weight to body weight (K) of newborn mice (CHAMP1+/+; n = 8, CHAMP1+/−; n = 14, CHAMP1−/−; n = 8). For box plots in (H, J, K), the bottom and top of the box show the lower and upper quartile values, respectively. The mean is indicated with a filled square and the median is indicated with a bar in the box. The bottom and top of the whiskers denote the 10th and 90th percentiles, respectively. P values were determined by Tukey–Kramer test. n.s., not statistically significant.
Mouse models have contributed to the understanding of the pathophysiology of ID and ASD.4 To explore the neuronal functions of CHAMP1 related to ID, we generated a CHAMP1 knockout mouse. We found that CHAMP1 homozygous knockout mice die soon after birth. Although brain structure of CHAMP1−/− mice was grossly normal, CHAMP1−/− neural stem cells (NSCs) showed a delay in neuronal differentiation in vitro, which was supported by CHAMP1 knockdown in vivo. In a behavioural test battery, CHAMP1 heterozygous knockout mice exhibited mild memory defects, altered social interaction and depression-like behaviours. Transcriptomic analysis revealed downregulation of neurotransmitter transport and NDD-related genes. Our study suggests that CHAMP1 plays a role in neuronal development, and CHAMP1 knockout mice contribute to understanding the pathophysiology of disorders seen in individuals with CHAMP1 mutations.
Materials and methods
Mice
CHAMP1 mutant mice (Accession No. CDB 1106K: https://large.riken.jp/distribution/mutant-list.html) were established as described (https://large.riken.jp/protocol/). To construct a targeting vector, a neomycin resistant gene, frt sequences and lox-P sites were inserted into the BAC clone containing Exon 2 of the CHAMP1 gene (RP24-377P6, BACPAC resources) (Fig. 1B). TT2 embryonic stem cells23 were electroporated with the linearized targeting vector and selected with G418 (Thermo Fisher Scientific). The homologous recombinant ES clones were identified by PCR and Southern blotting and were used to generate chimeric mice. The chimeric mice were crossed with C57BL/6J (wild-type) mice to obtain CHAMP1+/flox mice. The recombination event was confirmed by Southern blot analysis of BamHI-digested genomic DNA, which was performed at Phoenix Bio Co., Ltd (Japan). CHAMP1+/− mice were generated by breeding CHAMP1+/flox male mice with CAG-Cre female mice.24 CHAMP1 knockout mice were backcrossed into C57BL/6J for at least five generations. We bred the animals in groups of one or two under specific pathogen-free conditions (ad libitum access to food and water, 12:12 light:dark cycle, light on at 08:00 am). Mice were tagged using ear punch and randomly assigned to each experiment. All experimental procedures conformed to ‘Regulations for Animal Experiments and Related Activities at Tohoku University’ and ‘Guidelines for Conducting Animal Experiments at RIKEN Kobe Branch’ and were reviewed by the Institutional Laboratory Animal Care and Use Committee of Tohoku University and the Institutional Animal Care and Use Committee of RIKEN Kobe Branch and finally approved by the President of Tohoku University (2019AcA-024) and RIKEN Kobe Branch (A2001-03), and the same procedure was performed in Fujita Health University (AP16016).
Antibodies and PCR primers
Anti-CHAMP1 polyclonal antibody was purified using antigen-conjugated HiTrap NHS-activated HP Columns (GE Healthcare) from antiserum of rabbits immunized against mouse CHAMP1 aa515–646 (UNITECH) and used for immunofluorescence (IF) and immunoblot (IB) at 1:1000. Primary antibodies used in IF were as follows: mouse anti-Nestin (1:1000, Abcam), mouse anti-Tuj1 (βIII tubulin; 1:1000, BioLegend), rabbit anti-Pax6 (1:300, BioLegend), rabbit anti-Pax6 (1:1000, MBL), rabbit anti-Cux1 (1:1000, Santa Cruz Biotechnology), rat anti-Ctip2 (1:1000, Abcam), chicken anti-green fluorescent protein (GFP) (1:1000, Abcam), mouse anti-phospho-histone H3 Ser10 (1:1000, Cell Signalling), rabbit anti-PCNT (pericentrin; 1:1000, Abcam), rabbit anti-Ki67 (1:1000, Novusbio), mouse anti-glial fibrillary acidic protein (GFAP) (1:1000, Thermo Fisher Scientific), rabbit anti-Map2 (1:1000, BioLegend), mouse anti-Ankyrin-G (1:1000, Santa Cruz Biotechnology), and rabbit anti-POGZ (1:300).25 Polyclonal goat antibody against actin (Santa Cruz Biotechnology) was used in IB at 1:1000. Alexa Fluor 488-rabbit (Thermo Fisher Scientific), Alexa Fluor 568-mouse (Thermo Fisher Scientific), and Alexa Fluor 488-rat (Jackson Immunoresearch Laboratories) were used as secondary antibodies in IF at 1:1000. Alexa Fluor 488-chicken (1:500; Jackson Immunoresearch Laboratories), Cy3-anti-rabbit IgG (1:500; Jackson Immunoresearch Laboratories), Cy3-anti-rat IgG (1:500; Jackson Immunoresearch Laboratories) were used as secondary antibodies in IF at 1:500. Rabbit IgG-HRP (Santa Cruz Biotechnology) and goat-IgG-HRP (Santa Cruz Biotechnology) were used as secondary antibodies in IB at 1:1000. To stain DNA, 4′,6-diamidino-2-phenylindole (DAPI) was used at 1 μg/ml. The following primers were used for genotyping PCR: forward primer: 5′- CCTGAGAGGCCCCAAATAAAACTCCAAGTG - 3′; reverse primers: 5′- CTCGGTGTCTTTGCTCGGCTGCTCTTTCGG - 3′ (wild-type allele), 5′- TGAGCCCAGAAAGCGAAGGAGCAAAG −3′ (floxed allele), and 5′- CAGACCAGGATACACCACTACACACATACT - 3′ (knockout allele).
Cell culture
Mouse embryonic fibroblasts (MEFs) were isolated from embryonic (E) Day 14.5 embryos and maintained at 37°C in 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (DMEM, Nacalai Tesque) supplemented with 10% foetal bovine serum and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid. NSCs were cultured essentially as previously described.26 Briefly, brains from E14.5 mice were triturated in DMEM/F12, GlutaMAX supplement (Thermo Fisher Scientific) and cultured for 4 days in DMEM/F12, GlutaMAX supplement, containing N2 supplement (R & D Systems) added with 20 ng/ml EGF (Merck) and 20 ng/ml FGF2 (Merck) on dishes precoated with 15 μg/ml poly-L-ornithine (Merck) and 1 μg/ml fibronectin (Merck). Cells were then detached in DMEM/F12, GlutaMAX supplement and re-plated on dishes precoated as above, and cultured for 5 days in DMEM/F12, GlutaMAX supplement, containing N2 supplement without EGF and FGF2. In Supplementary Fig. 7, NSCs were cultured using neurosphere culture method.27 Briefly, cells were dissociated with neuron dissociation solutions (FUJIFILM Wako) according to the manufacturer’s instructions from the cortex in the E13.5 mouse brain and cultured for 4 days in the NSC culture medium as above on Costar Ultra-low attachment plate (Corning). Neurospheres were suspended and put on the dishes precoated with 15 μg/ml poly-L-ornithine and 1 μg/ml fibronectin. After 24 h, the neurospheres were transfected with a control or CHAMP1 siRNA using RNAiMAX (Thermo Fisher Scientific).. The sequences of siRNAs are as follows: CHAMP1 siRNA, AUAUGUCUGACGCUACCUUCGGUGA; control siRNA, UUCCUCUCCACGCGCAGUACAUUUA (Stealth; Thermo Fisher Scientific). Neurospheres were then differentiated for 5 days in the absence of EGF and FGF2 as described above. Primary cortical neurons were dissociated with neuron dissociation solutions (FUJIFILM Wako) according to the manufacturer’s instructions. Dissociated neurons were seeded on coverslips or dishes coated with 10 μg/ml poly-D-lysine (PDL; Merck) and 2 μg/ml Cultrex Mouse Laminin I (Trevigen) cultured in Neurobasal medium (Thermo Fisher Scientific) containing N21-MAX Media supplement (R & D Systems) and 0.5 mM L-glutamine (Nacalai Tesque).
Immunofluorescence staining and microscopic observation
Immunohistochemistry of frozen sections was performed according to our previous report.28 All samples were fixed in 1% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4, pH 7.4) for 12 h. Brains were cryoprotected in 10% and then 20% sucrose in PBS for 12 h each at 4°C and were frozen with Tissue-Tek O.C.T. compound (Sakura Finetek Japan). Coronal or sagittal sections were cut for 14 μm thickness using cryostat CM3050S (Leica Biosystems) and mounted onto MAS-coated glass slides (Matsunami Glass). Brain sections were masked with 3% albumin, from bovine serum, Fraction V pH7.0 (BSA, Fujifilm Wako) in TBST (403 mM Tris, pH 7.5, 137 mM NaCl, 27 mM KCl, and 0.1% Triton-X100) and were incubated with primary antibodies in 3% BSA/TBST or Can Get Signal immunostain solution A (TOYOBO) for 12 h at 4°C. After washes with TBST, sections were incubated with secondary antibodies in 3% BSA/TBST for 1 h at room temperature and were mounted with ProLong Gold (Thermo Fisher Scientific). Images were captured by a confocal laser microscope LSM800 (Zeiss) and BZ-9000 microscope (Keyence) using ×4, 0.20 numerical aperture (NA) Plan Apochromat air objective lens (Nikon) or ×10, 0.45 NA Plan Apochromat air objective lens (Nikon). Z-image stacks were captured in 0.2 μm increments on an IX-83 inverted microscope (Olympus) using ×60, 1.35 NA UPlan SApochromat oil objective lens (Olympus). Deconvolution was performed when necessary. Image stacks were projected and saved as TIFF files. Each experiment was successfully repeated at least three times.
Nissl staining
Cryosections after post-fixation with 4% PFA in PBS were washed in TBST, and incubated with NeuroTrace™ 500/525 Green Fluorescent Nissl Stain (1:300, Molecular Probes) in PBS for 30 min. After washing with TBST, the sections were mounted with VECTASHIELD antifade mounting medium (Vector Laboratories Inc).
In situ hybridization
In situ hybridization for frozen sections was performed as previously described.29,30 To obtain templates for cDNA fragment, full-length mouse CHAMP1 cDNA was amplified by PCR from a cDNA clone (mKIAA1802) in the ROUGE database (http://www.kazusa.or.jp/rouge/) and was inserted into EcoR1 and Xho1 sites in pcDNA3.1Zeo (−) expression vector (Thermo Fisher Scientific). A riboprobe was obtained from a CHAMP1 cDNA fragment (1542–2409 bp), which was cloned into a pBluescript IISK (−) (Agilent Technologies). POGZ riboprobe was obtained from a POGZ cDNA fragment (1–1000 bp), which was amplified from a cDNA clone (MMM1013-202761079, Horizon Discovery) and cloned into a pBluescript IISK (−).
Western blotting
Brains and MEFs were lysed in TNE-N buffer (1% NP-40, 150 mM NaCl, 20 mM Tris HCl, pH 7.5, and 2 mM EDTA). Protein concentration was measured by the Bio-Rad Protein assay kit (Bio-Rad). Extracted proteins were boiled for 5 min with 4× NuPAGE LDS (lithium dodecyl sulphate) sample buffer (Thermo Fisher Scientific). Proteins were separated by NuPAGE sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) Gel System (Thermo Fisher Scientific), electroblotted onto a polyvinylidene difluoride membrane (PVDF; Amersham Hybond-P, GE Healthcare). PVDF membrane was masked with 3% non-fat dry milk in TBST (150 mM NaCl, 0.1% Tween20, 25 mM Tris HCl pH7.5) for 30 min at room temperature, incubated with primary antibodies in Can Get Signal IB enhancer Solution I (TOYOBO) for 12 h at 4°C, washed with TBST and incubated with secondary antibodies in 3% BSA for 1 h in TBST at room temperature. After final washes, proteins were visualized using enhanced chemiluminescence, according to the manufacturer’s instructions. Chemiluminescence signals were detected using LAS4000mini Lumino Image Analyzer (GE Healthcare).
In utero electroporation
In utero electroporation of E14.5 C57BL/6J mouse cortex was performed as previously described with a minor modification.31,32 Specific siRNA for CHAMP1 or control, which is the same as the one used in in vitro culture, and pCAG-EGFP (kindly gifted from Prof. Tetsuichiro Saito, Chiba University, Japan) were injected into the lateral ventricle of the brain at concentrations of 2.0 μg/μl and 0.5 μg/μl, respectively. Injections were carried out using a glass capillary with the electric microinjector system (Narishige). The electroporation into progenitor cells in the ventricular zone (VZ) was performed with five pulses of 40 V using a square wave pulse electroporator (BEX). Electroporated embryos were fixed at E16.5 and E18.5.
Terminal deoxynucleotidyl transferase dUTP nick end labelling staining
The in situ cell death detection kit (Merck) was used to detect apoptotic cells according to the manufacturer’s protocol. Briefly, the sections were fixed in 4% PFA for 15 min at room temperature. The sections were incubated in 0.1% Triton X-100 for 2 min on ice. Transferase dUTP nick end labelling (TUNEL) mixture was added to brain sections and incubated in a humidified chamber for 60 min at 37°C.
Golgi-Cox staining and spine density analysis
Golgi-Cox staining was performed according to the manufacturer’s protocol of an FD Rapid Golgi Stain Kit (FD Neurotechnologies). Briefly, the unfixed brains were immersed in specific impregnation solution containing Solution A and B (1:1) in darkness at room temperature for 14 days and then incubated in solution C at 4°C in darkness for 3 days. After freezing with dry ice, the brains were sliced into 100 μm pieces using a vibratome (Leica VT1200; Leica Biosystems) and mounted on gelatin-coated slides with solution C. After drying at room temperature, the sections were stained with a mixed solution consisting of Solution C, Solution D and Milli-Q water at 1:1:2 for 5 min. After sections were rinsed in Milli-Q water, each section was dehydrated in graded alcohol solutions, cleared in xylene, and coverslipped. Spines were judged mushroom if the diametre of the head was much greater than the diametre of the neck, according to the previously described criteria.33
Sholl analysis
Image stacks of neurons differentiated from NSCs for 5 days immunostained with a Tuj1 antibody were traced and reconstructed as 2D binarized representations using ImageJ (National Institute of Health, http://rsb.info.nih.gov/ij/). Cells were analyzed by selecting the centre of their cell bodies, and the number of intersections at circles of increasing radii from the centre was counted using an ImageJ plugin for Sholl analysis.34
General protocol for behavioural tests
Mice used in this study had been backcrossed more than nine times to the C57BL/6J strain. Behavioural tests were performed with male mice that were at least 10-weeks-old. In each strain, we ran the experiments with a single batch (CHAMP1+/+ n = 20, CHAMP1+/− n = 20). Mice were group-housed in a room with a 12 h light/dark cycle (light on at 7:00 am) with access to food and water except for the period during which the olfactory test was being conducted. Room temperature was kept at 23 ± 2°C. Behavioural tests were performed between 08:00 am and 06:00 pm. After tests, all apparatus used was cleaned with diluted hypochlorite solution or 70% ethanol to prevent bias due to olfactory cues. The interval between tests was at least 1 day. Unless otherwise noted, behavioural tests were performed as previously described.35
T-maze forced alternation test
The forced alternation test was performed using an automatic T-maze apparatus with a video camera (O’HARA & Co.). It was constructed of 25 cm-high white plastic runways and partitioned off into six areas by sliding doors that can be opened downward. The stem of the T was comprised area S2 (12 × 24 cm), and the arms of the T comprise areas A1 and A2 (11.4 × 20.5 cm). Areas P1 and P2 were connecting passageways from respective arms (A1 and A2) to the start compartment area (S1). Mice were subjected to a forced alternation protocol for 7 days (one session consisting of 10 trials per day; cut-off time of 50 min). Each trial had first and second runs. On the first run, the mouse was forced to choose one of the arms of area (A1 or A2). After the mouse stayed for 10 s, the door that separated the arms of area (A1 or A2) and connecting passageway (P1 or P2) was opened to allow the mouse to return to the starting area (S1). The mouse was given a 3 s delay in area S1, followed by a free choice between both T arms. The correct response was the choice of the other arm that had not been chosen by the first trial. The location of the sample arm (left or right) was varied pseudo-randomly across trials. On Days 4–7, a delay (3, 30, 60 or 120 s) was applied after trial. The percentage of correct response was measured. Data acquisition, control of sliding doors and data analysis were performed by ImageTM software (see ‘Data analysis’).
Contextual and cued fear conditioning test
A contextual and cued fear conditioning test was conducted as previously described.36,37 This was performed using an automatic apparatus with a video camera (O’HARA & Co.). Each mouse was placed in an acrylic chamber (33 × 25 × 29 cm) consisting of white (side) and transparent (front, rear and top) walls with a stainless-steel grid floor (0.2 cm diametre, spaced 0.5 cm apart) (O’HARA & Co.) illuminated at 100 lux and allowed to explore freely for 2 min. White noise at 55 dB, which served as the conditioned stimulus (CS), was presented for 30 s, followed by a mild (2 s, 0.3 mA) foot shock, which served as the unconditioned stimulus (UCS). Two more CS–UCS pairings were presented with a 2 min interstimulus interval. Context test was conducted 1 day and 1 month after conditioning in the same chamber for 5 min on each mouse. Cued test with altered context was conducted after conditioning using a triangular box (31 × 31 × 32 cm) made of white opaque Plexiglas, which was located in a different room. The mice were allowed to explore the chamber for 180 s. Tone stimulus for the cued test (55 dB white noise) was applied for 3 min. In each test, freezing percentage and distance travelled (cm) were calculated automatically using ImageFZ software (see ‘Data analysis’).
Hot plate test
The hot plate test was used to evaluate sensitivity to a painful stimulus. Mice were placed on a hot plate (Columbus Instruments) at 55.0 ± 0.1°C. The latency to the first forepaw or hind paw response defined as either a foot shake or a paw lick, was recorded with 15 s cut-off time.
Barnes maze test
Barnes maze test was conducted as previously described.38 This test was performed on ‘dry land’, a white circular surface, 1.0 m in diametre, with 12 holes equally spaced around the perimetre in an automatic apparatus with a video camera (O’HARA & Co.). A black Plexiglas escape box (17 × 13 × 7 cm), which had paper cage chip on the bottom, was located under one of the holes. The hole above the escape box represented the target. The location of the target was consistent for a given mouse but was randomized across mice. The maze was rotated daily, with the spatial location of the target unchanged with respect to the distal visual room cues, to prevent a bias based on olfactory or proximal cues within the maze. During acquisition test, each trial ended when a mouse entered the escape box. If the mouse did not enter the target hole within 300 s, it was gently guided to the target hole by the experimenter. A probe trial was performed without the escape box 1 day after the last training session. The second probe trial was conducted 1 month after the last training session to evaluate memory retention. Latency, distance, and number of errors before first reaching the target hole, omission error to reach the target hole, and time spent around each hole were recorded with the use of ImageBM software (see ‘Data analysis’).
Social interaction test in a novel environment
In this test, two mice of identical genotypes that were previously housed in different cages were placed in a box together (40 × 40 × 30 cm) (O’HARA & Co.) and allowed to explore freely for 10 min. Behaviour was recorded and analyzed automatically using ImageSI software (see ‘Data analysis’). The total number of contacts, total duration of contact, mean duration per contact and total distance travelled were measured. If the two mice contacted each other and the distance travelled by either mouse was longer than 10 cm, the behaviour was classified as an ‘active contact’. Images were captured at three frames per second and distance travelled between two successive frames was calculated for each mouse.
Crawley’s sociability and social novelty preference test
This test is a method to investigate the effect of complex genetics on sociability and preference for social novelty.39 The testing apparatus consisted of a rectangular, three-chambered box and a lid with a video camera (O’HARA & Co.). Each chamber was 20 × 40 × 47 cm and the dividing walls were made from clear Plexiglas with small square openings (5 × 3 cm) allowing access into each chamber. The centre of the field was illuminated at 100 lux. A habituation session was performed in the apparatus for 10 min before the sociability test, and the wire cages in the lateral compartments were located in the corners of each compartment. In the sociability test, an unfamiliar C57BL/6J male mouse (stranger) that had no prior contact with the subject mice, was placed in one of the side chambers. The location of stranger mouse in the left or right side-chamber was systematically alternated between trials. The stranger mouse was enclosed in a small, circular wire cage, which allowed nose contact between the bars, but prevented fighting. The cage was 11 cm in height, with a bottom diametre of 9 cm, and vertical bars 0.5 cm apart. The subject mouse was first placed in the middle chamber and allowed to explore the entire test box for a 10 min session. The amount of time spent in each wire cage was measured by automatic apparatus with a video camera (O’HARA & Co.) fitted on top of the box. After the first 10 min, each mouse was tested in a second 10 min session to quantify social preference for a new stranger mouse. The new stranger mouse was enclosed in an identical small wire cage. The test mouse thus had a choice between the first, already-investigated mouse (familiar mouse) and the novel unfamiliar mouse (stranger mouse). The amount of time spent around each wire cage and chamber during the second 10 min session was recorded. Data acquisition and analysis were performed automatically using ImageCSI software (see ‘Data analysis’).
Twenty-four-hour home-cage monitoring
Two mice of the same genotype that had been housed separately were placed together in a home cage (15 × 25 × 30 cm) (O’HARA & Co.) with video camera attached. Their social behaviours and locomotor activities were monitored in a home cage for 1 week and the values for Days 4 through 6 were averaged. The counting of mice was designated as a ‘particle’ that was detected in each frame: two particles indicated that the mice were not in contact with each other and one particle indicated contact between the two mice. Analysis was performed automatically using ImageHA software (see ‘Data analysis’).
Rotarod test
This test, which uses an accelerating rotarod (UGO Basile), was performed on rotating drums (3 cm diametre) and latency to fall from the rotarod was recorded. The speed of the rotarod accelerated from 4 to 40 r.p.m. over a 5 min period. The test was performed three times per day.
Grip strength test
Grip strength was measured by a grip strength metre (O’HARA & Co.). After mice grasped a wire grid by the forelimbs, they were pulled backward until they release it. The peak force applied by the forelimbs of the mouse was recorded in Newtons (N). Each mouse was tested three times, and the greatest value obtained was used for further data analysis.
Wire-hang test
Wire-hang test apparatus was used to assess balance and grip strength. The apparatus consists of a box (21.5 × 22 × 30 cm) with a wire-mesh grid (10 × 10 cm) on top that can be inverted (O’HARA & Co.). The mice were placed on the wire mesh at the top of the apparatus, then the wire mesh was gently turned inside out. The latency to the mouse falling was recorded, with a 60 s cut-off time.
Beam test
Beam test was conducted as previously described.40 Motor coordination and balance were assessed with the test. This was performed by automatic apparatus with a video camera (O’HARA & Co.). The beam test was measuring the ability of mice to traverse a narrow beam to reach a dark box. The beams, with a rough painted surface, consisted of two different strips of iron. Each beam is 100 cm long, one was 2.8 cm (wide bar) and the other was 1.1 cm (narrow bar) placed horizontally 50 cm above the surface. One session of four trials was performed using the 2.8 cm beam and the other one of five trials was performed using the 1.1 cm beam. Mice were allowed up to 90 s to traverse each beam. The number of sideslips was recorded for each trial by the ImageBT programme (see ‘Data analysis’).
Porsolt forced swim test
Porsolt forced swim test was conducted as previously described,37 performed by automatic apparatus with a video camera (O’HARA & Co.). The apparatus was consisted of four Plexiglas cylinders (22 cm in height and 11 cm in diametre) that were filled with dilute sodium hypochlorite solution (∼23°C) up to a height of 7.5 cm of a white plastic chamber. The mice were placed in the cylinders, and the immobility and the distance travelled were recorded for 10 min. The test was performed for 2 days. Data acquisition and analysis were automatically performed. Immobility time (%) and distance travelled (cm) were measured by ImageTS software (see ‘Data analysis’).
Tail suspension test
Tail suspension test was conducted as previously described.41,42 Mice were suspended 30 cm above the floor of a white plastic chamber (O’HARA & Co.) and recorded for 10 min. This was performed by automatic apparatus with a video camera (O’HARA & Co.). Data acquisition and analysis were automatically performed. Immobility time (%) was measured by ImageTS software (see ‘Data analysis’).
Open field test
Each mouse was placed in the corner of the open field apparatus (40 × 40 × 30 cm) (Accuscan Instruments), which was illuminated at 100 lux. Total distance travelled (cm), vertical activity (rearing measured by counting the number of photobeam interruptions), time spent in the centre area (20 × 20 cm), and beam-break counts for stereotyped behaviours (defined by the number of breaks of the same beam) were recorded. Data were collected for 120 min.
Light/dark transition test
The apparatus for this test was conducted in a cage (21 × 42 × 25 cm) that was divided into two sections of equal size by a partition with a door (O’HARA & Co.). One chamber was made of white plastic and brightly illuminated, whereas the other was black and dark. Mice were placed in the dark side and allowed to move freely between two chambers with the door open for 10 min. The number of transitions between the two compartments, latency to first entry into the light chamber, distance travelled and time spent in each chamber were recorded with the use of ImageLD software (see ‘Data analysis’).
Elevated plus maze test
This test was conducted to assess anxiety-like behaviour. The elevated plus maze consisted of two open arms (25 × 5 cm) with 3 mm high transparent walls (O’HARA & Co.). The floors of the arms and the central square (5 × 5 cm) were made of white plastic plates and elevated to a height of 55 cm above the floor. The arms of the same type were arranged on opposite sides of the maze. The centre of the maze was illuminated at 100 lux. Each mouse was placed into the centre of the maze facing one of the closed arms and was recorded for 10 min. The distance travelled, number of total entries into the arms, percentage of entries into the open arms and percentage of time spent in the open arms were measured for 10 min using ImageEP software (see ‘Data analysis’).
Startle response and prepulse inhibition test
To measure acoustic startle response and prepulse inhibition (PPI), a startle reflex measurement system (O’HARA & Co.) was used. Before this test, mice were kept in a soundproof room separate from the testing room. A test session began by placing a mouse in a transparent PVC plastic cylinder where it was left undisturbed for 10 min. White noise (40 ms) was used as the startle stimulus for all trial types. The startle response was recorded for 400 ms starting with the onset of the startle stimulus. The background noise level in each chamber was 70 dB. A test session consisted of six trial types (i.e. two types for startle stimulus-only trials, and four types for PPI trials). The intensity of the startle stimulus was 110 or 120 dB. The prepulse sound was presented 100 ms before the startle stimulus, and its intensity was 74 or 78 dB. Four combinations of prepulse and startle stimuli were used (74–110, 78–110, 74–120 and 78–120 dB). Six blocks of the six trial types were presented in a pseudo-random order, such that each trial type was presented once within a block. The average inter-trial interval was 15 s (range: 10–20 s).
RNA sequence analysis
Total RNA was isolated from whole brain of male mouse embryos at embryonic Day 14.5 (E14.5, wild-type: n = 6, CHAMP1+/−; n = 4, CHAMP1−/−; n = 5) using RNeasy mini kit (QIAGEN), following the manufacturer’s instructions. Library construction and RNA-seq were performed using the Illumina sequencing platform (Annoroad Gene Technology). The short reads were mapped to the mm9 mouse reference genome using the STAR software package.43 The counts per million (CPM) were quantified from aligned reads by Cuffquant and Cuffnorm from the Cufflinks package and used for further analysis. The genes were annotated to the GENCODE annotation release M1 (NCBIM37), and the mitochondrial RNA and the ribosomal RNA were masked using handmade mtRNA/rRNA annotation list. Read counts per gene were obtained with HTSeq.44 Differentially expressed genes (DEGs) analysis and gene ontology (GO) annotation analysis were performed using iDEP.92.45 If none of the samples have the CPM (counts per million) values of more than 0.5 for a gene, the gene was discarded in the pre-process section in iDEP.92. The DEG was obtained under the minimum fold change >1.2 and false discovery rate (FDR) <0.1. Pre-ranked gene set enrichment analysis (GSEA) was performed using the GSEA software v.4.0.1 (Broad Institute).46 All (23 468) genes were ranked by fold change value in DEG analysis and used for pre-ranked GSEA. The gene set of 94 genes, in which damaging de novo mutations were found in NDD, were obtained from a previous report.6 To evaluate the significance of the enrichment of genes in the gene set in GSEA, random gene sets were created using Random Gene Set Generator (http://www.molbiotools.com/randomgenesetgenerator.html) and used for analysis together with the gene set.
Data analysis
Length in Fig. 3C, Supplementary Fig. 1C, H, 6C and D, and signal intensity in Fig. 4D were measured using Fiji.47 Angles in Fig. 3E were measured using MATLAB R2017b (Mathworks). The applications for behavioural studies (ImageTM, ImageFZ, ImageBM, ImageCSI, ImageHA, ImageBT, ImageTS, ImageLD, and ImageEP) were developed by Tsuyoshi Miyakawa (available through O’HARA & Co.) and are based on ImageJ. Statistical analysis according to paired t-test, Student’s t-test, one-way ANOVA, or two-way repeated-measures ANOVA was performed with the use of StatView 5.0.1 software (SAS Institute). Statistical analyses using the Tukey–Kramer post hoc test and the Steel–Dwass post hoc test were performed with Easy R (EZR),48 which is a graphical user interface for R [R Core Team, R: a language and environment for statistical computing, https://www.R-project.org/, (2018)]. More precisely, it is a modified version of R commander designed to add statistical functions frequently used in biostatistics. Data are represented as mean ± standard error of the mean (S.E.M.). If a P value was smaller than 0.05, the difference was considered statistically significant.
Figure 3.
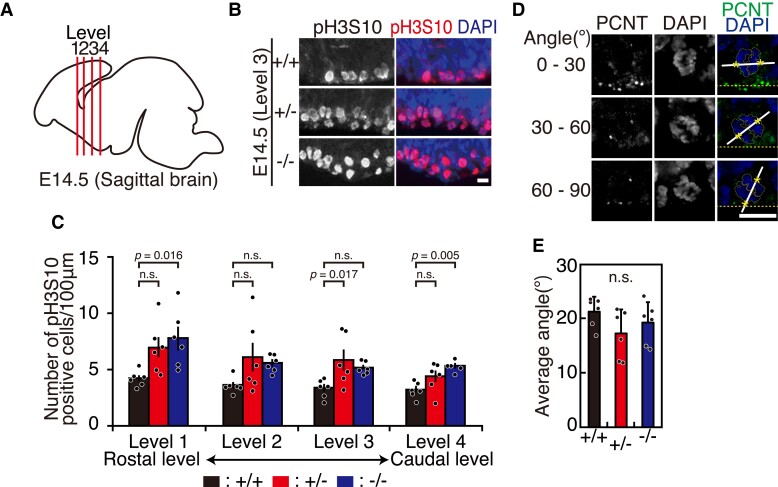
Mitotic cells are increased in CHAMP1 knockout mouse brains. (A) A schematic diagram of E14.5 brain depicting the levels of coronal sections, in which phospho-histone H3 at serine 10 was visualized. (B) Immunofluorescence staining of the VZ of E14.5 CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice at Level 3 in (A), using antibodies against phosphorylated histone H3 at serine 10 (pH3S10, red). Nuclei were stained with DAPI (blue). Scale bar: 10 μm. (C) Quantitative analysis of the number of pH3S10-positive cells in VZ of E14.5 mice at different levels shown in (A) (CHAMP1+/+; n = 6 (cortices), CHAMP1+/−; n = 6, CHAMP1−/−; n = 6). The data represent a minimum of 18 cells per cortex. Error bars represent S.E.M. P values were determined by Tukey–Kramer test. n.s., not statistically significant. (D) Immunofluorescence images of anaphase progenitor cells of E14.5 mice stained with antibodies against PCNT (green). Representative cells showing different orientation of the anaphase spindle axis to the ventricular surface are shown. Nuclei were stained with DAPI (blue). Centrosome positions, determined by PCNT signals, were shown by asterisks. Scale bar: 10 μm. (E) Average of the angle between anaphase spindle axis and the ventricular surface in CHAMP1+/+ (six cortices from three mice, at least 20 cells were counted per cortex), CHAMP1+/− (six cortices from three mice, at least 18 cells were counted per cortex), and CHAMP1−/− (six cortices from three mice, at least 16 cells were counted per cortex) mice. Error bars represent S.D. n.s. not statistically significant (Tukey–Kramer test).
Figure 4.
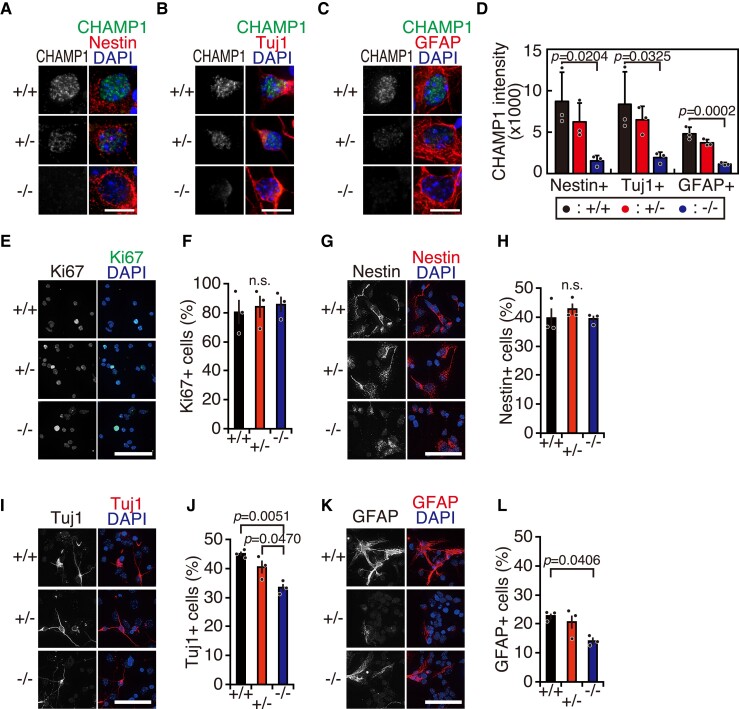
CHAMP1 is involved in NSC differentiation and axon outgrowth. (A) Immunofluorescence staining of NSCs isolated from the cortex of E14.5 CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice using antibodies against CHAMP1 (green) and Nestin (red). Nuclei were stained with DAPI (blue). Scale bar: 10 μm. (B) Immunofluorescence staining of neurons differentiated from NSCs of E14.5 CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice in vitro using antibodies against CHAMP1 (green) and Tuj1 (red). Nuclei were stained with DAPI (blue). Scale bar: 10 μm. (C) Immunofluorescence staining of astrocytes differentiated from NSCs of E14.5 CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice in vitro using antibodies against CHAMP1 (green) and GFAP (red). Nuclei were stained with DAPI (blue). Scale bar: 10 μm. (D) Quantification of CHAMP1 signal intensity in cultured NSCs (Nestin+), neurons (Tuj1+), and astrocytes (GFAP) isolated from E14.5 CHAMP1+/+ (n = 3), CHAMP1+/− (n = 3), and CHAMP1−/− (n = 3) mice. The data represent a minimum of 40 cells per mouse. Error bars represent S.D. P values were determined by Tukey–Kramer test. (E) Immunofluorescence staining of NSCs isolated from the cortex of E14.5 CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice using antibodies against Ki67 (green). Nuclei were stained with DAPI (blue). Scale bar: 50 μm. (F) Quantification of Ki67-positive NSCs (CHAMP1+/+; n = 3, CHAMP1+/−; n = 3, CHAMP1−/−; n = 3). The data represent a minimum of 1000 cells per mouse. Error bars represent S.E.M. P values were determined by Tukey–Kramer test. n.s., not statistically significant. (G, I, K) Immunofluorescence staining of NSCs isolated from the cortex of E14.5 CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice differentiated in vitro by growth factor withdrawal using antibodies against Nestin (G), Tuj1 (I), or GFAP (K) (red). Nuclei were stained with DAPI (blue). Scale bar: 50 μm. (H, J, L) Quantification of differentiated NSCs positive for Nestin (H), Tuj1 (J), or GFAP (L) (CHAMP1+/+; n = 3, CHAMP1+/−; n = 3, CHAMP1−/−; n = 3). The data represent a minimum of 1004 cells per mouse. Error bars represent S.E.M. P values were determined by Tukey–Kramer test. n.s., not statistically significant.
Data availability
The data that support the finding of this study are available from the corresponding author, upon reasonable request.
Results
CHAMP1 −/− mice were lethal at newborn
To explore the function of CHAMP1 in vivo, we generated a CHAMP1 knockout mouse. We first generated a transgenic mouse in which the CHAMP1 genomic region containing exon 2 was replaced with a gene cassette containing a neomycin resistant gene and lox-P sequences (Fig. 1B). We verified homologous recombination by Southern blot analysis (Fig. 1C). The CHAMP1 knockout mice were generated by excising the gene cassette by crossing with mice systemically expressing Cre recombinase under CAG promoter (CAG-Cre; Fig. 1B), which was confirmed by a polymerase chain reaction analysis (Fig. 1D), an IB analysis (Fig. 1E), as well as an immunofluorescence analysis of MEFs (Supplementary Fig. 1A). On C57BL/6J background, CHAMP1+/− mice were healthy and fertile, and CHAMP1−/− embryos were produced from heterozygous intercrosses at approximately the frequency predicted by Mendel’s law (Fig. 1F). However, CHAMP1−/− pups died within 2 days of birth (Fig. 1F). The newborn CHAMP1−/− mice looked superficially normal (Fig. 1G), although slightly smaller compared with wild-type mice (Fig. 1H). Absolute brain weight and ratios of brain weight to body weight showed no significant difference between wild-type and CHAMP1 knockout mice (Fig. 1I–K). Measurements using various parameters of cortical size, such as anteroposterior length, cortical length, and cortical width, also showed no difference between wild-type and CHAMP1 knockout mice (Supplementary Fig. 1B and C). We also measured these parameters in 13-week-old adult mice and found that there were no significant differences between wild-type and CHAMP1+/− mice (Supplementary Fig. 1D-H).
To explore the neonatal lethal phenotype of CHAMP1−/− mice further, we crossed CHAMP1+/− mice on C57BL/6 background with wild-type mice on Balb/c background and obtained CHAMP1+/− mice on mixed genetic background (Supplementary Fig. 2A). Intriguingly, when we crossed these CHAMP1+/− mice with mixed backgrounds, we could obtain CHAMP1−/− mice that survived to adulthood, although the frequency was lower than that predicted by Mendel’s law (Supplementary Fig. 2B). The CHAMP1−/− mice on mixed background were significantly smaller than their CHAMP1+/+ or CHAMP1+/− littermates (Supplementary Fig. 2C and D), and some of them exhibited kinked tails and kyphosis (Supplementary Fig. 2C). Expression of CHAMP1 and POGZ in adult brains was comparable between C57BL/6 and Balb/c backgrounds (Supplementary Fig. 2E). When we crossed the CHAMP1+/− mice on mixed background further with wild-type mice on Balb/c background to increase the contribution of Balb/c genetic background, the number of CHAMP1−/− mice obtained by crosses between these CHAMP1+/− mice decreased (Supplementary Fig. 2B), suggesting that CHAMP1−/− mice can survive to adulthood on mixed background, but not on pure genetic background. Systemic analysis of structural abnormalities of various organs in CHAMP1−/− mice using sagittal sections of newborn mice on C57BL/6 or mixed backgrounds did not reveal significant differences compared to CHAMP1+/+ or CHAMP1+/− mice (data not shown), leaving the cause of the neonatal death of CHAMP1−/− mice unspecified.
CHAMP1 localizes throughout the brain
We observed CHAMP1 expression in the brain. In an IB analysis, CHAMP1 expression increased at E17.5 and thereafter, i.e. the stages when neuronal production is almost completed (Fig. 2A), and it is expressed in each part of the neonatal brain in varying degrees (Fig. 2B). Expression of POGZ, a binding partner of CHAMP1, increased at E14.5-E15.5 and after birth (Fig. 2A, Supplementary Fig. 3A). POGZ showed a similar expression pattern in the neonatal brain as CHAMP1 (Fig. 2B, Supplementary Fig. 3B). By immunostaining of brain sections of E12.5, E14.5, neonatal, and adult mice, CHAMP1 was detected in each part of the brain, localizing in the nucleus of individual cells (Supplementary Fig. 3C-F). CHAMP1 was not detected in the brain sections of CHAMP1−/− mice, confirming the specificity of the fluorescent signal. In situ hybridization using brain sections obtained from E14.5 mice showed the expression of CHAMP1 mRNA throughout the brain, which was not detected in CHAMP1−/− mice (Supplementary Fig. 3G). We also examined POGZ mRNA in E14.5 mouse brains and found that its distribution is not altered in CHAMP1 knockout mice (Supplementary Fig. 4A). In E14.5, neonatal, and adult brains, POGZ localizes in the nucleus of the cells throughout the brain (Supplementary Fig. 4B–D), consistent with the previous reports.25,49,50 The nuclear localization of POGZ was detected in CHAMP1 knockout mice (Supplementary Fig. 4B–D), suggesting that CHAMP1 is not essential for POGZ localization in the nucleus. In immunostaining of cortical sections of E14.5 mice, CHAMP1 was detected in both NSCs and neurons, which were stained with Nestin and Tuj1, respectively (Fig. 2C and D). Overall, CHAMP1 localizes throughout the brain in both embryonic and adult mice, and the expression was increased during the embryonic period.
Figure 2.
CHAMP1 −/− mice do not show gross defects in the brain structure. (A) Expression of CHAMP1 and POGZ in brain during development. Whole-cell lysates prepared from mouse brains at indicated periods were subjected to IB analysis using antibodies as indicated. Quantification results of CHAMP1 are shown at the bottom (n = 3). Error bars represent S.D. (B) Expression of CHAMP1 and POGZ in whole brain, olfactory bulb (OB), cerebral cortex (CC), interbrain (IB), midbrain (MB), and cerebellum (CB) from newborn mice. Whole-cell lysate prepared from each part of the brain was subjected to IB analysis using antibodies as indicated. Quantification results of CHAMP1 are shown at the bottom (n = 3). Error bars represent S.D. (C, D) Immunofluorescence staining of a large dorsal cortical region of E14.5 CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice using antibodies against CHAMP1 (green) and Nestin (C) or Tuji1 (D) (red). Nuclei were stained with DAPI (blue). Scale bars: 100 μm. Scale bars in close-up images of boxed area: 10 μm. CP, cortical plate; SVZ, sub-ventricular zone; VZ, ventricular zone. (E) Nissl staining of coronal sections of newborn CHAMP1+/+ (upper), CHAMP1+/− (middle), and CHAMP1−/− (lower) mice. Scale bar: 1 mm. (F) Immunofluorescence staining of a large dorsal cortical region of newborn CHAMP1+/+, CHAMP1+/−, and CHAMP1−/− mice using antibodies against Pax6 (green) and Tuj1 (red). Nuclei were stained with DAPI (blue). Scale bar: 50 μm. (G) Thickness of the Pax6 (upper) and Tuj1 (lower)-positive cell layer of newborn mice shown in (F) (CHAMP1+/+; n = 5, CHAMP1+/−; n = 5, CHAMP1−/−; n = 5). Error bars represent S.D. P values were determined by Tukey–Kramer test. n.s., not statistically significant. (H) Immunofluorescence staining of a large dorsal cortical region of newborn CHAMP1+/+, CHAMP1+/−, and CHAMP1−/− mice using antibodies against Ctip2 (green) and Cux1 (red). Nuclei were stained with DAPI (blue). Scale bar: 50 μm. II〜VI, cortical layers. (I) Thickness of the Ctip2 (II–IV) and Cux1 (V)-positive cell layers of newborn mice shown in (H) (CHAMP1+/+; n = 5, CHAMP1+/−; n = 5, CHAMP1−/−; n = 5). Thickness of the layer VI was also measured. Error bars represent S.D. P values were determined by Tukey–Kramer test. n.s., not statistically significant.
CHAMP1 knockout mice do not show gross defects in brain structure
We morphologically and histologically analyzed the brain of neonatal CHAMP1 knockout mice. In Nissl staining of brain sections, no gross anatomical abnormalities were found between wild-type and CHAMP1 knockout mice (Fig. 2E). When we examined NSCs and neurons in the dorsal cortical area by Pax6 and Tuj1 immunofluorescence, respectively, no significant difference in the thickness of the Pax6 and Tuj1-positive cell layer were found between wild-type and CHAMP1 knockout newborn mice (Fig. 2F, G). The difference was also not apparent in E14.5 mice (Supplementary Fig. 5A). We also compared the thickness of the cortical layers. There was no significant difference in the thickness of the Ctip2 and Cux1-positive cell layers that represent layer V and layer II–IV, respectively (Fig. 2H and I), in newborn and adult brain of CHAMP1 knockout mice (Fig. 2H and I, Supplementary Fig. 5B). Collectively, the brain structure of CHAMP1 knockout mice appeared grossly normal.
Mitotic cells are increased in CHAMP1 knockout mouse brains
Since CHAMP1 is involved in chromosome segregation,11 we looked for mitotic defects in the brain of CHAMP1 knockout mice. We compared the number of mitotic progenitor cells in CHAMP1 knockout mice with that of wild-type mice by visualizing phospho-histone H3 at serine 10 (pH3S10), a mitotic phase marker, in E14.5 brains. To adjust the relative position along the longitudinal axis, we compared the number of cells positive for pH3S10 at four different levels along the longitudinal axis (Level 1–4, Fig. 3A). As shown in Fig. 3B and C, the number of pH3S10-positive cells tended to increase at the VZ in CHAMP1 knockout mice. Considering the role of CHAMP1 in chromosome segregation, the increase of mitotic cells may represent delay in mitotic progression rather than accelerated cell cycle. We did not find increase in apoptotic cells in the brain in E14.5 or neonatal CHAMP1 knockout mice compared with corresponding wild-type mice by terminal deoxynucleotidyl TUNEL staining (Supplementary Fig. 6A), suggesting that the mitotic delay is not severe enough to cause increased cell death. We further examined the orientation of the spindle axis in mitotic progenitor cells relative to the ventricular surface, because this has been proposed to influence cell fate determination.51 However, there was no significant difference between wild-type and CHAMP1 knockout E14.5 brains when we measured the orientation of the spindle axis (Fig. 3D and E).
Disruption of dendritic spines, membrane protrusions along dendrites, is found in patients with NDDs.52 We visualized and quantified the density of dendritic spines in the cerebral cortex and hippocampus of 13 week-old mice using Golgi-Cox staining, but found no difference between wild-type and CHAMP1+/− mice (Supplementary Fig. 6B and C). The density of mature spines, which exhibit mushroom-like morphologies, was also comparable between wild-type and CHAMP1+/− mice (Supplementary Fig. 6D).
CHAMP1 is involved in NSC differentiation in vitro
We enriched NSCs from the cortex of E14.5 mice by in vitro culture for 4 days in the presence of EGF and FGF2, which were then differentiated by growth factor withdrawal for 5 days. First, we addressed CHAMP1 expression in NSCs, neurons, and astrocytes, by immunofluorescence analysis with lineage-specific markers, Nestin, Tuj1 and GFAP, respectively. CHAMP1 was detected in cells expressing each of the respective markers, showing that CHAMP1 is expressed in NSCs, neurons and astrocytes, although expression is lower in astrocytes than in NSCs and neurons (Fig. 4A–D). The role of CHAMP1 in NSC proliferation was examined by measuring the rate of appearance of Ki67-positive cells in the presence of growth factors, and we found no difference between wild-type and CHAMP1 knockout mice (Fig. 4E and F). We also determined the role of CHAMP1 on NSC differentiation after growth factor withdrawal by quantifying the fraction of cells expressing lineage-specific markers. The percentages of Tuj1- and GFAP-positive cells were significantly reduced in CHAMP1−/− cells, whereas that of Nestin-positive cells was unaffected, compared with wild-type cells (Fig. 4G–L). These data suggest that CHAMP1 is involved in NSC differentiation in vitro to neuronal and astrocyte lineages.
We also performed CHAMP1 knockdown in NSCs from E14.5 mice and observed after differentiation for 5 days. In an IB analysis and immunofluorescence staining, we found that POGZ expression is reduced in CHAMP1 knockdown neurons (Supplementary Fig. 7A and B), suggesting that CHAMP1 is required for the stability of POGZ, which was also shown in a recent report.53 We also studied axon outgrowth in CHAMP1 knockdown neurons by measuring the number of axons stained with ankyrin-G, a marker of the axon initial segment, which were also positive for Map2, a dendritic marker. As shown in Supplementary Fig. 7D and E, the number of axons in CHAMP1 knockdown neurons was comparable with that in control neurons. Sholl analysis showed that the level of dendritic branching is not significantly different between CHAMP1 knockdown and control neurons (Supplementary Fig. 7F and G).
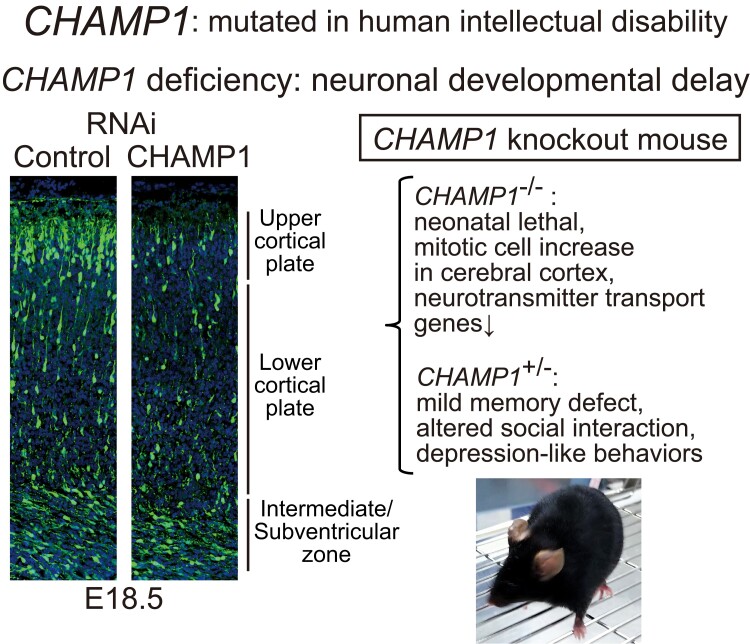
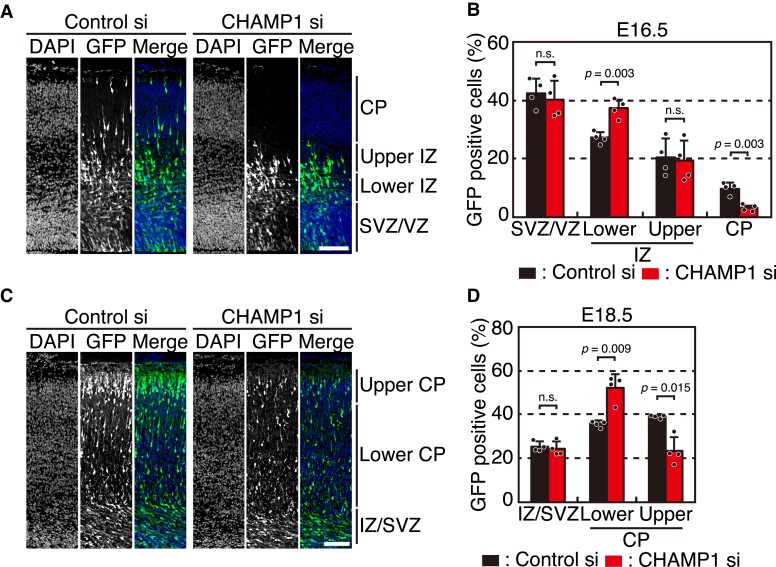
CHAMP1 knockdown leads to delayed cortical migration
To determine the role of CHAMP1 in NSCs of the developing cortex in vivo, we introduced an siRNA against CHAMP1 and an expression plasmid for GFP into the E14.5 mouse cortex by in utero electroporation. The embryos were analyzed for the localization of GFP-expressing cells in the developing cortex at E16.5 and E18.5. Some of the GFP-positive cells reached the cortical plate (CP) in the control embryos at E16.5, whereas most of the cells transfected with the CHAMP1 siRNA were localized in the VZ, sub-ventricular zone (SVZ) and intermediate zone (IZ) but not in the CP (Fig. 5A). The quantitative data showed that the fraction of GFP-positive cells was significantly increased in the lower IZ yet decreased in the CP in CHAMP1 knockdown embryos (Fig. 5B). Considering the results in vitro (Fig. 4G–L), these data suggest that CHAMP1 might be involved in promoting neuronal differentiation. At E18.5, the fraction of GFP-positive cells was increased in the lower CP and decreased in the upper CP (Fig. 5C and D), implicating that NSCs with CHAMP1 knockdown might be delayed in neuronal differentiation or have a defect in neuronal migration. The reason why the impairment of cortical cell localization was not detected in newborn mice (Fig. 2F and G) may be because the delay was small and only transiently detectable by acute CHAMP1 depletion by RNAi.
Figure 5.
CHAMP1 knockdown leads to delayed cortical migration. (A) Cell localization in E16.5 mouse cortices electroporated with mock or CHAMP1 siRNA at E14.5. Electroporated cells expressing GFP were shown in green. Nuclei were stained with DAPI (blue). Scale bar: 50 μm. CP, cortical plate; IZ, intermediate zone; SVZ, sub-ventricular zone; VZ, ventricular zone. (B) Quantification of GFP-positive cells treated with control or CHAMP1 siRNA in each layer shown in (A) (n = 4). Error bars represent S.D. P values were determined by Student’s t-test. n.s., not statistically significant. (C) Neuronal migration in E18.5 mouse cortices electroporated with mock or CHAMP1 siRNA at E14.5. Electroporated cells expressing GFP were shown in green. Nuclei were stained with DAPI (blue). Scale bar: 50 μm. (D) Quantification of GFP-positive cells treated with control or CHAMP1 siRNA in each layer shown in (C) (n = 4). Error bars represent S.D. P values were determined by Student’s t-test. n.s., not statistically significant.
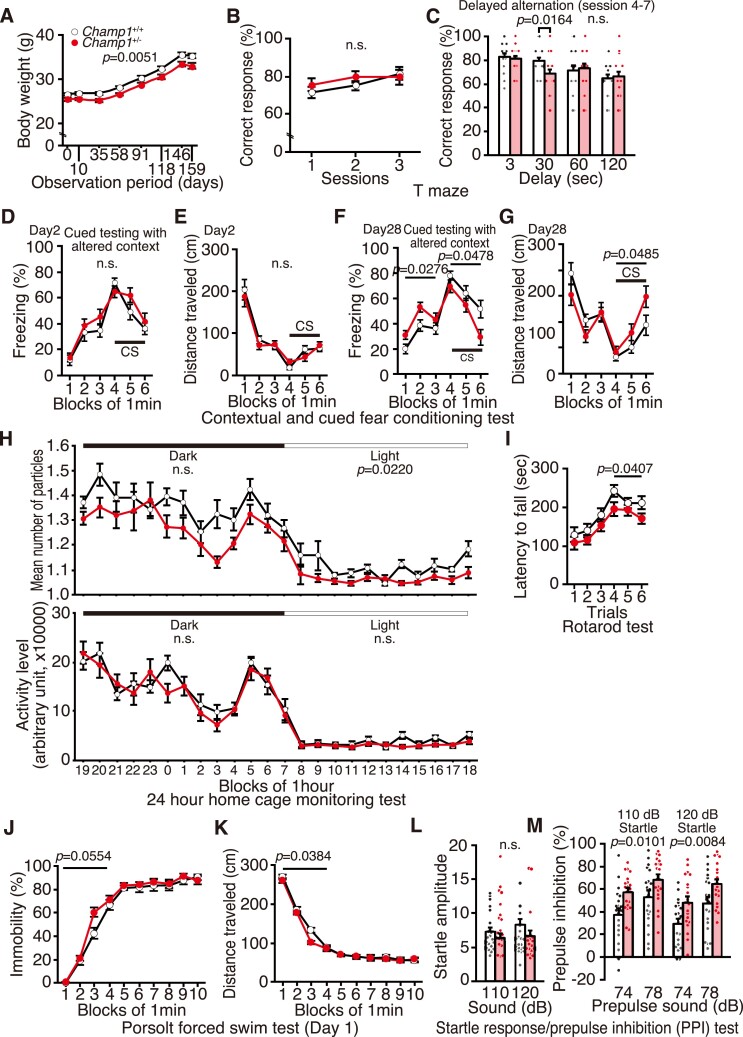
CHAMP1+/− mice have mild memory defects
The results so far show that neuronal defects in CHAMP1+/− mice are trending in the same direction, although not as strong, as that in CHAMP1−/− mice. Next, we performed a comprehensive battery of behavioural tests, which is available only for CHAMP1+/− mice, because CHAMP1−/− mice die soon after birth. We used 20 adult males of both wild-type and CHAMP1+/− mice at 10-weeks-old at test onset, which were backcrossed at least nine generations from the original C57BL/6N × CBA/J mix genetic background to generate C57BL/6J genetic background. CHAMP1+/− mice used for the behavioural test battery showed a small but significant decrease in body weight throughout the observation period (Fig. 6A, Supplementary Fig. 8A), which was not apparent in mice that were not used for the behavioural test (Supplementary Fig. 1D). The small difference in body weight may become apparent only in the strictly controlled rearing conditions.
Figure 6.
CHAMP1+/− mice show mild memory defects, altered social interaction and depressive behaviours. (A) Body weight of CHAMP1+/+ (white circle; n = 20) and CHAMP1+/− (red circle; n = 20) mice during 10–37 weeks-old, plotted against observation period of the behavioural test battery. (B, C) T-maze forced alteration test. Percentage of correct response in the first three training (B) and the four consecutive sessions with various delay times (C) of CHAMP1+/+ (white bars) and CHAMP1+/− (red bars) mice. (D-G) Cued and contextual fear conditioning test: (D) percentage of freezing time and (E) total distance moved in the chamber on Day 2, and (F) percentage of freezing time and (G) total distance moved in the chamber after 1 month of CHAMP1+/+ (white circle) and CHAMP1+/− (red circle) mice. CS: conditioned stimulus. (H) Social interaction test in home cage: mean number of particles, which represents the contact between two mice (upper graph, one particle: two mice were together, two particles: they were separating), and mean activities (lower graph) of CHAMP1+/+ (white circle) and CHAMP1+/− (red circle) mice. (I) Latency to fall of CHAMP1+/+ (white circle) and CHAMP1+/− (red circle) mice in the rotarod test. (J, K) Porsolt forced swim test: (J) percentage of immobility and (K) total distance moved on Day 1 by CHAMP1+/+ (white circle) and CHAMP1+/− (red circle) mice. (L, M) Startle response and PPI test. Acoustic startle response to stimuli of 110 or 120 dB (L), and inhibition of the startle response by prepulse of 74 or 78 dB (M) in CHAMP1+/+ (white bars) and CHAMP1+/− (red bars) mice. For all graphs, twenty male mice of both genotypes were used, and error bars represent S.E.M. P values for genotype effects were determined by two-way repeated measurement ANOVA. In (C) at 30 s, Student’s t-test was used. In (F) and (G), data before CS (1–3 min) and during CS (4–6 min) were analyzed separately. In (I), data for the 1st to 3rd trials and the 4th to 6th trials were analyzed separately. In (L) and (M), data for early time points (1–4 min) and late time points (5–10 min) were analyzed separately.
To evaluate working memory, a T-maze test was performed according to the forced alteration protocol. The percentage of correct responses of CHAMP1+/− mice was not significantly different from that of wild-type mice (Fig. 6B and C). However, there was a difference only in 30 s delay (P = 0.0164, Fig. 6C), which may reflect a slight impairment in working memory in CHAMP1+/− mice.
Next we performed a cued and contextual fear conditioning test to assess the involvement of CHAMP1 in fear memory. In the conditioning period, there was no significant difference between wild-type and CHAMP1+/− mice in the percentage of freezing or distance travelled caused by the foot shock delivered as the UCS following the white noise as the conditioned stimulus (CS; Supplementary Fig. 8B) but the distance travelled during the 1st electrical foot shock was greater in CHAMP1+/− mice (Supplementary Fig. 8E, foot shock 1, P = 0.0278). In a hot plate test, latency to the first hind paw response did not differ between wild-type and CHAMP1+/− mice (Supplementary Fig. 8F), excluding the possibility that CHAMP1+/− mice have defective sensitivity to a painful stimulus. During Day 2 of context testing, there was no difference between wild-type and CHAMP1+/− mice in the percentages of freezing or distance travelled (Supplementary Fig. 8C). In cued testing with altered context, in which CS was presented without UCS, the percentages of freezing and distance travelled were comparable between wild-type and CHAMP1+/− mice (Fig. 6D and E). In the context testing 1 month after the fear conditioning, there was no difference between wild-type and CHAMP1+/− mice in the percentages of freezing and distance travelled (Supplementary Fig. 8D). However, in the cued testing after 1 month, the percentage of freezing of CHAMP1+/− mice was significantly increased before CS (P = 0.0276) and decreased during CS (P = 0.0478, Fig. 6F). Accordingly, total distance travelled by CHAMP1+/− mice were longer during CS (P = 0.0485, Fig. 6G). Decreased freezing in response to CS despite the increase in generalized freezing before CS only after a month might be explained by a problem in remote, but not recent, cued memory in CHAMP1+/− mice.
We then used the Barnes maze test to measure spatial learning and memory. There were no significant differences in latency, number of errors, distance travelled and number of omission errors before reaching the target hole between wild-type and CHAMP1+/− mice (Supplementary Fig. 9A). In probe trials, in which the escape box was removed, performed either 1 day or 1 month after training, there was no significant difference between wild-type and CHAMP1+/− mice in the time spent around the target (Supplementary Fig. 9B), showing that spatial memory is not affected. Collectively, these results suggest that CHAMP1+/− mice have a cued memory defect but not a spatial memory defect.
CHAMP1+/− mice show altered social interaction
To evaluate social interaction, a 24 h home-cage monitoring test was conducted. As shown in Fig. 6H, where mean number of particles represents the contact between two mice (one particle: two mice were together, two particles: they were separating), CHAMP1+/− mice exhibited increased interaction compared with wild-type mice, which was statistically significant in inactive periods (‘light’, P = 0.0220, Fig. 6H). Activity level was comparable between CHAMP1+/− and wild-type mice. These data suggest that CHAMP1+/− mice show altered social interaction.
In the social interaction test, which measures social behaviour in a novel environment, there was no difference between wild-type and CHAMP1+/− mice in all the parameters (Supplementary Fig. 9C). There was also no difference between wild-type and CHAMP1+/− mice in a Crawley’s sociability and social novelty preference test (Supplementary Fig. 9D and E), suggesting that social recognition is not impaired.
CHAMP1+/− mice present depression-like behaviours
In a rotarod test that assesses motor coordination, latency to fall in the 4th to 6th trials were significantly shorter in CHAMP1+/− mice compared with that in wild-type mice (P = 0.0407, Fig. 6I). Neuromuscular strength did not differ between wild-type and CHAMP1+/− mice, which was shown by a grip strength test and a wire-hung test (Supplementary Fig. 10A and B). The finding that body weight was slightly smaller in adult CHAMP1+/− mice (Fig. 6A) also makes it unlikely that the result of the rotarod test was due to physical problems. Motor function was also assessed in a beam test. Number of slips, moving speed, and number of move episodes were no different between wild-type and CHAMP1+/− mice (Supplementary Fig. 10C). These results suggest that CHAMP1+/− mice have a tendency to give up easily through psychological but not physical reason.
Next, we examined depression-like behaviour in a Porsolt forced swim test performed on two consecutive days. Total immobility time tended to be longer in CHAMP1+/− mice only in early time points in the first day trial (P = 0.0554, Fig. 6J). Accordingly, total distance travelled was significantly shorter in CHAMP1+/− mice only in early time points in the first day trial (P = 0.0380, Fig. 6K). Total immobility time and total distance travelled did not differ between wild-type and CHAMP1+/− mice in the second day trial (Supplementary Fig. 10D). In a tail suspension test, which is also for detecting depression-like behaviour, total immobility time was not different between wild-type and CHAMP1+/− mice (Supplementary Fig. 10E). Collectively, these results imply that CHAMP1+/− mice have a minor tendency to present depression-like behaviour.
To evaluate anxiety, an open field test, a light/dark transition test, and an elevated plus maze test were performed but none showed any statistical difference between wild-type and CHAMP1+/− mice (Supplementary Fig. 10F–H). When we examined acoustic startle response and PPI, the acoustic startle response was not different between wild-type and CHAMP1+/− mice (Fig. 6L). However, CHAMP1+/− mice exhibited increased PPI for 74 dB or 78 dB prepulse before 110 dB or 120 dB startle stimulus (P = 0.0101, 0.0084, Fig. 6M, see Discussion).
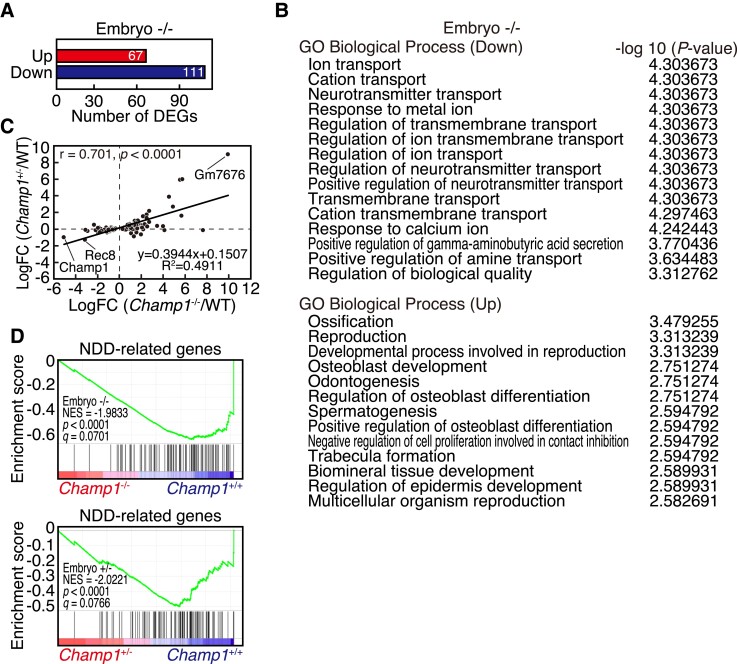
Gene expression changes in CHAMP1 knockout mice
To explore the alteration of gene expression profiles in CHAMP1 knockout mice, we performed RNA-seq analysis of whole brains of wild-type, CHAMP1+/−, and CHAMP1−/− mouse embryos at E14.5. We found 178 DEGs in embryonic brain of CHAMP1−/− mice compared with wild-type mice at cut-offs corresponding to false discovery rate (FDR) <0.1 and fold change (FC) >1.2, among which 111 were downregulated and 67 were upregulated (Fig. 7A, Supplementary Fig. 11A). A GO enrichment analysis of DEGs downregulated in CHAMP1−/− embryonic brain showed enrichment for genes related to neurotransmitter transport (Fig. 7B), implicating the role of CHAMP1 in brain function. Among the DEGs related to neurotransmitter transport, Slc6a1, a gamma-aminobutyric acid (GABA) transporter associated with ID,54 was included (Supplementary Fig. 11A). For DEGs upregulated in CHAMP1−/− embryonic brain, genes involved in regulation of ossification were enriched, which may be related to skeletal abnormalities found in CHAMP1−/− mice on mixed background (Supplementary Fig. 2C). Expression changes of most of the DEGs found in CHAMP1−/− embryonic brain were not significant in CHAMP1+/− embryonic brain, but they showed similar expression changes, which was demonstrated by a correlation analysis (Fig. 7C).
Figure 7.
Gene expression changes in CHAMP1 knockout mice. (A) Number of DEGs in embryonic brain of CHAMP1−/− mice. Genes with FDR <0.1 and fold change (FC) >1.2 were selected as DEGs. (B) GO of biological processes enriched in DEGs in embryonic brain of CHAMP1−/− mice. (C) Correlation between fold changes of gene expression in embryonic brain of CHAMP1−/− and CHAMP1+/− mice for 178 DEGs of CHAMP1−/− embryonic brain. Spearman’s rank correlation coefficients (r) and P values are shown. (D) GSEA plots of DEGs between wild-type and CHAMP1−/− (upper) or CHAMP1+/− (lower) embryonic brain for the list of NDD-related genes. Nominal P values and FDR (q) are shown.
To address the relationship of CHAMP1 with ID, we performed GSEA against the 94 genes, in which damaging de novo mutations were found in NDD.6 The expression of these genes was significantly downregulated not only in CHAMP1−/−, but also in CHAMP1+/− embryonic brain (Fig. 7D). We judged significant enrichment of NDD-related genes by testing the gene set together with 1000 random gene sets with comparable sizes (Supplementary Fig. 11B). Collectively, these data suggest that CHAMP1 plays a role in the expression of neurotransmitter transport and NDD-related genes, dysregulation of which may be related to ID.
Discussion
In this study, we report the first characterization of a CHAMP1 knockout mouse. We found that CHAMP1−/− mice are smaller in size and neonatally lethal. Mitotic cells were increased in CHAMP1 knockout embryonic cerebral cortex, and CHAMP1−/− NSCs showed a delay in neuronal differentiation in vitro, which was also suggested in vivo by CHAMP1 knockdown. In a behavioural test battery, CHAMP1+/− mice exhibited mild memory defects, altered social interaction, and depression-like behaviours. Transcriptomic analysis revealed downregulation of neurotransmitter transport and NDD-related genes.
Newborn CHAMP1−/− mice were neonatally lethal but often survived the first day after birth. As milk was detected in their stomach (data not shown), severe breathing and suckling problems are unlikely to be the cause of death. Apparent defects in organ structures were not found in CHAMP1−/− mice through systemic analysis, leaving the cause of neonatal death of CHAMP1−/− mice unspecified. Interestingly, CHAMP1−/− mice can survive to adulthood on mixed genetic background, suggesting that the neonatal lethality is somehow rescued on the mixed background.
CHAMP1 deficiency in NSCs resulted in delayed neuronal differentiation, suggesting that CHAMP1 plays a role in neuronal development. Similar neuronal developmental delay was also reported in NSCs depleted of POGZ,49 which may reflect the close relationship between them. Mitotic cells were increased in the cerebral cortex of CHAMP1 knockout mice. CHAMP1 functions in the regulation of kinetochore–microtubule attachment,11 and defective chromosome segregation and multipolar spindle formation were seen in lymphoblast cells isolated from an individual with a CHAMP1 mutation.12 It is known that genes related to spindle formation and chromosome segregation are mutated in severe ID and/or microcephaly, causing cortical development failure.55 Therefore, mitotic defects caused by CHAMP1 deficiency may lead to delay in mitotic progression and neuronal development. However, the delay is supposed to be small and transiently detectable, as impairment of cortical layering was not detected in newborn mice.
Recently, a comprehensive evaluation of the neurobehavioural phenotype of individuals with CHAMP1 mutations was reported.10 In our behavioural test battery, CHAMP1+/− mice showed a relatively mild phenotype in a subset of tests. However, a slightly cued memory impairment in the cued and contextual fear conditioning test, as well as a slight working memory impairment in the T-maze test are consistent with ID, as memory deficits are commonly seen in mouse models of ID and ASD genes, such as Fmr1, Shank and Mecp2.4 In addition, altered social interaction, which was found in the 24 h home-cage monitoring test, is also a common feature of NDD.56 It is of note that depression-like behaviour was found in CHAMP1+/− mice in a Porsolt forced swim test. Abnormal findings in a rotarod test may also be related to depression-like behaviour. Interestingly, anxiety and depression were frequently manifested in individuals with CHAMP1 mutations,10 suggesting that depression-like behaviour is a characteristic phenotype of CHAMP1 deficiency. Pathophysiological significance of increased PPI in CHAMP1+/− mice is currently unknown. PPI is a common psychophysiological index of sensorimotor gating, and reduction of PPI is typically seen in schizophrenia and also in other disorders.57 On the other hand, relationship between increased PPI levels and diseases are relatively unfamiliar. It is noteworthy that mice mutated in CHD8, a gene affected in ASD, also exhibit an increased PPI level,58 implying a relationship between increased PPI and NDD.
To understand the role of CHAMP1, findings on POGZ, an interaction partner of CHAMP1 that is also related to ID and ASD,18–21 are informative. Similarly to CHAMP1, depletion of POGZ was reported to cause a chromosome segregation defect.59 POGZ is also involved in DNA double-strand break (DSB) repair,60 and we recently reported that CHAMP1 functions in DSB repair together with POGZ.53 POGZ is widely expressed in neurons throughout the brain.25,49,50 POGZ homozygous knockout mice die during embryonic development or shortly after birth.61,62 To assess neuronal defects by avoiding embryonic lethality, a brain-specific conditional POGZ knockout (POGZ cKO) mouse was generated.50 In behavioural tests, POGZ cKO−/− mice showed motor learning deficits and increased social interaction.50 Notably, significant differences were only seen between wild-type and POGZ cKO−/− mice but not between wild-type and POGZ cKO+/− mice, implying that CHAMP1 deficiency phenotype may become more apparent in a brain-specific conditional CHAMP1 homozygous knockout mouse. In contrast, a model mouse heterozygous for a de novo POGZ mutation found in an ASD patient (POGZWT/Q1038R) showed NDD-related behavioural abnormalities, suggesting a dominant-negative effect of the POGZ mutant.49 Reduced body weight seen in CHAMP1 knockout mice was also observed in both POGZ cKO and POGZWT/Q1038R mice.49,50 Both studies, as well as other studies suggest that POGZ plays a role in transcriptional regulation.49,50,61,63 The possibility that CHAMP1 also functions in transcriptional regulation is worth considering, which can explain the downregulation of neurotransmitter transport and NDD-related genes in embryonic CHAMP1−/− brains.
It has not been shown whether truncated CHAMP1 transcripts are expressed in individuals with CHAMP1 mutations. Even if they are expressed, they will not function properly considering that truncated CHAMP1 proteins cannot localize to chromatin to form a complex with POGZ.8 This suggests that individuals with CHAMP1 mutations show haplo-insufficient phenotype. The observation that severity of ID and other symptoms is not correlated to the size of truncation supports this notion.8 However, relatively mild phenotypes reported in individuals with 13q34 microdeletions that include the CHAMP1 locus16 leaves the possibility that truncated CHAMP1 plays a dominant role in the pathogenesis of CHAMP1 individuals. Further study is required to obtain insights into the pathophysiology of the disorders in humans with CHAMP1 mutations and to develop therapeutic interventions.
Supplementary Material
Acknowledgements
We thank members of the K.T. laboratory for discussions, Y. Yoshizaki for his critical reading of the manuscript and A. Harata, K. Yokoyama, S. Makino and Y. Nishimura for technical assistance. We acknowledge The CHAMP1 Research Foundation (https://champ1foundation.org/) for continuing support of our study.
Abbreviations
- ADHD =
attention deficit hyperactivity disorder
- ASD =
autism spectrum disorder
- CAMP =
chromosome alignment-maintaining phosphoprotein
- CDK1 =
cyclin-dependent kinase 1
- CP =
cortical plate
- CS =
conditioned stimulus
- DAPI =
4′,6-diamidino-2-phenylindole
- DEG =
differentially expressed gene
- DSB =
double-strand break
- FC =
fold change
- FDR =
false discovery rate
- GFAP =
glial fibrillary acidic protein
- GFP =
green fluorescent protein
- GO =
gene ontology
- GSEA =
gene set enrichment analysis
- IB =
immunoblot
- ID =
intellectual disability
- IF =
immunofluorescence
- IZ =
intermediate zone
- MEF =
mouse embryonic fibroblast
- NDD =
neurodevelopmental disorder
- NSC =
neural stem cell
- POGZ =
pogo transposable element-derived protein with zinc-finger domain
- PPI =
prepulse inhibition
- SVZ =
sub-ventricular zone
- UCS =
unconditioned stimulus
- VZ =
ventricular zone
Contributor Information
Masayoshi Nagai, Department of Molecular Oncology, Institute of Development, Aging and Cancer (IDAC), Tohoku University, Sendai, Miyagi 980-8575, Japan.
Kenji Iemura, Department of Molecular Oncology, Institute of Development, Aging and Cancer (IDAC), Tohoku University, Sendai, Miyagi 980-8575, Japan.
Takako Kikkawa, Department of Developmental Neuroscience, United Centers for Advanced Research and Translational Medicine (ART), Tohoku University Graduate School of Medicine, Sendai, Miyagi 980-8575, Japan.
Sharmin Naher, Department of Developmental Neuroscience, Tohoku University Graduate School of Life Sciences, Sendai, Miyagi 980-8575, Japan.
Satoko Hattori, Division of Systems Medical Science, Institute for Comprehensive Medical Science (ICMS), Fujita Health University, Toyoake, Aichi 470-1192, Japan.
Hideo Hagihara, Division of Systems Medical Science, Institute for Comprehensive Medical Science (ICMS), Fujita Health University, Toyoake, Aichi 470-1192, Japan.
Koh-ichi Nagata, Department of Molecular Neurobiology, Institute of Developmental Research, Aichi Developmental Disability Center, Kasugai, Aichi 480-0392, Japan; Department of Neurochemistry, Nagoya University Graduate School of Medicine, Nagoya, Aichi 466-8550, Japan.
Hayato Anzawa, Department of Applied Information Sciences, Graduate School of Information Sciences, Tohoku University, Sendai 980-8579, Japan.
Risa Kugisaki, Department of Molecular Oncology, Institute of Development, Aging and Cancer (IDAC), Tohoku University, Sendai, Miyagi 980-8575, Japan.
Hideki Wanibuchi, Department of Molecular Pathology, Osaka City University Graduate School of Medicine, Osaka 545-8585, Japan.
Takaya Abe, Laboratory for Animal Resources and Genetic Engineering, RIKEN Center for Biosystems Dynamics Research, Kobe, Hyogo 650-0047, Japan.
Kenichi Inoue, Laboratory for Animal Resources and Genetic Engineering, RIKEN Center for Biosystems Dynamics Research, Kobe, Hyogo 650-0047, Japan.
Kengo Kinoshita, Department of Applied Information Sciences, Graduate School of Information Sciences, Tohoku University, Sendai 980-8579, Japan; Division of Integrated Genomics, Tohoku Medical Megabank Organization, Tohoku University, Sendai, 980-8573, Japan; Department of In Silico Analysis, Institute of Development, Aging and Cancer, Tohoku University, Sendai, 980-8575, Japan.
Tsuyoshi Miyakawa, Division of Systems Medical Science, Institute for Comprehensive Medical Science (ICMS), Fujita Health University, Toyoake, Aichi 470-1192, Japan.
Noriko Osumi, Department of Developmental Neuroscience, United Centers for Advanced Research and Translational Medicine (ART), Tohoku University Graduate School of Medicine, Sendai, Miyagi 980-8575, Japan.
Kozo Tanaka, Department of Molecular Oncology, Institute of Development, Aging and Cancer (IDAC), Tohoku University, Sendai, Miyagi 980-8575, Japan.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS KAKENHI Grant Numbers 24370078, 19K22410); Ministry of Education, Culture, Sports, Science and Technology (MEXT KAKENHI Grant Numbers 26116501, 16H01296, JP16H06276 (AdAMS)); and grants from the Takeda Science Foundation, Princess Takamatsu Cancer Research Fund (10-24210) to K.T., Ministry of Education, Culture, Sports, Science and Technology (MEXT Grant-in-Aid for Scientific Research on Innovative Areas Grant Number 16H06530) to N.O., Japan Society for the Promotion of Science (Grant-in-Aid for JSPS Research Fellow Grant Number 17J02608) and a Grant Programme Research from the Division for Interdisciplinary Advanced Research and Education, Tohoku University, to M.N., Grant-in-Aid for Joint Research by Young Researchers, Tohoku University, Ensemble Grants for Early Career Researchers, Tohoku University, to K.I. Behavioural analysis, which was carried out at Institute for Comprehensive Medical Science, Fujita Health University, was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT Promotion of Distinctive Joint Research Centre Programme Grant Number JPMXP0618217663).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain Communications online.
References
- 1. Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–187. [DOI] [PubMed] [Google Scholar]
- 2. Flore LA, Milunsky JM. Updates in the genetic evaluation of the child with global developmental delay or intellectual disability. Semin Pediatr Neurol. 2012;19(4):173–180. [DOI] [PubMed] [Google Scholar]
- 3. American Psychiatric Association . Diagnostic and statistical manual of mental disorders; DSM-V. American Psychiatric Association; 2013. [Google Scholar]
- 4. Verma V, Paul A, Amrapali Vishwanath A, Vaidya B, Clement JP. Understanding intellectual disability and autism spectrum disorders from common mouse models: Synapses to behaviour. Open Biol. 2019;9(6):180265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deciphering Developmental Disorders Study . Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519(7542):223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deciphering Developmental Disorders Study . Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hempel M, Cremer K, Ockeloen CW, et al. De Novo mutations in CHAMP1 cause intellectual disability with severe speech impairment. Am J Hum Genet. 2015;97(3):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Isidor B, Kury S, Rosenfeld JA, et al. De Novo truncating mutations in the kinetochore-microtubules attachment gene CHAMP1 cause syndromic intellectual disability. Hum Mutat. 2016;37(4):354–358. [DOI] [PubMed] [Google Scholar]
- 9. Tanaka AJ, Cho MT, Retterer K, et al. De novo pathogenic variants in CHAMP1 are associated with global developmental delay, intellectual disability, and dysmorphic facial features. Cold Spring Harb Mol Case Stud. 2016;2(1):a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levy T, Lerman B, Halpern D, et al. CHAMP1 Disorder is associated with a complex neurobehavioral phenotype including autism, ADHD, repetitive behaviors and sensory symptoms. Hum Mol Genet. 2022;31(15):2582–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Itoh G, Kanno S, Uchida KS, et al. CAMP (C13orf8, ZNF828) is a novel regulator of kinetochore-microtubule attachment. EMBO J. 2011;30(1):130–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Okamoto N, Tsuchiya Y, Kuki I, et al. Disturbed chromosome segregation and multipolar spindle formation in a patient with CHAMP1 mutation. Mol Genet Genomic Med. 2017;5(5):585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ben-Haim R, Heyman E, Benyamini L, et al. CHAMP1 Mutations cause refractory infantile myoclonic epilepsy. J Pediatr Neurol. 2020;18:27–32. [Google Scholar]
- 14. Dong Y, Shi X, Du K, et al. First Chinese patient with mental retardation-40 due to a de novo CHAMP1 frameshift mutation: Case report and literature review. Exp Ther Med. 2021;22(2):907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Asakura Y, Osaka H, Aoi H, Mizuguchi T, Matsumoto N, Yamagata T. Intellectual disability and microcephaly associated with a novel CHAMP1 mutation. Hum Genome Var. 2021;8(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reinstein E, Liberman M, Feingold-Zadok M, Tenne T, Graham JM Jr. Terminal microdeletions of 13q34 chromosome region in patients with intellectual disability: Delineation of an emerging new microdeletion syndrome. Mol Genet Metab. 2016;118(1):60–63. [DOI] [PubMed] [Google Scholar]
- 17. Sagi-Dain L, Goldberg Y, Peleg A, et al. The rare 13q33-q34 microdeletions: Eight new patients and review of the literature. Hum Genet. 2019;138(10):1145–1153. [DOI] [PubMed] [Google Scholar]
- 18. Ye Y, Cho MT, Retterer K, et al. De novo POGZ mutations are associated with neurodevelopmental disorders and microcephaly. Cold Spring Harb Mol Case Stud. 2015;1(1):a000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White J, Beck CR, Harel T, et al. POGZ Truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stessman HAF, Willemsen MH, Fenckova M, et al. Disruption of POGZ is associated with intellectual disability and autism Spectrum disorders. Am J Hum Genet. 2016;98(3):541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan B, Zou Y, Zhang Y, et al. A novel de novo POGZ mutation in a patient with intellectual disability. J Hum Genet. 2016;61(4):357–359. [DOI] [PubMed] [Google Scholar]
- 22. Assia Batzir N, Posey JE, Song X, et al. Phenotypic expansion of POGZ-related intellectual disability syndrome (white-sutton syndrome). Am J Med Genet A. 2020;182(1):38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yagi T, Tokunaga T, Furuta Y, et al. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214(1):70–76. [DOI] [PubMed] [Google Scholar]
- 24. Matsumura H, Hasuwa H, Inoue N, Ikawa M, Okabe M. Lineage-specific cell disruption in living mice by cre-mediated expression of diphtheria toxin A chain. Biochem Biophys Res Commun. 2004;321(2):275–279. [DOI] [PubMed] [Google Scholar]
- 25. Ibaraki K, Hamada N, Iwamoto I, et al. Expression analyses of POGZ, A responsible gene for neurodevelopmental disorders, during mouse brain development. Dev Neurosci. 2019;41(1–2):139–148. [DOI] [PubMed] [Google Scholar]
- 26. Yanagisawa M, Nakashima K, Arakawa H, et al. Astrocyte differentiation of fetal neuroepithelial cells by interleukin-11 via activation of a common cytokine signal transducer, gp130, and a transcription factor, STAT3. J Neurochem. 2000;74(4):1498–1504. [DOI] [PubMed] [Google Scholar]
- 27. Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12(11):4565–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kikkawa T, Sakayori N, Yuuki H, et al. Dmrt genes participate in the development of cajal-retzius cells derived from the cortical hem in the telencephalon. Dev Dyn. 2020;249(6):698–710. [DOI] [PubMed] [Google Scholar]
- 29. Osumi N, Hirota A, Ohuchi H, et al. Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development. 1997;124(15):2961–2972. [DOI] [PubMed] [Google Scholar]
- 30. Kikkawa T, Obayashi T, Takahashi M, Fukuzaki-Dohi U, Numayama-Tsuruta K, Osumi N. Dmrta1 regulates proneural gene expression downstream of Pax6 in the mammalian telencephalon. Genes Cells. 2013;18(8):636–649. [DOI] [PubMed] [Google Scholar]
- 31. Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240(1):237–246. [DOI] [PubMed] [Google Scholar]
- 32. Tsunekawa Y, Britto JM, Takahashi M, Polleux F, Tan SS, Osumi N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 2012;31(8):1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12(7):2685–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ferreira TA, Blackman AV, Oyrer J, et al. Neuronal morphometry directly from bitmap images. Nat Methods. 2014;11(10):982–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakao A, Miki T, Shoji H, et al. Comprehensive behavioral analysis of voltage-gated calcium channel beta-anchoring and -regulatory protein knockout mice. Front Behav Neurosci. 2015;9:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shoji H, Takao K, Hattori S, Miyakawa T. Contextual and cued fear conditioning test using a video analyzing system in mice. J Vis Exp. 2014;85:50871.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shoji H, Takao K, Hattori S, Miyakawa T. Age-related changes in behavior in C57BL/6J mice from young adulthood to middle age. Mol Brain. 2016;9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maeta K, Hattori S, Ikutomo J, et al. Comprehensive behavioral analysis of mice deficient in Rapgef2 and Rapgef6, a subfamily of guanine nucleotide exchange factors for rap small GTPases possessing the ras/rap-associating domain. Mol Brain. 2018;11(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moy SS, Nadler JJ, Perez A, et al. Sociability and preference for social novelty in five inbred strains: An approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3(5):287–302. [DOI] [PubMed] [Google Scholar]
- 40. Kanetake T, Sassa T, Nojiri K, et al. Neural symptoms in a gene knockout mouse model of sjogren-larsson syndrome are associated with a decrease in 2-hydroxygalactosylceramide. FASEB J. 2019;33(1):928–941. [DOI] [PubMed] [Google Scholar]
- 41. Steru L, Chermat R, Thierry B, Simon P. The tail suspension test: A new method for screening antidepressants in mice. Psychopharmacology (Berl). 1985;85(3):367–370. [DOI] [PubMed] [Google Scholar]
- 42. Onouchi T, Kobayashi K, Sakai K, et al. Targeted deletion of the C-terminus of the mouse adenomatous polyposis coli tumor suppressor results in neurologic phenotypes related to schizophrenia. Mol Brain. 2014;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anders S, Pyl PT, Huber W. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ge SX, Son EW, Yao R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-seq data. BMC Bioinf. 2018;19(1):534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48(3):452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matsumura K, Seiriki K, Okada S, et al. Pathogenic POGZ mutation causes impaired cortical development and reversible autism-like phenotypes. Nat Commun. 2020;11(1):859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Suliman-Lavie R, Title B, Cohen Y, et al. Pogz deficiency leads to transcription dysregulation and impaired cerebellar activity underlying autism-like behavior in mice. Nat Commun. 2020;11(1):5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18(1):4–11. [DOI] [PubMed] [Google Scholar]
- 52. Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fujita H, Ikeda M, Ui A, et al. CHAMP1-POGZ Counteracts the inhibitory effect of 53BP1 on homologous recombination and affects PARP inhibitor resistance. Oncogene. 2022;41(19):2706–2718. [DOI] [PubMed] [Google Scholar]
- 54. Johannesen KM, Gardella E, Linnankivi T, et al. Defining the phenotypic spectrum of SLC6A1 mutations. Epilepsia. 2018;59(2):389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Poirier K, Lebrun N, Broix L, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45(6):639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crawley JN. Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin Neurosci. 2012;14(3):293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Takahashi H, Hashimoto R, Iwase M, Ishii R, Kamio Y, Takeda M. Prepulse inhibition of startle response: Recent advances in human studies of psychiatric disease. Clin Psychopharmacol Neurosci. 2011;9(3):102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Katayama Y, Nishiyama M, Shoji H, et al. CHD8 Haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537(7622):675–679. [DOI] [PubMed] [Google Scholar]
- 59. Nozawa RS, Nagao K, Masuda HT, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010;12(7):719–727. [DOI] [PubMed] [Google Scholar]
- 60. Baude A, Aaes TL, Zhai B, et al. Hepatoma-derived growth factor-related protein 2 promotes DNA repair by homologous recombination. Nucleic Acids Res. 2016;44(5):2214–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gudmundsdottir B, Gudmundsson KO, Klarmann KD, et al. POGZ Is required for silencing mouse embryonic beta-like hemoglobin and human fetal hemoglobin expression. Cell Rep. 2018;23(11):3236–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Markenscoff-Papadimitriou E, Binyameen F, Whalen S, et al. Autism risk gene POGZ promotes chromatin accessibility and expression of clustered synaptic genes. Cell Rep. 2021;37(10):110089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun X, Cheng L, Sun Y. Autism-associated protein POGZ controls ESCs and ESC neural induction by association with esBAF. Mol Autism. 2022;13(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the finding of this study are available from the corresponding author, upon reasonable request.