Abstract
The extraction of phenolic compounds from defatted hempseeds was optimized using a simplex lattice mixture design with three solvents (water, methanol, and acetone). The response variables were total phenolic content (TPC) and antioxidant activity evaluated by different spectrophotometric tests. The results showed that the binary acetone-water mixture in equal proportions is the optimal combination to achieve the maximum TPC (53.65 mg GAE per g extract) with higher antioxidant activities (265.53, 36.25, 119.03, 69.46, and 68.91 mg TE g−1 extract for the TAC, DPPH, ABTS, FRAP, and CUPRAC tests respectively). In addition, the phenolic profile analysis of defatted hemp seeds by HPLC-DAD/ESI-MS2 techniques showed the predominance of hydroxycinnamic acid amides and lignanamides. It allowed visualizing the effect of each solvent mixture on the relative extracted amount of each identified phenolic compound. This study suggests that N-trans-caffeoyltyramine, cannabisin A, and cannabisin B might contribute strongly to the potent antioxidant activity of hempseed extracts. Thus, it encourages the use of defatted hemp seeds as a source of antioxidants with added value for pharmaceutical and cosmetic applications.
The extraction of phenolic compounds from hemp seeds was optimized using a mixture design. 50% aqueous acetone was the optimal solvent to extract 33 phenolic compounds with a richness in hydroxycinnamic acid amides and lignanamides.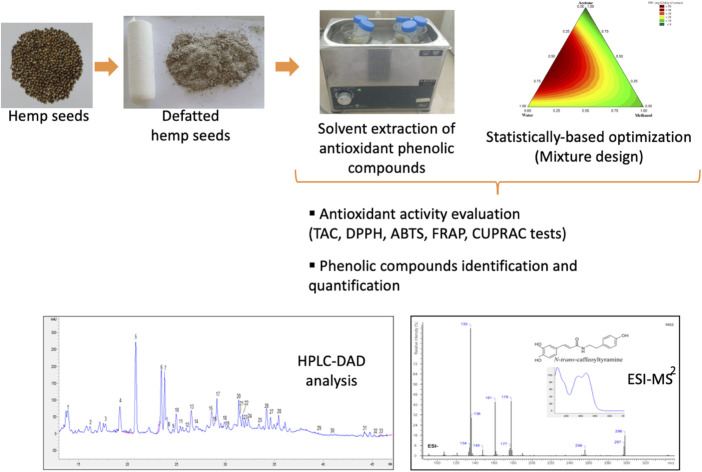
1. Introduction
Hemp (Cannabis sativa L.) is an annual plant of the Cannabaceae family. It is widespread in several regions of the world and is cultivated for industrial, therapeutic, recreational, and nutritional purposes. This plant had been banned and classified on the list of risky narcotics for a long time due to the psychotropic Δ9-tetrahydrocannabinol molecule. Recently, some countries have started easing restrictions to allow its cultivation and production, as well as its import and export for therapeutic, cosmetic, or industrial purposes, provided the used varieties have a low THC content (e.g., inferior to 0.2% w/w for European regulations).1 In Morocco, hemp was unlawfully cultivated for centuries in the mountainous region of the Rif for recreational use. The recent legalization of cannabis in Morocco is an excellent opportunity to take advantage of this culture known for its countless benefits.
Over the last years, hemp seeds and their oil have found a niche in the human food market owing to their nutritional and nutraceutical potential. Hemp seeds are a good source of protein (25–30%), fiber (30–40%), and oil (25–35%).2 Hemp seed oil is well known for its richness in polyunsaturated fatty acids.3 It is also a good source of antioxidants, such as γ-tocopherol,4 which could potentially prevent oxidative stress-related diseases.5 Furthermore, several studies showed the richness of hemp seeds in bioactive compounds mainly located in the hulls6 and remaining in the cake after oil extraction.7,8 Therefore, this by-product of hemp seed oil extraction is an interesting, inexpensive, and phenol-rich matrix, which can be used as a raw material for bioactive compounds extraction.
Bioactive compounds are secondary metabolites of self-defense produced by the plant in response to biotic or abiotic stress. They are of several classes, such as phenolic compounds, alkaloids, or terpenes, and are rising to prominence in several fields.9 Hemp seed phenolic compounds belong essentially to the phenylpropionamides class comprising phenylanamides and lignanamides.10 Hemp seeds are particularly rich in caffeoyltyramine, cannabisin A, and cannabisin B molecules. Several phenylpropionamides were isolated from hemp seeds (whole seeds, hulls, or cake) and showed interesting biological activities with important health virtues, including antioxidant, anti-inflammatory, anti-cancer, and anti-neuroinflammatory properties.6,11–13
The content of bioactive compounds varies according to the plant species, the organ studied, the environment, the genotype, and the method and conditions of extraction.14–16 Several techniques are available today to extract bioactive compounds from plant materials, but organic solvent-based extraction is the most widely used method.17 Nevertheless, the choice of solvent always remains a challenging determinant, affecting extraction selectivity. Usually, pure solvents cannot ensure the complete extraction of phenolic compounds with their distinct chemical structures and polarities. Solvent mixtures are frequently employed to improve the extraction selectivity of bioactive compounds.18
Mixture design is a statistical method based on regression analysis highlighting the relationship between the response and the studied factors. It reduces the use of raw materials, solvents, and time while promoting the selectivity and optimization required for targeted plant and food analysis.19 Mixture designs, such as simplex-lattice and simplex-centroid designs, have proven effective in numerous areas. Several studies have used this statistical tool to optimize the extraction of phenolic compounds from different plant matrices.20,21 The results of these studies open up promising perspectives for studying the solvent effect on the extraction of phenolic compounds from the cake of hemp seeds to obtain antioxidant-rich extracts.
To our knowledge, this work is the first one that optimizes the extraction of antioxidant compounds from defatted hemp seeds and evaluates the solvent effect on the phenolic profile of different solvent extracts. In addition, no previous study was interested in the characterization of phenolic compounds of Moroccan hemp seeds or the evaluation of their antioxidant potential. The works in the literature primarily concerned the evaluation of hemp seed oil composition.3,4 We hypothesized in this study that Moroccan hemp seeds would exhibit high antioxidant phenolic content, and the application of response surface analysis could optimize their extraction. Therefore, the objectives of this work were (i) to characterize the phenolic profile of Moroccan hemp seeds and (ii) to optimize the extraction of phenolic compounds with a higher antioxidant activity using a mixture design approach.
2. Materials and methods
2.1. Chemicals and reagents
Acetonitrile and formic acid were of LC-MS grade, while the other solvents used were of analytical grade. All reagents, solvents, and standards employed were purchased from Sigma-Aldrich (St-Louis, MO., USA), except for the ESI Tuning mix from Agilent Technologies (Santa Clara, CA., USA).
2.2. Plant material
Hemp seeds of a local ecotype were kindly provided by the National Agency of Medicinal and Aromatic Plants (ANPMA), Taounate, Morocco. The cultivation of cannabis and the production of seeds were carried out in 2020 at the ANPMA experimental station in northern Morocco. After cleaning the seeds, they were finely ground using a laboratory grinder. The comminuted seeds were defatted in a Soxhlet apparatus using petroleum ether at 45 °C for 5 hours. The cakes were stored in plastic bags at −20 °C until further use.
2.3. Experimental design
A simplex lattice mixture design, which is a variant of response surface methodology, was used in this study. This experimental design allows studying the solvent effect on phenolic content and antioxidant activities by modeling these responses according to all the possible combinations of three solvents while minimizing the number of experiments. Three solvents were investigated: water, methanol, and acetone. These solvents were chosen based on preliminary experiments (data not shown) to select three solvents capable of extracting the maximum of phenolic compounds among five solvents (water, ethanol, methanol, acetone, and isopropanol).
In total, 14 different extractions were performed (Table 1). The three solvents of the mixture design were studied at six levels through binary and ternary combinations with a duplicate at the central point (ternary mixture with equal proportions).
Simplex-lattice mixture design experiments for extraction of antioxidant compounds from defatted hemp seed and the correspondent responses.
| Run | Independent variables | Responses | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Water | Methanol | Acetone | TPCa | TACb | DPPHb | ABTSb | FRAPb | CUPRACb | |
| 1 | 1/3 | 2/3 | 0 | 22.56 | 166.38 | 13.32 | 75.83 | 31.02 | 33.38 |
| 2 | 2/3 | 1/3 | 0 | 22.20 | 178.52 | 8.48 | 77.83 | 23.91 | 19.28 |
| 3 | 1 | 0 | 0 | 27.51 | 125.70 | 2.70 | 64.13 | 15.54 | 17.92 |
| 4 | 2/3 | 1/6 | 1/6 | 33.35 | 191.39 | 20.74 | 93.99 | 39.43 | 38.62 |
| 5 | 1/6 | 1/6 | 2/3 | 36.46 | 194.46 | 18.41 | 81.29 | 48.25 | 63.65 |
| 6 | 0 | 1 | 0 | 14.10 | 151.51 | 7.81 | 31.25 | 29.80 | 41.51 |
| 7 | 0 | 0 | 1 | 6.86 | 41.24 | 0.92 | 19.27 | 12.21 | 22.13 |
| 8 | 1/3 | 1/3 | 1/3 | 38.80 | 217.66 | 25.18 | 117.47 | 45.38 | 65.97 |
| 9 | 0 | 1/3 | 2/3 | 13.52 | 119.61 | 5.71 | 30.33 | 20.67 | 37.21 |
| 10 | 1/3 | 0 | 2/3 | 45.81 | 247.89 | 34.15 | 122.37 | 61.01 | 84.92 |
| 11 | 2/3 | 0 | 1/3 | 50.19 | 231.42 | 26.91 | 117.41 | 51.65 | 56.33 |
| 12 | 1/6 | 2/3 | 1/6 | 27.79 | 163.40 | 15.31 | 79.44 | 38.57 | 43.88 |
| 13 | 0 | 2/3 | 1/3 | 20.01 | 172.27 | 9.52 | 49.81 | 17.83 | 43.29 |
| 14 | 1/3 | 1/3 | 1/3 | 41.89 | 237.60 | 25.61 | 105.41 | 48.34 | 56.27 |
Total phenolic content (TPC) is expressed in mg gallic acid equivalent per g of extract (mg GAE per g).
Total antioxidant capacity (TAC), cupric reducing antioxidant capacity (CUPRAC), Ferric reducing antioxidant power (FRAP), ABTS-, and DPPH- radical scavenging are expressed in mg torolox equivalent per g of extract (mg TE g−1).
The selected-response parameters (Yi dependent variables) were total phenolic content (Y1), total antioxidant capacity (Y2), DPPH radical scavenging activity (Y3), ABTS radical cation scavenging activity (Y4), ferric reducing antioxidant power (Y5), and cupric reducing antioxidant capacity (Y6). Linear (eqn. (1)), quadratic (eqn. (2)), special cubic (eqn. (3)), and full cubic (eqn. (4)) mathematical regression models were evaluated to express responses as a function of independent variables.
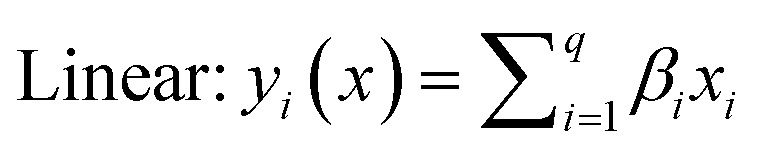 |
1 |
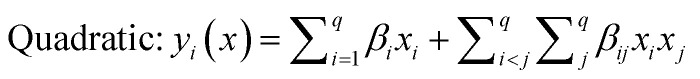 |
2 |
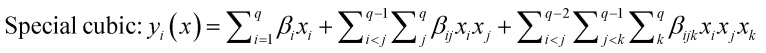 |
3 |
Full cubic:
| 4 |
β i represents the linear regression coefficient, βij the binary interaction coefficient, βijk the ternary interaction coefficient.
2.4. Extract preparation
Each extraction was carried out by mixing 6 ml of the solvent mixture with 0.6 g of defatted hemp seeds. The tubes were vortexed for 5 min, sonicated for 45 min in an ultrasound bath (Transonic T460, Germany) in a darkened cold room, and then centrifuged for 10 min at 4800 rpm. After repeating the extraction process twice, the supernatants were combined and evaporated using a rotary evaporator. The resulting dry extract was resuspended in 2 ml of methanol, filtered (0.45 μm), and stored at 4 °C until analysis. Fourteen extracts were produced according to the experimental design presented in Table 1. All extractions were realized in triplicate.
2.5. Total phenolic content of defatted hemp seed extracts
The total phenolic content (TPC) of hemp seed extracts was quantified according to Moccia et al.7 with modifications. 100 μl of the Folin-Ciocalteu reagent were added to 100 μl of the extract and 1.3 ml of water. The mixture was incubated for 10 min, mixed with 1 ml of 10% sodium carbonate (Na2CO3) solution, and re-incubated for 1 h 20 min in the dark. The absorbance was read at 765 nm against a blank using a UV-visible spectrophotometer (Jenway 7305, France). A calibration curve was established using the gallic acid standard (0.0156–0.25 mg ml−1), and the results were expressed as mg gallic acid equivalent per g of extract (mg GAE per g extract). The limits of detection (LOD) and quantification (LOQ) were 14 and 43 μg ml−1, respectively.
2.6. Antioxidant activity of defatted hemp seed extracts
Antioxidant activities of hemp seed extracts were assessed using several tests based on either scavenging free radicals or reducing capacity. For all these spectrophotometric tests, the absorbance measurement was carried out using a UV-visible spectrophotometer (Jenway 7305, France), and the results were presented as mg Trolox Equivalent per g of extract (mg TE g−1 extract) by referring to a Trolox (2,5,7,8-tetramethylchroman-2-carboxylic acid) calibration curve (20–280 μg ml−1).
2.6.1. Total antioxidant capacity assay
The Total Antioxidant Capacity (TAC) was evaluated based on the phosphomolybdenum method,22 consisting of reducing molybdenum(vi) to molybdenum(v). Briefly, the phosphomolybdenum reagent was prepared by mixing 4 mM ammonium molybdate, 28 mM sodium phosphate, and 0.6 M sulfuric acid in equivalent proportions. The test was performed by adding 1.5 ml of the reagent to 50 μl of the extract. The tubes were well vortexed and incubated in a water bath at 95 °C for 90 min. The absorbance was measured immediately at 695 nm against a blank.
2.6.2. DPPH free radical scavenging activity
DPPH• (2,2-diphenyl-1-picrylhydrazyl radical) is a purple-colored free radical that turns yellow when neutralized by antioxidants. 100 μl of the extract were mixed with 2 ml of a DPPH methanolic solution (1.3 × 10−4 M). The mixture was vortexed and incubated for 30 min in the dark. The absorbance was then measured at 517 nm.
2.6.3. ABTS radical cation scavenging assay
The ABTS radical cation scavenging assay was performed as described by Grochowski and his co-authors with some modifications.22 This test is based on the decolorization of the blue-green colored ABTS˙+ in the presence of antioxidants. The ABTS˙+ was generated by mixing a methanolic solution of 7 mM ABTS (2,2-azino-bis(3-ethylbenzo-thiazoline-6-sulfonic acid)) with an aqueous solution of 2.45 mM potassium persulfate (1 : 1; v/v). The mixture was incubated at room temperature in darkness for 16 h, and the absorbance (at 734 nm) was adjusted to 1 by dilution. Then, 50 μl of the extract were mixed with 2 ml of the ABTS•+ solution. After vortexing, the mixture was incubated for 30 min at room temperature in dark conditions, and the absorbance was measured at 734 nm.
2.6.4. Ferric reducing antioxidant power assay
The ferric reducing antioxidant power (FRAP) test relies on reducing Fe3+/ferricyanide complex to Fe2+based on electron-donating capacity. It was carried out according to the method reported in the literature.23 100 μl of the extract were mixed with 0.65 ml of phosphate buffer (0.2 M, pH 6.6) and 0.65 ml of 1% ferricyanide. After 20 min of incubation at 50 °C in a water bath, 0.65 ml of 10% trichloroacetic acid was added to stop the reaction. Then, 0.65 ml was collected from each tube and mixed with 0.65 ml of water and 0.25 ml of 0.1% FeCl3. The absorbance was read at 700 nm against a blank.
2.6.5. Cupric reducing antioxidant capacity assay
The cupric reducing antioxidant capacity (CUPRAC) test is based on reducing Cu2+ to Cu1+ by antioxidants. It was performed following Kadriye's method,23 which consists of adding 50 μl of extract to 1.5 ml of a reagent (10 mM CuCl2, 1 M ammonium acetate, 7.5 mM neocuproine (1 : 1 : 1)). After 30 min of incubation, the absorbance of the chromophore Cu(i)–neocuproine chelate was recorded at 450 nm.
2.7. HPLC-DAD and MS analyses of phenolic compounds
Phenolic compounds separation was performed on an Eclipse XDB-C18 column (3.5 μm particle size, 150 × 4.6 mm internal diameter; Agilent Technologies, USA) using an Agilent 1260 Infinity II high-performance liquid chromatography system (HPLC, Agilent Technologies, USA), equipped with a diode array detector (DAD). The mobile phase consisted of water (A) and acetonitrile (B), with 1% formic acid. The separation was carried out using the following gradient: 2–12.5% B (0–6 min), 12.5–30% B (6–23 min), 30–45% B (23–33 min), 45–75% B (33–38 min), 75% B (38–42 min), 75–100% B (42–47 min), 100% B (47–49 min), 100–2% B (49–50 min), and 2% B (50–51 min). 10 μl of the sample were injected with a flow rate of 0.6 ml min−1. The wavelengths 254, 280, 300, and 340 nm were used to monitor the separation of extracts. Thus, the UV-visible spectra of the compounds were recorded between 190-800 nm. After their separation, each peak was collected, at the output of the HPLC system, in 2 ml vials and then identified by mass spectrometry.
The mass spectra of the phenolic compounds were recorded in negative and positive modes by direct infusion (at 500 μl h−1) of the peaks collected from the HPLC system on an ion trap mass spectrometer (Esquire HCT mass spectrometer, Bruker Daltonics, Germany) equipped with electrospray ionization (ESI) source. ESI operating parameters were set as follows: spray voltage 4500 V, dry gas temperature 200 °C, nebulizer 10 psi, and dry gas 4 L min−1, using smart mode with a target mass of 400, 500, and 600 m/z. MS2 mass spectra were then produced for each mass scan by isolating the precursor ion inside the ion trap and using a collision energy of 1–10% arbitrary unit. A mass range of 50–1000 m/z was used at a speed of 26 000 m/z/s to acquire mass spectra in ultra-scan mode. The instrument was calibrated using the ESI Tuning mix. Esquire Control software was used for instrument control and data acquisition, while mass data processing was performed using ACDlabs 2021.2.1 software. The error between observed and calculated masses was expressed as parts per million (ppm). Phenolic compounds were identified by comparing their MS, MS2 fragments, and UV spectra with published data.
The chromatograms were taken at 220, 254, 280, 320, and 340 nm. The peak areas obtained from the HPLC-DAD profile at 280 nm were used to semi-quantify the identified phenolic compounds because it is the common maximum absorption wavelength among most of these compounds. This wavelength has been described as adequate for quantifying phenolic compounds in various vegetable matrices,21,24,25 including those analyzed in hemp seeds.10 The semi-quantification was performed using an external N-trans-caffeoyltyramine standard curve (60–980 μg ml−1, LOD = 34 μg ml−1, LOQ = 102 μg ml−1) due to the limited availability of the majority of the phenolic compounds identified in this work. Results are expressed as mg N-trans-caffeoyltyramine equivalent per g of extract (CTE per g extract). Only benzoic acid (60–980 μg ml−1, LOD = 48 μg ml−1, LOQ = 146 μg ml−1), p-coumaric acid (75–1200 μg ml−1, LOD = 24 μg ml−1, LOQ = 73 μg ml−1), ferulic acid (60–980 μg ml−1, LOD = 34 μg ml−1, LOQ = 104 μg ml−1), and sinapic acid (60–980 μg ml−1, LOD = 32 μg ml−1, LOQ = 95 μg ml−1) were quantified using their commercial standards. The HPLC profiles were visualized and analyzed by the Agilent Chemstation 32 software.
2.8. Statistical analysis
The mixture design experiments, regression analysis, results of ANOVA, and contour plots were generated and processed using Statistica software version 10.0 (StatSoft Inc., USA). Data were recorded as mean ± SD, and p-values < 0.05 were held as statistically significant.
3. Results and discussion
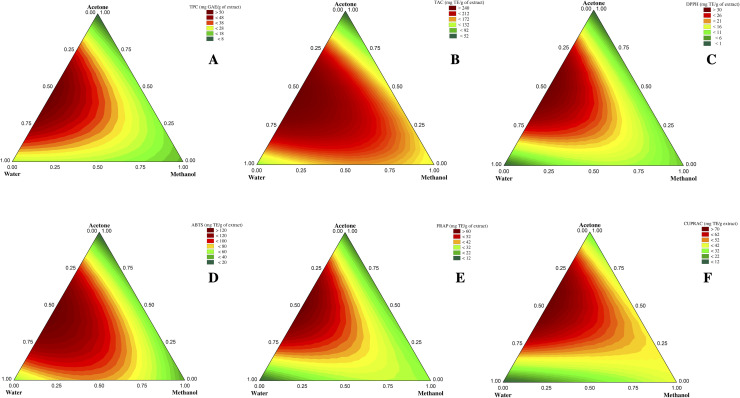
The mixture design used in this study allows mathematical modeling of the experiment. The model represents the relationship between the response and solvent proportions and allows predicting the response at any point within the experimental domain of interest even if no experiment was performed. The R2 coefficient reflects the variance proportion explained by the model. The best mathematical model is represented graphically using contour plots which allow visualizing the evolution of each response studied as a function of solvent mixtures within the experimental domain. The responses (TPC, TAC, DPPH, ABTS, FRAP, and CUPRAC) obtained for each experiment are presented in Table 1.
3.1. Solvent effect on TPC
Hemp is rich in various bioactive compounds. Several studies have focused on the unique cannabinoid profile specific to this species.26 Over the last years, more importance has been devoted to hempseed phenolic compounds. These substances have a broad spectrum of biological activities (antibacterial, anti-carcinogenic, anti-inflammatory, antioxidant) linked to their reducing nature and affinity for proteins and metal ions.27 The optimization of their extraction is therefore of great interest.
The total phenolic content (TPC) of defatted hempseeds was investigated for different solvent proportions. As shown in Table 1, TPC varies considerably from 6.86 to 50.19 mg GAE per g of extract, depending on the solvent mixtures. The best binary mixture was acetone-water (runs 10 and 11), while the best ternary mixture was water-acetone-methanol (1/3 : 1/3 : 1/3).
Several regression models were tested to determine which model best fits the experimental data (ESI Table S1†). The results showed that only the quadratic model was significant (p = 0.00006) and explained a large part of the variation of phenolic content in response to changing solvent proportions (R2 = 0.95). The model's goodness was confirmed by the insignificant lack of fit (p > 0.05).
Table 2 presents the regression coefficients used to construct the TPC prediction equation. Analyzing these coefficients is very useful in understanding the effect of each variable on the studied response. In fact, the coefficient in absolute value reflects each variable's weight, while its positive or negative sign indicates whether there is a positive or negative effect on the response. For the TPC prediction equation, we notice that although all the coefficients are positive, only those corresponding to water (β1), acetone–water (β13), and acetone–methanol (β23) mixtures are significant (p < 0.05), which proves the synergistic effect of these solvent combinations on the extraction of phenolic compounds. In addition, β13 corresponding to the acetone–water mixture is the greatest indicating that this solvent combination contributes more to improving the extraction of phenolic compounds. This finding corroborates other studies which reported the effectiveness of moderately polar mixtures such as acetone-water in extracting phenolic compounds from protein-rich plant matrices (which is the case of hemp seeds), thanks to its ability to degrade polyphenol–protein complexes.21
Regression equation coefficients and model fitting of the quadratic model for each studied response.
| Responsesa (Yi) | Coefficients | p-values | R 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β 1 | β 2 | β 3 | β 12 | β 13 | β 23 | Model | Lack of fit | ||
| TPC | 23.99b | 15.14 | 6.82 | 15.51 | 150.81b | 37.23b | 0.000064 | 0.402 113 | 0.95 |
| TAC | 118.95b | 147.06b | 50.6b | 146.51 | 694.36b | 208.11b | 0.000200 | 0.516 828 | 0.93 |
| DPPH | 0.67 | 8.21b | 1.05 | 32.48b | 135.35b | 15.88 | 0.000010 | 0.093201 | 0.97 |
| ABTS | 54.97b | 35.11b | 18.03 | 153.19b | 378.33b | 80.86 | 0.000072 | 0.567 471 | 0.95 |
| FRAP | 11.41b | 28.53b | 15.33b | 40.53 | 202.45b | 3.16 | 0.000209 | 0.286 987 | 0.93 |
| CUPRAC | 10.25 | 41.20b | 26.83b | 2.69 | 240.38b | 39.54 | 0.001257 | 0.558 710 | 0.89 |
Y i = β1 Water+ β2 Methanol+ β3 Acetone+ β12 Water × Methanol+ β13 Water × Acetone+ β23 Methanol × Acetone.
Significant at p < 0.05.
Plant matrices contain several classes of bioactive compounds, which vary in terms of polarity. The extraction efficiency of phenols conspicuously depends on the degree of dissolution of these compounds in a given solvent, related to their respective polarities.28 Generally, phenolic compounds have an affinity for polar solvents. Their solubility mainly depends on their structure,15 including their molecular size, the presence of hydroxyl groups, the length of the constituent hydrocarbon chains, and the degree of methoxylation.
Contour plots showed that binary and ternary solvents appeared particularly enriched in phenols than pure solvents (Fig. 1A). Indeed, pure acetone extract recorded the lowest TPC value among all tested mixtures (Table 1), corroborating the fact that using pure acetone promotes its self-association (acetone–acetone self-associated molecules) and therefore decreases the chances of its association with the plant matrix compounds for their extraction.29
Fig. 1. Contour plots of TPC (a), TAC (b), DPPH (c), ABTS (d), FRAP (e), and CUPRAC (f). The vertices correspond to pure solvents; the points on the edges correspond to binary mixtures, while the points inside the triangle correspond to ternary mixtures.
Previous studies have highlighted the suitability of mixtures between organic solvents (e.g., acetone, ethanol, methanol) and water for phenolic compounds extraction from different plant matrices and have shown their synergistic effect.19,21 This can be explained by the ability of water to swell the cells and disintegrate the walls, enhancing the intracellular penetration of organic solvents.30 Furthermore, some phenolic compounds are often conjugated with other molecules, such as polysaccharides and proteins, easily dissolved in water.21
Also, some physical characteristics of solvents, such as density and viscosity, could massively impact their extraction capacities. In general, solvents with low density and viscosity have a good extraction power because their diffusivity is high, and the movement of solvent and solute molecules is important, thus improving the extraction efficiency.31 Combining organic solvents with water positively affects phenolic compounds extraction as it decreases the density and viscosity values of the solvent system, thereby increasing its diffusivity and facilitating the extraction process.
3.2. Solvent effect on antioxidant activity
Antioxidants are compounds that can delay or stop the oxidation process generated by free radicals. Indeed, when the production rate of free radicals exceeds antioxidants, the organism can be subjected to oxidative stress that can damage physiological functions.32 Antioxidants are natural substances belonging to several chemical classes, such as phenolic compounds. These latter are generally the most prevalent class and are known to be powerful antioxidants due to their ability to donate electrons, hydrogens, or chelate metals.27
Five different spectrophotometric tests were performed to fully understand the solvent effect on the antioxidant activity of defatted hemp seeds. According to their mechanisms of action (H-donating capacity/Redox properties/metal-chelating), each test involves a type of antioxidant. The antioxidant activity of the studied extracts was in the range of 0.92–34.15, 19.27–122.37, 12.21–61.01, 17.92–84.92, and 41.24–247.89 mg TE g−1 extract for DPPH, ABTS, FRAP, CUPRAC, and TAC, respectively. The highest antioxidant activity for all the tests carried out was recorded in the run 10, corresponding to acetone : water mixture (2/3 : 1/3). The lowest activity was observed in water for the CUPRAC test and pure acetone for all the other tests (Table 1).
Upon investigating the generated regression models, the quadratic model was chosen for all antioxidant tests because the special cubic and full cubic models do not significantly improve the fit to the surface (ESI Table S1†). All quadratic models were statistically significant and showed no significant lack of fit at the 95% confidence level, with an R2 ranging from 0.89 to 0.97. Regression coefficients to build the equation of prediction of each antioxidant test are listed in Table 2. No antagonistic effect appeared for the different mixtures of solvents. However, some mixtures were more synergistic than others. Pure acetone was the solvent with the lowest antioxidant activity, while its combination with water gave the higher activity. This could be explained by the synergistic effect of the acetone-water mixture, which showed the highest regression coefficient for all the antioxidant activity tests. As previously reported, pure solvent extracts recorded lower antioxidant activity than their aqueous mixture.6,20
According to the contour plots of antioxidant activity tests (Fig. 1B–F), we notice that they all roughly followed the same trend observed for phenolic compounds (Fig. 1A). Indeed, correlation results have shown a significant (p < 0.05) and strong correlation between the content of phenolic compounds and antioxidant activities. Correlation coefficients (r) between TPC and antioxidant capacity tests are 0.73 for CUPRAC, 0.88 for both FRAP and TAC, 0.91 for DPPH, and 0.95 for ABTS. Some structural features of phenolic compounds have a direct impact on their antioxidant capacity, such as the number and position of hydroxyl groups, the extent of conjugation, the distance between the carbonyl group and the aromatic ring, the degree of methoxylation, in addition to the number of aromatic rings.33 For the TAC test (Fig. 1B), it is noted from contour plots that methanol and some aqueous methanolic and acetone-methanolic extracts have moderate antioxidant activity, albeit they have a low capacity for extracting phenols (Fig. 1A). Antioxidants other than phenolic compounds could be extracted, contributing thus to this activity, or the nature of the phenolic compounds extracted by these two mixtures could result in a more effective antioxidant activity.
As a visualization of the predictive model, these ternary graphs show that the acetone-water mixture (50–50) is responsible for the highest TPC value and the highest activity for all the antioxidant activity tests performed. Chen et al.,6 studying hempseed kernels and hull, also found that 50% and 75% acetone extracts had the highest TPC values and the best DPPH scavenging activity compared to absolute or aqueous methanolic and ethanolic extracts.
3.3. Solvent effect on the phenolic profile
The phenolic profile of each extract was analyzed for an in-depth study of the solvent effect on the individual extraction of phenols. After separating phenolic compounds on HPLC-DAD (ESI Fig. S1†), the peaks obtained were analyzed by ESI-MS2. The identification of the compounds detected in UV was carried out by comparing the precursor ion mass (MS) and the mass fragments (MS2) in both negative and positive modes with those found in the literature7,10–12 and mass spectrum databases available online (MassBank of North America). In total, 33 phenolic compounds were identified in defatted hemp seeds, including 5 hydroxycinnamic acid amides, 20 lignanamides, 4 cannabinoids, and 4 phenolic acids. These compounds are listed in Table 3 with their retention time (HPLC-DAD), molecular formulas, UV λmax, experimental and calculated m/z, error (ppm), MS2 fragments, and the proposed assignment. The results showed that defatted hemp seeds are rich in phenylpropionamides, a phenolic class including lignanamides and hydroxycinnamic acid amides (HCA). These latter, also called phenolamides, are associations between phenolic acids and aliphatic or aromatic amines.34 The compounds 5, 6, 9,11, and 18 were assigned to HCA, specifically hydroxycinnamoyl amides with tyramine or spermidine as amine moiety. Some studies have also detected the presence of other HCA in hempseeds with octopamine moiety, such as caffeoyl-octopamine and coumaroyl-octopamine.12,35 MS2 spectrum of compounds 4 and 5 showed, respectively, the deprotonated molecular ions [M − H]−at m/z 298.1161 and 298.1159, and the protonated molecular ions [M + H]+ at m/z 300.1155 and 300.1160. Their fragmentations in [M−H]− gave the same intensive ions at m/z 135, 161, and 178 (ESI Fig. S2†). The fragments at m/z 135 (base peak) and 178 could be formed, respectively, as a consequence of tyramine moiety cleavage and N–Cα bond breakage.12 In addition, the UV spectra of compounds 4 and 5 mostly resembled those previously reported for N-trans-caffeoyltyramine. These compounds were tentatively identified as N-trans-caffeoyltyramine geometrical isomers. Similarly, compound 10, exhibiting an experimental mass in [M−H]−atm/z 312.1322 with the fragments 178, 135, 297, 312, and 148, was likely N-feruloyltyramine.
Identified compounds by HPLC-DAD/ESI-MS2 in defatted hemp seeds.
| N° peak | RT (min) | Compound name | Molecular formula | UV λmax (nm) | Theoretical mass (m/z) | Experimental mass (m/z) | Error (ppm) | Mass fragments (%intensity) |
|---|---|---|---|---|---|---|---|---|
| Negative mode [M − H] − | ||||||||
| 1 | 13.9 | Benzoic acid | C7H6O2 | 222, 285 | 122.0373 | 121.0375 | 1.65 | Not fragmented |
| 2 | 16.1 | p-Coumaric acid | C9H8O3 | 230, 295, 310 | 164.0478 | 163.0481 | 1.84 | 119 (100), 93 (8) |
| 3 | 17.7 | Ferulic acid | C10H10O4 | 225, 292, 315 | 194.0584 | 193.0588 | 2.07 | 134 (100), 178 (37), 149 (13) |
| 4 | 19.2 | N-trans-caffeoyltyramine isomer | C17H16NO4 | 220, 280, 315 | 299.1158 | 298.1161 | 1.01 | 135(100), 178(46), 298(45), 161(25), 136(12), 284(6) |
| 5 | 20.8 | N-trans-caffeoyltyramine | C17H16NO4 | 220, 290, 320 | 299.1158 | 298.1159 | 0.34 | 135(100), 178(43), 161(42), 136(30), 298(16), 256(5), 148(5) |
| 6 | 23.4 | Cannabisin A | C34H30O8N2 | 255 | 594.2002 | 593.2014 | 2.02 | 593(100), 454(27), 639(21), 523(16), 536(13), 428(11), 482(11) |
| 7 | 23.7 | Cannabisin B | C34H32O8N2 | 225, 250, 285, 310, 335 | 596.2159 | 595.2148 | −1.85 | 432(100), 595(93), 485(68), 269(33), 322(30) |
| 8 | 24.2 | N-trans-coumaroyltyramine | C17H17O3N | 224, 290, 310 | 283.1208 | 282.1218 | 3.54 | 145(100), 119(85), 282(54), 162(46), 134(14), 240(10) |
| 9 | 24.6 | Cannabisin B isomer 1 | C34H32O8N2 | 255, 310 | 596.2159 | 595.2151 | −1.34 | 416(100), 595(99), 269(46), 432(45), 458(30), 485(29), 295(23) |
| 10 | 24.9 | N-feruloyltyramine | C18H19O4N | 220, 292, 318 | 313.1314 | 312.1322 | 2.56 | 178(100), 135(45), 297(46), 312(45), 148(12) |
| 11 | 25.3 | Cannabisin B isomer 2 | C34H32O8N2 | 224, 250, 290, 335 | 596.2159 | 595.2164 | 0.84 | 485(100), 432(40), 322(33), 269(20), 348(9), 456(8), 595 (6) |
| 12 | 25.9 | Demethylgrossamide | C35H34N2O8 | 225, 250, 285, 330 | 610.2315 | 609. 2324 | 1.47 | 283(100), 446(66), 268(26), 609(9), 377(4) |
| 13 | 26.4 | Cannabisin C | C35H34O8N2 | 220, 245, 292, 320 | 610.2315 | 609. 2321 | 0.98 | 499(100), 609(95), 446(70), 447(27), 336(20), 269(5) |
| 14 | 26.9 | Cannabisin C isomer | C35H34O8N2 | 220, 280, 335, 410 | 610.2315 | 609.2332 | 2.79 | 446(100), 485(66), 609(21), 322(19), 472(7), 279(6), 499(6) |
| 15 | 28.5 | Cannabisin D | C36H36N2O8 | 225, 250, 285, 340 | 624.2472 | 623.2479 | 1.12 | 460(100), 623(94), 283(35), 268(7), 444(9), 499(4) |
| 16 | 28.9 | 3,3-Didemethylgrossamide | C34H32N2O8 | 225, 290, 324 | 596.2159 | 595.2168 | 1.51 | 432(100), 269(99), 458(36), 595(22), 295(10), 338(7), 250(2) |
| 17 | 29.1 | Tri-p-coumaroylspermidine | C34H37N3O6 | 255 | 583.2688 | 582.2691 | 0.52 | 462(100), 582(87), 342(76), 316(10), 436(11), 299(4), 217(2), 533(2) |
| 18 | 29.8 | Cannabisin E | C36H38N2O9 | 220, 283, 316 | 642.2577 | 641.2579 | 0.31 | 623(100), 489(90), 281(65), 431(40), 641(30), 591(15), 460(11), 312(12) |
| 19 | 30.2 | Grossamide K | C28H29NO7 | 225, 288, 325 | 491.1949 | 490.1957 | 1.63 | 472(100), 490(43), 460(34), 488(2) |
| 20 | 31.3 | Cannabisin M | C34H32N2O8 | 223, 285, 315 | 596.2159 | 595.2147 | −2.04 | 298(100), 595(57), 430(17), 427(5), 547(4) |
| 21 | 31.5 | 3,3′-demethyl-heliotropamide | C34H32N2O8 | 223, 285, 315 | 596.2159 | 595.2168 | 1.51 | 107(100), 298(49), 595(23) |
| 22 | 31.7 | Unnamed condensed trilignanamide | C51H47N3O12 | 222, 278, 315 | 893.3160 | 892.3164 | 0.44 | 430(100), 595(85), 593(55), 727(24), 485(11), 322(11), 892(9) |
| 23 | 32.0 | Cannabisin Q | C34H32N2O8 | 290, 320 | 596.2159 | 595.2154 | −0.84 | 298(100), 595(42), 296(7), 178(1) |
| 24 | 32.3 | Cannabisin F | C36H36N2O8 | 225, 290 | 624.2472 | 623.2478 | 0.96 | 460(100), 623(61), 297(35), 486(29), 352(5) |
| 25 | 33.4 | Isocannabisin N | C35H34N2O9 | 225, 294, 312 | 610.2315 | 609.2298 | −2.79 | 609(100), 296(41), 312(25), 417(17), 446(5), 176(4), 581(3) |
| 26 | 34.2 | Grossamide | C36H36N2O8 | 226, 285, 322 | 624.2472 | 623.2485 | 2.08 | 623(100), 460(77), 591(47), 297(32), 471(30), 551(23), 432(17), 486(15), 428(11), 282(11) |
| 27 | 34.5 | Cannabisin O | C54H53N3O12 | 229, 315 | 935.3629 | 934.3641 | 1.28 | Not fragmented |
| 28 | 35.4 | Unnamed lignanamide | — | 228, 312 | — | 589.2366 | — | 426 (100), 261(10), 443(8), 589(7), 255(7), 279(5), 163(5) |
| 29 | 39.4 | Dihydrocannabinol | C21H28O2 | 272 | 312.2089 | 311.2092 | 0.96 | 293(100), 311(82), 223(59), 275(41), 201(26), 235(20), 171(15) |
| 30 | 40.7 | Cannabidiol (CBD) | C21H30O2 | 280 | 314.2246 | 313.2257 | 3.51 | 313(100), 201(96), 171(36), 295(34), 277(26), 202(14), 165(6), 172(5), 183(2), 129(0,5) |
| 31 | 44.2 | Cannabielsoic acid | C22H29O5 | 280 | 374.2099 | 373.2105 | 1.61 | 205(100), 329(94), 311(84), 373(56), 271(35), 173(16), 259(8) |
| 32 | 45.2 | Sinapic acid | C11H12O5 | 275 | 224.0690 | 223.0697 | 3.13 | 225(100), 223(34), 195(36), 125(35), 179(24), 221(20), 163(18), 206(16), 164(12), 155(17) |
| 33 | 45.9 | Cannabidiolic acid (CBDA) | C22H30O4 | 224, 270, 310 | 358.2144 | 357.2141 | −0.84 | 339(100), 357(21), 340(19), 341(11), 311(7), 313(5), 289(3), 179(2), 271(1), 245(1) |
| Positive mode [M + H]+ | ||||||||
| 1 | 13.9 | Benzoic acid | C7H6O2 | 222, 285 | 122.0373 | 123.0376 | 2.44 | 123(100), 95(74), 122(20), 79(17), 97(15), 107(14), 96(12), 20(7) |
| 4 | 19.2 | N-trans-caffeoyltyramine isomer | C17H16NO4 | 220, 280, 315 | 299.1158 | 300.1155 | −0.99 | 163(100), 138(18), 300(16), 121(10), 145(4) |
| 5 | 20.8 | N-trans-caffeoyltyramine | C17H16NO4 | 220, 290, 320 | 299.1158 | 300.1160 | 0.67 | 163(100), 300(18), 138(15), 121(7), 145(5) |
| 6 | 23.4 | Cannabisin A | C34H30N2O8 | 255 | 594.2002 | 595.2011 | 1.51 | 458(100), 595(18), 459(5) |
| 7 | 23.7 | Cannabisin B | C34H32N2O8 | 225, 250, 285, 310, 335 | 596.2159 | 597.2167 | 1.34 | 460(100), 597(11), 432(5), 350(3), 295(1) |
| 8 | 24.2 | N-trans-coumaroyltyramine | C17H17NO4 | 224, 290, 310 | 283.1208 | 284.1214 | 2.11 | 147(100), 284(30), 148(14), 119(2) |
| 9 | 24.6 | Cannabisin B isomer 1 | C34H32N2O8 | 255, 310 | 596.2159 | 597.2151 | −1.34 | 460(100), 432(32), 597(25), 295(13), 350(7), 418(5), 279(5) |
| 10 | 24.9 | N-feruloyltyramine | C17H17NO4 | 220, 292, 318 | 313.1314 | 314.1319 | 1.59 | 177(100), 314(28), 145(13), 178(13), 313(9), 117(1) |
| 11 | 25.3 | Cannabisin B isomer 2 | C17H17NO4 | 224, 250, 290, 335 | 596.2159 | 597.2164 | 0.84 | 460(100), 597(7), 350(3), 187(3), 131(1), 295(2), 323(2) |
| 12 | 25.9 | Demethylgrossamide | C35H34N2O8 | 225, 250, 285, 330 | 610.2315 | 611.2318 | 0.49 | 474(100), 611(51), 446(20), 350(16), 309(7), 591(5), 187(3) |
| 13 | 26.4 | Cannabisin C | C35H34N2O8 | 220, 245, 292, 320 | 610.2315 | 611.2297 | −2.94 | 474(100), 611(30), 446(21), 364(12), 309(5), 201(4) |
| 14 | 26.9 | Cannabisin C isomer | C35H34N2O8 | 220, 280, 335, 410 | 610.2315 | 611.2317 | 0.33 | 474(100), 611(48), 446(21), 472(12), 350(7), 337(4), 309(4), 454(3) |
| 15 | 28.5 | Cannabisin D | C36H36N2O8 | 225, 250, 285, 340 | 624.2472 | 625.2479 | 1.12 | 488(100), 460(31), 625(20),364(19), 323(9), 201(5), 297(2), 244(1), 439(1) |
| 16 | 28.9 | 3,3-Didemethylgrossamide | C34H32N2O8 | 225, 290, 324 | 596.2159 | 597.2170 | 1.84 | 434(100), 297(49), 597(44), 279(41), 323(25), 251(15), 460(14), 233(6), 271(5) |
| 17 | 29.1 | Tri-p-coumaroylspermidine | C34H37N3O6 | 255 | 583.2688 | 584.2691 | 0.51 | 438(100), 420(73), 584(17), 204(14), 275(7), 565(4), 534(3) |
| 18 | 29.8 | Cannabisin E | C36H38N2O9 | 220, 283, 316 | 642.2577 | 643.2586 | 1.40 | 643(100), 462(80), 282(17), 625(15), 338(10), 341(9), 325(7), 489(7) |
| 19 | 30.2 | Grossamide K | C28H29NO7 | 225, 288, 325 | 491.1949 | 492.1929 | −4.06 | 462(100), 493(57), 325(45), 463(13), 337(9), 307(7), 293(1) |
| 20 | 31.3 | Cannabisin M | C34H32N2O8 | 223, 285, 315 | 596.2159 | 597.2164 | 0.84 | 297(100), 434(82), 597(52), 300(30), 337(17), 187(17), 533(16), 460(15), 279(14) |
| 21 | 31.5 | 3,3′-demethyl-heliotropamide | C34H32N2O8 | 223, 285, 315 | 596.2159 | 597.2167 | 1.34 | 523(100), 297(45), 597(42), 458(33), 337(21), 853(20) |
| 22 | 31.7 | Unnamed condensed trilignanamide | C51H47N3O12 | 222, 278, 315 | 893.3160 | 894.3171 | 1.23 | 894(00), 757(99), 484(30), 729(28), 374(10), 647(10), 594(10), 429(4) |
| 23 | 32.0 | Cannabisin Q | C34H32N2O8 | 290, 320 | 596.2159 | 597.2162 | 0.50 | 297(100), 597(31), 460(24), 434(21), 187(21), 279(11), 233(5), 251(5) |
| 24 | 32.3 | Cannabisin F | C36H36N2O8 | 225, 290 | 624.2472 | 625.2453 | −3.03 | 462(100), 625(70), 325(44), 307(12), 351(8), 293(5), 201(2) |
| 25 | 33.4 | Isocannabisin N | C35H34N2O9 | 225, 294, 312 | 610.2315 | 611.2319 | 0.65 | 448(100), 611(80), 311(64), 314(35), 187(33), 474(25), 293(14), 177(13), 591(8), 409(6) |
| 26 | 34.2 | Grossamide | C36H36N2O8 | 226, 285, 322 | 624.2472 | 625.2481 | 1.44 | 625(100), 462(93), 325(57), 351(42), 307(30), 488(25), 293(15),292(5), 626(5) |
| 27 | 34.5 | Cannabisin O | C54H53N3O12 | 229, 315 | 935.3629 | 936.3609 | −2.13 | 799(100), 538(36), 634(30), 771(21), 512(15), 936(12), 675(8), 388(6) |
Several lignanamides were also detected and characterized, including cannabisin A (6), B (7, 9, and 11), C (13 and 14), D (15), E (18), M (20), Q (23), F (24), O (27), isocannabisin N (25), demethylgrossamide (12), 3,3-didemethylgrossamide (16), grossamide K (19), 3,3-demethyl-heliotropamide (21), and grossamide (26). Lignanamides are complex structures resulting from the oxidation of phenolamides.1 Cannabisin A and B were reported as the major lignanamides in hempseeds.7 In the present study, three isomers (7, 9, and 11) showed the molecular ions [M − H]−at m/z 595.2148, 595.2151, and 595.2164, and intensive fragments [M − H]− at m/z 485 and 432, corresponding to the loss of the catechol unit (−110 amu) and tyramine moiety (−163 amu). Based on MS, MS2, UV-DAD spectra, and the literature previously reported on hemp seeds, these compounds were tentatively identified as cannabisin B isomers. In a similar way, two isomers (13 and 14) showed an identical molecular formula (C35H34O8N2), with a similar fragmentation pattern by losing a catechol unit (−110 amu) and tyramine moiety (−163 amu), giving, respectively, fragment ions [M − H]− at m/z 499 and 446 as product ions. Both compounds were assigned as cannabisin C isomers. Compound 6 showed [M − H]− and [M + H]+ ions at m/z 593.2014 and 595.2011, respectively, supporting the formula C34H30N2O8 (calculated mass 594.2002). Its MS2 fragment ions in negative and positive modes and UV-DAD spectra (ESI Fig. S2†) were comparable to those previously reported for cannabisin A.12 Another unnamed lignanamide (compound 28) has been detected but not yet characterized. This compound showed the molecular ion [M − H]−at m/z 589.2366, with intensive MS2 fragment at m/z 426 (base peak), corresponding to a possible loss of the tyramine moiety (−163 amu). This suggests that this compound could be a tyramine-type hydroxycinnamoyl amide, characterized by the tyramine moiety.12
Some cannabinoids, namely, dihydrocannabinol, cannabidiol (CBD), cannabidiolic acid (CBDA), and cannabielsoic acid, which is a photo-oxidation product of CBD and CBDA,36 were detected and assigned for the compounds 29, 30, 31, and 33. These compounds have already been noticed in hempseed extracts.7,35,36
The compounds identified in defatted hemp seeds are extracted in a solvent-dependent manner. Table 4 shows the content of each identified compound in the studied extracts. Depending on the proportion used from each solvent in the mixture design, the relative content of the identified compounds varied more or less noticeably. In other words, some compounds were present in one solvent and absent in another, while others, even if they were present in all the solvents studied, their quantity extracted was different.
Relative content of the identified compounds in different solvent extracts of defatted hemp seedsa (mean ± SD).
| Compounds | Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | Run 6 | Run 7 | Run 8 | Run 9 | Run10 | Run11 | Run12 | Run13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenolic acids | |||||||||||||
| Benzoic acid | 11.46 ± 0.127 | 7.05 ± 0.750 | 2.02 ± 0.080 | 10.13 ± 0.040 | 9.27 ± 0.064 | 10.12 ± 0.150 | 4.19 ± 0.095 | 8.71 ± 0.006 | 4.31 ± 0.019 | 15.73 ± 0.041 | 10.12 ± 0.070 | 5.41 ± 0.015 | 10.01 ± 0.078 |
| p-Coumaric acid | 1.01 ± 0.120 | 0.95 ± 0.049 | 2.37 ± 0.370 | 1.25 ± 0.055 | 0.53 ± 0.015 | 0.52 ± 0.012 | Nd | 0.86 ± 0.009 | Tr | 1.41 ± 0.085 | 1.37 ± 0.035 | 0.016 ± 0.005 | 0.54 ± 0.011 |
| Ferulic acid | 3.12 ± 0.417 | 2.14 ± 0.068 | 8.57 ± 1.180 | 3.84 ± 0.029 | Nd | 1.90 ± 0.034 | Nd | 2.75 ± 0.053 | Nd | 3.87 ± 0.313 | 3.92 ± 0.180 | 1.49 ± 0.112 | 1.45 ± 0.018 |
| Sinapic acid | 1.16 ± 0.009 | 0.49 ± 0.003 | Tr | 0.68 ± 0.011 | 0.89 ± 0.011 | 1.02 ± 0.011 | 0.67 ± 0.004 | 0.92 ± 0.003 | 0.72 ± 0.002 | 1.19 ± 0.004 | 0.76 ± 0.002 | 0.83 ± 0.04 | 0.96 ± 0.015 |
| Total phenolic acids | 16.69 | 11.08 | 12.96 | 15.9 | 10.69 | 13.56 | 4.86 | 13.24 | 5.03 | 22.2 | 16.17 | 7.75 | 12.96 |
| Hydroxycinnamic acid amides b | |||||||||||||
| N-trans-caffeoyltyramine isomer | 8.16 ± 0.098 | 1.11 ± 0.101 | Nd | 3.24 ± 0.005 | 4.29 ± 0.361 | 6.58 ± 0.125 | 2.29 ± 0.012 | 5.84 ± 0.220 | 2.48 ± 0.036 | 12.23 ± 0.080 | 5.15 ± 0.152 | 2.34 ± 0.023 | 6.93 ± 0.161 |
| N-trans-caffeoyltyramine | 8.70 ± 0.277 | 1.25 ± 0.010 | Nd | 7.17 ± 0.265 | 14.07 ± 0.330 | 7.60 ± 0.283 | 2.08 ± 0.022 | 24.95 ± 0.083 | 2.80 ± 0.081 | 33.83 ± 0.054 | 9.16 ± 0.166 | 8.01 ± 0.039 | 10.87 ± 0.437 |
| N-trans-coumaroyltyramine | Nd | Nd | Nd | Nd | Nd | Nd | Nd | Nd | Nd | Tr | Nd | Nd | Nd |
| N-feruloyltyramine | 3.21 ± 0.009 | 1.31 ± 0.003 | 0.87 ± 0.038 | 2.63 ± 0.060 | 4.42 ± 0.086 | 2.91 ± 0.140 | 0.96 ± 0.012 | 5.48 ± 0.021 | 1.36 ± 0.021 | 7.65 ± 0.043 | 3.51 ± 0.026 | 3.02 ± 0.060 | 3.74 ± 0.074 |
| Tri-p-coumaroylspermidine | 3.39 ± 0.134 | Nd | Nd | 5.59 ± 0.124 | 8.63 ± 0.342 | 3.80 ± 0.061 | Nd | 10.08 ± 0.083 | 2.45 ± 0.047 | 11.02 ± 0.648 | 6.64 ± 0.030 | 7.47 ± 0.053 | 6.49 ± 0.114 |
| Total HCA | 23.46 | 3.66 | 0.87 | 18.63 | 31.41 | 20.88 | 5.33 | 46.36 | 9.09 | 64.72 | 24.46 | 20.85 | 28.03 |
| Lignanamides b | |||||||||||||
| Cannabisin A | 10.36 ± 0.307 | 2.41 ± 0.003 | Nd | 5.63 ± 0.025 | 10.89 ± 0.531 | 9.04 ± 0.197 | 3.94 ± 0.004 | 14.27 ± 0.045 | 3.55 ± 0.009 | 21.65 ± 0.004 | 9.63 ± 0.021 | 6.05 ± 0.375 | 10.92 ± 0.071 |
| Cannabisin B | 6.07 ± 0.415 | Nd | Nd | 2.46 ± 0.030 | 7.74 ± 0.286 | 4.69 ± 0.168 | 2.50 ± 0.023 | 11.18 ± 0.050 | 1.96 ± 0.010 | 18.63 ± 0.052 | 4.98 ± 0.138 | 4.25 ± 0.291 | 5.68 ± 0.017 |
| Cannabisin B isomer 1 | 0.93 ± 0.026 | 0.34 ± 0.296 | Nd | 0.71 ± 0.008 | 0.96 ± 0.026 | 0.75 ± 0.049 | 0.55 ± 0.007 | 1.26 ± 0.004 | 0.57 ± 0.005 | 1.75 ± 0.005 | 0.94 ± 0.007 | 0.78 ± 0.019 | 0.94 ± 0.048 |
| Cannabisin B isomer 2 | 1.51 ± 0.053 | 0.73 ± 0.022 | Nd | 1.25 ± 0.078 | 1.32 ± 0.052 | 1.00 ± 0.285 | 0.61 ± 0.002 | 1.97 ± 0.006 | 0.67 ± 0.005 | 2.62 ± 0.011 | 1.52 ± 0.001 | 0.99 ± 0.066 | 1.38 ± 0.008 |
| Demethylgrossamide | 1.35 ± 0.024 | Nd | Nd | 0.91 ± 0.025 | 1.01 ± 0.054 | 1.04 ± 0.017 | 0.57 ± 0.005 | 1.57 ± 0.009 | 0.59 ± 0.003 | 1.98 ± 0.018 | 1.16 ± 0.107 | 0.82 ± 0.066 | 1.30 ± 0.015 |
| Cannabisin C | 5.24 ± 0.032 | 0.89 ± 0.063 | Nd | 2.96 ± 0.017 | 5.02 ± 0.420 | 3.75 ± 0.335 | 1.29 ± 0.023 | 6.23 ± 0.078 | 1.50 ± 0.017 | 9.26 ± 0.283 | 4.81 ± 0.011 | 2.95 ± 0.105 | 4.01 ± 0.011 |
| Cannabisin C isomer | 1.68 ± 0.082 | 0.65 ± 0.094 | Nd | Nd | 0.88 ± 0.009 | 1.12 ± 0.141 | Nd | 1.40 ± 0.060 | 0.59 ± 0.008 | 1.78 ± 0.259 | 1.70 ± 0.003 | 0.93 ± 0.089 | 0.88 ± 0.011 |
| Cannabisin D | 5.43 ± 0.213 | 1.22 ± 0.042 | Nd | 4.67 ± 0.201 | 3.95 ± 0.242 | 4.11 ± 0.460 | 0.78 ± 0.006 | 5.58 ± 0.143 | 2.13 ± 0.032 | 6.57 ± 0.702 | 6.39 ± 0.032 | 4.56 ± 0.029 | 3.81 ± 0.557 |
| 3,3-Didemethylgrossamide | 1.20 ± 0.091 | Nd | Nd | Nd | 1.72 ± 0.143 | 1.22 ± 0.295 | Nd | 1.73 ± 0.100 | Nd | 3.28 ± 0.694 | Nd | Nd | 1.47 ± 0.398 |
| Cannabisin E | 1.85 ± 0.082 | Nd | Nd | 1.68 ± 0.027 | 1.98 ± 0.082 | 1.28 ± 0.016 | 0.53 ± 0.004 | 1.84 ± 0.259 | 0.69 ± 0.009 | 2.89 ± 0.115 | 1.90 ± 0.011 | 1.53 ± 0.038 | 1.45 ± 0.038 |
| Grossamide K | Nd | Nd | Nd | Nd | Nd | Nd | 0.39 ± 0.001 | Nd | Nd | Nd | Nd | Nd | Nd |
| Cannabisin M | 3.38 ± 0.162 | 0.90 ± 0.018 | Nd | 2.47 ± 0.133 | 4.36 ± 0.129 | 3.23 ± 0.107 | 1.29 ± 0.012 | 5.38 ± 0.017 | 1.71 ± 0.022 | 8.17 ± 0.007 | 3.47 ± 0.030 | 3.18 ± 0.031 | 4.05 ± 0.033 |
| 3,3′-demethyl-heliotropamide | 1.30 ± 0.104 | 0.61 ± 0.026 | Nd | 0.86 ± 0.124 | 1.33 ± 0.174 | 1.30 ± 0.076 | 0.67 ± 0.009 | 1.66 ± 0.009 | 0.68 ± 0.092 | 2.47 ± 0.007 | 1.12 ± 0.024 | 1.01 ± 0.039 | 1.24 ± 0.227 |
| Unnamed condensed trilignanamide | 1.95 ± 0.113 | 0.62 ± 0.026 | Nd | 1.09 ± 0.102 | 2.12 ± 0.046 | 2.02 ± 0.048 | 0.77 ± 0.018 | 2.71 ± 0.004 | 0.95 ± 0.077 | 3.61 ± 0.019 | 1.69 ± 0.022 | 1.53 ± 0.031 | 2.03 ± 0.232 |
| Cannabisin Q | 1.34 ± 0.010 | Nd | Nd | 0.86 ± 0.035 | 1.50 ± 0.045 | 1.10 ± 0.021 | 0.70 ± 0.007 | 1.83 ± 0.096 | 0.76 ± 0.001 | 2.68 ± 0.027 | 1.27 ± 0.012 | 1.18 ± 0.062 | 1.34 ± 0.032 |
| Cannabisin F | 2.74 ± 0.048 | 0.89 ± 0.042 | Nd | 1.28 ± 0.082 | 1.80 ± 0.101 | 2.04 ± 0.014 | 0.96 ± 0.004 | 2.26 ± 0.301 | 1.18 ± 0.075 | 3.42 ± 0.007 | 2.11 ± 0.010 | 1.35 ± 0.179 | 2.08 ± 0.033 |
| Isocannabisin N | 1.23 ± 0.015 | Nd | Nd | 0.71 ± 0.017 | 1.21 ± 0.032 | 0.80 ± 0.008 | Nd | 1.47 ± 0.056 | 0.57 ± 0.003 | 1.99 ± 0.117 | 1.15 ± 0.011 | 1.00 ± 0.029 | 0.96 ± 0.006 |
| Grossamide | 3.68 ± 0.071 | 0.87 ± 0.015 | Nd | 2.21 ± 0.159 | 4.79 ± 0.089 | 2.91 ± 0.080 | 0.96 ± 0.011 | 6.04 ± 0.021 | 1.49 ± 0.024 | 8.94 ± 0.023 | 3.30 ± 0.024 | 3.63 ± 0.022 | 3.98 ± 0.070 |
| Cannabisin O | 2.28 ± 0.207 | Nd | Nd | 0.61 ± 0.060 | 3.08 ± 0.025 | 1.12 ± 0.018 | Nd | 3.67 ± 0.040 | 0.68 ± 0.005 | 4.53 ± 0.029 | 1.84 ± 0.086 | 2.79 ± 0.038 | 1.57 ± 0.034 |
| Unnamed lignanamide | 3.57 ± 0.227 | Nd | Nd | 0.99 ± 0.022 | 3.62 ± 0.014 | 1.80 ± 0.013 | Nd | 4.70 ± 0.075 | 0.92 ± 0.009 | 5.58 ± 0.011 | 3.07 ± 0.024 | 3.33 ± 0.055 | 2.22 ± 0.038 |
| Total lignanamides | 57.09 | 10.13 | — | 31.34 | 59.30 | 44.31 | 16.54 | 76.72 | 21.18 | 111.81 | 52.05 | 41.87 | 51.32 |
| Total phenylpropanoids | 80.55 | 13.79 | 0.87 | 49.96 | 90.71 | 65.19 | 21.87 | 123.09 | 30.27 | 176.53 | 76.51 | 62.72 | 79.35 |
| Cannabinoids b | |||||||||||||
| Dihydrocannabinol | 0.67 ± 0.031 | 0.51 ± 0.003 | Nd | 0.62 ± 0.002 | 0.59 ± 0.001 | 0.65 ± 0.002 | Nd | 0.60 ± 0.007 | 0.52 ± 0.003 | 0.64 ± 0.002 | 0.65 ± 0.002 | 0.57 ± 0.017 | 0.62 ± 0.006 |
| Cannabidiol (CBD) | 0.56 ± 0.005 | Nd | Nd | 0.50 ± 0.002 | 0.56 ± 0.002 | 0.57 ± 0.007 | Nd | 0.54 ± 0.003 | 0.53 ± 0.012 | 0.63 ± 0.040 | 0.52 ± 0.004 | 0.53 ± 0.008 | 0.61 ± 0.006 |
| Cannabielsoic acid | 1.67 ± 0.028 | 0.60 ± 0.002 | Nd | 1.04 ± 0.016 | 1.53 ± 0.016 | 1.86 ± 0.022 | 0.98 ± 0.020 | 1.58 ± 0.033 | 1.16 ± 0.012 | 1.85 ± 0.055 | 1.08 ± 0.002 | 1.37 ± 0.024 | 1.81 ± 0.024 |
| Cannabidiolic acid (CBDA) | 1.12 ± 0.007 | Nd | Nd | 0.57 ± 0.004 | 1.04 ± 0.021 | 1.33 ± 0.037 | 0.97 ± 0.005 | 1.05 ± 0.016 | 0.90 ± 0.009 | 0.98 ± 0.004 | 0.61 ± 0.003 | 0.88 ± 0.024 | 1.16 ± 0.052 |
| Total cannabinoids | 4.03 | 1.12 | — | 2.73 | 3.72 | 4.40 | 1.95 | 3.77 | 3.10 | 4.10 | 2.86 | 3.34 | 4.21 |
Nd, not detected; Tr, traces.
Hydroxycinnamic acid amides, lignanamides, and cannabinoids are expressed in mg caffeoyltyramine equivalent per g of extract (CTE per g extract).
In general, the studied defatted hemp seeds are especially rich in hydroxycinnamic acid amides and lignanamides (especially cannabisins) which are present in greater quantities than phenolic acids (Table 4). The most abundant HCA was N-trans-caffeoyltyramine, while the most representative lignanamides were cannabisin A and cannabisin B. However, cannabinoids were detected in small quantities. These results are in perfect agreement with several studies which have determined the particular abundance of caffeoyltyramine and cannabisins in hempseeds.12,16 Furthermore, it is noteworthy that the three identified phenolic acids (compounds 2, 3, and 32) are cinnamic acid derivatives. Benzoic acid was present in all solvent extracts, while no phenolic hydroxybenzoic acid was detected, contrary to several studies which proved their presence in hemp seeds such as 4-hydroxybenzoic acid, salicylic acid, vanillic acid, and protocatechuic acid.10,14,35
Aside from these identified phenolic classes, Rea Martinez et al.35 has also detected the presence of some flavonoids in defatted hempseeds, such as quercetin, rutin, vitexin, isovitexin, genistin, naringenin, apigenin, and diosmetin. In the same line, Nigro and his collaborators have reported quercetin and Kaempferol derivatives in hempseed extracts.12
The HPLC-DAD analysis of the thirteen extracts showed that the binary mixture corresponding to the run 10 (2/3 acetone 1/3 water) as well as the ternary mixture corresponding to the run 8 (1/3 water 1/3 acetone 1/3 methanol) have the best capacity to extract phenolic compounds from defatted hempseeds which are in agreement with the Folin-Ciocalteu method. For instance, the run 10 showed that 33.83, 21.65, and 18.63 mg CTE per g extract were recorded for N-trans-caffeoyltyramine, cannabisin A, and cannabisin B respectively. The run 8 recorded 24.95, 14.27, and 11.18 mg CTE per g extract for the same compounds, respectively.
It is readily noted that some phenolic acids, such as p-coumaric and ferulic, were more abundant in pure water extract (2.37 mg g−1 of p-coumaric acid and 8.57 mg g−1 of ferulic acid). In contrast, they were moderately present in the mixtures of aqueous organic solvents and absent in pure acetone (Table 4). A similar result was obtained in a previous work investigating the solvent effect on phenols extraction from chia seeds.21 Conversely, phenylpropionamides were almost absent in the water extract and were extracted in large quantities in the aqueous acetone mixture (2/3 acetone 1/3 water), reaching a value of 176.53 mg CTE per g extract.
It is noteworthy that methanol was reported in several studies to be effective for extracting low molecular weight compounds, while aqueous acetone is effective for extracting higher molecular weight compounds.17,30 This agrees with our results since phenylpropionamides, the abundant compounds in hemp seeds, have a high molecular weight.
Using the Folin assay, the 100% water extract (run 3) and binary mixtures with 66% water extracts (runs 2 and 11) recorded considerable values of phenols compared to binary mixtures with 33% water extracts (run 1 vs. run 2 and run 11 vs. run 10). However, HPLC-DAD analysis showed that 100% water extract is almost ineffective for extracting phenols, particularly phenylpropionamides, and 66% of water mixtures extracted small quantities. This could be explained by the fact that water, when present in large proportions, may extract other non-phenolic interfering compounds with reducing behavior, such as sugars, aromatic amines, and organic acids.37
On the other hand, some studies have pointed out that acetone extraction has the disadvantage of extracting chlorophyll which may interfere with phenols and bias the Folin phenol assay result.6,38 However, our HPLC-DAD results comparing the extraction of the compounds assert that aqueous acetone (2/3 acetone 1/3 water) is the ideal mixture to extract phenols from the hemp seed matrix effectively. This solvent mixture allows the extraction of 64.72 mg CTE per g extract of hydroxycinnamic acid amides, 111.81 mg CTE per g extract of lignanamides, and 4.10 mg CTE per g extract of cannabinoids.
Phenylpropionamides arouse much interest due to their biological activities. Several review papers peculiarly tackled their considerable pharmacological potential.34,39 Indeed, N-trans-caffeoyltyramine and cannabisin B were isolated from hemp seeds and revealed predominant radical scavenging activity and protective effect against in vitro oxidation of human low-density lipoprotein.6 Hempseed lignanamides have also shown effective antioxidant and acetylcholinesterase inhibitory activities13 and anti-neuroinflammatory activity.11 In addition, hemp seed lignanamides rich fraction demonstrated an anticancer effect against U-87 cancer cell proliferation.12
3.4. Multi-response optimization and model validation
Optimization of all responses (TPC, TAC, DPPH, ABTS, FRAP, CUPRAC) simultaneously was performed based on maximizing the overall desirability function. The result showed that the binary acetone-water mixture in equal proportions (50–50) is the optimal combination to extract more phenols and have potent antioxidant activity. A repeatable study was conducted using these optimal conditions to assess the model's predictive capacity. The observed values for all responses were close to the predicted values (R2 > 0.99) and belonged to the confidence interval (Table 5). Therefore, the model has a satisfactory accuracy and can be validated.
Observed and predicted values for total phenolic content (TPC) and antioxidant tests (DPPH, TAC, ABTS, FRAP, and CUPRAC) at optimized extraction conditions (acetone–water 50–50%).
| Responses | Observed values (Mean ± SD) | Predicted values | Confidence intervals at 95% |
|---|---|---|---|
| TPC (mg GAE per g extract) | 53.65 ± 2.98 | 53.27 | [47.15–59.38] |
| TAC (mg TE per g extract) | 265.53 ± 8.69 | 258.96 | [229.08–288.84] |
| DPPH (mg TE per g extract) | 36.25 ± 1.48 | 34.68 | [30.91–38.45] |
| ABTS (mg TE per g extract) | 119.03 ± 1.14 | 131.41 | [115.18–147.63] |
| FRAP (mg TE per g extract) | 69.46 ± 3.68 | 63.93 | [55.59–72.27] |
| CUPRAC (mg TE per g extract) | 68.91 ± 3.29 | 78.45 | [65.16–91.73] |
This result meets several studies that tested the solvent effect on extracting phenolic compounds from hempseeds6,38 or other different plant matrices.40,41 These studies found that the binary combination between acetone and water is the most adequate but not necessarily with 50–50% proportions. However, other researchers have proven the effectiveness of other solvent combinations, such as the ternary mixture of methanol–acetone–water 7 : 7 : 6.18 It is worth considering that these reported studies did not optimize the extraction using a mixture design as in our case, but they only compared the solvent effect at punctual solvents proportions. Only one study applying a mixture design approach to hemp was found in the literature.19 This author investigated the effect of methanol, ethanol, and water on the extraction of phenolic compounds from whole plant residues that remain after the production of cannabis resin and found that the optimal combination is 30% water-70% ethanol. Similarly to our result, 50% acetone −50% water was the optimal solvent for phenolic antioxidants recovery from white and black mustard grains20 and Jambolan fruits42 applying mixture design analysis.
In our study, optimization of the solvent mixture allows extracting an amount of total phenolic content reaching 53.65 mg GAE per g of extract, which corresponds to 2.95 mg GAE per g of defatted seeds. This result is quite similar to that of Rea Martinez (56.7 mg GAE per g extract) obtained using 75% ethanol with subsequent fractioning with ethyl acetate35 and higher than that obtained using 80% ethanol (2.21 mg GAE per g seeds) from Futura cultivar.36
The common solvent in several other studies that determined hempseed cake's phenolic content was 80% methanol. In a comparative study of raw and extruded hempseed cakes from the CRS1 variety, the TPC achieved varies from 0.385 to 0.906 mg GAE per g sample.16 In the same context (80% methanolic extracts), Moccia et al. found a value of 1.71 mg GAE per g of hempseed flour,7 while Siano et al. recorded 0.744 mg GAE per g of hempseed flour,8 both from Fedora cultivar. All these reported TPC values were lower than our result, affirming optimization processes' utility and importance. However, another study also using 80% methanol reported higher TPC values than ours, reaching 381.8 to 779.8 mg GAE per 100 g of hempseed cake of seven industrial hemp cultivars.14 These differences in phenols' content might be due to the cultivar, the environmental conditions, the extraction method, or the solvent used.
Regarding antioxidant activities, the results of the optimal solvent mixture were 265.53 mg TE g−1 extract, 36.25 mg TE g−1 extract, 119.03 mg TE g−1 extract, 69.46 mg TE g−1 extract, and 68.91 mg TE g−1 extract, respectively, for the TAC, DPPH, ABTS, FRAP, and CUPRAC tests. It was reported in a previous work a value of 4.7 mg TE g−1 of extract of hempseed flour for the DPPH assay,8 which is lower than our results. Irakli and his research group recorded a value of 458.8–1066.3 mg TE per 100 g and 338.4–806.8 mg TE per 100 g of hempseed flour, respectively, for ABTS and FRAP tests depending on the genotype and the growing year.14 Various other authors investigated the antioxidant power of hempseeds or defatted hempseeds,7,36 and they used different ways to express their antioxidant results, such as the percentage of inhibition of free radicals or using different standards (ascorbic acid or quercetin). Therefore, no comparison can be made between them.
It is worth mentioning that extracts showing important antioxidant activities have a particular abundance of N-trans-caffeoyltyramine, cannabisin A, and cannabisin B, suggesting the antioxidant potential of these compounds. Indeed, some previous studies have isolated these phenylpropionamides from hemp seeds and demonstrated their antioxidant capacity.6,13
Overall, this important antioxidant activity suggests that defatted hempseeds, a by-product of the hempseed oil industry, can be sustainably recycled and potentially used in pharmacological and cosmetic applications.
4. Conclusion
The extraction of phenolic compounds and antioxidants from defatted hemp seeds was optimized using a mixture design of three solvents (water, methanol, and acetone). The optimal combination of solvents is 50% aqueous acetone which achieved maximum phenolic compounds and better antioxidant activity in all the tests performed (DPPH, ABTS, FRAP, TAC, CUPRAC). Also, HPLC-DAD and ESI-MS2 analyses revealed the abundance of hydroxycinnamic acid amides and lignanamides in defatted hemp seed extracts, thus demonstrating the effect of solvent mixtures on each identified phenolic compound.
The results obtained in this study could support future research on hemp seeds and their valorization since this is the first work that reported optimizing the extraction of hemp seed bioactive compounds. Besides, it encourages using defatted hemp seeds as a source of antioxidants with added value for pharmaceutical and cosmetic applications.
Author contributions
Chaymae Benkirane: experiment, formal analysis, software, writing-original draft. Abdessamad Ben Moumen: formal analysis, validation. Marie-Laure Fauconnier: project administration, visualization, writing – review & editing. Kamal Belhaj: investigation, data curation. Malika Abid: investigation, validation, supervision. Hana Serghini Caid: visualization, writing – review & editing. Ahmed Elamrani: funding acquisition, project administration, visualization. Farid Mansouri: conceptualization, methodology, writing-original draft, software, supervision.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This research was supported by: the Moroccan Ministry of Higher Education, Scientific Research and innovation, and the National Agency of Medicinal and Aromatic Plants through the VPMA2/ref 2020/1 project; the Wallonia Brussels International (WBI).
Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra04081f
References
- Isidore E. Karim H. Ioannou I. Antioxidants. 2021;10:942. doi: 10.3390/antiox10060942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W. Zhang P. Ying D. Fang Z. Compr. Rev. Food Sci. Food Saf. 2020;19:282–308. doi: 10.1111/1541-4337.12517. [DOI] [PubMed] [Google Scholar]
- Taaifi Y. Benmoumen A. Belhaj K. Aazza S. Abid M. Azeroual E. Elamrani A. Mansouri F. Serghini-Caid H. Int. J. Food Sci. Technol. 2021;56:5931–5947. [Google Scholar]
- Stambouli H. El Bouri A. Bouayoun T. Bellimam A. Ann. Toxicol. Anal. 2006;18:119–125. [Google Scholar]
- Jiang Q. Christen S. Shigenaga M. K. Ames B. N. Am. J. Clin. Nutr. 2001;74:714–722. doi: 10.1093/ajcn/74.6.714. [DOI] [PubMed] [Google Scholar]
- Chen T. He J. Zhang J. Li X. Zhang H. Hao J. Li L. Food Chem. 2012;134:1030–1037. doi: 10.1016/j.foodchem.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Moccia S. Siano F. Russo G. L. Volpe M. G. La Cara F. Pacifico S. Piccolella S. Picariello G. Int. J. Food Sci. Nutr. 2020;71:410–423. doi: 10.1080/09637486.2019.1666804. [DOI] [PubMed] [Google Scholar]
- Siano F. Moccia S. Picariello G. Russo G. L. Sorrentino G. Di Stasio M. La Cara F. Volpe M. G. Molecules. 2018;24:1–13. doi: 10.3390/molecules24010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twaij B. M. Hasan N. Int. J. Plant Biol. 2022;13:4–14. [Google Scholar]
- Leonard W. Zhang P. Ying D. Xiong Y. Fang Z. Food Chem. 2021;346:128606. doi: 10.1016/j.foodchem.2020.128606. [DOI] [PubMed] [Google Scholar]
- Zhou Y. Wang S. Ji J. Lou H. Fan P. ACS Omega. 2018;3:15988–15995. doi: 10.1021/acsomega.8b02250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro E. Crescente G. Formato M. Pecoraro M. T. Mallardo M. Piccolella S. Daniele A. Pacifico S. Molecules. 2020;25:1–25. doi: 10.3390/molecules25051049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X. Tang J. Dos Santos Passos C. Nurisso A. Simoes-Pires C. A. Ji M. Lou H. Fan P. J. Agric. Food Chem. 2015;63:10611–10619. doi: 10.1021/acs.jafc.5b05282. [DOI] [PubMed] [Google Scholar]
- Irakli M. Tsaliki E. Kalivas A. Kleisiaris F. Sarrou E. Cook C. M. Antioxidants. 2019;8:491. doi: 10.3390/antiox8100491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alara O. R. Abdurahman N. H. Ukaegbu C. I. Curr. Res. Food Sci. 2021;4:200–214. doi: 10.1016/j.crfs.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W. Zhang P. Ying D. Xiong Y. Fang Z. J. Food Sci. 2021;86:3159–3175. doi: 10.1111/1750-3841.15813. [DOI] [PubMed] [Google Scholar]
- Brglez Mojzer E. Knez Hrnčič M. Škerget M. Knez Ž. Bren U. Molecules. 2016;21:901. doi: 10.3390/molecules21070901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh S. S. El-Din Bekhit A. Birch J. Antioxidants. 2014;3:67–80. doi: 10.3390/antiox3010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aazza S. Journal of Chemistry. 2021;2021:9738656. [Google Scholar]
- Boscariol Rasera G. Hilkner M. H. de Alencar S. M. de Castro R. J. S. Ind. Crops Prod. 2019;135:294–300. [Google Scholar]
- Alcântara M. A. de Lima Brito Polari I. de Albuquerque Meireles B. R. L. de Lima A. E. A. da Silva Junior J. C. de Andrade Vieira É. dos Santos N. A. de Magalhães Cordeiro A. M. T. Food Chem. 2019;275:489–496. doi: 10.1016/j.foodchem.2018.09.133. [DOI] [PubMed] [Google Scholar]
- Grochowski D. M. Uysal S. Aktumsek A. Granica S. Zengin G. Ceylan R. Locatelli M. Tomczyk M. Phytochem. Lett. 2017;20:365–372. [Google Scholar]
- Kadriye I. Demirata B. Apak R. Food Analytical Methods. 2012;5:1150–1158. [Google Scholar]
- Ben Moumen A. Mansouri F. Richard G. Abid M. Fauconnier M. L. Sindic M. El Amrani A. Serghini Caid H. Int. J. Food Sci. Technol. 2015;50:804–810. [Google Scholar]
- Mansouri F. Ben Moumen A. Belhaj K. Richard G. Fauconnier M. L. Sindic M. Caid H. S. Elamrani A. Emirates Journal of Food and Agriculture. 2018;30:549–562. [Google Scholar]
- Pattnaik F. Nanda S. Mohanty S. Dalai A. K. Kumar V. Ponnusamy S. K. Naik S. Chemosphere. 2022;289:133012. doi: 10.1016/j.chemosphere.2021.133012. [DOI] [PubMed] [Google Scholar]
- Li A. N. Li S. Zhang Y. J. Xu X. R. Chen Y. M. Bin Li H. Nutrients. 2014;6:6020–6047. doi: 10.3390/nu6126020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeel A. Jan S. A. Ullah I. Shinwari Z. K. Xu M. PeerJ. 2019:1–19. doi: 10.7717/peerj.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav D. L. Karthick N. K. Kannan P. P. Shanmugam R. Elangovan A. Arivazhagan G. J. Mol. Struct. 2017;1130:497–502. [Google Scholar]
- Cabajar A. A. Siacor F. D. C. Lim K. J. A. Lobarbio C. F. Y. Taboada E. B. Applied Science and Engineering Progress. 2021;14:156–164. [Google Scholar]
- Serna-Vázquez J. Ahmad M. Z. Boczkaj G. Castro-Muñoz R. Molecules. 2021;26:5037. doi: 10.3390/molecules26165037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisoschi A. M. Pop A. Eur. J. Med. Chem. 2015;97:55–74. doi: 10.1016/j.ejmech.2015.04.040. [DOI] [PubMed] [Google Scholar]
- Chen J. Yang J. Ma L. Li J. Shahzad N. Kim C. K. Sci. Rep. 2020;10:2611. doi: 10.1038/s41598-020-59451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumani M. Duval R. E. Ropars A. Risler A. Robin C. Larbat R. Biomed. Pharmacother. 2020;131:110762. doi: 10.1016/j.biopha.2020.110762. [DOI] [PubMed] [Google Scholar]
- Rea Martinez J. Montserrat-De La Paz S. De La Puerta R. García-Giménez M. D. Fernández-Arche M. Á. Food Funct. 2020;11:4057–4066. doi: 10.1039/d0fo00066c. [DOI] [PubMed] [Google Scholar]
- Frassinetti S. Moccia E. Caltavuturo L. Gabriele M. Longo V. Bellani L. Giorgi G. Giorgetti L. Food Chem. 2018;262:56–66. doi: 10.1016/j.foodchem.2018.04.078. [DOI] [PubMed] [Google Scholar]
- Rover M. R. Brown R. C. J. Anal. Appl. Pyrolysis. 2013;104:366–371. [Google Scholar]
- Liang J. Zago E. Nandasiri R. Khattab R. Eskin N. A. M. J. Am. Oil Chem. Soc. 2018;95:1319–1327. [Google Scholar]
- Pollastro F. Minassi A. Fresu L. G. Curr. Med. Chem. 2018;25:1160–1185. doi: 10.2174/0929867324666170810164636. [DOI] [PubMed] [Google Scholar]
- Nasr A. Zhou X. Liu T. Yang J. Zhu G.-P. Turk. J. Biochem. 2019;44:231–239. [Google Scholar]
- Van Ngo T. Scarlett C. J. Bowyer M. C. Ngo P. D. Van Vuong Q. J. Food Qual. 2017;2017:9305047. [Google Scholar]
- De Morais Sousa M. De Lima A. Araujo B. Q. Dos Santos Rocha M. dos Santos Monção Filho E. De Sousa R. P. Das Graças Lopes Citó A. M. Sattler J. A. G. De Almeida-Muradian L. B. Do Nascimento Nogueira N. Food Analytical Methods. 2021;15:34–45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.