Abstract
DNA origami has created complex structures of various spatial dimensions. However, their versatility in terms of function is limited due to the lower number of the intrinsic building blocks, i.e. nucleotides, compared with the number of amino acids. Therefore, protein origami has been proposed and demonstrated to precisely fabricate artificial functional nanostructures. Despite their hierarchical folded structures, chain-like peptides and DNA share obvious similarities in both structures and properties, especially in terms of chain hybridization; therefore, replacing DNA with peptides to create bioactivities not only has high theoretical feasibility but also provides a new bottom-up synthetic strategy. However, designing functionalities with tens to hundreds of peptide chains using the similar principle of DNA origami has not been reported, although the origami strategy holds great potential to generate more complex bioactivities. In this perspective review, we have reviewed the recent progress in and highlighted the advantages of peptide assembly and origami on the orientation of artificially created bioactivities. With the great potential of peptide origami, we appeal to develop user-friendly softwares in combination with artificial intelligence.
Chain-like peptides and DNA share obvious similarities in terms of chain hybridization; thus, replacing DNA with peptides to create bioactivities not only has high theoretical feasibility but also provides a new bottom-up synthetic strategy.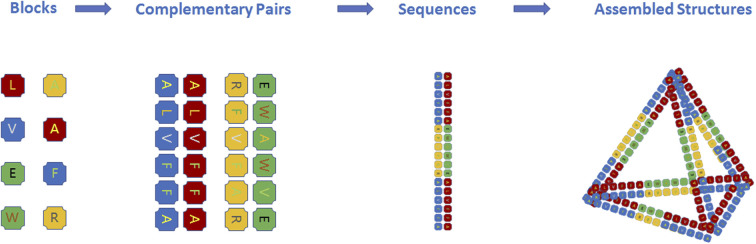
In cells, peptides form versatile nanostructures with different bioactivities through self-assembly and co-assembly. Proteins such as collagen,1 actin2 and the peptides/proteins that are associated with amyloid diseases3 can self-assemble into fibrous structures. Helicase4 and nuclease5 are naturally ring-shaped structures. TMV capsid protein,6 α-hemolysin,7 and the Bacillus anthracis protective antigen8 are tubular structures. Citrate synthase,9 lysyl oxidase,10 RecR proteins,11 ribonucleotide reductase I,12 and hydrolase CS2 13 can self-assemble into the lariat structures. Transferrin,14 vault protein,15 clathrin,16 chaperonin,17 and the capsids of viruses18 can form cage structures. These ordered nanostructures formed by the peptide assembly play essential roles in life.
One of the ultimate goals of synthetic biology is to create artificial cells, and assembly is an important approach. DNA origami has been used to create versatile nanostructures since its origin19 and proof-of-concept reports20 published by Seeman and coworkers. The nanostructures from DNA origami can act as scaffolds to synthesize inorganic particles or units to conduct multi-component and multi-functional assembly. Recently, de novo designed proteins have been investigated in various fields such as catalysis,21 energy (i.e., light harvesting22), medicine (i.e., vaccine R&D,23 gene and drug delivery24), and environmental science (i.e., water treatment25). Related review papers have been published by Guang et al.26 and Yan et al.27 However, the current artificial protein design is normally folded from one to a few long single-peptide chains (as the subunits of tertiary/quaternary structures), and designing complex artificial nanostructures with tens to hundreds of peptide chains using the similar principle of DNA origami (Fig. 1) has not been reported yet.
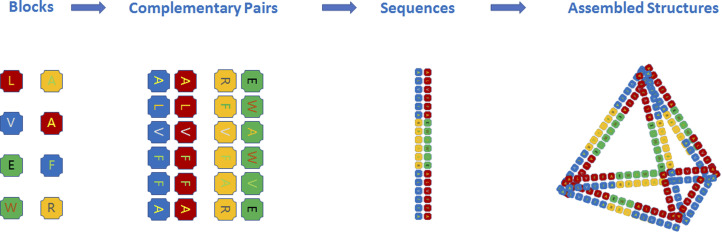
Fig. 1. Peptide assembly with the principle of DNA origami. All amino acids are blocks for peptide assembly, and short peptide sequences can be designed as complementary pairs similar to base pairs (A T, G C) for peptide origami. Peptide sequences containing such complementary pairs can further assemble into nanostructures.
Origami: a special type of assembly
The concept of protein origami was proposed by Jerala et al.28 Similar to DNA origami, protein origami employs the complementary pairing capability of coiled-coil secondary structures; for example, the first artificial tetrahedron was fabricated using six pairs of coiled-coil sequences, which were all set in a single chain. Significantly, unlike DNA origami, the current so-called protein origami has not employed stapling sequences, which are very short sequences normally over ten in number used in DNA origami. In this regard, current protein origami is more like protein folding from an artificial peptide chain using the complementary pairing capability of coiled-coil sequences. Single peptide chains, termed as the primary sequence of proteins, not only contain all the information for intramolecular folding and intermolecular self-assembly29 but also determine the final chemical properties and bioactivities of proteins. Generally, peptides are defined as either protein fragments or the amino acid sequence of proteins before folding, while proteins are the 3-D structures after the intramolecular folding and intermolecular self-assembly of peptides. In order to include most current techniques to create desirable bioactivities, especially for peptide/protein origami, self-assembly and co-assembly with more than a single peptide chain, hereafter we suggest calling them a peptide assembly (Fig. 1).
Despite advances in protein design and engineering, precisely fabricating multiprotein architectures with both structural and functional sophistication is still challenging. Considering the combination number of the basic building blocks, the huge difference between the twenty kinds of amino acids and four types of bases undoubtedly endows the peptide assembly with a great advantage. Similar to antibodies produced by B cells, the enormous number of peptide sequence or amino acid combinations can create numerous bioactivities. Because two amino acid residues cannot form a robust conjugation like that of base pairs, the origami-like strategy of peptide assembly is hard to design due to the orthogonal issue. In addition, both synergistic effects and long-distance interactions among amino acids make it more difficult than DNA origami.
The success of DNA origami is mainly ascribed to the two A T and G C pairings. However, the limitation of both the permutation and combination number and chemical activities of these DNA bases significantly restrict their potential to produce enough bioactivities, which might be the reason for life to use DNA as a conserved genetic material and use peptides to create bio-functionalities. Therefore, to employ the origami strategy for peptide assembly, the most important requirement is to realize orthogonal complementary pairing between short peptide motifs. Here, we introduce a new idea of using short peptide sequences or motifs instead of single amino acids to form conjugations with both specificity and affinity. Fortunately, leucine zipper sequences or so-called coiled-coils (a short α-helix peptide sequence consisting of ∼35 amino acids) can perfectly realize this idea,30–32 which has been successfully demonstrated by Jerala et al.33,34 If one could consider these special peptide sequences similar to DNA base pairs, the principle of origami can naturally extend from DNA to peptides. Regarding the regulation capabilities of both specificity and affinity, peptide motifs show more advantages than DNA sequences due to the various interactions among amino acids. Moreover, such orthogonal motifs could be derived from some natural proteins, i.e., prions,35 α-synuclein,36,37 insulin,38 A-β,39 the core sequence of natural silk proteins40 and the artificially designed ionic-complementary peptides,41 which could be a giant bank for developing orthogonal pairs for peptide assembly.
The increasing protein data bank can also provide a resource pool for orthogonal pairs. For example, motifs like β-sheets and coiled-coils coexist in proteins, and normally form natural orthogonal pairs. Simply with coiled-coils, scientists have already designed complex nanostructures such as cages for diverse applications in biomedicine.42 Secondly, short peptides with less than 100 amino acid residues can be easily synthesized in the solid phase, while longer peptides can be obtained through genetic engineering, i.e., fused protein expression. Thirdly, the rapid development of molecular dynamics simulation makes the theoretical prediction of peptide spatial structures increasingly accurate.43 There are already several powerful software such as Gromacs, NAMD, RoseTTAfold and α-Fold that not only allow scientists to understand the dynamic process of peptide assembly but also help perfect the design of peptide sequences. Prof. Susan Lindquist from MIT wrote, “Humans have been domesticating plants and animals about 10 000 years ago, and now it's time to domesticate molecules”. Now it is time to develop peptide origami.
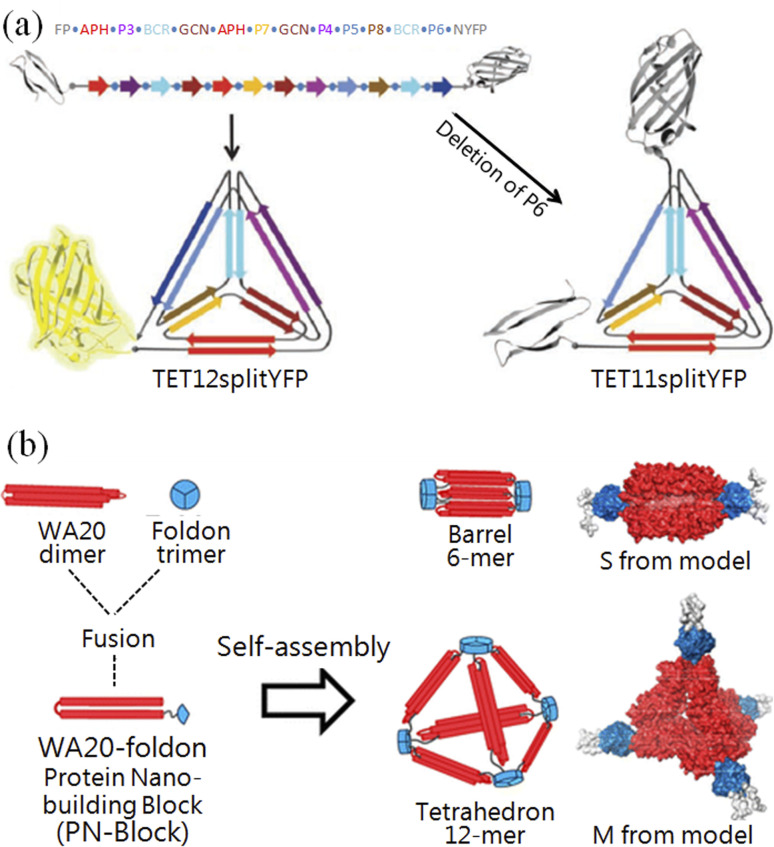
Woolfson and co-workers constructed a single-layered protein grid using two coiled-coil orthogonal complementary pairs, which further self-assembled into hollow balls with a diameter of 100 nm.44 Jerala and co-workers designed an artificial peptide containing six coiled-coil orthogonal complementary pairs (Fig. 2a), which was produced by the genetic engineering method and can fold into a tetrahedral structure with a 10 nm length.45 Ryadnov and co-workers constructed the smallest artificial viruses using coiled-coil orthogonal complementary pairs.46 Baker and co-workers successfully constructed 2-D47 and 3-D nanostructures48 using a theoretical calculation that has been published online.49,50 Recently, they also precisely controlled the β-sheet curvature of proteins at the nanoscale.51 Ryoichi et al. constructed versatile nanostructures with one dimer of WA20 and one trimer of foldon conjugated peptides52 (Fig. 2b). These results indicate that peptide origami is already comparable to DNA origami, and has ushered a new era for creating novel nanostructures and bioactivities.
Fig. 2. Creating pre-designed nanostructures with peptide origami. (a) A tetrahedron was folded by a single peptide chain, which was fused with a fluorescent protein.45 (b) Constructing nanostructures with genetically fused peptide WA20–foldons.52.
Superiority of peptide origami
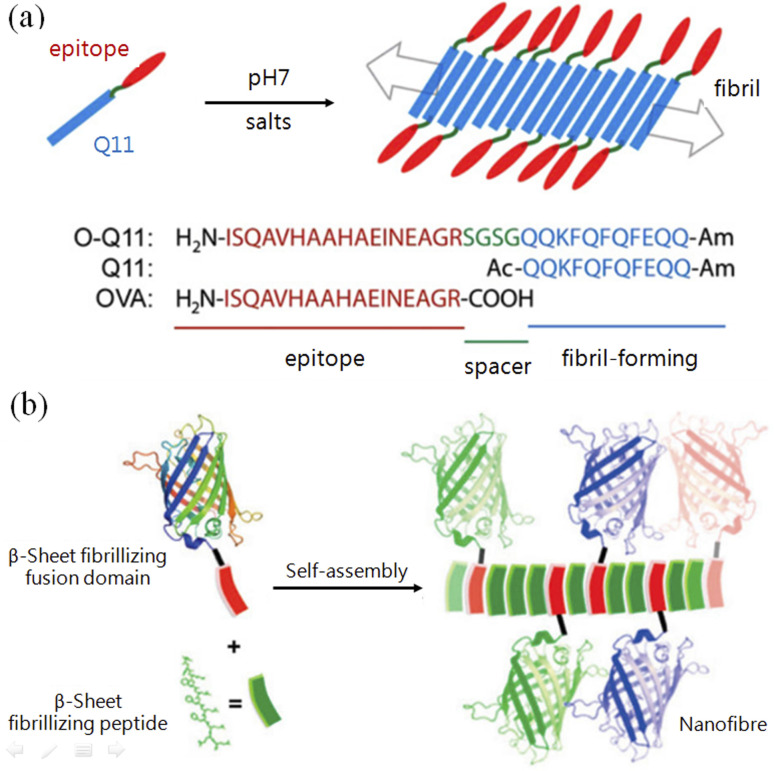
In living systems, efficient functions are normally achieved by nanostructures that are subtly arranged with complex molecules, such as molecular machines (2016 Nobel Prize in Chemistry) and light-harvesting complexes, both of which were listed as the top ten research hotspots in biology. In contrast, DNA origami involves covalently conjugating functional groups to complementary DNA sequences in advance to realize pre-designed functionalities.53 There are a few approaches to realize site-specific conjugation to proteins, such as the unnatural amino acid-based conjugation invented by Schultz and co-workers,54 but these are highly skilled. Due to the orthogonality of the complementary pairs, coiled-coil motifs could produce more complex nanostructures than β-tails. The development of new vaccines requires the following effects: long-term, highly effective immune-stimulating enhancement with minimal or complete elimination of adjuvant additions. With both protein fusion and peptide assembly/origami, high-efficiency polyvalent vaccines or combined vaccines of multiple diseases can be achieved by displaying different types and quantities of epidemic antigens on the surface of predesigned nanostructures such as nanofibers55 (Fig. 3a). In this way, catalytic performance can also be enhanced by clustering proteinases or antigens on the surface of fibrils.56 With the same principle, Collier and co-workers constructed fibrils using β-tail-fused fluorescent proteins, which not only retain the structure's fluorescence but can also emit different fluorescence by mixing fluorescent proteins in different ratios56 (Fig. 3b).
Fig. 3. Application demonstrations of peptide origami with β-tails. (a) Immune stimulating action was enhanced by conjugating antigen epitopes with fibril-forming β-tail peptides.55 (b) Replacing the antigen epitope with fluorescent proteins (of different colors) to construct nanofibrils with β-tail-based peptide origami.56.
Compared with DNA origami, the central dogma endows more advantages to peptide origami, i.e., bioactivities created by peptide origami hold the potential to be replicated in vivo. Although DNA self-replicates, DNA origami requires temperature ramping to finish the annealing process, which is hard to achieve in vivo. Peptides can be excreted from cells after being produced from the protein synthesizing machinery, which inspires us to yield bioactivities en masse using cells. Yeates and co-workers reported a fusion protein including two orthogonal complementary pair subunits, and this protein can further self-assemble into protein cages with a diameter of 16 nm.57 Afterwards, they created another kind of porous cube with a pore diameter of 130 nm using the same peptide origami approach.58 Our group overcame the shortcomings of the poor photostability and complex synthetic process of organic molecular probes and used the natural tripeptide GYK (G, glycine, Y, tyrosine, K, lysine) to generate fluorescence.59 The red shift of about 100 nm in the fluorescence of GYK assemblies already overcame the obstacle of natural amino acids.60 Such peptide assemblies hold great promise for developing novel fluorescent probes for cell labeling and imaging.61
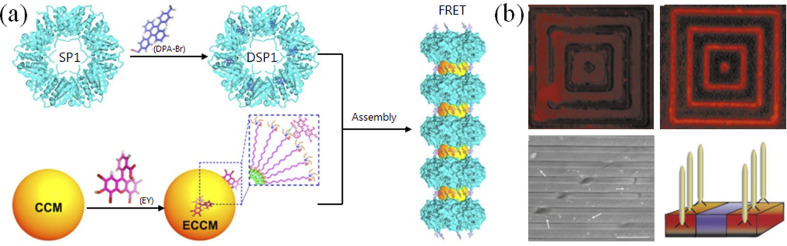
Due to the intrinsic chemical complexity based on amino acid sequences, peptides can be screened out or designed to specifically respond to metal ions62 or recognize inorganic materials with relatively high affinity (Fig. 4). Liu and co-workers constructed protein-based nanorings and nanorods using the recognition between metal ions and proteins, and demonstrated their application value in artificial light-harvesting systems (Fig. 4a).63 In combination lab experiments with theoretical simulations, Zhou and co-workers conducted a systematic study on controlling GAV peptide origami on a liquid–solid interface.64 In general, thiol group-modified DNA can form robust conjugation with some materials (such as gold and silver), while such functional groups are naturally contained in peptides. Fan and co-workers reported an interesting method wherein polyA has strong affinity to gold nanoparticles, whereby gold–DNA hybrid nanostructures were constructed by DNA origami.65 Systematic evolution of ligands by exponential enrichment (SELEX) could screen out DNA aptamers binding to different molecules and materials,66 but so far these aptamers cannot be compared to antibodies in terms of affinity. However, peptide aptamers have a higher affinity than DNA aptamers and can be selected using a bio-evolution system. For example, Belcher and co-workers successfully applied the phage display technique to screen out peptides with high affinity to semiconductors (Fig. 4b).67 Hence, regarding their diversity and affinity, we believe that there is plenty of room at the bottom of peptide origami.
Fig. 4. Peptide–inorganic hybrid functional nanomaterials. (a) Artificial light-harvesting hybrid nanostructures by co-assembling proteins and nanoparticles.63 Stable protein one (SP1) forms ring-like nanostructures, DSP1, which were modified by a thiol-reactive chromophore 9-[4-(bromomethyl)phenyl]-10-(4-methylphenyl)anthracene (DPA-Br, as the donor chromophore). EY (eosin Y, as the acceptor chromophore)-modified CCMs (core-cross-linked micelles), ECCMs, can co-assemble with DSP1 into antenna-like hybrid nanostructures for FRET (Förster resonance energy transfer). (b) Screening out semiconductor-recognizing peptides using the phage display technique. GaAs, but not SiO2, was specifically recognized by phages that were fluorescently labeled with tetramethyl rhodamine (TMR), which was verified by scanning electron microscopy.67.
Outlook
Although the current nanostructures created by peptide origami are less complex than those made by DNA origami, and the computational assisted programs are also limited to a few peptide chains that are much less than the chain number used in current DNA origami, peptide origami has already demonstrated its feasibility and superiority, and holds great potential to combine other material blocks for fabricating hybrid origami structures, i.e., DNA–peptide hybrid origami structures.68
Our group previously discovered strong physical adsorption between serum albumin and amphiphilic polymer-encapsulated nanoparticles,69 and proposed a “mortise–tenon joint” mechanism.70 This mortise–tenon joint is independent of the properties of the material encapsulated inside the polymer, and with this advantage, nanostructures with a discrete number of ligands (i.e., peptides) can be obtained by gel electrophoresis.71 Such peptide/protein–nanoparticle72 hybrid building blocks hold great potential for bottom-up origami-like nanofabrication, with which Parak and co-workers demonstrated the significant differences of nanoparticles conjugated with a different number of antibodies in tumor-targeting therapy.73 Artificial light-harvesting complexes have been a hot spot to use “infinite” light energy due to the depleting fossil fuels such as coal, oil and natural gas. However, improving the current light energy conversion rate is still challenging, and the delicate design of photosynthetic nanostructures is key; therefore, peptide origami could be the most effective way to solve this problem.
Consent for publication
All authors agree to be published.
Author contributions
Feng Zhang and Liping Wang conceived the concept and supervised the writing of the manuscript. Feng Zhang, Liping Wang, Yuxing Ma, Ruoyang Zhao, and Xiaofang Li collected the literature, prepared the figures and drafted the manuscript. Feng Zhang, Liping Wang, Yuxing Ma, Ruoyang Zhao, Xiaofang Li, Enqi Wu, Qiqige Du, and Jun Guo read and approved the final manuscript.
Conflicts of interest
The authors declare that they have no competing interests.
Supplementary Material
Acknowledgments
This work was supported by the National Key Research and Development Program of China (No. 2021YFA1200402), the National Natural Science Foundation of China (No. T2241002, U1832125 and 51763019), the start-up funding from Wenzhou Institute, University of Chinese Academy of Sciences (No. WIUCASQD2021003 and WIUCASQD2021011), the Program Funded by the University for Fostering Distinguished Young Scholars, the Grassland Talents Program of Inner Mongolia Autonomous Region, the Distinguished Young Scholars Foundation of Inner Mongolia Autonomous Region, and the Young Leading Talents of Science and Technology Program of Inner Mongolia Autonomous Region.
Biographies
Biography
Yuxing Ma.

Yuxing Ma received her Master degree at Inner Mongolia Agricultural University. Then worked as a research fellow at Hetao College and a research assistant in Wenzhou Institute, University of Chinese Academy of Sciences. Her research interests focused on the biomaterials and biomedical application.
Biography
Xiaofang Li.

Xiaofang Li is currently pursuing a PhD degree under the supervision of Professor Feng Zhang in Inner Mongolia Agricultural University, and meantime working as a research assistant in Wenzhou Institute, University of Chinese Academy of Sciences and University of Shanghai for Science and Technolgoy. Xiaofang Li received her Master of Engineering degree from Tianjin Polytechnic University in 2012, and now her research interests focus on computational biology and its applications in biological medicine.
Biography
Ruoyang Zhao.

Ruoyang Zhao received her PhD degree from Inner Mongolia Agricultural University. She is now working as a postdoctoral fellow in Professor Feng Zhang's group, Wenzhou Institute, UCAS. Her research interests in self-assembly of peptides and its applications in biological medicine.
Biography
Jun Guo.

Jun Guo received his PhD degree from Inner Mongolia Agricultural University. He worked as a postdoctoral fellow in Professor Feng Zhang and Professor Bo Song's groups, University of Shanghai for Science and Technology. His research interests include the design of biomimetic materials from self-assembling peptides.
Biography
Liping Wang.

Liping Wang received her PhD degree at East China Normal University, and finished her postdoctoral training and worked as a research fellow at Shanghai Jiao Tong University. She has been working as an associated professor at Wenzhou Institute, University of Chinese Academy of Sciences since 2021. Her research interests focused on the drug delivery, bioactive materials and biomedical application.
Biography
Feng Zhang.

Feng Zhang is a Professor at University of Shanghai for Science and Technology. He received his PhD degree from Shanghai Institute of Applied Physics, Chinese Academy of Sciences. After two PostD trainings in Germany and USA, he started his independent career. His research interests are focused on the nanofabrication of bioactivities through peptide assembly (especially peptide origami) and the development of new quantum biological methodologies for analytical and biomedical applications.
References
- Darvish D. M. Mater. Today Bio. 2022;15:100322. doi: 10.1016/j.mtbio.2022.100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q. Li W. Yuan M. Gong P. Zhang Y. Zhang F. Luminescence. 2017;32:618–624. doi: 10.1002/bio.3229. [DOI] [PubMed] [Google Scholar]
- Li D. Liu C. Biochemistry. 2020;59:639–646. doi: 10.1021/acs.biochem.9b01069. [DOI] [PubMed] [Google Scholar]
- Li K. Zheng J. Wirawan M. Trinh N. M. Fedorova O. Griffin P. R. Pyle A. M. Luo D. Nucleic Acids Res. 2021;49:9978–9991. doi: 10.1093/nar/gkab712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S. A. Zhu W. Graham S. Rambo R. White M. F. Gloster T. M. Nat. Commun. 2020;11:500. doi: 10.1038/s41467-019-14222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Wang X. Zhou K. Chen G. Wang Q. ACS Nano. 2018;12:1673–1679. doi: 10.1021/acsnano.7b08316. [DOI] [PubMed] [Google Scholar]
- Fiaschi L. Di Palo B. Scarselli M. Pozzi C. Tomaszewski K. Galletti B. Nardi-Dei V. Arcidiacono L. Mishra R. P. Mori E. Pallaoro M. Falugi F. Torre A. Fontana M. R. Soriani M. Bubeck Wardenburg J. Grandi G. Rappuoli R. Ferlenghi I. Bagnoli F. Clin. Vaccine Immunol. 2016;23:442–450. doi: 10.1128/CVI.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D. Krantz B. A. J. Mol. Biol. 2017;429:753–762. doi: 10.1016/j.jmb.2017.01.015. [DOI] [PubMed] [Google Scholar]
- Lee S. H. Kim K. J. Biochem. Biophys. Res. Commun. 2019;509:722–727. doi: 10.1016/j.bbrc.2018.12.172. [DOI] [PubMed] [Google Scholar]
- Vallet S. D. Gueroult M. Belloy N. Dauchez M. Ricard-Blum S. ACS Omega. 2019;4:8495–8505. doi: 10.1021/acsomega.9b00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q. Liu Y. P. Yan X. X. Liang D. C. DNA Repair. 2014;24:10–14. doi: 10.1016/j.dnarep.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Kwak Y. Jiang W. Dassama L. M. Park K. Bell 3rd C. B. Liu L. V. Wong S. D. Saito M. Kobayashi Y. Kitao S. Seto M. Yoda Y. Alp E. E. Zhao J. Bollinger Jr J. M. Krebs C. Solomon E. I. J. Am. Chem. Soc. 2013;135:17573–17584. doi: 10.1021/ja409510d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C.-Y. Zlotnick A. Mecinovi J. Chem. Commun. 2014;50:10281–10283. doi: 10.1039/c4cc04650a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. He J. Fan K. Yan X. Nanoscale. 2019;11:12449–12459. doi: 10.1039/c9nr03823j. [DOI] [PubMed] [Google Scholar]
- Kato K. Tanaka H. Sumizawa T. Yoshimura M. Yamashita E. Iwasaki K. Tsukihara T. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:525–531. doi: 10.1107/S0907444908004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halebian M. Morris K. Smith C. Subcell. Biochem. 2017;83:551–567. doi: 10.1007/978-3-319-46503-6_20. [DOI] [PubMed] [Google Scholar]
- Bracher A. Paul S. S. Wang H. Wischnewski N. Hartl F. U. Hayer-Hartl M. PLoS One. 2020;15:e0230090. doi: 10.1371/journal.pone.0230090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. de Ruiter M. V. Maassen S. J. Cornelissen J. Methods Mol. Biol. 2018;1798:1–9. doi: 10.1007/978-1-4939-7893-9_1. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- Chen J. H. Seeman N. C. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- Song R. Wu X. Xue B. Yang Y. Huang W. Zeng G. Wang J. Li W. Cao Y. Wang W. Lu J. Dong H. J. Am. Chem. Soc. 2019;141:223–231. doi: 10.1021/jacs.8b08893. [DOI] [PubMed] [Google Scholar]
- Bai Y. Luo Q. Liu J. Chem. Soc. Rev. 2016;45:2756–2767. doi: 10.1039/c6cs00004e. [DOI] [PubMed] [Google Scholar]
- Purcell A. W. McCluskey J. Rossjohn J. Nat. Rev. Drug Discovery. 2007;6:404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- Dai B. Li D. Xi W. Luo F. Zhang X. Zou M. Cao M. Hu J. Wang W. Wei G. Zhang Y. Liu C. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2996–3001. doi: 10.1073/pnas.1416690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolisetty S. Mezzenga R. Nat. Nanotechnol. 2016;11:365–371. doi: 10.1038/nnano.2015.310. [DOI] [PubMed] [Google Scholar]
- Wei G. Xi W. Nussinov R. Ma B. Chem. Rev. 2016;116:6516–6551. doi: 10.1021/acs.chemrev.5b00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Liu K. Xing R. Yan X. Chem. Soc. Rev. 2016;45:5589–5604. doi: 10.1039/c6cs00176a. [DOI] [PubMed] [Google Scholar]
- Ljubetic A. Lapenta F. Gradisar H. Drobnak I. Aupic J. Strmsek Z. Lainscek D. Hafner-Bratkovic I. Majerle A. Krivec N. Bencina M. Pisanski T. Velickovic T. C. Round A. Carazo J. M. Melero R. Jerala R. Nat. Biotechnol. 2017;35:1094–1101. doi: 10.1038/nbt.3994. [DOI] [PubMed] [Google Scholar]
- Joshi K. B. Verma S. J. Pept. Sci. 2008;14:118–126. doi: 10.1002/psc.955. [DOI] [PubMed] [Google Scholar]
- Huang C. C. Ravindran S. Yin Z. George A. Biomaterials. 2014;35:5316–5326. doi: 10.1016/j.biomaterials.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M. Bentley D. Ghosh I. J. Am. Chem. Soc. 2004;126:734–735. doi: 10.1021/ja036634y. [DOI] [PubMed] [Google Scholar]
- Shen W. Lammertink R. G. H. Sakata J. K. Kornfield J. A. Tirrell D. A. Macromolecules. 2005;38:3909–3916. [Google Scholar]
- Majerle A. Hadzi S. Aupic J. Satler T. Lapenta F. Strmsek Z. Lah J. Loris R. Jerala R. Proc. Natl. Acad. Sci. U. S. A. 2021;118:e2021899118. doi: 10.1073/pnas.2021899118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic Abram S. Gradisar H. Aupic J. Round A. R. Jerala R. ACS Chem. Biol. 2021;16:310–315. doi: 10.1021/acschembio.0c00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R. Angew. Chem., Int. Ed. Engl. 2018;57:6086–6089. doi: 10.1002/anie.201800197. [DOI] [PubMed] [Google Scholar]
- Zhang F. Lin X. J. Ji L. N. Du H. N. Tang L. He J. H. Hu J. Hu H. Y. Biochem. Biophys. Res. Commun. 2008;368:388–394. doi: 10.1016/j.bbrc.2008.01.091. [DOI] [PubMed] [Google Scholar]
- Zhang F. Ji L. N. Tang L. Hu J. Hui H. Y. Xu H. J. He J. H. Acta Biochim. Biophys. Sin. 2005;37:113–118. [PubMed] [Google Scholar]
- Zhou X. Zhang Y. Zhang F. Pillai S. Liu J. Li R. Dai B. Li B. Zhang Y. Nanoscale. 2013;5:4816–4822. doi: 10.1039/c3nr00886j. [DOI] [PubMed] [Google Scholar]
- Li H. Zhang F. Zhang Y. Ye M. Zhou B. Tang Y. Z. Yang H. J. Xie M. Y. Chen S. F. He J. H. Fang H. P. Hu J. J. Phys. Chem. B. 2009;113:8795–8799. doi: 10.1021/jp903446g. [DOI] [PubMed] [Google Scholar]
- Humenik M. Scheibel T. ACS Nano. 2014;8:1342–1349. doi: 10.1021/nn404916f. [DOI] [PubMed] [Google Scholar]
- Zhang S. G. Marini D. M. Hwang W. Santoso S. Curr. Opin. Chem. Biol. 2002;6:865–871. doi: 10.1016/s1367-5931(02)00391-5. [DOI] [PubMed] [Google Scholar]
- Lapenta F. Aupic J. Strmsek Z. Jerala R. Chem. Soc. Rev. 2018;47:3530–3542. doi: 10.1039/c7cs00822h. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov S. Park H. Varghese N. Huang P. S. Pavlopoulos G. A. Kim D. E. Kamisetty H. Kyrpides N. C. Baker D. Science. 2017;355:294–298. doi: 10.1126/science.aah4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. M. Harniman R. L. Barnes F. R. Boyle A. L. Collins A. Mantell J. Sharp T. H. Antognozzi M. Booth P. J. Linden N. Miles M. J. Sessions R. B. Verkade P. Woolfson D. N. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradisar H. Bozic S. Doles T. Vengust D. Hafner-Bratkovic I. Mertelj A. Webb B. Sali A. Klavzar S. Jerala R. Nat. Chem. Biol. 2013;9:362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble J. E. De Santis E. Ravi J. Lamarre B. Castelletto V. Mantell J. Ray S. Ryadnov M. G. J. Am. Chem. Soc. 2016;138:12202–12210. doi: 10.1021/jacs.6b05751. [DOI] [PubMed] [Google Scholar]
- Gonen S. DiMaio F. Gonen T. Baker D. Science. 2015;348:1365–1368. doi: 10.1126/science.aaa9897. [DOI] [PubMed] [Google Scholar]
- King N. P. Sheffler W. Sawaya M. R. Vollmar B. S. Sumida J. P. Andre I. Gonen T. Yeates T. O. Baker D. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga N. Tatsumi-Koga R. Liu G. Xiao R. Acton T. B. Montelione G. T. Baker D. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N. P. Bale J. B. Sheffler W. McNamara D. E. Gonen S. Gonen T. Yeates T. O. Baker D. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos E. Basanta B. Chidyausiku T. M. Tang Y. Oberdorfer G. Liu G. Swapna G. V. Guan R. Silva D. A. Dou J. Pereira J. H. Xiao R. Sankaran B. Zwart P. H. Montelione G. T. Baker D. Science. 2017;355:201–206. doi: 10.1126/science.aah7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N. Yanase K. Sato T. Unzai S. Hecht M. H. Arai R. J. Am. Chem. Soc. 2015;137:11285–11293. doi: 10.1021/jacs.5b03593. [DOI] [PubMed] [Google Scholar]
- Urban M. J. Dutta P. K. Wang P. Duan X. Shen X. Ding B. Ke Y. Liu N. J. Am. Chem. Soc. 2016;138:5495–5498. doi: 10.1021/jacs.6b00958. [DOI] [PubMed] [Google Scholar]
- Chin J. W. Martin A. B. King D. S. Wang L. Schultz P. G. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra J. S. Tian Y. F. Jung J. P. Collier J. H. Proc. Natl. Acad. Sci. U. S. A. 2010;107:622–627. doi: 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudalla G. A. Sun T. Gasiorowski J. Z. Han H. Tian Y. F. Chong A. S. Collier J. H. Nat. Mater. 2014;13:829–836. doi: 10.1038/nmat3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y. T. Cascio D. Yeates T. O. Science. 2012;336:1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- Lai Y. T. Reading E. Hura G. L. Tsai K. L. Laganowsky A. Asturias F. J. Tainer J. A. Robinson C. V. Yeates T. O. Nat. Chem. 2014;6:1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Ramachandran S. Zhong R. Lal R. Zhang F. Chembiochem. 2019;20:2324–2330. doi: 10.1002/cbic.201900166. [DOI] [PubMed] [Google Scholar]
- Guo J. Li X. Lian J. Gao F. Zhao R. Song B. Zhang F. ACS Macro Lett. 2021;10:825–830. doi: 10.1021/acsmacrolett.1c00251. [DOI] [PubMed] [Google Scholar]
- Guo J. Zhao R. Gao F. Li X. Wang L. Zhang F. ACS Macro Lett. 2022;11:875–881. doi: 10.1021/acsmacrolett.2c00180. [DOI] [PubMed] [Google Scholar]
- Singh N. Singh R. Joshi K. B. Verma S. ChemPlusChem. 2020;85:2001–2009. doi: 10.1002/cplu.202000464. [DOI] [PubMed] [Google Scholar]
- Sun H. Zhang X. Miao L. Zhao L. Luo Q. Xu J. Liu J. ACS Nano. 2016;10:421–428. doi: 10.1021/acsnano.5b05213. [DOI] [PubMed] [Google Scholar]
- Kang S. G. Huynh T. Xia Z. Zhang Y. Fang H. Wei G. Zhou R. J. Am. Chem. Soc. 2013;135:3150–3157. doi: 10.1021/ja310989u. [DOI] [PubMed] [Google Scholar]
- Yao G. B. Pei H. Li J. Zhao Y. Zhu D. Zhang Y. N. Lin Y. F. Huang Q. Fan C. H. NPG Asia Mater. 2015;7:e159. [Google Scholar]
- Fang X. Tan W. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley S. R. English D. S. Hu E. L. Barbara P. F. Belcher A. M. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- Udomprasert A. Bongiovanni M. N. Sha R. Sherman W. B. Wang T. Arora P. S. Canary J. W. Gras S. L. Seeman N. C. Nat. Nanotechnol. 2014;9:537–541. doi: 10.1038/nnano.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R. Liu Y. Zhang P. Liu J. Zhao G. Zhang F. ACS Appl. Mater. Interfaces. 2014;6:19465–19470. doi: 10.1021/am506497s. [DOI] [PubMed] [Google Scholar]
- Liu Y. Zhong R. Zhang P. Ma Y. Yun X. Gong P. Wei J. Zhao X. Zhang F. ACS Appl. Mater. Interfaces. 2016;8:2478–2485. doi: 10.1021/acsami.5b08386. [DOI] [PubMed] [Google Scholar]
- Guo J. Zhong R. B. Li W. R. Liu Y. S. Bai Z. J. Yin J. Liu J. R. Gong P. Zhao X. M. Zhang F. Appl. Surf. Sci. 2015;359:82–88. [Google Scholar]
- Tripathi S. K. Kesharwani K. Kaul G. Akhir A. Saxena D. Singh R. Mishra N. K. Pandey A. Chopra S. Joshi K. B. ChemMedChem. 2022;17:e202200251. doi: 10.1002/cmdc.202200251. [DOI] [PubMed] [Google Scholar]
- Colombo M. Fiandra L. Alessio G. Mazzucchelli S. Nebuloni M. De Palma C. Kantner K. Pelaz B. Rotem R. Corsi F. Parak W. J. Prosperi D. Nat. Commun. 2016;7:13818. doi: 10.1038/ncomms13818. [DOI] [PMC free article] [PubMed] [Google Scholar]