Abstract
Bacteriophage φYeO3-12 is a lytic phage of Yersinia enterocolitica serotype O:3. The phage receptor is the lipopolysaccharide O chain of this serotype that consists of the rare sugar 6-deoxy-l-altropyranose. A one-step growth curve of φYeO3-12 revealed eclipse and latent periods of 15 and 25 min, respectively, with a burst size of about 120 PFU per infected cell. In electron microscopy φYeO3-12 virions showed pentagonal outlines, indicating their icosahedral nature. The phage capsid was shown to be composed of at least 10 structural proteins, of which a protein of 43 kDa was predominant. N-terminal sequences of three structural proteins were determined, two of them showing strong homology to structural proteins of coliphages T3 and T7. The phage genome was found to consist of a double-stranded DNA molecule of 40 kb without cohesive ends. A physical map of the phage DNA was constructed using five restriction enzymes. The phage infection could be effectively neutralized using serum from a rabbit immunized with whole φYeO3-12 particles. The antiserum also neutralized T3 infection, although not as efficiently as that of φYeO3-12. φYeO3-12 was found to share, in addition to the N-terminal sequence homology, several common features with T3, including morphology and nonsubjectibility to F exclusion. The evidence conclusively indicated that φYeO3-12 is the first close relative of phage T3 to be described.
Yersinia enterocolitica is a Gram-negative species which contains several serotypes, some of which are pathogenic to humans. The major pathogens in Europe, Canada, Japan, and South Africa belong to serotypes O:3 and O:9, and those in the United States belong to serotype O:8 (11). The main reservoir in nature for Y. enterocolitica is pigs (15), and human infections usually take place after ingestion of contaminated foodstuffs.
A number of yersiniophages have been described, but only a few have been characterized by electron microscopy and to our knowledge none have been studied in detail. In our laboratory a number of Yersinia-specific bacteriophages have been isolated, all originating from the raw incoming sewage of the Turku City sewage treatment plant, and the phages have been used as genetic tools (32). One of the phages, φYeO3-12, was isolated as specific to Y. enterocolitica serotype O:3. The phage could infect Escherichia coli C600 expressing the cloned O antigen of Y. enterocolitica serotype O:3 and spontaneous phage-resistant Y. enterocolitica serotype O:3 strains were missing the O antigen, indicating that the O antigen is the phage receptor (4, 5). The serotype O:3 specificity makes the phage φYeO3-12 a potential biotechnological tool, and therefore we have initiated its detailed characterization. Here we present the biological and physical properties of the phage and evidence suggesting that φYeO3-12 is closely related to coliphages T3 and T7.
MATERIALS AND METHODS
Culture conditions.
Bacterial strains, bacteriophages and plasmids used in this study are listed in Table 1. Virulence plasmid-cured Y. enterocolitica serotype O:3 strain 6471/76-c (31) was the usual host for propagation of phage φYeO3-12. The φYeO3-12 and its host are available under accession no. HER 249 and 1249, respectively, at the Felix d'Herelle Reference Center for Bacterial Viruses. Bacterial strains were grown in tryptone soya broth medium (TSB; Oxoid), and incubations were done at room temperature (RT; 22 to 25°C) unless specified otherwise. E. coli strains were grown in Luria broth (LB) at 37°C, and ampicillin (100 μg/ml) was added when required. Solid medium was obtained by adding 2% (wt/vol) agar to LB, and soft agar was obtained by adding 0.5% (wt/vol) agar to TSB (7, 30).
TABLE 1.
Bacterial strains, bacteriophages, and plasmids used in this study
| Bacterial strain, bacteriophage, or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Yersiniae | ||
| 6471/76 (YeO3) | Y. enterocolitica serotype O:3; patient isolate; wild type | 31 |
| 6471/76-c (YeO3-c) | Virulence plasmid-cured derivative of 6471/76 | 31 |
| IP23047 | Y. frederiksenii serotype O:3 | Elisabeth Carniel, Institut Pasteur |
| IP22404 | Y. mollaretii serotype O:3 | Elisabeth Carniel, Institut Pasteur |
| IP22828 | Y. kristensenii serotype O:3 | Elisabeth Carniel, Institut Pasteur |
| Several species and serotypes | See Table 2 | Laboratory collection |
| E. coli K-12 | ||
| C600 | thi thr leuB tonA lacY supE | 6 |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 λ− Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | 36 |
| IJ511 | ΔlacX74 galK2 galT22 supE44 hsdS | Ian Molineux |
| IJ512 | F42 (F′ lac) derivative of IJ511 | Ian Molineux |
| IJ855 | ara Δ(lac-proAB) thi supD hsrD | Ian Molineux |
| IJ1419 | lacY1 or Δ(lac)6 supE44 galK2 galT22 λ− rfbD1 metB1 mcrA1 hsdR2 rpoC319 (tsnB) | Ian Molineux |
| IJ1420 | lacY1 or Δ(lac)6 supE44 galK2 galT22 λ− rfbD1 metB1 mcrA1 hsdR2 rpoC320 (BR3) | Ian Molineux |
| S. sonnei D2 371-48 | Abortively infects T7+ (R. Hausmann) | Ian Molineux |
| Bacteriophages | ||
| φYeO3-12 | Y. enterocolitica serotype O:3 specific; wild type; isolated from Turku City sewage | 4 |
| ΔPK | Protein kinase deletion derivative of φYeO3-12 | This study |
| T7+ | Wild type from F. W. Studier | Ian Molineux |
| T3+ | Wild type from F. W. Studier (Luria strain) | Ian Molineux |
| Plasmid pAY100 | The O antigen gene cluster of Y. enterocolitica serotype O:3 cloned into pBR322 | 4 |
Propagation and purification of phage φYeO3-12.
Bacteriophage φYeO3-12 (Table 1) was stored at −70°C in TSB supplemented with 7% dimethyl sulfoxide (DMSO). Large-scale purification of φYeO3-12 virions was done as described elsewhere (30). Briefly, an overnight culture of Y. enterocolitica serotype O:3 strain 6471/76-c was diluted 10-fold in TSB in a total volume of 1 liter divided into four 2-liter Erlenmeyer flasks and infected with φYeO3-12 at a multiplicity of infection (MOI) of 1. The infected cultures were incubated at 25°C with vigorous aeration (250 rpm), until, usually after 2.5 h, the bacterial lysis took place. The lysed culture was treated with DNase I (1.2 μg/ml; Roche Molecular Biochemicals) and RNase A (1 μg/ml; Sigma Chemicals, St. Louis, Mo.) at RT for 30 min. Sodium chloride (final concentration, 1 M) was added to the treated lysate and incubated on ice for 1 h, and then the solution was centrifuged at 11,000 × g at 4°C for 20 min in a Sorvall GS-3 fixed-angle rotor to remove the precipitated bacterial debris. The phage was recovered from the supernatant by precipitating with polyethylene glycol (PEG) 8000 (10%, wt/vol; ≥60 min; 0°C) and was resuspended into SM buffer (30). The phage was further purified by chloroform extraction and one to three rounds of discontinuous glycerol density gradient ultracentrifugation at 35,000 rpm at 4°C for 4 h in a Sorvall TH-641 swing-out rotor. After ultracentrifugation the phages were resuspended in SM buffer containing 8% sucrose to yield a typical concentration of ca. 1014 PFU/ml, as determined by phage titration (30).
Host range determination.
The host range of φYeO3-12 was determined by pipetting 20-μl droplets of serial dilutions of concentrated phage stocks (up to 1014 PFU/ml) on lawns of different bacterial strains prepared on LB plates. The formation of plaques of lysis, i.e., the plating efficiency, was tested both at RT and at 37°C.
Electron microscopy.
Phage particles were negatively stained with 2% (wt/vol) phosphotungstic acid, pH 7. Prior to examination, the particles were sedimented at about 25,000 × g for 60 min in a Beckman (Palo Alto, Calif.) J2-21 centrifuge, using a JA-18.1 fixed-angle rotor. This was followed by two washes in 0.1 M ammonium acetate, pH 7.2. Stained particles were observed in a Philips EM 300 electron microscope operated at 60 kV. Magnification was monitored with catalase crystals (Worthington, Freehold, N.J.) (24). Dimensions were measured on photographic prints at a final magnification of ×297,000.
Density and fatty acid analysis.
A PEG-precipitated bacteriophage suspension was loaded on a continuous CsCl density gradient and ultracentrifuged to equilibrium at 40,000 rpm (Sorvall TST 60.4) for 24 h at 4°C. After centrifugation, fractions were collected for phage titration and density measurement. For fatty acid analysis the phage particles were treated with sodium dodecyl sulfate (SDS) (0.5%) and proteinase K (0.2 mg/ml) for 70 min at 56°C, extracted, and converted into methylester derivatives which were analyzed by gas chromatography (18).
One-step growth curve.
A mid-exponential-phase culture (10 ml) of Y. enterocolitica serotype O:3 strain 6471/76-c (optical density at 600 nm [OD600], 0.4 to 0.5) was harvested by centrifugation and resuspended in 0.25 volume of fresh TSB (ca. 109 CFU/ml). Phage was added at an MOI of 0.0005 and allowed to adsorb for 5 min at RT. The mixture was then centrifuged, pelleted cells were resuspended in 10 ml of TSB, and incubation was continued at RT. Samples were taken at 5-min intervals. The first set of samples was immediately diluted and plated for phage titration. A second set of samples was treated with 1% (vol/vol) chloroform to release intracellular phages in order to determine the eclipse period before phage titration (9).
Immune sera.
Immunization of rabbits with bacteriophage φYeO3-12 was performed as follows. Preimmunization serum samples were collected from the rabbits prior to immunization. Three young rabbits were immunized with purified phages (ca. 1013 PFU/rabbit) in Freund's complete adjuvant (Difco 0638) in phosphate-buffered saline (PBS) by subcutaneous injection of four sites (0.25 ml each) on the back of the rabbit. Booster immunizations were done by injecting phages in Freund's incomplete adjuvant (Sigma F-5506) every 3rd week for three times. Humoral immune responses were monitored by analyzing serum samples by enzyme immunoassay (EIA) for specific antibodies on the 5th and 8th weeks of immunization (see below). The rabbits were killed and the blood was collected after 10 weeks of immunization. The sera were separated from the blood after clotting and were stored frozen at −20°C (or −70°C for longer periods).
EIA.
The wells of a 96-well microtiter plate (Nunc-Immuno plate; MaxiSorp surface) were coated with heat-inactivated φYeO3-12 (140 min at 80°C followed by 65 min at 95°C) in PBS (100 μl per well containing about 1010 phage particles [ca. 1 μg]) overnight at RT. After three washes with PBS, the wells were blocked with 150 μl of 5% skim milk powder (wt/vol) in water for 2 h at 37°C and then washed again three times with PBS. The rabbit sera were diluted in 1% normal sheep serum (NSS) in PBS to obtain twofold dilutions between 1:8,000 and 1:256,000, and 75 μl of the dilutions was incubated in the wells for 1.5 h at 37°C, after which the wells were washed three times with PBS. Then 75 μl of swine horseradish peroxidase-conjugated immunoglobulin against rabbit immunoglobulins (P217; Dako A/S, Colostrup, Denmark) diluted 1:1,000 in 1% NSS-PBS was added and incubated for 1 h at 37°C. The plates were washed three times with PBS, and 75 μl of a substrate solution (3 mg of 1,2-phenylenediamine/ml and 0.02% H2O2 in citrate buffer [per liter, 4.97 g of citric acid × H2O and 9.9 g of Na2HPO4 × 2H2O]) was added and incubated for 10 min at RT. The reactions were terminated by adding 125 μl of 1 M HCl to the wells. Optical absorbances were measured at 492 nm with a Labsystems Multiskan MCC/340 Photometer. Each sample was analyzed in duplicate. For negative controls, wells with 1:8,000 diluted preimmune sera from the rabbits and wells without any rabbit serum were included.
The sensitivity of the strongest rabbit serum for the phage particles was determined essentially as described above with the exception that wells were coated with a 100-μl volume of different amounts (1 μg, 0.5 μg, 0.1 μg, 50 ng, 10 ng, 5 ng, 1 ng, 0.5 ng, and 0.1 ng) of native φYeO3-12 particles, the rabbit serum was diluted 1:2,000 and 1:10,000 in 1% NSS-PBS, and 75 μl of the dilutions was incubated in the wells for 1.5 h at 37°C. After a wash step, the antibodies that had bound to the phage particles were detected as described above with the same negative controls. Each sample was analyzed in triplicate.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Samples of purified virions of φYeO3-12 were heated at 95°C for 15 min in SDS gel-loading buffer (30). Electrophoresis was conducted at a constant current of 13 mA in SDS–10% polyacrylamide gels using the Hoefer SE 600 device (Amersham Pharmacia Biotech) as described by Laemmli (23). After electrophoresis the gels were either stained with Coomassie brilliant blue R-350 (Pharmacia) or blotted to a nitrocellulose membrane (BAS 83; Schleicher & Schuell, Dassel, Germany) using the Trans-Blot Semi-Dry Transfer Cell (Bio-Rad), according to the manufacturer's instructions. The membrane was blocked overnight at RT in 5% skim milk powder in PBS. After a wash, it was incubated with the strongest rabbit serum (diluted 1:20,000 in 5% skim milk powder in PBS) overnight at RT. The membrane was washed and then incubated with 1:2,000 diluted P217 for 2 h at RT. After four washes, the bound peroxidase was detected using the ECL Western blotting kit (RPN 2106; Amersham International plc, Little Chalfont, England).
Affinity purification of antibodies against φYeO3-12 proteins.
Antibodies monospecific for phage proteins were affinity purified from the rabbit antiserum as described elsewhere (29). SDS-PAGE samples of purified virions of φYeO3-12 were electrophoresed using a 10% polyacrylamide separation gel and then transferred to a nitrocellulose membrane. The membrane was stained with Ponceau S (2%, wt/vol) for 5 min to visualize the protein bands, and the membrane was excised into 27 horizontal strips. The strips were incubated overnight at RT in 5% skim milk powder in PBS to block nonspecific binding sites. After a wash step, each strip was incubated overnight at RT with 1 ml of the strongest rabbit serum diluted 1:50 in 5% skim milk powder in PBS. The strips were washed four times with PBS. Specific anti-protein antibodies were eluted by treatment with 1,050 μl of 0.2 M HCl-glycine buffer (pH 2.2) at RT for 15 min. The pH of the eluate was immediately neutralized by the addition of 450 μl of 1 M K2HPO4, and then it was dialyzed overnight at 4°C against PBS. The antibodies were stored at 4°C.
Neutralization.
The crude rabbit serum, heat-inactivated (56°C for 30 min) rabbit serum, and affinity-purified antibodies were tested for their abilities to neutralize φYeO3-12, T3, and T7 infections. The crude rabbit serum was also preincubated 10 min at RT with an excess of T3 (107 PFU) before being tested for its ability to neutralize φYeO3-12 infection. The sera were diluted in PBS to obtain twofold dilutions, and 50 μl of the dilutions was incubated with a constant amount (ca. 100 PFU) of different phages for 10 min at RT. Then 150 μl of an overnight culture of indicator bacteria (Y. enterocolitica serotype O:3 strain 6471/76-c or IJ855) was added, the mixture was plated, and plaques were counted.
N-terminal amino acid analysis.
SDS-PAGE of purified phage particles was run in a Mini-PROTEAN II gel electrophoresis apparatus (Bio-Rad). The electrophoresis and subsequent electroblotting onto Problott (ABI Foster City, Calif.) were performed according to the work of Moos et al. (28). Amino-terminal amino acid sequencing of the immobilized protein was performed with an Applied Biosystems 477A protein sequencer (ABI, Ltd.) equipped with an on-line ABI 120A PTH (phenylthiohydantoin)-amino acid analyzer. Standard cycle parameters provided by the manufacturer were used. Protein similarity searches were done at the European Bioinformatics Institute site (http://www.ebi.ac.uk/fasta3) using the Fasta3 search with default parameters against the SwissProt All library.
DNA techniques.
Phage DNA was obtained from high-titer phage preparations as described by Sambrook et al. (30). Plasmid DNA was extracted from E. coli by alkaline lysis according to the procedure of Birnboim and Doly (10). All enzymatic treatments of DNA were performed as recommended by the manufacturers. DNA electrophoresis was carried out in agarose gels using standard TAE buffer (30). A physical map of the DNA of φYeO3-12 was determined using five restriction enzymes (BclI, EcoRV, PmlI, SnaBI, and XbaI; New England Biolabs, Beverly, Mass.) by analysis of single and double digests. The existence of cohesive ends was assayed by comparing the restriction patterns of phage DNA with and without prior treatment with T4 DNA ligase. Aliquots of phage DNA were heated at 70°C for 10 min in order to melt the cohesive ends and then were either slowly cooled down to RT or immediately placed on ice. After ligation and digestion with DraI or StuI, the samples were heated again at 70°C and then kept on ice until loading in an agarose gel (19). Lambda DNA was used as a positive control for cohesive ends.
RESULTS AND DISCUSSION
Host range of φYeO3-12.
Previous work established that the φYeO3-12 receptor is a homopolymer of 6-deoxy-l-altropyranose, the O antigen of Y. enterocolitica serotype O:3 (4). The host range of φYeO3-12 was assayed using almost 300 strains belonging to eight Yersinia species (Table 2). A large number of Y. enterocolitica serotype O:3 strains were included in order to test O:3 strains isolated from different origins. The results (Table 2) showed that φYeO3-12 could form plaques only on Y. enterocolitica serotypes O:1, O:2, and O:3, i.e., serotypes with an O antigen known to contain 6-deoxy-l-altropyranose. No difference in sensitivity was found between Y. enterocolitica serotype O:3 strains of human and animal origin. Also, Yersinia frederiksenii serotype O:3 and Yersinia mollaretii serotype O:3 were found to be phage sensitive. No plaques were produced on strains of serotypes of Y. enterocolitica or of other Yersinia species where 6-deoxy-l-altropyranose is not present in the O antigen. Similar phage propagation in sensitive strains was observed at RT and 37°C.
TABLE 2.
Bacteriophage φYeO3-12 sensitivity of Yersinia species
| Yersinia species | φYeO3-12-sensitive serotypesa | φYeO3-12-resistant serotypesa |
|---|---|---|
| Y. enterocolitica | O:1 (2), O:2 (2), O:3 (224) | O:1,2,3 (1), O:4 (1), O:4,32 (1), O:5 (9), O:5,27 (4), O:6 (2), O:6,30 (5), O:6,31 (3), O:7,8 (3), O:8 (7), O:9 (28), O:10 (5), O:13 (1), O:13,7 (2), O:13a,13b (1), O:13,18 (1), O:14 (1), O:20 (2), O:21 (3), O:25 (1), O:25,26,44 (1), O:26,44 (1), O:28,50 (1), O:34 (1), O:35,36 (1), O:35,52 (1), O:41(27),42 (1), O:41,43 (1), O:41(27),43 (2), O:41(27)K1 (1), O:50 (1), K1 nontypeable (2), nontypeable (3) |
| Y. pseudotuberculosis | I, IA, III (7) | |
| Y. intermedia | O:52,54, O:16,21 (2) | |
| Y. kristensenii | O:12,25, O:16, O:3, nontypeable (6) | |
| Y. frederiksenii | O:3 (1) | O:16, O:35, O:48, O:58,16, K1 nontypeable, nontypeable (7) |
| Y. mollaretii | O:3 (1) | O:59(20,36,7) (1) |
| Y. bercovieri | O:58,16 (1) | |
| Y. ruckerii | Nontypeable (1) |
The number of strains studied is given in parentheses.
The single Y. kristensenii serotype O:3 strain tested was resistant to φYeO3-12. To find out whether this was due to absence of the biosynthetic genes of the O:3 O antigen, the Y. kristensenii genomic DNA was isolated and used as a template in PCR. As a positive control, template DNA from Y. enterocolitica O:3 was used. Two sets of primers from the Y. enterocolitica O:3 O-antigen gene cluster (accession no. Z18920) were used. Identical PCR products were obtained from both species (data not shown). This suggested that in the Y. kristensenii O:3 strain studied, the O-antigen expression was somehow inactivated or its surface exposure was blocked.
Morphology and physical properties of φYeO3-12 particles.
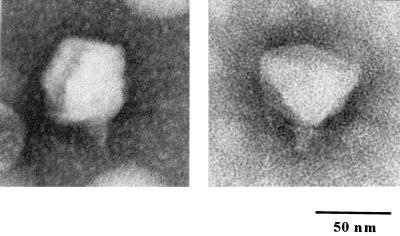
Electron microscopy of phosphotungstic acid-stained φYeO3-12 virions (Fig. 1) revealed particles with approximate dimensions of 57 nm for the head and 15 by 8 nm for the tail. Extended tail fibers were not seen. Normal capsids sometimes showed pentagonal outlines, indicating their icosahedral nature. Based on its morphology, φYeO3-12 belongs to the family Podoviridae (25) and to type C in Bradley's classification (12); furthermore, it resembles a typical member of the T7 group (H.-W. Ackermann, personal communication) (1). Other Y. enterocolitica phages characterized to date by electron microscopy have been of type A in Bradley's classification (22) or have been classified into the families Myoviridae or Podoviridae (2).
FIG. 1.
Electron micrograph of negatively stained φYeO3-12 virions.
In CsCl gradients the density of the phage was found to be ca. 1.3 g/ml. We noticed that φYeO3-12 lost infectivity very rapidly when CsCl was present. Fatty acid analysis failed to reveal any host-derived fatty acids from the phage particle.
Size and structure of the φYeO3-12 genome.
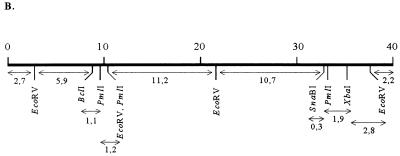
The phage genome was shown to consist of double-stranded DNA after its digestion with different restriction endonucleases (Fig. 2A). The size of the full-length phage genome was estimated to be about 40 kb, i.e., close to that of coliphages T3 and T7 (8, 17). Comparison of restriction patterns of unligated and ligated phage DNA showed similar DNA fragment patterns, indicating that φYeO3-12 DNA does not have cohesive ends (data not shown). A restriction map of the φYeO3-12 DNA was determined by analysis of single and double digests with restriction enzymes BclI, EcoRV, PmlI, SnaBI, and XbaI (Fig. 2).
FIG. 2.
Characterization of the φYeO3-12 genome. (A) Agarose gel electrophoresis of φYeO3-12 DNA after single and double digestions with restriction enzymes BclI, EcoRV, PmlI, SnaBI, and XbaI. (B) Restriction map of phage φYeO3-12 DNA (scale in kilobases) constructed on the basis of the data in panel A. The restriction fragments are indicated by double-headed arrows, below which the fragment sizes in kilobases are given.
Studier (34) proposed that HpaI restriction patterns could be easily testable and precisely informative in comparing T7-related phages. However, comparison of the HpaI patterns of T3, T7, and φYeO3-12 did not clearly indicate whether φYeO3-12 is more closely related to T7 or to T3 (data not shown).
One-step growth curve.
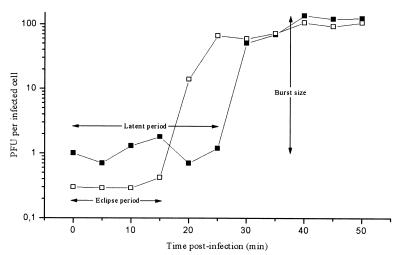
The one-step growth curve of φYeO3-12 propagated on Y. enterocolitica serotype O:3 strain 6471/76-c was determined and is shown in Fig. 3. Eclipse and latent periods of 15 and 25 min, respectively, were observed, followed by a short growth period of 10 min; the burst size was 100 to 140 PFU per infected cell. These values fit into the range observed with T7 group phages.
FIG. 3.
One-step growth curve of bacteriophage φYeO3-12 on Y. enterocolitica serotype O:3 strain 6471/76-c at RT. Shown are the PFU per infected cell in chloroform-treated cultures (□) and in untreated cultures (■) at different time points. Each data point is a mean from three experiments.
T7 group features of φYeO3-12.
It is known that most members of the T7 phage group abortively infect the host strain Shigella sonnei D2 371-48, displaying the same pattern of breakdown of newly synthesized phage DNA (21), while T3 plates normally on this host. S. sonnei D2 was transformed with pAY100, which encodes the φYeO3-12 receptor. φYeO3-12 plated normally on this host (data not shown). In addition to this, T7 and most of its relatives, T3 being the exception, fail to grow in cells harboring the F plasmid (26). Plating assays of φYeO3-12 on isogenic female and male hosts, IJ511 (F−) and IJ512 (F+), transformed with pAY100, showed that φYeO3-12 plated on IJ512/pAY100 (F+) with essentially normal efficiency, and the plaques appeared normal in every way (data not shown).
To further characterize the host requirements of φYeO3-12, we looked at the plating efficiencies of φYeO3-12 on E. coli RNA polymerase rpoC319 and rpoC320 mutants, again transformed with pAY100. T7+ does not form plaques on rpoC319 cells but plates with near-normal efficiency on rpoC320 cells (13), whereas T3+ productively infects rpoC319 cells (14), suggesting that the T3 gp2 (gene product) interaction with E. coli RNA polymerase differs from that of T7 gp2. In line with the T3-like F plasmid results above, φYeO3-12 plated normally on the rpoC319/pAY100 cells (data not shown).
A very common type of variation encountered among different T7 group phage strains is a deletion of some portion of the gene 0.7 region, and plating efficiency on rpoC320 cells gives a good preliminary indication whether a given phage strain carries a deletion (34). T3 and T7 phages defective in the 0.7 gene, which codes for a protein kinase that is involved in host shutoff, are unable to plate on rpoC320 cells (13); however, rpoC320 is known to be less restrictive to 0.7 mutants of T3 than it is to 0.7 mutants of T7 (Ian Molineux, personal communication). Furthermore, it is known that 0.7 deletion mutants grow better (in the laboratory) than wild-type phages and that gene 0.7 is important for growth in poor media and at elevated temperatures (20, 27). As we had serendipitously encountered a faster-growing mutant of φYeO3-12, we wondered whether this mutant was defective in the 0.7 gene. The φYeO3-12 mutant (designated ΔPK) showed no differences when compared to φYeO3-12 in plating efficiencies on rpoC319 and rpoC320 cells (data not shown). φYeO3-12 and ΔPK DNA were digested with the HpaI and EcoRV restriction enzymes. Evident in the HpaI digestion was a ca. 0.7-kb difference in one of the fragments, indicating a putative 1.75% deletion, and in the EcoRV digestion one of the restriction sites was missing, and the 2.7- and 8.2-kb fragments (see Fig. 2) were joined together, giving a ca. 10.2-kb fragment (data not shown), placing the deletion close to one end of the phage genome. Nucleotide sequence analysis of the phages (unpublished data) confirms that ΔPK indeed is a gene 0.7 deletion derivative of φYeO3-12, as suggested by the above results.
Antiserum specific for φYeO3-12 particles.
All three of the immunized rabbits developed good humoral immune responses against φYeO3-12 particles. In EIA analysis using phage particles as the antigen, the mean absorbance values of the preimmune rabbit sera at a 1:8,000 dilution were <0.1. The mean absorbance values of the three rabbit sera at a 1:64,000 dilution were 0.211, 0.260, and 0.463, respectively, when heat-inactivated φYeO3-12 particles were used as the antigen. The absorbance value of the strongest serum at a dilution of 1:256,000 was still significantly higher than the background (0.208), and therefore it was used in later experiments.
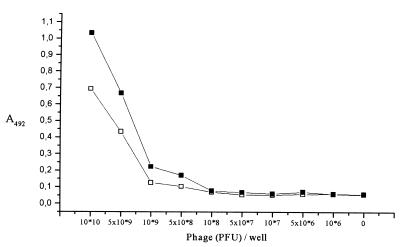
EIA analysis was also used to characterize the antigen-antibody relationships. When decreasing numbers of phage particles were used as the antigen, the sensitivity of the rabbit serum (Fig. 4) was about 5 × 108 PFU (corresponding to ca. 50 ng [total mass] of phage particles) at a 1:2,000 dilution and 109 PFU (ca. 0.1 μg) at a 1:10,000 dilution when native φYeO3-12 particles were used as the antigen, and the mean absorbance values of two times background were considered significant. The sensitivity of 50 ng of antigen (phage in our case) is very typical for an assay of this type (35).
FIG. 4.
Characterization of the rabbit antiserum specific for φYeO3-12 particles. The sensitivity of the antiserum in detection of immobilized native phage particles was assessed using EIA. Shown are the absorbance values obtained using antiserum dilutions of 1:10,000 (□) and 1:2,000 (■). Each data point is a mean from three experiments.
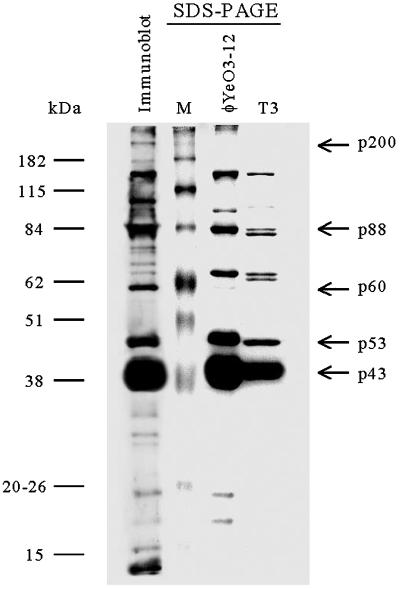
Analysis of the phage structural proteins.
SDS-PAGE of the virion proteins (Fig. 5) revealed one predominant polypeptide with a molecular mass of about 43 kDa (p43), which most likely is the major capsid protein (see the discussion of N-terminal sequences below). From extensively purified phage particles (four to five rounds of ultracentrifugation), at least 10 other polypeptide bands ranging from ca. 15 to >200 kDa could also be detected by Coomassie blue staining. Immunogenic phage proteins were identified using rabbit antiserum specific for φYeO3-12 (Fig. 5). The immunostained protein bands were essentially identical to those seen in the Coomassie-stained gel (Fig. 5), although some qualitative differences were evident: p60, p100, and p200 were overrepresented in the immunoblot. Also, some low-molecular-weight proteins gave relatively stronger signals in the immunoblot than in Coomassie staining. The tail fiber protein of T7 (gp17; Mr, 61,441) forms a trimer, with an average mass of 166 kDa for the trimer (33). It is possible that p60 could be monomeric gp17 of φYeO3-12, and subsequently p200 could be a gp17 trimer of φYeO3-12.
FIG. 5.
SDS-PAGE of the φYeO3-12 and T3 particles and immunoblot of the φYeO3-12 proteins developed with rabbit antiserum. The phage proteins discussed in the text are indicated by arrows at the right. Protein molecular size markers are shown at the left.
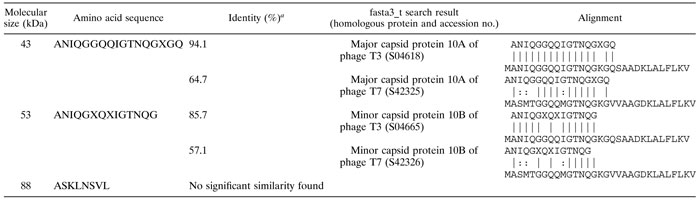
N-terminal amino acid analysis of phage proteins.
N-terminal sequences of the most abundant proteins were determined (Table 3). This analysis indicated that the p43 N terminus was highly similar to that of the major capsid protein 10A (gp10A) of phages T3 and T7 (Table 3). The N terminus of p53 was identical to that of p43. This indicated that, as in T3 and T7, a translational frameshift (−1) may take place close to the end of the p43 gene of φYeO3-12, giving rise to a minor capsid protein (16). The N-terminal sequence of p88 revealed no significant similarities in the databases, indicating that it is a novel protein.
TABLE 3.
N-terminal sequences of φYeO3-12 structural proteins
Computed with respect to query.
Inhibition and neutralization of φYeO3-12 infection.
To characterize the host specificity mechanisms of phage φYeO3-12, inhibition and neutralization tests were performed. The phage infection could be inhibited with purified lipopolysaccharide (LPS) (10 to 100 μg/ml) or with glycerol (1%). The effect of glycerol was noticed by chance after purification of the phage by discontinuous glycerol density gradients. Inhibition by glycerol was reversible when it was diluted below 1%. The mechanism of inhibition by glycerol is not known, but we assume that glycerol in high concentrations can occupy the 6-deoxy-l-altropyranose binding site of the phage adsorption complex. In addition, we found that φYeO3-12 did not degrade the Y. enterocolitica O:3 O antigen when LPS samples from bacteria exposed to the phage were compared to samples from intact bacteria (data not shown).
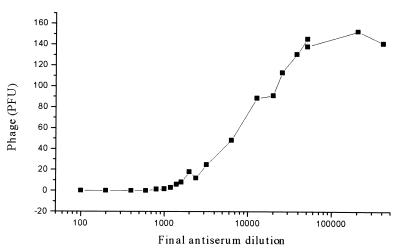
Specific antisera are used to neutralize viruses and phages. The neutralization of φYeO3-12 by the strongest rabbit antiserum was efficient; at a final dilution of 1:10,000, the antiserum inhibited 50% of the ca. 150 PFU of φYeO3-12 (Fig. 6). At a final dilution of 1:20, up to 5 × 106 PFU of φYeO3-12 was totally neutralized, showing the high capacity of the antiserum. Heat treatment of the antiserum at 56°C for 30 min did not affect its efficiency (data not shown), indicating that activation of serum complement did not play a role in neutralization. T7 was not inhibited by the antiserum, in contrast to T3, which was totally neutralized by a 1:2 final dilution and 50% neutralized by a 1:600 final dilution of the antiserum when ca. 100 PFU of T3 particles was used in the assay. On the other hand, T3 is inactivated by anti-T7 serum at about 10% of the homologous rate (3).
FIG. 6.
Neutralization efficiency of the rabbit antiserum specific for φYeO3-12 particles. Shown are obtained PFU values of φYeO3-12 at different antiserum dilutions. Each data point is a mean from one to five experiments.
We wondered whether we could affinity purify neutralizing antibodies from the antiserum using electroblotted phage proteins as the solid phase in preparing monospecific antibodies to different phage proteins. The antibodies were prepared as described in Materials and Methods and then assayed for neutralizing ability against φYeO3-12. Only monospecific anti-p43 antibodies could neutralize φYeO3-12 with about 95% efficiency; the other monospecific antibodies showed no neutralizing activity. The latter finding could be due to one or more of several different factors: (i) the only anti-p43 antibodies are neutralizing, (ii) blotted adsorptive proteins have the wrong conformation and the neutralizing antibodies do not bind to them, and/or (iii) the amount of neutralizing antibodies obtained by the strip affinity purification method was too small. In line with the last point was the fact that anti-p53 antibodies did not show neutralizing activity, but p53 was immunostained when anti-p43 antibodies were used as the primary antibody in immunoblotting (data not shown). The monospecific anti-p43 antibodies were also able to neutralize T3 with about 50% efficiency (data not shown). Further work is needed to elucidate the mechanism behind the neutralizing activity of the anti-p43 antibodies.
Conclusions.
In electron microscopy φYeO3-12 virions showed pentagonal outlines, indicating their icosahedral nature, and thus φYeO3-12 was classified as a typical member of the T7 group. In line with the morphology results, φYeO3-12 showed T7 group features when plating was tested on an F-plasmid-containing host. On E. coli RNA polymerase mutants, φYeO3-12 plated like T3. N-terminal sequences of three structural proteins were determined, and two of them showed strong homology to structural proteins of coliphages T3 and T7. The sequence identity percentages were higher for T3 than for T7, suggesting that φYeO3-12 is more closely related to T3 than to T7. This was further supported by the fact that φYeO3-12-specific antiserum neutralized T3 infection but not T7 infection. The evidence conclusively indicated that φYeO3-12 is the first close relative of phage T3 to be described. The nucleotide sequence of φYeO3-12 (unpublished data) does confirm these conclusions.
ACKNOWLEDGMENTS
Turku Graduate School for Biomedical Sciences, the National Technology Agency, and the Academy of Finland are thanked for financial support.
Jukka Hellman is acknowledged for N-terminal sequencing, and Hans-Wolfgang Ackermann is acknowledged for the electron microscopy of the φYeO3-12 particles. We thank Erkki Eerola and Kirsti Tuomela for HPLC analysis of the fatty acids, Elise Ervelä for LPS degradation studies, Anne Peippo for phage purifications and DNA isolations, and Jyri Kurkinen for digital art images. Ian Molineux's help with bacterial strains, phages, and personal communication is greatly appreciated.
REFERENCES
- 1.Ackermann H-W, DuBow M S. Family Podoviridae. In: Ackermann H-W, DuBow M S, editors. Viruses of prokaryotes. II. Natural groups of bacteriophages. Boca Raton, Fla: CRC Press; 1987. pp. 85–100. [Google Scholar]
- 2.Ackermann H-W, DuBow M S, Gershman M, Karska-Wysocki B, Kasatiya S S, Loessner M J, Mamet-Bratley M D, Regué M. Taxonomic changes in tailed phages of enterobacteria. Arch Virol. 1997;142:1381–1390. [PubMed] [Google Scholar]
- 3.Adams M H, Wade E. Classification of bacterial viruses: the relationship of two Serratia phages to coli-dysentery phages T3, T7, and D44. J Bacteriol. 1954;68:320. doi: 10.1128/jb.68.3.320-325.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Hendy A, Toivanen P, Skurnik M. Expression cloning of the Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb Pathog. 1991;10:47–59. doi: 10.1016/0882-4010(91)90065-i. [DOI] [PubMed] [Google Scholar]
- 5.Al-Hendy A, Toivanen P, Skurnik M. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect Immun. 1992;60:870–875. doi: 10.1128/iai.60.3.870-875.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appleyard R K. Segregation of new lysogenic types during growth of doubly lysogenic strain derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 8.Bailey J N, Dembinski D R, McAllister W T. Derivation of a restriction map of bacteriophage T3 DNA and comparison with the map of bacteriophage T7 DNA. J Virol. 1980;35:176–183. doi: 10.1128/jvi.35.1.176-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birge E A. Bacterial and bacteriophage genetics: an introduction. 2nd ed. New York, N.Y: Springer-Verlag Inc.; 1988. [Google Scholar]
- 10.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley D E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967;31:230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchstein S R, Hinkle D C. Genetic analysis of two bacterial RNA polymerase mutants that inhibit the growth of bacteriophage T7. Mol Gen Genet. 1982;188:211–218. doi: 10.1007/BF00332677. [DOI] [PubMed] [Google Scholar]
- 14.Chamberlin M. Isolation and characterization of prototrophic mutants of Escherichia coli unable to support the intracellular growth of T7. J Virol. 1974;14:509–516. doi: 10.1128/jvi.14.3.509-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen S G. Yersinia enterocolitica in Danish pigs. J Appl Bacteriol. 1980;48:377–382. doi: 10.1111/j.1365-2672.1980.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 16.Condron B G, Gesteland R F, Atkins J F. An analysis of sequences stimulating frameshifting in the decoding of gene 10 of bacteriophage T7. Nucleic Acids Res. 1991;19:5607–5612. doi: 10.1093/nar/19.20.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn J J, Studier F W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 18.Eerola E, Lehtonen O P. Optimal data processing procedure for automatic bacterial identification by gas-liquid chromatography of cellular fatty acids. J Clin Microbiol. 1988;26:1745–1753. doi: 10.1128/jcm.26.9.1745-1753.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero M, de los Reyes-Gavilán C G, Caso J L, Suárez J E. Characterization of φ393-A2, a bacteriophage that infects Lactobacillus casei. Microbiology. 1994;140:2585–2590. [Google Scholar]
- 20.Hirsch-Kauffmann M, Herrlich P, Ponta H, Schweiger M. Helper function of T7 protein kinase in virus propagation. Nature. 1975;255:508–510. doi: 10.1038/255508a0. [DOI] [PubMed] [Google Scholar]
- 21.Hyman R W, Brunovskis I, Summers W C. A biochemical comparison of the related bacteriophages T7, φI, φII, W31, H, and T3. Virology. 1974;57:189–206. doi: 10.1016/0042-6822(74)90120-2. [DOI] [PubMed] [Google Scholar]
- 22.Kawaoka Y, Otsuki K, Tsubokura M. Characteristics of Yersinia enterocolitica bacteriophages. Zentbl Bakteriol Hyg Abt 1 Orig A. 1982;253:102–109. [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Luftig R. An accurate measurement of the catalase crystal period and its use as an internal marker for electron microscopy. J Ultrastruct Res. 1967;20:91–102. doi: 10.1016/s0022-5320(67)80038-8. [DOI] [PubMed] [Google Scholar]
- 25.Matthews R E F. The classification and nomenclature of viruses. Intervirology. 1981;16:53–60. [Google Scholar]
- 26.Molineux I J. Host-parasite interactions: recent developments in the genetics of abortive phage infections. New Biol. 1991;3:230–236. [PubMed] [Google Scholar]
- 27.Molineux I J. The T7 family of bacteriophages. In: Creighton T E, editor. Encyclopedia of molecular biology. New York, N.Y: John Wiley and Co; 1999. pp. 2495–2507. [Google Scholar]
- 28.Moos M, Jr, Nguyen N Y, Liu T Y. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988;263:6005–6008. [PubMed] [Google Scholar]
- 29.Salamitou S, Lemaire M, Fujino T, Ohayon H, Gounon P, Beguin P, Aubert J P. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J Bacteriol. 1994;176:2828–2834. doi: 10.1128/jb.176.10.2828-2834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Skurnik M. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J Appl Bacteriol. 1984;56:355–363. doi: 10.1111/j.1365-2672.1984.tb01362.x. [DOI] [PubMed] [Google Scholar]
- 32.Skurnik M. Molecular genetics of Yersinia lipopolysaccharide. In: Goldberg J, editor. Genetics of bacterial polysaccharides. Boca Raton, Fla: CRC Press; 1999. pp. 23–51. [Google Scholar]
- 33.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A D, Wall J S, Hainfeld J F, Studier F W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 34.Studier F W. Relationships among different strains of T7 and among T7-related bacteriophages. Virology. 1979;95:70–84. doi: 10.1016/0042-6822(79)90402-1. [DOI] [PubMed] [Google Scholar]
- 35.Voller A, Bidwell D. Enzyme-linked immunosorbent assay. In: Rose N R, Friedman H, Fahey J L, editors. Manual of clinical laboratory immunology. Washington, D.C.: American Society for Microbiology; 1986. pp. 99–109. [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]