Abstract
Rapid point-of-care (POC) diagnosis of bacterial infection diseases provides clinical benefits of prompt initiation of the antimicrobial therapy and reduction of the overuse/misuse of unnecessary antibiotics for nonbacterial infections. We present here a point-of-care compatible method for rapid bacterial infection detection in 10 minutes. We use a large volume solution scattering imaging (LVSi) system with low magnifications (1–2X) to visualize bacteria in clinical samples, thus eliminating the need for culture-based isolation and enrichment. We tracked multiple intrinsic phenotypic features of individual cells in a short video. By clustering these features with a simple machine learning algorithm, we can differentiate Escherichia coli from similar-sized polystyrene beads, distinguish bacteria with different shapes, and distinguish E. coli from urine particles. We applied the method to detect urinary tract infections in 104 patient urine samples with a 30-second LVSi video, and the results showed 92.3% accuracy compared with the clinical culture results. This technology provides opportunities for rapid bacterial infection diagnosis at the point-of-care settings.
Keywords: Bacteria detection, UTI screening, multiple phenotypic features, solution scattering imaging, machine learning
Graphical Abstract
A point-of-care compatible UTI screening approach performed directly on a urine sample in less than 10 minutes based on large volume solution scattering imaging (LVSi) tracking of multiple intrinsic bacterial phenotypic features for rapid infection detection (referred to as LVSi-RD).
Every year, over 2.8 million people suffer bacterial infections in the United States, leading to approximately 223,900 hospital admissions and 35,000 deaths. Urinary tract infections (UTI) are the third most common type of bacterial infection (after respiratory and gastrointestinal infections), affecting half of the population during their lifetime and accounting for considerable morbidity and healthcare expenditure with an estimated annual cost of ~$3.5 billion in the US.1–3 Although many UTIs are uncomplicated, they can develop into life-threatening infections, such as sepsis, particularly with the rising incidence of antimicrobial resistance to commonly used antibiotics. The problem of antimicrobial resistance is aggravated by the overuse and misuse of antibiotics in healthcare facilities,4–6 as current diagnostic timelines often necessitate empirical treatments. The pathogens responsible for UTIs pose a high threat of developing antimicrobial resistance, and thus are critical targets for the advancement of rapid diagnostic methods.
Standard diagnostic methods for bacterial identification and antibiotic susceptibility testing (AST) are lab-based and typically take 2–4 days for results to be reported to the provider.7, 8 Rapid point-of-care (POC) urinary tract infection detection methods could help to reduce use of unnecessary antibiotics, to improve the choice of therapy when needed, and to optimize clinical laboratory resources. Traditionally, to identify bacteria at a species/strain level, they are isolated, purified, and identified with a series of observations of cellular and colony morphology along with biochemical tests.9, 10 While faster UTI screening methods like dipstick tests and manual microscopy are available, they are less reliable than the standard culture-based methods that require 24 h or more to produce results.11–14 For high throughput analysis, automated devices based on light scattering analysis or particle imaging recognition show lower sensitivity rates (30–70%) than manual microscopy or culturing for bacterial identification.15, 16 Modern methods, such as Matrix Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry, also require culture-based enrichment and isolation.9, 17 To bypass bacterial culturing, attempts have been made to identify pathogens using real-time Polymerase Chain Reaction (PCR),18, 19 Raman scattering,20 colorimetric detection,21 and imaging methods based on machine22–24 and deep learning25–27. However, these methods have their limitations: real-time PCR is expensive, Raman scattering has limited throughput, the colorimetric detection requires concentration and physiologic-based staining of the bacteria, and traditional machine learning and deep learning-based techniques require extensive datasets for robust generalization capacities and are currently limited to a small number of species and cases.
Single particle analysis is important in characterizing particle analytes, including single cells28. To fully characterize particles and increase specificity, multi-parameter detection and tracking of particles at single particle resolution are preferred29–31. Using light scattering imaging, such as dark-field microscopy, one can obtain the trajectory of particles by capturing and recording the scattering intensity and temporal information of particles31. The analysis of small particle trajectories at high spatial and temporal resolution is a powerful tool that characterizes the mechanisms of particle motion in living cells and other systems22, 32–35. Many existing single cell analysis methods rely on extrinsic markers and biochemical labels that target specific molecules on or in cells, such as nanoparticles, fluorochromes, quantum dots, magnetic beads, and stable isotopes10,11,36. However, labeling requires additional sample processing and usually affects cell viability and downstream analysis37. Cells possess intrinsic features, including size, shape, density, optical, mechanical, and electrical properties, that are detectable without the need for labeling. These intrinsic cellular features function as markers and allow for label-free analysis that can be universally applied to all cells22, 38–40. A digital electrical impedance method has been developed to sensitively measure the metabolic activity and growth of single bacterial cells by reducing the culture volume to nanoliter scale39. However, the method is still focused on the pure cultured bacterial samples and suffers from evaporation issues due to the small sample volume.
Current methods for single bacterial cell tracking involve high magnification imaging (eg., dark-field microscopy) for detailed single bacterial cell analysis34,35 or external labeling for pure bacterial sample detection36, making real sample application in clinical settings difficult. A recent advance in microfluidics introduced an adaptable mechanism to enable fast AST with clinical urine samples. This method flows the clinical samples along microfluidic channels and images cell elongation with high-resolution microscopy.41 However, trapping of bacterial cells requires prior knowledge of bacterial size, and the loading time was long for samples with low bacterial concentrations. This method also cannot differentiate normal bacterial growth from those antibiotics that promotes cell elongation. In our previous work, we achieved direct AST results on clinical urine samples in 60–90 min with large volume solution scattering imaging (LVSi) by tracking single cell division42 and by object scattering intensity detection43. Here we present a point-of-care compatible UTI screening approach that directly analyzes urine samples via LVSi tracking of multiple intrinsic bacterial phenotypic features. This rapid detection (RD) screening method detects bacteria in less than 10 minutes, reducing unnecessary antibiotic use and clinical laboratory resources. LVSi-RD is compatible with POC settings since it allows direct analysis of clinical samples without bacterial enrichment and isolation. Sufficient volume of the specimen can be observed in a single field of view with LVSi for direct detection of clinically relevant concentrations of bacteria. Intrinsic phenotypic features of single particles, including particle size, shape, and motion, can be tracked with scattering light intensities and position profiles. By tracking these intrinsic phenotypic features of single cells in a short video and clustering with a machine learning algorithm, we can differentiate E. coli from similarly-sized polystyrene beads, discriminate between common bacterial morphologies, accurately determine the presence of E. coli and/or similarly-shaped bacteria in urine, and differentiate bacteria from other particles commonly found in urine. We demonstrated the diagnosis of clinical UTIs in patient urine samples with our method and validated the results using traditional culture-based methods and clinical lab testing.
Results and Discussion
Principle
Light scattering is a label-free and non-invasive analytical method for real-time detection and classification of small particle analytes, such as bacteria and cells44–46. Flow cytometry systems, such as the FDA-approved UF-1000i (Sysmex), use a combination of light scattering and fluorescence to rapidly screen for the presence of bacteria in urine44. However, flow cytometry requires expensive and complex instrumentation and needs regular calibration, maintenance, and personnel training. Here, we present a cuvette-based LVSi system with forward scattering detection and associated multi-phenotypic feature tracking algorithms for rapid detection and characterization of bacterial cells (Fig. 1A and Supporting Information Fig. S1). Unlike the flow cytometry system that collects snapshots of information from single particles, LVSi records time-lapsed video of multiple particles and measures the temporal light scattering profile with single cell resolution. As shown in Fig. 1A, LVSi images particles (bacterial cells and impurity particles) in the sample solution as individual bright spots moving over time due to physiological movement, Brownian motion, and thermal reflux. To extract the phenotypic features of individual particles, an automated image processing algorithm is implemented. First, common background noise and image drift are corrected with temporal local minimal subtraction, which improves the image contrast. Second, a Laplace of Gaussian (LOG) filter is used to detect individual cells and particles (bright spots in the image). Third, directional linking of spots in adjacent frames is performed using the Kalman filter to obtain single cell tracking trajectories (Fig. 1A). Finally, the image features, including time profiles of particle scattering intensity and position, are extracted from each trajectory for detailed particle characterization.
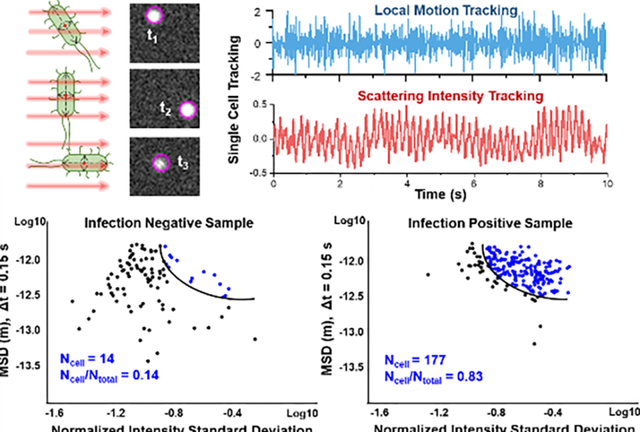
Figure 1. Principle of single cell phenotypic feature tracking for rapid E. coli detection and differentiation from 1 μm polystyrene beads.
(A) Schematic illustration of the experimental setup for single E. coli imaging and temporal trajectory tracking. (B) E. coli rotation-induced scattering intensity fluctuation tracking compared to 1 μm beads. (C) Sub-pixel motion tracking of single E. coli compared to 1 μm beads. (D) Characterization of E. coli cell population from single cell phenotypic features including intensity fluctuation (Normalized Intensity Standard Deviation, NISD) (from panel B) and micro-motion (MSD, mean square displacement) (from panel C).
The scattering intensity provides particle size information as it is proportional to the scattering cross sections of the particle. The intensity fluctuation patterns also depend on particle shape and orientation for non-sphere particles, such as rod-shaped bacterial cells. One example is E. coli, which scatters maximum light when the illumination is perpendicular to the long axis of the cell, and scatters minimal light when illumination is in parallel with the long axis of the cell (Fig. 1B)47. The intensity fluctuates with the free rotation of the bacterial cells in a solution, which can be used for cell size and shape characterization47. To compare the intensity results of different particles, scattering intensity values (It) are normalized to the mean intensity (μ) and the detection accuracy of the normalized intensity is ~ 0.002 (Supporting Information Fig. S2). To quantify the scattering intensity fluctuation of single particles, the normalized intensity standard deviation (NISD) is calculated by:
| [1] |
where It is the scattering intensity profile over time t, μ is the mean intensity, and σ is the intensity standard deviation.
Particles in a solution move due to Brownian motion and thermal drift. For live bacterial cells, the movement also includes intrinsic metabolic activity (ATP-dependent diffusive-like motion) with all viable cells, as well as flagella motor-driven activity with motile bacterial species. 48–51 By carefully tracking the movement of single particles, the intrinsic Brownian motion and cell activity can be extracted for both size and cell viability characterization. In the present LVSi system, the thermal flux induced drift dominates a particle’s overall motion in the 30 s duration video. To remove the low frequency drift effects and extract the micro motion of particles, local movement in the minimal time period was used. Images were recorded at 65 frames per second (fps), so the minimum time for local movement tracking is ~15 ms. The sub-pixel displacement of single cells is measured with a 2-D Laplace of Gaussian (LOG) fitting, from which the center position is derived at each time point (xt, yt), and the local movement is defined as the center position’s displacement between two adjacent frames (Δxt=xt+1-xt, Δyt=yt+1-yt, Fig. 1C). With a tracking accuracy of ~40 nm for local displacement (Supporting Information Fig. S2), the local micro motion of single particles is tracked over time for Brownian motion and cell activity characterization. To quantify the micro motion, the mean square displacement (MSD), 〈[Δx(t)2]〉, for a given duration, lag t, is calculated from the trajectory of each particle, which can be fit to extract physical information, such as the diffusion coefficient (D) of each particle for size estimation. According to the Einstein’s equation:
| [2] |
| [3] |
where t is time, T is Temperature, η is the viscosity of the fluid, and r is the radius of the particle49. Thus, the MSD is inversely proportional to the particle size at a given condition. With both the NISD and MSD obtained, the particle population in the 30 s solution scattering video was clustered for particle classification. Fig. 1D shows one representative classification of E. coli cells.
Differentiate E. coli cells from spherical polystyrene beads
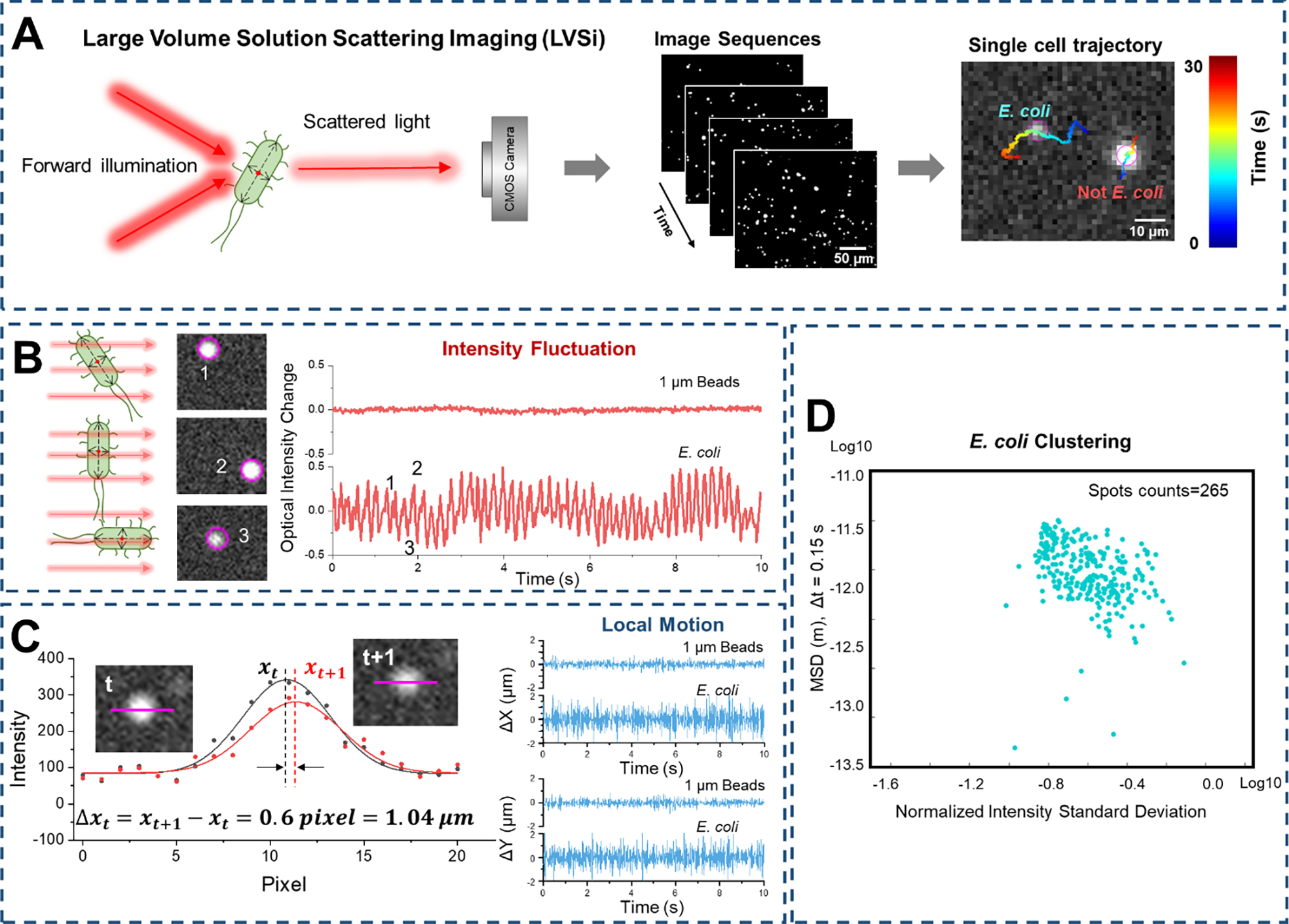
To establish the method, E. coli cultures (see Materials and Methods) and 1 μm polystyrene beads were tested for rapid differentiation and classification. First, pure E. coli cells and 1 μm polystyrene beads in solution were imaged separately with LVSi at 65 fps for 30 s. For each particle in the video, single-cell trajectory was tracked and both the intensity fluctuation and the x-y local movement profile were mapped. One representative result is shown in Figure 2A, in which the E. coli cell shows higher intensity fluctuation and larger micro motion than the abiotic bead. For more accurate comparisons, the local movement along the x axis (Δx, similar for y direction movement) and intensity profile (normalized to the average intensity value) were plotted in Figure 2B. E. coli shows continuous motion with ~ 1 μm oscillation amplitude, while ~ 0.2 μm micro motion is observed from the spherical beads. Also, E. coli shows greater intensity variation compared to 1 μm polystyrene beads. To classify the E. coli cells and beads, a traditional machine learning method, support vector machine (SVM)52, was employed for efficient unsupervised clustering. Figure 2C shows the training result with an accuracy of 98.2% for E. coli and 98.5% for bead classification. To test the trained model, a sample with both E. coli cells and 1 μm polystyrene beads at an approximately 4:1 ratio was measured (Fig. 2D). The classification of particles in the sample using extracted intensity fluctuation (NISD) and local movement (MSD) information confirmed ~80% of the mixed sample as bacterial cells (Fig. 2D).
Figure 2. Differentiation of E. coli from polystyrene beads by phenotypic features tracking.
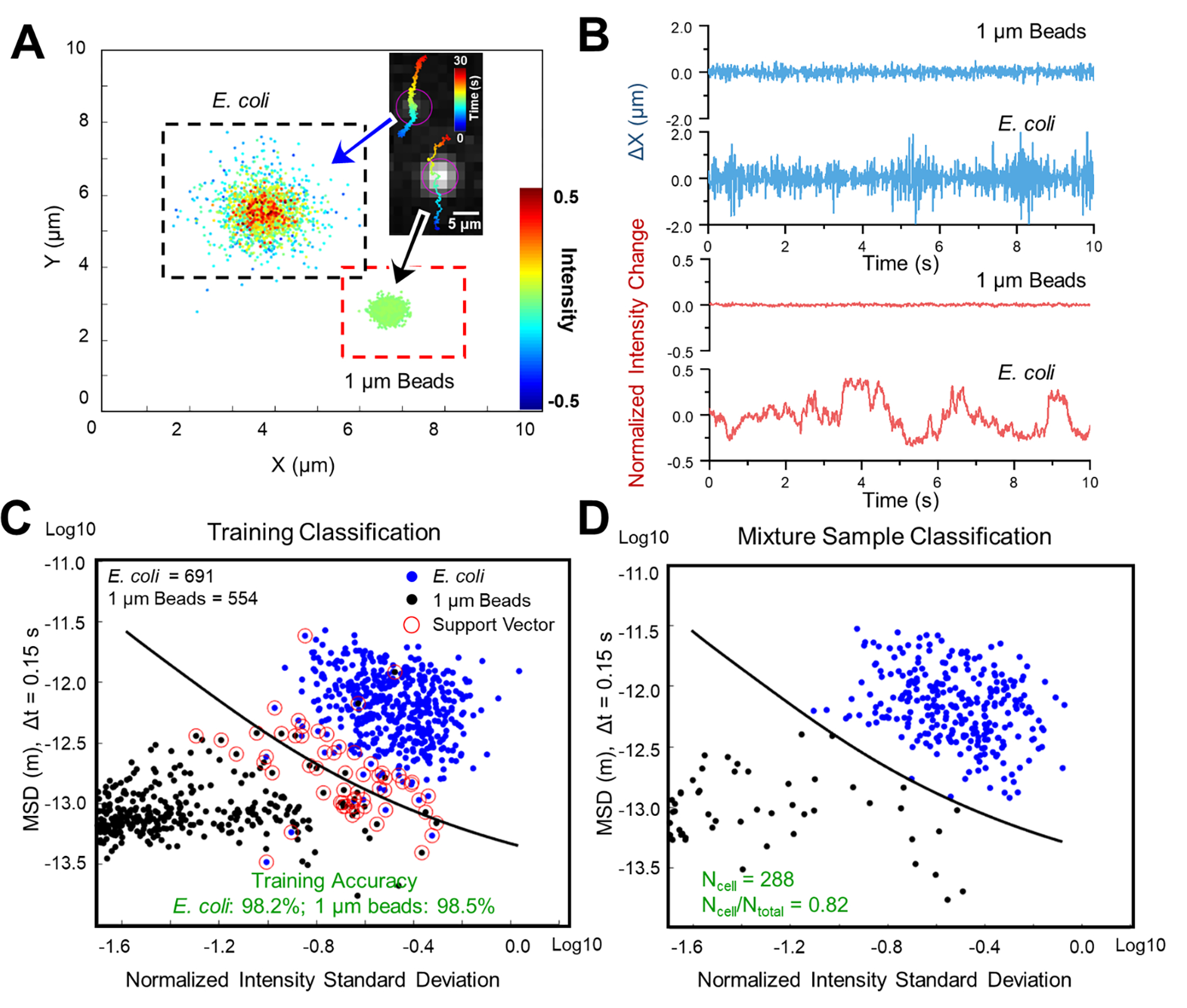
(A) Single cell motion and intensity mapping for E. coli and 1 μm polystyrene beads. (B) Comparison of the corresponding micro motion (top panel) and intensity fluctuation (lower panel) of single E. coli cell and 1 μm polystyrene bead. (C) Training results with machine learning classification (Support Vector Machine, SVM) based on mean squared displacement (MSD) of single cell motion and normalized intensity standard deviation (NISD) of single cell intensity. (D) Classification of a 4:1 mixed sample of E. coli: polystyrene beads with the trained SVM model. Scale bar: 5 μm.
The particle information extracted from the video of their forward light scattering, the x-y locations and intensity as a function of time, are correlated with three key intrinsic particle phenotypic features – size, shape, and activity – and thus can be used to differentiate bacteria from abiotic particles. For spherical abiotic beads, the intensity fluctuations and the local movements were small, due to the homogeneous shape and pure Brownian motion. In contrast, for E. coli cells, the scattering intensity variation over time provides information about the size, rod shape, and the activity state of the cell. The peak intensity correlates with the length of the cell, the valley intensity reflects its width, and the intensity fluctuation pattern indicates the bacterial activities (e.g., metabolic or motor-driven activity35,36). The tracked micro motion of each bacterial cell also reflected the size and activity (e.g., metabolic or motor-driven activity) status of the cell, especially for the motile cells, which showed periodic intensity fluctuation with a certain frequency and regular movement patterns (Supporting Information Fig. S3). Thus, multiple phenotypic features tracking allows for the rapid differentiation of E. coli cells from the homogeneous spherical beads with the LVSi-RD system. Note, in the current LVSi study, most of the bacterial cells lacked large, obvious motor-driven movements (e.g., swimming, running). Therefore, the observed intensity fluctuation and micro motion features were likely related to minimal motile movements, thermal Brownian motion, and/or metabolic activities, providing a more universal detection approach for all live pathogens.
Differentiate rod-shaped bacteria (e.g., E. coli) from spherical-shaped bacteria (e.g., Staphylococcus saprophyticus)
Bacteria are classified, in part, according to their cell shape, such as bacilli (rods), cocci (spheres), and others (spiral, curved). As this information can be useful in determining empirical treatment, we evaluated the capability of LVSi-RD to differentiate bacilli and cocci bacteria. E. coli and S. saprophyticus are used as models for rod- and sphere-shaped cells, respectively, as they are microorganisms frequently associated with UTIs. Both bacterial cultures were prepared according to the Materials and Methods and subject to LVSi-RD with the protocol described above. The intensity-motion mapping obtained from representative cell trajectories for each organism is shown in Figure 3A. E. coli exhibited approximately double the motion in both x and y direction, and broader intensity range compared to S. saprophyticus. Figure 3B plots the time profile of x direction motion and normalized intensity. The intensity profile of E. coli shows more frequent intensity fluctuations (Fig. 3B, upper panel), indicating the rotational motion of the rod-shaped cell. However, the NISD for E. coli and S. saprophyticus cells is similar (Fig. 3B, lower panel), perhaps due to the movement of S. saprophyticus cell clusters. The NISD and MSD of the local movement of the bacterial cell are calculated and trained with the SVM algorithm. The training accuracies were ~81.3% and 81.5% for E. coli and S. saprophyticus, respectively (Fig. 3C). To test the trained model, we obtained another set of data from individual pure cultures of both E. coli cells (n = 109) and S. saprophyticus cells (n = 96) and classified them with the trained model (Fig. 3D). The pure culture testing accuracies were 84.4% and 88.5% for E. coli and S. saprophyticus, respectively (Fig. 3D). These results show that with LVSi and machine learning classification, we can distinguish bacilli from cocci bacteria, providing valuable information for initial bacterial differentiation. Our method mainly relies on the size and shape differences, with the potential to differentiate bacterial cells with similar shape, such as E. coli and Klebsiella pneumoniae (Supporting Information Fig. S4). Although the current classification accuracies were less than 70% for differentiating E. coli and Klebsiella pneumoniae, with optimization of the imaging system and additional data for training and calibration of a deep learning algorithm, we anticipate LVSi-RD can include additional phenotypic features (such as active movement and aspect ratio) to aid comprehensive interpretation of the urine sample.
Figure 3. Differentiation of rod-shaped bacteria (E. coli) and spherically-shaped bacteria (S. saprophyticus) by single-cell phenotypic features tracking.
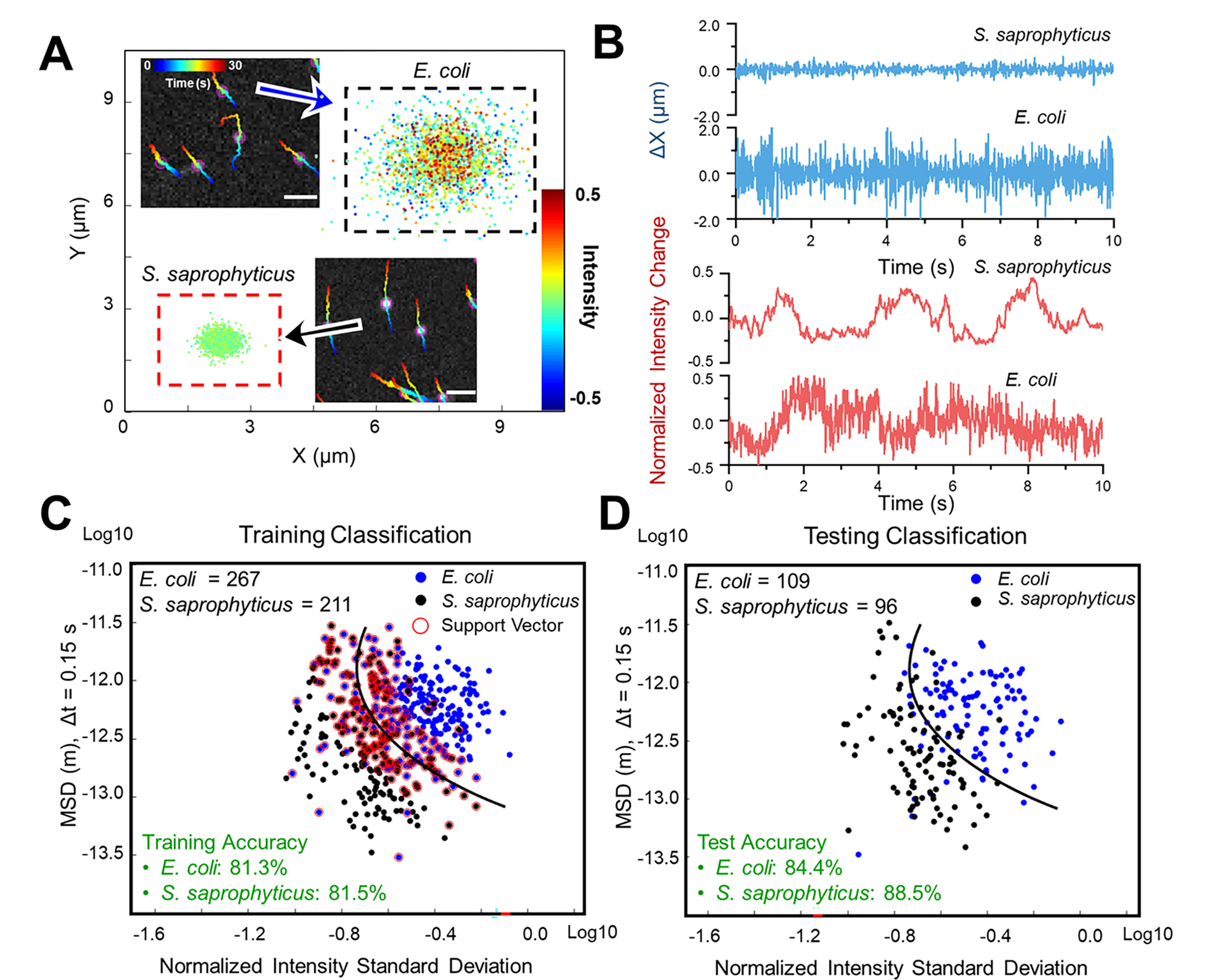
(A) Single cell motion and intensity mapping for E. coli and S. saprophyticus. (B) Comparison of the corresponding micro motion (upper panel) and intensity fluctuation (lower panel) for single E. coli and S. saprophyticus cells. (C) Training results obtained from individual pure cultures of E. coli (n = 267) and S. saprophyticus (n = 211) with machine learning classification (Support Vector Machine, SVM) based on mean squared displacement (MSD) of single cell motion and normalized intensity standard deviation (NISD) of single cell intensity. (D) Testing results obtained from individual pure cultures of both E. coli cells (n = 109) and S. saprophyticus cells (n = 96) with the trained SVM model. Scale bar: 20 μm.
Differentiate E. coli from urine particles
We have demonstrated that our LVSi and machine learning classification can be used to differentiate E. coli from polystyrene beads and distinguish between rod-shaped and spherically-shaped bacteria. However, to diagnose UTIs, the method must differentiate bacterial cells and non-bacterial particles in urine samples. Nonbacterial particles in urine originate from various sources and are heterogeneous in size, shape, and concentration. This dramatic heterogeneity makes them inherently difficult to classify as some particles may display structural features that are similar to bacterial cells. To test LVSi-RD for urine samples, we obtained LVSi video of pure E. coli cultures and pooled, healthy urine samples with particles (no bacteria present) for NISD and MSD tracking and SVM model training. Figure 4A shows the trajectory and intensity-motion mapping of both E. coli and urine particles with very small differences observed directly. By further plotting the x direction motion and intensity fluctuation profile, we observe some minimal intensity amplitude differences, while the x direction motion patterns are indistinguishable (Fig. 4B). We proceeded to train the SVM algorithm with the NISD and MSD data and found the accuracy for E. coli classification remains high (94.1%), while the accuracy for urine particles was 63.9% (Fig. 4C). These results imply that E. coli cells showed consistent features while urine particle characteristics varied considerably. With the trained model, we tested another set of data with 80 E. coli cells and 66 urine particles and obtained testing accuracies of 90% for E. coli detection and 62.1% for urine particles classification (Fig. 4D). Thus, LVSi-RD can detect the presence of E. coli cells in a urine sample with high sensitivity. Although urine particles may contribute to some false positive bacterial counts, it may not affect the accuracy for signifying a bacterial-positive sample with an optimized infection threshold, as described in the next section.
Figure 4. Differentiation of bacterial cells from urine particles by phenotypic features tracking.
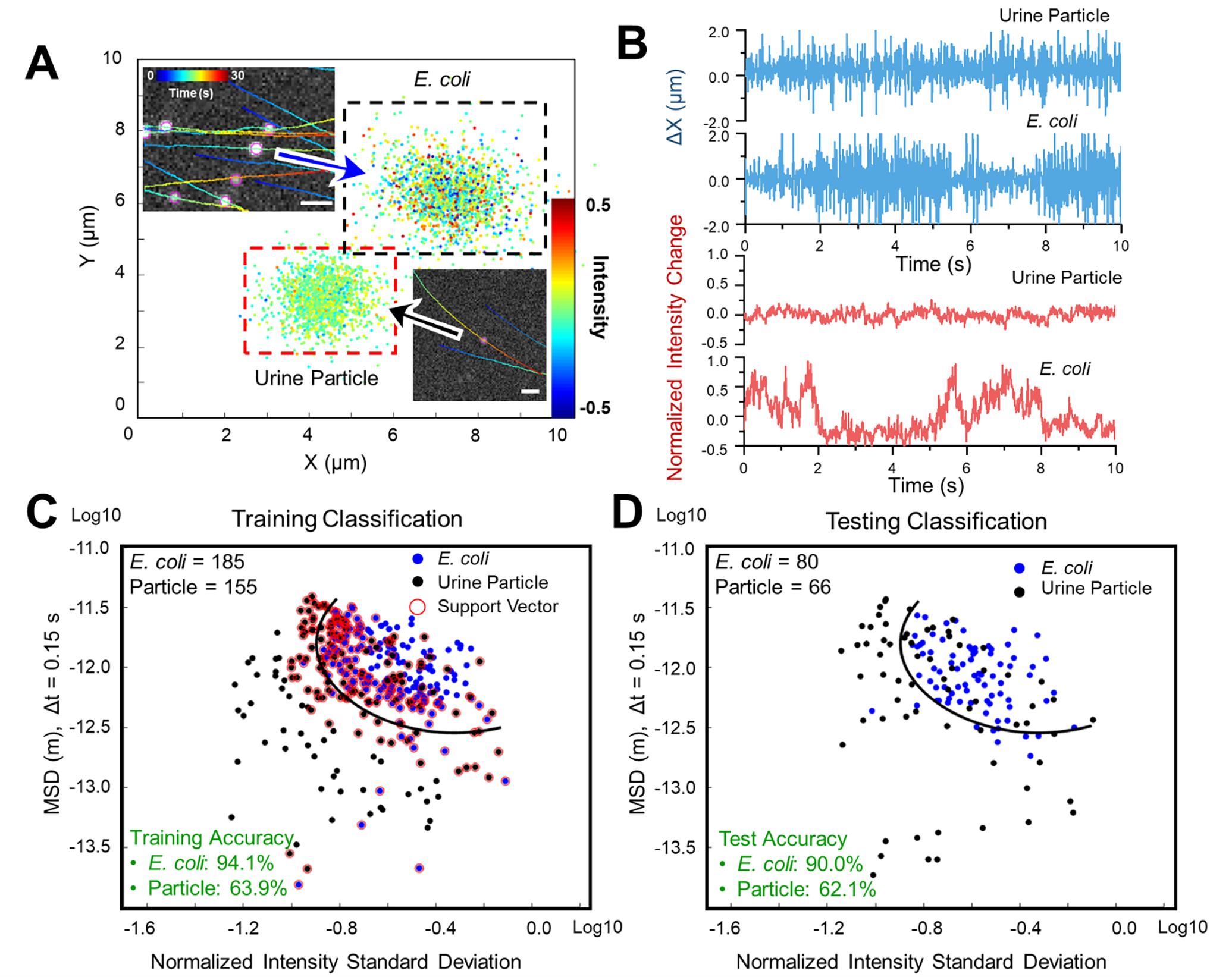
(A) Single cell motion and intensity mapping for cultured E. coli cells and urine particles. (B) Comparison of the corresponding micro motion (top panel) and intensity fluctuation (lower panel) of a single E. coli cell and a single urine particle. (C) The corresponding training results of E. coli (n = 185) and urine particles (n = 155) with machine learning classification (Support Vector Machine, SVM) based on mean squared displacement (MSD) of single cell motion and normalized intensity standard deviation (NISD) of single cell intensity. (D) The corresponding testing results of E. coli (n = 80) and urine particles (n = 66) with the trained SVM model. Scale bar: 20 μm.
UTI detection with clinical samples
To clinically assess LVSi-RD, we first determined an infection threshold and then used it to test 104 clinical urine samples to screen samples for UTI detection (Fig. 5). Since E. coli is the most predominant pathogen causing 80–90% of the community-acquired UTIs, the trained model of distinguishing E. coli from urine particles was used for clinical UTI screening. To determine a proper infection threshold, the receiver operating characteristic (ROC) curve was constructed using the relative amounts of cells/all particles (NCell/NTotal) as a predictor, and results were evaluated to determine the infection threshold for UTIs. To obtain a proper threshold of this trained model for real clinical samples, we validated the model with 20 identified clinical samples (10 were positive and 10 were negative; determined by the clinical microbiological culture), which contain mixtures of bacterial cells and urine particles with different ratios. From the ROC curve of these 20 samples, we determined the infection threshold of 0.5 with a sensitivity of 90% and a specificity of 100% (Supporting Information Fig. S5). The determined threshold was above the false positive rate (<0.4) caused by the urine particles in the trained model (Fig. 4D). Note, with the determined infection threshold (TI) of 0.5, the trained model for E. coli differentiation from urine particles could also apply to the similarly sized and shaped bacteria of the Enterobacteriaceae family, such as K. pneumoniae (Supporting Information Fig. S6). Based on the infection threshold (TI = 0.5), the sample was classified as UTI positive when the NCell/NTotal > TI. A representative negative sample (Sample #11), shown in Fig. 5A, exhibits the SMV model particle characterization in which a small relative proportion of particles were determined to be bacterial cells, while more than 80% of the particles were classified as bacterial cells in a positive sample (Sample #12), shown in Figure 5B. Using a threshold of 0.5, 61 of the 104 clinical samples exhibited low ratio of bacterial cells to total particles (NCell/NTotal) and were categorized as LVSi-RD detection negative, while the remaining 43 samples revealed E. coli or other similar-sized, rod-shaped bacterial cells (e.g., K. pneumoniae and other Gram-negative bacilli) and were categorized as infection LVSi-RD detection positive (Supporting Information Table S1). For cross validation, these results were compared with the gold standard culture-based method results obtained in the Microbiology Lab at Mayo Clinic, where the samples were collected (Fig. 5C, Supporting Information Table S1). The classification results were also evaluated using the ROC curve constructed using NCell/NTotal of all clinical samples (Fig. 5D). From the ROC curve of 104 samples, with the infection threshold of 0.5, we detected positive UTI infections with a sensitivity of 84% and a specificity of 100% (Supporting Information Table S1). Eight false negative samples were determined from the 104 samples tested, demonstrating an accuracy of 92.3% for rapid UTI detection (Supporting Information Fig. S7). The parallel microbiological agar plating results showed that most of these false negative samples had bacterial concentrations below 1,000 CFU/mL after sample preparation (Supporting Information Fig. S8), which is below the commonly used UTI clinical thresholds of 10,000 to 100,000 CFU/mL as a concerning level of bacteria 53.
Figure 5. Rapid infection detection with the trained SVM model.
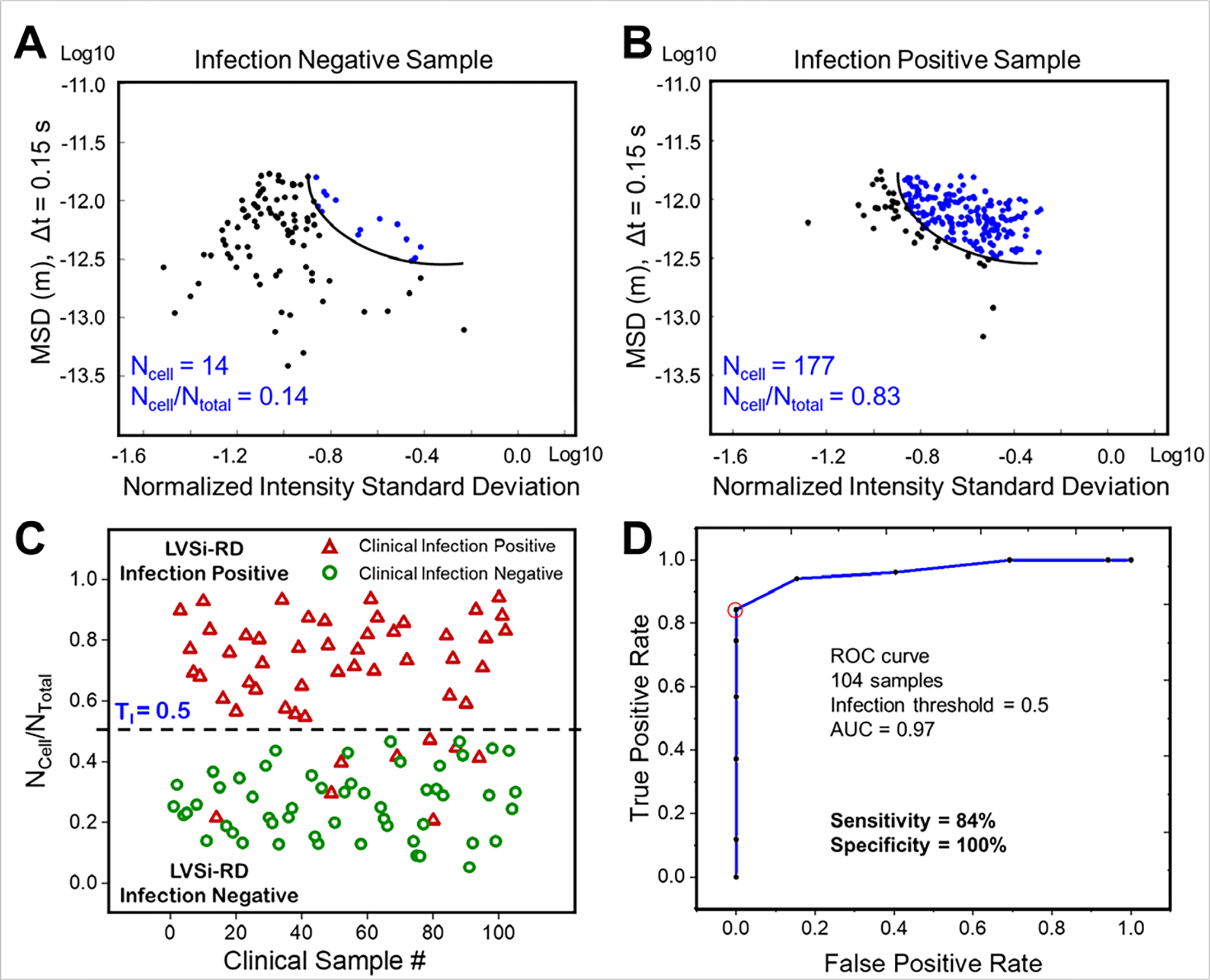
(A) SVM classification result of one representative infection negative sample (Sample #11). (B) SVM classification result of one representative infection positive sample (Sample #12). (C) Comparison of culture-based detection and phenotypic tracking for LVSi-RD of UTIs. TI indicates the determined infection threshold. (D) The ROC curve for UTI diagnosis evaluation from LVSi-RD. At the threshold of 0.5, the sensitivity and specificity were 84% and 100%, respectively.
The present LVSi-RD system has an imaging volume of ~5 μL, allowing imaging of sufficient numbers of bacterial cells simultaneously in urine samples at clinically-relevant concentrations (>10,000 CFU/mL). However, when the bacterial concentration is below 1,000 cells/mL and less than 5 cells can be detected, the current classification protocol does not accurately detect bacterial infections. However, this bacterial concentration is likely an appropriate level for detection accuracies to decrease, since the clinical value of these levels is unclear 53. Each sample underwent initial on-site microbiological plating validation with overnight culturing after the samples were transported to the research lab. Among the eight false negative samples, one sample (#94; Supporting Information Table S1) tested negative with the initial on-site plating validation. It is likely due to the viability lost during the cold storage and transportation, which could be avoided when evaluated on-site with fresh urine. Another five of the remaining seven samples (#14, #52, #79, #80, and #87; Supporting Information Table S1) were validated on-site with concentrations between 104-105 CFU/mL, while the remaining two false negative samples (#49 and #69; Supporting Information Table S1) displayed bacterial concentrations higher than 105 CFU/mL (Supporting Information Fig. S7). After all sample handling, including prewarming, filtration, and dilution steps, parallel microbiological plating validation results revealed low counts of bacterial cells (below 1,000 cells/mL, Supporting Information Fig. S8) for five of the false negative samples. Therefore, most of the false negative results were due to a combination of low initial bacterial concentration and sample handling procedures, such as filtration or dilution. We anticipate the rate of false negative results can be reduced after optimization of the sample handling protocol.
The current sample handling protocol is outlined here: Clinical samples are collected, refrigerated for 12–24 h, and transported on ice to the research lab. To mimic a fresh and warm urine sample, a 30-min prewarming incubation at 37°C was performed prior to testing. Next, to remove large particles, the sample was filtered, and then diluted with microbiological media. To enable single particle tracking, a quick check of total particle concentration was performed in LVSi after the initial 10X dilution of the sample. Additional dilutions of the sample were carried out to achieve a concentration of ~5 × 104 particles/mL. These simple steps averaged ~ 2 minutes for each sample. Then, the sample was subjected to LVSi for 30 s video detection. The final tracking of multi-phenotypic features and SVM clustering with a trained model took ~ 5 min. Therefore, the current total assay time for LVSi-RD of a clinical sample includes 30 min sample pre-warming, 2 min sample pre-treatment (filtration/dilution), 30 s LVSi detection, and 5 min multi-phenotypic features tracking and SVM clustering for a total of 37.5 min. Ultimately, in a point-of-care setting, the prewarming step would not be necessary for fresh urine samples and the sample pre-treatment time could be reduced with an improved sample collection device that integrates a filter. Thus, the total time for LVSi-RD method of infection determination can be as short as ~5–6 min, superior to 2–3 days of the gold standard urine culture tests and 3 hours of FDA approved UTI POCT devices (eg. BacterioScan) for UTI screening.
Conclusion
We demonstrated that a time-lapse large-volume solution scattering imaging system can be used for rapid UTI detection in clinical samples via label-free tracking of multiple phenotypic features. By real-time tracking of local motion, intensity, and intensity fluctuation of individual bacteria and particles in solution, different particles (including cells and polystyrene beads), bacterial cells with different shapes, and bacterial cells and urine particles can be distinguished and classified with a simple machine learning algorithm. By applying the classification model to 104 clinical urine samples, the model accurately predicted 92.3% of the clinically identified infection-positive samples containing rod-shaped bacterial cells including E. coli, K. pneumoniae, and Gram-negative bacilli, which dominate UTI pathogens. In summary, LVSi-RD can test clinical urine samples without overnight culturing and detect rod-shaped bacterial cells in a total assay time of less than 10 minutes.
Materials and Methods
Materials.
E. coli ATCC 25922, S. saprophyticus ATCC 15305, and K. pneumoniae ATCC 25955 were purchased from American Type Culture Collection (ATCC) and stored at −80°C in 5% glycerol. Polystyrene beads (1 μm) were purchased from Bangs Laboratories, Inc and suspended in PBS. Pooled, healthy human urine purchased from BioereclamationIVT (Lot number: BRH1311635) was filtered using 5 μm syringe filters.
Sample preparation.
E. coli, S. saprophyticus, and K. pneumoniae were grown overnight (~15 h) in Luria−Bertani (LB) broth (per liter: 10 g peptone 140, 5 g yeast extract, and 5 g sodium chloride) or Mueller Hinton Broth (MHB, per liter: 2.0 g beef infusion solids, 1.5 g starch, and 17.5 g casein hydrolysate) at 37°C with agitation. E. coli and S. saprophyticus cultures were diluted in fresh LB broth or MHB, respectively, to concentrations of ~5 × 104 cells/mL. Bacterial suspensions (70 μL) were transferred into a cuvette at 37 °C for imaging. Similarly, pooled, healthy human urine purchased from BioereclamationIVT (Lot number: BRH1311635) was filtered using 5 μm syringe filters and diluted in fresh LB broth to concentrations of ~5 × 104 particles/mL. Urine particle suspensions (70 μL) were transferred into a cuvette at 37 °C for imaging.
Clinical urine samples.
De-identified excess and residual clinical urine samples were obtained from the clinical microbiology laboratory at Mayo Clinic Hospital, Phoenix, Arizona (Approved by Mayo Clinic Biospecimen Subcommittee BIO00015462). Clinical urine samples were stored at 4°C and transported in an insulated box with ice packs. Prior to processing, urine samples were pre-warmed for 30 min at 37°C for cell recovery. In the current protocol (Supporting Information Fig. S9), the clinical sample was filtered with a 5 μm pore size filter to remove large substances and then diluted with microbiological media to supply nutrients. The sample dilution process includes an initial 10-fold dilution with microbiological media to provide bacterial nutrients. Then, the sample is measured in the LVSi imaging system and a 10 s video was processed (the first 4 steps of video processing in Supporting Information Fig. S10) for a quick count of particle concentration. If the concentration is greater than 105 particles/mL, additional dilutions are performed to ensure the final concentration of detectable particles is between 5 × 104 and 105 particles/mL. The sample dilution process takes about 2–5 minutes for imaging, counting, and dilutions. After mixing, diluted samples (70 μL) were transferred to cuvettes (Uvette, Eppendorf, Germany), and subjected to LVSi. A total of 104 clinical urine samples were tested in a blinded and independent fashion with 1) standard microbiological plating in the Microbiological Lab at Mayo Clinic Arizona, 2) our LVSi-RD testing, and 3) the parallel agar plating for on-site cross validation in the research lab. Upon completion of each experimental batch, direct comparison was performed between LVSi-RD and the gold standard from both clinical site and on-site validation (Supporting Information Table S1)
LVSi.
The large volume scattering imaging system (Fig. 1A and Supporting Information Fig. S1) consists of an 800 mW, 780 nm infrared (IR, Bandwidth: 30 nm) LEDs (M780LP1, Thorlabs, Inc., USA), with collimating and focusing lens and a central blocking aperture to focus a ring-shaped illumination through the sample or the reference cuvettes. Wide-view and deep field depth scattering images were recorded by a CMOS camera (BFS-U3–16S2M-CS, Point Grey Research Inc., Canada) at 65 fps through the variable zoom lenses (NAVITAR 12X, Navitar, USA) with zoom factors set at 2.0X for the sample cuvettes. The image volume was determined by the viewing size and focal depth of the optics. For the experiments described in this study, the viewing volume of 2.5 mm × 1.9 mm × 1.0 mm was equivalent to 4.8 μL at 2.0X magnifying power. The sample cuvette was placed on a temperature-controlled breadboard (PTC1, Thorlabs, Inc., USA) to maintain temperature at 37 °C.
Biosafety.
All sample preparations and measurements were performed in biosafety level 2 (BSL2) laboratories following an IBC-approved BSL2 protocol.
Single particle tracking.
Individual cells recorded by LVSi are resolved as bright spots, and the image sequences were taken as stacks and analyzed by ImageJ plug-in TrackMate.54 First, each spot was detected with a Laplacian of Gaussian (LoG) filter with a defined radius and threshold. Then, the spots from adjacent time frames were connected with a Karman filter for directional linking, so that each bright spot became a single-cell trajectory with both intensity and x, y position information at each frame (Supporting Information Fig. S10).
Support Vector Machines (SVM).
For each single particle trajectory, we extracted a set of handcrafted features based on intensity and position information. Starting with the intensity features, we obtained the statistical features over 30 sec. Statistic features, including mean, standard deviation, normalized intensity standard deviation (NISD), kurtosis, skewness, quartiles, and motion features, including local mean displacement, MSD, kurtosis, and skewness, were extracted. SVM were utilized for their ability to find optimal separation hyperplanes in binary classification, with radial basis function (RBF) kernels. A list with all 10 features was tested, among which NISD of the intensity and MSD of the local displacement provided the most accurate classification results. Therefore, these two features were used in this study and paper for LVSi-RD (Supporting Information Fig. S10).
Supporting Information
Supporting Information Available: the prototype LVSi system; evaluation of tracking accuracy; comparison of intensity fluctuation; differentiation of E. coli and K. pneumoniae by phenotypic features tracking; the ROC curve for threshold determination with the first 20 clinical urine samples; classification of different samples with the trained model of distinguishing E. coli from urine particles; clinical urine sample rapid detection results; initial sample validation results; initial and parallel plating validation result of 8 false negative samples; flow chart for clinical urine sample preparation; flow chart for LVSi video processing and machine learning.
Supplementary Material
ACKNOWLEDGMENT
Financial support from NIH (R01AI138993) is acknowledged.
Footnotes
The authors declare no competing financial interest.
Reference
- 1.Griebling TL, Urologic diseases in america project: trends in resource use for urinary tract infections in men. J Urol 2005, 173 (4), 1288–94. [DOI] [PubMed] [Google Scholar]
- 2.Griebling TL, Urologic diseases in America project: trends in resource use for urinary tract infections in women. J Urol 2005, 173 (4), 1281–7. [DOI] [PubMed] [Google Scholar]
- 3.Foxman B, The epidemiology of urinary tract infection. Nat Rev Urol 2010, 7 (12), 653–60. [DOI] [PubMed] [Google Scholar]
- 4.The antibiotic alarm. Nature 2013, 495 (7440), 141. [DOI] [PubMed] [Google Scholar]
- 5.Gross M, Antibiotics in crisis. Curr Biol 2013, 23 (24), R1063–5. [DOI] [PubMed] [Google Scholar]
- 6.Van Boeckel TP; Gandra S; Ashok A; Caudron Q; Grenfell BT; Levin SA; Laxminarayan R, Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 2014, 14 (8), 742–750. [DOI] [PubMed] [Google Scholar]
- 7.Liu C; Bayer A; Cosgrove SE; Daum RS; Fridkin SK; Gorwitz RJ; Kaplan SL; Karchmer AW; Levine DP; Murray BE; M JR; Talan DA; Chambers HF; Infectious Diseases Society of, A., Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin Infect Dis 2011, 52 (3), e18–55. [DOI] [PubMed] [Google Scholar]
- 8.Mammina C, The global crisis of multidrug resistance: how to face healthcare associated infections without effective antibiotics? Iran J Microbiol 2013, 5 (2), 99–101. [PMC free article] [PubMed] [Google Scholar]
- 9.Davenport M; Mach KE; Shortliffe LMD; Banaei N; Wang TH; Liao JC, New and developing diagnostic technologies for urinary tract infections. Nat Rev Urol 2017, 14 (5), 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kshikhundo RI, S., Bacterial species identification. World News of Natural Sciences 2016, 3, 26–38. [Google Scholar]
- 11.Mori R; Yonemoto N; Fitzgerald A; Tullus K; Verrier-Jones K; Lakhanpaul M, Diagnostic performance of urine dipstick testing in children with suspected UTI: a systematic review of relationship with age and comparison with microscopy. Acta Paediatr 2010, 99 (4), 581–4. [DOI] [PubMed] [Google Scholar]
- 12.Whiting P; Westwood M; Watt I; Cooper J; Kleijnen J, Rapid tests and urine sampling techniques for the diagnosis of urinary tract infection (UTI) in children under five years: a systematic review. BMC Pediatr 2005, 5 (1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vazouras K; Velali K; Tassiou I; Anastasiou-Katsiardani A; Athanasopoulou K; Barbouni A; Jackson C; Folgori L; Zaoutis T; Basmaci R; Hsia Y, Antibiotic treatment and antimicrobial resistance in children with urinary tract infections. J Glob Antimicrob Resist 2020, 20, 4–10. [DOI] [PubMed] [Google Scholar]
- 14.Wilson ML; Gaido L, Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis 2004, 38 (8), 1150–8. [DOI] [PubMed] [Google Scholar]
- 15.Shayanfar N; Tobler U Fau - von Eckardstein A; von Eckardstein A Fau - Bestmann L; Bestmann L, Automated urinalysis: first experiences and a comparison between the Iris iQ200 urine microscopy system, the Sysmex UF-100 flow cytometer and manual microscopic particle counting. (1434–6621 (Print)). [DOI] [PubMed] [Google Scholar]
- 16.van den Broek D; Keularts IM; Wielders JP; Kraaijenhagen RJ, Benefits of the iQ200 automated urine microscopy analyser in routine urinalysis. Clin Chem Lab Med 2008, 46 (11), 1635–40. [DOI] [PubMed] [Google Scholar]
- 17.Kaleta EJ; Clark AE; Cherkaoui A; Wysocki VH; Ingram EL; Schrenzel J; Wolk DM, Comparative analysis of PCR-electrospray ionization/mass spectrometry (MS) and MALDI-TOF/MS for the identification of bacteria and yeast from positive blood culture bottles. Clin Chem 2011, 57 (7), 1057–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dark PM; Dean P; Warhurst G, Bench-to-bedside review: the promise of rapid infection diagnosis during sepsis using polymerase chain reaction-based pathogen detection. Crit Care 2009, 13 (4), 217–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niemz A; Ferguson TM; Boyle DS, Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol 2011, 29 (5), 240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosch P; Harz M; Schmitt M; Peschke KD; Ronneberger O; Burkhardt H; Motzkus HW; Lankers M; Hofer S; Thiele H; Popp J, Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl Environ Microbiol 2005, 71 (3), 1626–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael I; Kim D; Gulenko O; Kumar S; Kumar S; Clara J; Ki DY; Park J; Jeong HY; Kim TS; Kwon S; Cho YK, A fidget spinner for the point-of-care diagnosis of urinary tract infection. Nat Biomed Eng 2020, 4 (6), 591–600. [DOI] [PubMed] [Google Scholar]
- 22.Jo Y; Jung J; Kim M.-h.; Park H; Kang S-J; Park Y, Label-free identification of individual bacteria using Fourier transform light scattering. Opt. Express 2015, 23 (12), 15792–15805. [DOI] [PubMed] [Google Scholar]
- 23.He Y; Xu W; Zhi Y; Tyagi R; Hu Z; Cao G, Rapid bacteria identification using structured illumination microscopy and machine learning. Journal of Innovative Optical Health Sciences 2017, 11 (01), 1850007. [Google Scholar]
- 24.Liu H; Li Z; Shen R; Li Z; Yang Y; Yuan Q, Point-of-Care Pathogen Testing Using Photonic Crystals and Machine Vision for Diagnosis of Urinary Tract Infections. Nano Lett 2021, 21 (7), 2854–2860. [DOI] [PubMed] [Google Scholar]
- 25.John AQ; Rose N; Pius KBM; Patrick B; William L; Alfred A, Deep Convolutional Neural Networks for Microscopy-Based Point of Care Diagnostics. PMLR: 2016; pp 271–281. [Google Scholar]
- 26.Liang Y; Kang R; Lian C; Mao Y, An End-to-End System for Automatic Urinary Particle Recognition with Convolutional Neural Network. J Med Syst 2018, 42 (9), 165. [DOI] [PubMed] [Google Scholar]
- 27.Hay EA; Parthasarathy R, Performance of convolutional neural networks for identification of bacteria in 3D microscopy datasets. Plos Comput Biol 2018, 14 (12), e1006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cong Y; Ludtke SJ, Single particle analysis at high resolution. Methods Enzymol 2010, 482, 211–35. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y; Yu H; Shan X; Wang W; Liu X; Wang S; Tao N, Label-Free Tracking of Single Organelle Transportation in Cells with Nanometer Precision Using a Plasmonic Imaging Technique. Small (Weinheim an der Bergstrasse, Germany) 2015, 11 (24), 2878–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taute KM; Gude S; Tans SJ; Shimizu TS, High-throughput 3D tracking of bacteria on a standard phase contrast microscope. Nat Commun 2015, 6, 8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian H; Sheetz MP; Elson EL, Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys J 1991, 60 (4), 910–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adan A; Alizada G; Kiraz Y; Baran Y; Nalbant A, Flow cytometry: basic principles and applications. Crit Rev Biotechnol 2017, 37 (2), 163–176. [DOI] [PubMed] [Google Scholar]
- 33.Babick F, Chapter 3.2.1 - Dynamic light scattering (DLS). In Characterization of Nanoparticles, Hodoroaba V-D; Unger WES; Shard AG, Eds. Elsevier: 2020; pp 137–172. [Google Scholar]
- 34.Yoon J; Lee K; Park Y. J. a. Q. M., A simple and rapid method for detecting living microorganisms in food using laser speckle decorrelation. 2016. [Google Scholar]
- 35.Macnab RM, Examination of Bacterial Flagellation by Dark-Field Microscopy. Journal of Clinical Microbiology 1976, 4 (3), 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai M; Mine K; Tomonari H; Uchiyama J; Matuzaki S; Niko Y; Hadano S; Watanabe S, Dark-Field Microscopic Detection of Bacteria using Bacteriophage-Immobilized SiO2@AuNP Core-Shell Nanoparticles. Anal Chem 2019, 91 (19), 12352–12357. [DOI] [PubMed] [Google Scholar]
- 37.Hunt HK; Armani AM, Label-free biological and chemical sensors. Nanoscale 2010, 2 (9), 1544–59. [DOI] [PubMed] [Google Scholar]
- 38.Freudiger CW; Min W; Saar BG; Lu S; Holtom GR; He C; Tsai JC; Kang JX; Xie XS, Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science 2008, 322, 1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scherer B; Surrette C; Li H; Torab P; Kvam E; Galligan C; Go S; Grossmann G; Hammond T; Johnson T; St-Pierre R; Nelson JR; Potyrailo RA; Khire T; Hsieh K; Wang TH; Wong PK; Puleo CM, Digital electrical impedance analysis for single bacterium sensing and antimicrobial susceptibility testing. Lab Chip 2021, 21 (6), 1073–1083. [DOI] [PubMed] [Google Scholar]
- 40.Zhang F; Wang S; Yang Y; Jiang J; Tao N, Imaging Single Bacterial Cells with Electro-optical Impedance Microscopy. ACS Sens 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H; Torab P; Mach KE; Surrette C; England MR; Craft DW; Thomas NJ; Liao JC; Puleo C; Wong PK, Adaptable microfluidic system for single-cell pathogen classification and antimicrobial susceptibility testing. Proc Natl Acad Sci U S A 2019, 116 (21), 10270–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F; Jiang J; McBride M; Yang Y; Mo M; Iriya R; Peterman J; Jing W; Grys T; Haydel SE; Tao N; Wang S, Direct Antimicrobial Susceptibility Testing on Clinical Urine Samples by Optical Tracking of Single Cell Division Events. Small (Weinheim an der Bergstrasse, Germany) 2020, n/a (n/a), 2004148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang F; Jiang J; McBride M; Zhou X; Yang Y; Mo M; Peterman J; Grys T; Haydel SE; Tao N; Wang S, Rapid Antimicrobial Susceptibility Testing on Clinical Urine Samples by Video-Based Object Scattering Intensity Detection. Anal Chem 2021, 93 (18), 7011–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broeren MA; Bahceci S; Vader HL; Arents NL, Screening for urinary tract infection with the Sysmex UF-1000i urine flow cytometer. J Clin Microbiol 2011, 49 (3), 1025–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fouchet P; Jayat C; Hechard Y; Ratinaud MH; Frelat G, Recent advances of flow cytometry in fundamental and applied microbiology. Biol Cell 1993, 78 (1–2), 95–109. [DOI] [PubMed] [Google Scholar]
- 46.Steen HB, Light scattering measurement in an arc lamp-based flow cytometer. Cytometry 1990, 11 (2), 223–30. [DOI] [PubMed] [Google Scholar]
- 47.Jo Y; Jung J; Lee JW; Shin D; Park H; Nam KT; Park JH; Park Y, Angle-resolved light scattering of individual rod-shaped bacteria based on Fourier transform light scattering. Sci Rep 2014, 4, 5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pottash AE; McKay R; Virgile CR; Ueda H; Bentley WE, TumbleScore: Run and tumble analysis for low frame-rate motility videos. BioTechniques 2017, 62 (1), 31–36. [DOI] [PubMed] [Google Scholar]
- 49.Mörters P; Peres Y, Brownian Motion. Cambridge University Press: Cambridge, 2010. [Google Scholar]
- 50.Lane MC; Alteri CJ; Smith SN; Mobley HLT, Expression of flagella is coincident with uropathogenic <em>Escherichia coli</em> ascension to the upper urinary tract. Proceedings of the National Academy of Sciences 2007, 104 (42), 16669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parry BR; Surovtsev IV; Cabeen MT; O’Hem CS; Dufresne ER; Jacobs-Wagner C, The Bacterial Cytoplasm Has Glass-like Properties and Is Fluidized by Metabolic Activity. Cell 2014, 156 (1–2), 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winters-Hilt S; Merat S, SVM clustering. BMC Bioinformatics 2007, 8 Suppl 7, S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burd EM; Kehl KS, A Critical Appraisal of the Role of the Clinical Microbiology Laboratory in the Diagnosis of Urinary Tract Infections. 2011, 49 (9_Supplement), S34–S38. [Google Scholar]
- 54.Tinevez JY; Perry N; Schindelin J; Hoopes GM; Reynolds GD; Laplantine E; Bednarek SY; Shorte SL; Eliceiri KW, TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


