Abstract
Objective:
Determine the time to positivity (TTP) of blood cultures among infants with late-onset bacteremia and predictors of TTP >36 hours.
Design:
Retrospective cohort study
Setting:
16 birth centers in 2 health care systems
Patients:
Infants with positive blood cultures obtained >72 hours after birth.
Outcome:
The main outcome was TTP, defined as the time interval from specimen collection to when a neonatal provider was notified of culture growth. TTP analysis was restricted to the first positive culture per infant. Patient and infection-specific factors were analyzed for association with TTP >36 hours.
Results:
Of 10,235 blood cultures obtained from 3,808 infants, 1,082 (10.6%) were positive. Restricting to bacterial pathogens and the first positive culture, the median TTP (25th–75th percentile) for 428 cultures was 23.5 hours (18.4–29.9); 364 (85.0%) resulted in 36 hours. Excluding coagulase-negative staphylococci (CoNS), 275/294 (93.5%) cultures were flagged positive by 36 hours. In a multivariable model, CoNS isolation and antibiotic pretreatment were significantly associated with increased odds of TTP >36 hours. Projecting a 36-hour empiric duration at one site, and assuming that all negative evaluations were associated with an empiric course of antibiotics, we estimated that 1,164 doses of antibiotics would be avoided in 629 infants over 10 years, while delaying a subsequent antibiotic dose in 13 infants with bacteremia.
Conclusions:
Empiric antibiotic administration in late-onset infection evaluations (not targeting CoNS) can be stopped at 36 hours. Longer durations (48 hours) should be considered when there is pretreatment or antibiotic therapy is directed at CoNS.
Keywords: Neonatology, Sepsis, Microbiology, Therapeutics
INTRODUCTION
Optimizing antibiotic use in health care settings is a key strategy to combat the global rise of antimicrobial resistance.1 In neonatal intensive care units (NICUs), empiric treatment for suspected infection is a large contributor of antibiotic use.2 The duration of empiric antibiotic therapy should be based on the time to positivity (TTP) for the majority of confirmed cases. Traditionally, empiric antibiotic duration ranges from 48 to 72 hours.3,4 Widespread use of automated blood culture detection systems has shortened the time in which most pathogens are reported5 that could allow shorter empiric durations and reduce antibiotic utilization in the NICU. In early-onset sepsis (EOS) evaluations, a 36-hour empiric antibiotic duration has been proposed to capture the majority of cases while minimizing antibiotics among the ultimately uninfected.6–8 National guidelines now suggest that a 36-hour empiric duration may be appropriate in EOS evaluations.9,10 However, it is unclear whether a 36-hour empiric duration can also be applied to late-onset sepsis evaluations. Prior studies report that TTP in late-onset sepsis is longer than in EOS,6,11 but few studies have evaluated the impact of patient characteristics or antibiotic pretreatment on TTP in late-onset sepsis.
The objectives of this study were to determine TTP in late-onset bacteremia cases and factors associated with a TTP >36 hours, and to estimate the probability of pathogen isolation after 36 hours. We also estimated difference in antibiotic use with a 36-hour versus 48-hour empiric antibiotic duration.
METHODS
Study setting
This was a retrospective cohort study of infants admitted to NICUs in two health care systems who had a blood culture obtained >72 hours after birth. Study centers and time periods included: Pennsylvania Hospital (PAH, infants born from 1/1/2009-12/31/2020, excluding infants born 1/1/2015-9/30/2016 due to lack of available data), and 15 birth facilities affiliated with Kaiser Permanente Northern California (KPNC, infants born from 1/1/2010-12/31/2020). The study was approved by the Institutional Review Board of both centers, PAH (#828920) and KPNC (#1262699-33), with a waiver of informed consent. Patient demographics, antibiotic information, and blood culture results were obtained from electronic medical records. Assessments of contaminants, TTP, incubation time, and central line presence were determined by chart review. Data for central line and incubation time were available for KPNC infants and a subset of PAH infants born after 10/1/2016. Analyses that included these variables were restricted to this subcohort.
Blood culture processing
All blood cultures obtained >72 hours after birth with detectable microbial growth were included in the analysis. At KPNC, 1 ml of blood was inoculated in an aerobic bottle, followed by incubation at an off-site centralized microbiology laboratory using BacT/Alert Pediatric FAN. PAH used an on-site microbiology laboratory and the BD BACTEC system. Between 2009–2014 at PAH, 1 ml of blood was inoculated in an aerobic bottle. After 10/1/2016, 2 ml of blood was inoculated, divided equally between an aerobic and an anaerobic bottle. Microbiology staff at both sites call and inform the neonatal provider in real-time when the automated microbial detection system alarms to signal microbial growth.
Study Definitions
TTP was defined as the duration in hours from when the culture specimen was obtained to the time that laboratory staff reported growth to a neonatal provider. Pre-analytic time was defined as the duration in hours from when the culture specimen was obtained to the time it was incubated in the automated detection system.
Known commensal organisms such as diphtheroids, Micrococcus species, etc. were classified as contaminants. Coagulase-negative staphylococci (CoNS) were classified as pathogens when managed accordingly by the clinical team with ≥5 days of antibiotics. For infants with multiple positive blood cultures, a new bacteremia episode was defined as isolation of a different organism or isolation of the same organism >2 days after completion of antibiotic course for previous bacteremia. Contaminants and fungi were excluded from the primary TTP analysis. For all TTP analyses, only the first episode of bacteremia for each infant was used to avoid nonindependence associated with repeated measures. All episodes of bacteremia were used to calculate rates of positivity.
Pretreatment with antibiotics was defined as parenteral antibiotic administration 1 to 24 hours prior to drawing the culture specimen.
Central line-associated bloodstream infection (CLABSI) was defined as pathogen isolation from a culture obtained with a central line in place for ≥2 days or ≤2 days of its removal.
Analysis
We compared the median TTP and interquartile range (IQR), and the proportion of positive bacterial cultures by 24, >24 to 36, >36 to 48 and >48 hours in the following groups: (A) pathogens versus contaminants; (B) pathogens excluding CoNS versus CoNS managed as pathogen; (C) Gram-positive (excluding CoNS) versus Gram-negative pathogens; (D) cultures obtained prior to antibiotic administration versus pretreated cultures; (E) CLABSI episodes versus non-line associated infections; and (F) PAH versus KPNC. We generated Kaplan-Meier curves for TTP in these groups to graphically present the differences. We determined the probability of a culture growing a bacterial pathogen after 36 hours using the formula provided by Lambregts et al , where TTPe is the proportion of positive blood cultures growing ≤36 hours after birth, and X is the rate of positivity for blood cultures obtained >72 hours after birth.12
In the subcohort with all variables available, we tested for associations between TTP >36 hours and gestational age at birth, postnatal age when culture was obtained, sex, presence of central line, pathogen type, study center, and antibiotic pretreatment. For the multivariable logistic regression model, we included variables significantly associated with the outcome in bivariable analysis and forced study center into the model.
To estimate the impact of a 36-hour versus a 48-hour empiric antibiotic duration, we used PAH data that contained information on all sepsis evaluations including those with negative blood culture results. A new evaluation was defined as a blood culture obtained >2 days after completion of antibiotic course for a previous bacteremia, or >2 days after a previous negative culture. We assumed that all evaluations were associated with empiric antibiotic initiation consisting of an 8-hourly dose for Gram-positive coverage (e.g., oxacillin) and a 24-hourly dose for Gram-negative coverage (e.g., aminoglycoside). Further, we assumed that these antibiotics would be discontinued after 48 hours if cultures resulted as sterile at that time. We estimated that a 36-hour empiric duration compared to 48-hour would avoid one antibiotic dose for Gram-positive coverage at the 40th hour.
All analyses were performed using Stata 16 (StataCorp, College Station, TX).
RESULTS
A total of 10,235 blood cultures were obtained from 3,808 infants >72 hours after birth, with 77.9% of the cultures obtained at KPNC. Supplemental Figure delineates derivation of the study population. A total of 1,082 blood cultures were positive (10.6%). After excluding cultures that were repeat positive due to persistent bacteremia/fungemia (n=441, 40.8%) and contaminants (n=145, 13.4%), there were 496 unique episodes of late-onset bacteremia or fungemia in 447 infants. Median gestational age for the cohort was 26 weeks (IQR 24–29 weeks). Median postnatal age at the first episode was 14 days (IQR 9–24 days) (Supplemental Table 1). Of 496 episodes, antibiotic pretreatment was observed in 62 (12.5%) episodes. Data for central line was available for 378 episodes, and 252 (66.7%) of those were identified as CLABSI.
Of the 496 episodes of late-onset bacteremia or fungemia, Gram-positive organisms accounted for 58.1% of all pathogens isolated. CoNS was the predominant organism, representing 28.8% of all pathogens. Gram-negative organisms were second most common (33.3%), followed by fungi (6.1%) and polymicrobial infections (2.6%) (Table 1). Organisms excluded as contaminants are shown in Supplemental Table 2.
Table 1.
Blood Culture Pathogens Isolated from all Late-Onset Infection Evaluation Episodes
| Pathogens Isolated | N = 496 | Median TTP (Q1, Q3) | Positive by 36 hours1 |
|---|---|---|---|
| Gram-positive, n (%) | 288 (58.1) | ||
| CoNS | 143 | 30.4 (24.3, 39.2) | 98 (68.5) |
| Staphylococcus aureus2 | 99 | 23.9 (19.5, 29.0) | 89 (89.9) |
| Streptococcus agalactiae | 27 | 15.0 (11.0, 19.0) | 27 (100.0) |
| Enterococcus sp. | 11 | 23.1 (18.3, 27.6) | 10 (90.9) |
| Other Streptococcus sp. | 7 | 19.3 (9.6, 28.0) | 7 (100.0) |
| Bacillus cereus | 1 | 24.0 | 1 (100.0) |
| Gram-negative, n (%) | 165 (33.3) | ||
| Escherichia coli | 82 | 19.8 (16.9, 23.1) | 77 (93.9) |
| Klebsiella sp. | 38 | 17.4 (12.5, 21.0) | 38 (100.0) |
| Pseudomonas aeruginosa | 18 | 24.1 (22.1, 28.8) | 16 (88.9) |
| Enterobacter sp. | 11 | 17.6 (17.4, 22.7) | 9 (81.8) |
| Serratia sp. | 10 | 24.5 (17.1, 25.7) | 10 (100.0) |
| Citrobacter sp. | 2 | 25.3 (23.2, 27.5) | 2 (100.0) |
| Morganella sp. | 2 | 19.8 (19.0, 20.5) | 2 (100.0) |
| Proteus mirabilis | 1 | 20.8 | 1 (100.0) |
| Stenotrophomonas maltophilia | 1 | 37.5 | 0 |
| Fungi, n (%) | 30 (6.1) | 38.0 (30.6, 47.1) | 14 (46.7) |
| Poly-microbial, n (%) | 13 (2.6) | 21.7 (15.0, 23.9) | 13 (100.0) |
Row percentages shown
Includes 16 methicillin-resistant Staphylococcus aureus.
CoNS, coagulase-negative staphylococci; Q1, first quartile; Q3, third quartile; sp., species; TTP, time to positivity.
TTP
Restricting to the first episode of pathogenic bacteremia (n=428), the median TTP was 23.5 hours (IQR 18.4─29.9), with 54.2% detected by 24 hours, 85.1% by 36 hours, and 94.9% by 48 hours. The probability of detecting a bacterial pathogen after 36 hours was 1.8% (95% CI 1.4–2.2) and the probability of detecting a non-CoNS pathogen after 36 hours was 0.5% (95% CI 0.3–0.7).
Figure 1 shows TTP curves by specific characteristics. Pathogens had a significantly shorter TTP compared to contaminants (Figure 1A). CoNS had a longer TTP compared to other pathogens (Figure 1B). Median TTP of non-CoNS pathogens was 21.2 hours (IQR 17.1–25.7) with 93.5% detected within 36 hours (Table 2). Excluding CoNS, the median TTP remained longer for other Gram-positive organisms compared to Gram-negative organisms. However, the proportion detected by 36 hours did not differ (91.9% vs. 94.5%, P=0.38) (Figure 1C).
Figure 1. Kaplan-Meier Curves of TTP for Late-Onset Bacteremia: Subgroup Comparisons.
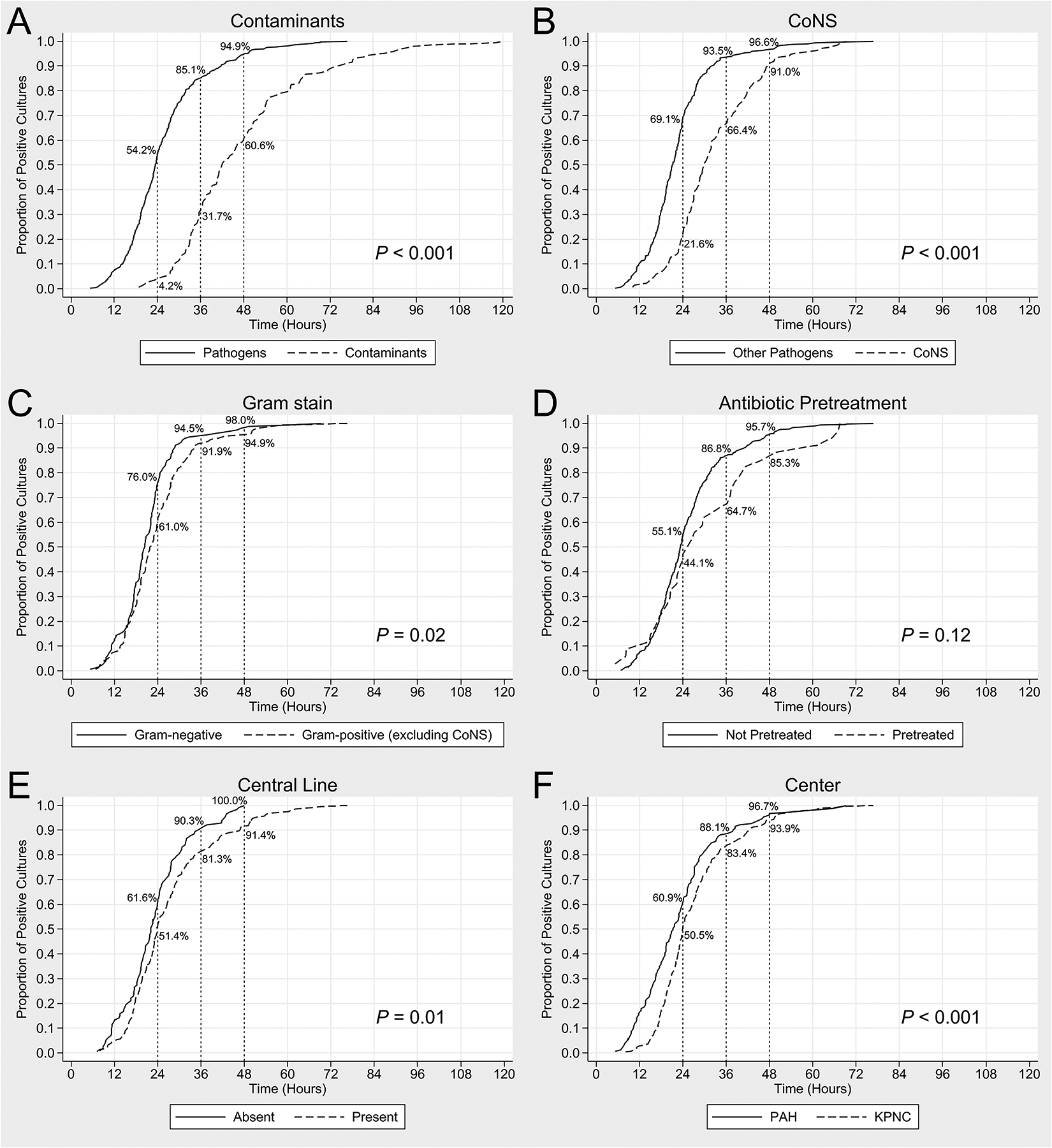
Figure 1 demonstrates the TTP of positive cultures among the following subgroups: (A) pathogens versus contaminants; (B) pathogens excluding CoNS versus CoNS managed as pathogen; (C) Gram-positive (excluding CoNS) versus Gram-negative pathogens; (D) cultures obtained prior to antibiotic administration versus pretreated cultures; (E) CLABSI episodes versus non-line associated infections; and (F) PAH versus KPNC. The P values shown demonstrate the difference in median TTP (by Mann-Whitney U Test) of the respective comparison groups. CoNS, coagulase-negative staphylococci; CLABSI, central line-associated blood stream infections; KPNC, Kaiser Permanente Northern California; PAH, Pennsylvania Hospital; TTP, time to positivity.
Table 2.
TTP for Neonatal Late-Onset Bacteremia by Culture Characteristics
| Characteristics | TTP, median (Q1, Q3) | P 1 | TTP2 | |||
|---|---|---|---|---|---|---|
| 24 hours, n (%) | >24–36 hours, n (%) | >36–48 hours, n (%) | ≥48 hours, n (%) | |||
| All bacterial pathogens (n=428) | 23.5 (18.4, 29.9) | – | 232 (54.2) | 132 (30.8) | 42 (9.8) | 22 (5.1) |
| Pathogens vs. Contaminants 3 | ||||||
| Pathogens (n=428) | 23.5 (18.4, 29.9) | <0.001 | 232 (54.2) | 132 (30.8) | 42 (9.8) | 22 (5.1) |
| Contaminants (n=142) | 42.1 (34.1, 54.3) | 6 (4.2) | 39 (27.5) | 41 (28.9) | 56 (39.4) | |
| CoNS (managed as pathogens) vs. Other Pathogens | ||||||
| CoNS (n=134) | 30.0 (24.6, 40.3) | <0.001 | 29 (21.6) | 60 (44.8) | 33 (24.6) | 12 (9.0) |
| Other Pathogens (n=294) | 21.2 (17.1, 25.7) | 203 (69.1) | 72 (24.5) | 9 (3.1) | 10 (3.4) | |
| Gram-positive (excluding CoNS) vs. Gram-negative | ||||||
| Gram-positive (n=136) | 22.1 (17.7, 27.6) | 0.02 | 83 (61.0) | 42 (30.9) | 4 (2.9) | 7 (5.2) |
| Gram-negative (n=146) | 20.4 (17.1, 23.9) | 111 (76.0) | 27 (18.5) | 5 (3.4) | 3 (2.1) | |
| Pretreatment with antibiotics | ||||||
| Pretreated cultures (n=34) | 25.8 (19.4, 38.6) | 0.12 | 15 (44.1) | 7 (20.6) | 7 (20.6) | 5 (14.7) |
| Cultures obtained prior to antibiotics (n=394) | 23.3 (18.4, 29.4) | 217 (55.1) | 125 (31.7) | 35 (8.9) | 17 (4.3) | |
| Central Line 4 | ||||||
| Present (n=208) | 23.8 (19.0, 31.7) | 0.01 | 107 (51.4) | 62 (29.8) | 21 (10.1) | 18 (8.7) |
| Absent (n=113) | 22.1 (17.5, 27.6) | 69 (61.1) | 33 (29.2) | 11 (9.7) | 0 | |
| Study Center | ||||||
| KPNC (n=277) | 24.0 (19.7, 31.1) | <0.001 | 140 (50.5) | 91 (32.9) | 29 (10.5) | 17 (6.1) |
| PAH (n=151) | 21.3 (15.1, 27.3) | 92 (60.9) | 41 (27.2) | 13 (8.6) | 5 (3.3) | |
P values demonstrate difference in median TTP (by Mann-Whitney U Test) between the respective subgroups.
Row percentages shown.
Excludes 3 cases (2 Staphylococcus aureus and 1 Candida albicans) managed as contaminants by clinical team; infants remained well without antibiotic treatment.
Only shown for infants with central line data available.
CoNS, coagulase-negative staphylococci; KPNC, Kaiser Permanente Northern California; PAH, Pennsylvania Hospital; Q1, first quartile; Q3, third quartile; TTP, time to positivity.
CoNS accounted for 45/64 (70.3%) of the organisms detected after 36 hours. The remaining 19 organisms included Staphylococcus aureus (n=10), Escherichia coli (n=4), Pseudomonas aeruginosa (n=2), Enterobacter cloacae (n=1), Enterococcus faecalis (n=1), and Stenotrophomonas maltophila (n=1). Antibiotic pretreatment was observed in 5/19 (26.3%) of these non-CoNS cultures.
While antibiotic pretreatment was not associated with a lower median TTP (Figure 1D), pretreated cultures were less likely to be positive by 36 hours compared to non-pretreated cultures (64.7% vs. 86.8%; P=0.001). Central line presence was associated with a longer median TTP (Figure 1E). PAH cultures (on-site microbiology facility) had a significantly shorter median TTP compared to KPNC cultures (centralized, off-site facility) (Figure 1F). Median pre-analytic time was significantly higher for KPNC (8 hours, IQR 5–10 hours) compared to PAH (1 hour, IQR 0–2 hours; P<0.001). However, the proportion of positive cultures by 36 hours was similar between the two centers (88.1% vs. 83.4%; P=0.19).
Factors associated with TTP >36 hours
In the bivariable analysis (Table 3), central line presence, antibiotic pretreatment, and CoNS were significantly associated with TTP >36 hours. In multivariable analysis, CoNS and pretreatment remained significantly associated with a TTP >36, but central line presence did not.
Table 3.
Bivariable and Multivariable Association between Patient and Culture Characteristics and TTP >36 hours1
| Characteristics | Bivariable | Multivariable | ||
|---|---|---|---|---|
| OR (95% CI) | P | aOR (95% CI) | P | |
| Gestational age (weeks) | 0.96 (0.88, 1.04) | 0.35 | – | – |
| Sex (male vs. female) | 0.90 (0.49, 1.66) | 0.74 | – | – |
| Postnatal age (days) at positive culture | 1.00 (0.98, 1.01) | 0.68 | – | – |
| Central line (present vs. absent) | 2.21 (1.08, 4.51) | 0.03 | 1.34 (0.60, 3.02) | 0.48 |
| Pretreated vs. not pretreated | 6.07 (2.16, 17.06) | 0.001 | 5.31 (1.46, 19.27) | 0.01 |
| CoNS vs. other pathogens | 14.34 (7.10, 28.96) | <0.001 | 14.60 (6.98, 30.58) | <0.001 |
| Gram-positive vs. Gram-negative2 | 1.67 (0.56, 4.97) | 0.36 | – | – |
| KPNC vs. PAH | 1.71 (0.58, 5.04) | 0.33 | 2.26 (0.68, 7.55) | 0.19 |
Includes first positive episode of late-onset sepsis growing a pathogen (except fungus and poly-microbial) for infants who had data available for central line (N=313).
Excludes CoNS (N=237).
aOR, adjusted odds ratio; CI, confidence intervals; CoNS, coagulase-negative staphylococci; KPNC, Kaiser Permanente Northern California; PAH, Pennsylvania Hospital; OR, odds ratio; TTP, time to positivity.
Antibiotic use with empiric duration of 36 versus 48 hours
At PAH, there were 174 episodes of late-onset bacteremia in 152 infants and 1,164 episodes of negative evaluations in 629 infants. Stopping antibiotics at 36 hours would have avoided 1,164 antibiotic doses for Gram-positive coverage. It also would have resulted in an interruption of antibiotic therapy for <8 hours in 7 infants and >8 hours in 6 infants with an eventual positive culture. The longest TTP was for a culture growing Enterobacter cloacae (83.6 hours). For this episode, a 36-hour empiric duration would have interrupted the 40th hour antibiotic dose and delayed subsequent dose by 44 hours, whereas a 48-hour empiric duration would have interrupted the 48th hour antibiotic dose and delayed subsequent dose by 36 hours. For every case with an interruption of antibiotic therapy of >8 hours, 194 antibiotic doses would be avoided.
DISCUSSION
In a large cohort of infants admitted to NICUs in 2 health care systems, 85.1% of late-onset bacteremia cases, and 93.5% of non-CoNS pathogens, were identified by 36 hours. CoNS bacteremia and antibiotic pretreatment were associated with TTP >36 hours. We found a low probability (1.8%) for a culture that is negative at 36 hours to signal growth thereafter, and an even lower probability (0.5%) that it will detect a non-CoNS organism.
Based on TTP data, recent reports have proposed shortening the empiric antibiotic duration for EOS evaluation from 48–72 hours to 24–36 hours.7,11,13 Our study highlights some additional considerations when contemplating similarly limiting empiric antibiotic duration for late-onset infection evaluations. First, we found that a longer pre-analytic time correlated with a significant center-specific difference in TTP. Studies reporting TTP in late-onset bacteremia often calculate TTP from the time of incubation6,14–17 to detection of bacterial growth by the culture machine.11,16,17 In real-world practice, TTP determining empiric antibiotic duration also includes the time from blood collection to incubation, and the time from detection of growth to provider notification.3 TTP calculated from the time of incubation can give shorter durations both by excluding the pre-analytic time, and by not accounting for the growth that potentially occurred during that time. Samples from KPNC, processed at an off-site microbiology facility, had a significantly longer pre-analytic time, which corresponded to longer median TTP and a lower percentage of positive cultures at 24 hours. However, by 36 hours, a similar proportion were detected at both centers, suggesting a potentially generalizable 36-hour cut-off.
Second, we found that pretreated cultures had significantly longer TTP compared to cultures drawn before antibiotic administration, a finding reported by others as well.18 Since only 64.7% of eventual positive pretreated cultures resulted by 36 hours, a shortened empiric duration may not be sufficient. These cases highlight the role for culture-independent techniques such as polymerase chain reaction-based methods in supplementing culture-based methods. While blood cultures remain the best way to determine susceptibility for a broad group of pathogens, culture independent techniques offer specific advantages in that they are less sensitive to pre-treatment19 can detect organisms not easily cultured.20 While challenges remain with widespread availability and lack of guidance on how best to integrate them in clinical practice21 these tests may allow both earlier identification of pathogens and increase clinician confidence in a sterile result.
Lastly, the positivity rate of blood cultures in suspected late-onset infection evaluations can be as high as 30%,3,6,14,22–24 substantially greater than the 2–4% positivity rates seen in EOS evaluation.6,7,23 This higher prior probability of detecting bacteremia may change how clinicians view the risks and benefits of early antibiotic discontinuation. Consistent with other studies, we found that the probability of detecting bacteremia after 36 hours was low.6,11,12 In a study of over 700 late-onset blood cultures, most pathogens were detected by 24–36 hours;11 and almost all Gram-negative organisms were detected by 24 hours. The authors suggested that aminoglycoside coverage could be stopped at 24 hours. In our study, only 76.0% of Gram-negative pathogens were detected by 24 hours. These differences may be due to differing patient characteristics or longer pre-analytic time for some samples in our study.
While the majority of late-onset bacteremia cases were detected in 36 hours, many pretreated cultures and cases of CoNS bacteremia were not. If considering a 36-hour empiric duration, these 2 scenarios could be resolved differently. Since pretreatment is known to the ordering provider, they may elect to choose a longer empiric antibiotic duration. For CoNS cases, a more general risk-benefit assessment is required. Management of CoNS bacteremia is challenging, as definitive criteria for excluding contamination are rarely met in neonates. It is also associated with lower mortality and it is unclear whether a few hours’ interruption of antibiotic therapy impacts morbidity.14,25 Studies on vancomycin reduction initiatives in the NICU have previously discussed the risk-benefit of empiric CoNS coverage.26,27 Many NICU providers conclude that the benefits of using oxacillin/nafcillin (less nephrotoxic antibiotic; superior activity toward methicillin-sensitive Staphylococcus aureus; avoids selection pressure against vancomycin) outweigh the risks of delaying CoNS coverage. Aligned with previous studies, we conclude that empiric coverage not targeting CoNS can be stopped at 36 hours.11 If antibiotics are targeted at CoNS, empiric coverage for 48 hours may be warranted.
A strength of the study is a large sample size and inclusion of multiple study centers with differing microbiology processing systems, making the results more generalizable. Limitations include the lack of information on clinical presentation, site of blood draw, and details of blood inoculant volume as a predictor of TTP. Lastly, our estimates of antibiotic doses reduced using a 36-hour convention for “rule out sepsis” would differ depending on what proportion of infants were ultimately treated in the face of sterile blood cultures for conditions such as necrotizing enterocolitis, urinary tract infection, cellulitis, and depending on center practice regarding treatment for “culture-negative” sepsis.
CONCLUSIONS
Empiric antibiotic coverage for late-onset infection evaluations (not targeting CoNS) can be stopped at 36 hours. Longer durations (48 hours) should be considered when there is pretreatment or antibiotic therapy is directed at CoNS.
Supplementary Material
What is already known on this topic
The duration of empiric antibiotic therapy in sepsis evaluation is conventionally based on the time to positivity (TTP) for the majority of confirmed cases.
Automated blood culture detection systems can shorten the time to detection of pathogens and allow shorter empiric antibiotic durations in the NICU.
A 36-hour empiric duration is being used in early-onset sepsis evaluations. It is unclear whether 36-hour empiric duration can be applied to late-onset sepsis evaluations.
What this study adds
Among 428 infants with late-onset bacteremia, 85% of all cultures, and 94% of non-CoNS pathogens were detected by 36 hours.
CoNS isolation and pretreatment with antibiotics were significantly associated with TTP >36 hours in multivariable model adjusting for patient- and culture-specific factors.
How this study might affect research, practice or policy
Empiric antibiotic coverage for late-onset sepsis evaluations not targeting CoNS (such as regimens using oxacillin for Gram-positive coverage) can be stopped at 36 hours.
ACKNOWLEDGEMENTS
The authors would like to thank Suyi Zhu, RN, MSN for assisting with data collection.
FUNDING
Dr. Flannery receives funding from the Agency for Healthcare Research and Quality (K08HS027468). Dr. Mukhopadhyay receives funding from Eunice Kennedy Shriver National Institute of Child Health and Human Development from the National Institutes of Health grant (K23HD088753). Dr. Coggins receives funding from the National Heart, Lung and Blood Institute of the National Institutes of Health (T32HL007891). None of the authors has conflicts of interest to declare relevant to this study.
Abbreviations
- CoNS
coagulase-negative staphylococci
- EOS
early-onset sepsis
- IQR
interquartile range
- KPNC
Kaiser Permanente Northern California
- NICU
neonatal intensive care unit
- PAH
Pennsylvania Hospital
- TTP
time to positivity
Footnotes
CONTRIBUTORSHIP STATEMENT
Dr. Mukhopadhyay and Dr. Kuzniewicz conceptualized and designed the study, contributed to the interpretation of the results, drafted the initial manuscript, and reviewed and revised the final manuscript.
Dr. Flannery and Ms. Briker contributed to study design, data acquisition, interpretation of results, drafted the initial manuscript, critically reviewed the manuscript, and approved the final manuscript as submitted.
Dr. Dhudasia contributed to study design and data acquisition, performed statistical analyses, critically reviewed the manuscript, and approved the final manuscript as submitted.
Dr. Coggins contributed to the interpretation of the results, critically reviewed the manuscript, and approved the final manuscript as submitted.
Ms. Woodford, Ms. Walsh and Ms. Li contributed to data collection and verification, critically reviewed the manuscript, and approved the final manuscript as submitted.
Dr. Puopolo contributed to the interpretation of the results, critically reviewed the manuscript, and approved the final manuscript as submitted.
COMPETING INTERESTS
Authors have no conflicts of interest to declare relevant to this study.
REFERENCES
- 1.Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Healthcare Quality Promotion (DHQP). Biggest Threats and Data: 2019 AR Threats Report. Available at: https://www.cdc.gov/drugresistance/biggest-threats.html. Updated 2021. Accessed Oct 21, 2021.
- 2.Cantey JB, Wozniak PS, Sánchez PJ. Prospective surveillance of antibiotic use in the neonatal intensive care unit: Results from the SCOUT study. Pediatr Infect Dis J. 2015;34(3):267–272. doi: 10.1097/INF.0000000000000542 [doi]. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser JR, Cassat JE, Lewno MJ. Should antibiotics be discontinued at 48 hours for negative late-onset sepsis evaluations in the neonatal intensive care unit? J Perinatol. 2002;22(6):445–447. doi: 10.1038/sj.jp.7210764 [doi]. [DOI] [PubMed] [Google Scholar]
- 4.Rubin LG, Sánchez PJ, Siegel J, et al. Evaluation and treatment of neonates with suspected late-onset sepsis: A survey of neonatologists’ practices. Pediatrics. 2002;110(4):e42. doi: 10.1542/peds.110.4.e42 [doi]. [DOI] [PubMed] [Google Scholar]
- 5.Opota O, Croxatto A, Prod’hom G, Greub G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin Microbiol Infect. 2015;21(4):313–322. doi: S1198–743X(15)00183–4 [pii]. [DOI] [PubMed] [Google Scholar]
- 6.Jardine L, Davies MW, Faoagali J. Incubation time required for neonatal blood cultures to become positive. J Paediatr Child Health. 2006;42(12):797–802. doi: JPC980 [pii]. [DOI] [PubMed] [Google Scholar]
- 7.Kuzniewicz MW, Mukhopadhyay S, Li S, Walsh EM, Puopolo KM. Time to positivity of neonatal blood cultures for early-onset sepsis. Pediatr Infect Dis J. 2020;39(7):634–640. doi: 10.1097/INF.0000000000002632 [doi]. [DOI] [PubMed] [Google Scholar]
- 8.Meyers JM, Tulloch J, Brown K, Caserta MT, D’Angio CT, GOLISANO CHILDREN’S HOSPITAL NICU ANTIBIOTIC STEWARDSHIP TEAM. A quality improvement initiative to optimize antibiotic use in a level 4 NICU. Pediatrics. 2020;146(5):e20193956. doi: 10.1542/peds.2019-3956. Epub 2020 Oct 14. doi: e20193956 [pii]. [DOI] [PubMed] [Google Scholar]
- 9.National Institute for Health and Care Excellence. Neonatal infection: antibiotics for prevention and treatment (NG195). Available at: https://www.nice.org.uk/guidance/ng195. Accessed Oct 21, 2021. [PubMed]
- 10.Puopolo KM, Benitz WE, Zaoutis TE, COMMITTEE ON FETUS AND NEWBORN, COMMITTEE ON INFECTIOUS DISEASES. Management of neonates born at ≥35 0/7 weeks’ gestation with suspected or proven early-onset bacterial sepsis. Pediatrics. 2018;142(6):e20182894. doi: 10.1542/peds.2018-2894. doi: e20182894 [pii]. [DOI] [PubMed] [Google Scholar]
- 11.Ur Rehman Durrani N, Rochow N, Alghamdi J, Pelc A, Fusch C, Dutta S. Minimum duration of antibiotic treatment based on blood culture in rule out neonatal sepsis. Pediatr Infect Dis J. 2019;38(5):528–532. doi: 10.1097/INF.0000000000002182 [doi]. [DOI] [PubMed] [Google Scholar]
- 12.Lambregts MMC, Bernards AT, van der Beek MT, Visser LG, de Boer MG. Time to positivity of blood cultures supports early re-evaluation of empiric broad-spectrum antimicrobial therapy. PLoS One. 2019;14(1):e0208819. doi: 10.1371/journal.pone.0208819 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marks L, de Waal K, Ferguson JK. Time to positive blood culture in early onset neonatal sepsis: A retrospective clinical study and review of the literature. J Paediatr Child Health. 2020;56(9):1371–1375. doi: 10.1111/jpc.14934 [doi]. [DOI] [PubMed] [Google Scholar]
- 14.Abdelhamid SM. Time to positivity and antibiotic sensitivity of neonatal blood cultures. J Glob Infect Dis. 2017;9(3):102–107. doi: 10.4103/jgid.jgid_1_17 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Prats JA, Cooper TR, Schneider VF, Stager CE, Hansen TN. Rapid detection of microorganisms in blood cultures of newborn infants utilizing an automated blood culture system. Pediatrics. 2000;105(3 Pt 1):523–527. doi: 10.1542/peds.105.3.523 [doi]. [DOI] [PubMed] [Google Scholar]
- 16.Huggard D, Powell J, Kirkham C, Power L, O’Connell NH, Philip RK. Time to positivity (TTP) of neonatal blood cultures: A trend analysis over a decade from ireland. J Matern Fetal Neonatal Med. 2021;34(5):780–786. doi: 10.1080/14767058.2019.1617687 [doi]. [DOI] [PubMed] [Google Scholar]
- 17.Vamsi SR, Bhat RY, Lewis LE, Vandana KE. Time to positivity of blood cultures in neonates. Pediatr Infect Dis J. 2014;33(2):212–214. doi: 10.1097/INF.0000000000000018 [doi]. [DOI] [PubMed] [Google Scholar]
- 18.Scheer CS, Fuchs C, Gründling M, et al. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: A prospective clinical cohort study. Clin Microbiol Infect. 2019;25(3):326–331. doi: S1198–743X(18)30449-X [pii]. [DOI] [PubMed] [Google Scholar]
- 19.Gies F, Tschiedel E, Felderhoff-Müser U, Rath PM, Steinmann J, Dohna-Schwake C. Prospective evaluation of SeptiFast multiplex PCR in children with systemic inflammatory response syndrome under antibiotic treatment. BMC Infect Dis. 2016;16:378–9. doi: 10.1186/s12879-016-1722-9 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinha M, Jupe J, Mack H, Coleman TP, Lawrence SM, Fraley SI. Emerging technologies for molecular diagnosis of sepsis. Clin Microbiol Rev. 2018;31(2):e00089–17. Print 2018 Apr. doi: 10.1128/CMR.00089-17 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rub DM, Dhudasia MB, Healy T, Mukhopadhyay S. Role of microbiological tests and biomarkers in antibiotic stewardship. Semin Perinatol. 2020;44(8):151328. doi: S0146–0005(20)30111–7 [pii]. [DOI] [PubMed] [Google Scholar]
- 22.Gowda H, Norton R, White A, Kandasamy Y. Late-onset neonatal sepsis-A 10-year review from North Queensland, Australia. Pediatr Infect Dis J. 2017;36(9):883–888. doi: 10.1097/INF.0000000000001568 [doi]. [DOI] [PubMed] [Google Scholar]
- 23.Guerti K, Devos H, Ieven MM, Mahieu LM. Time to positivity of neonatal blood cultures: Fast and furious? J Med Microbiol. 2011;60(Pt 4):446–453. doi: 10.1099/jmm.0.020651-0 [doi]. [DOI] [PubMed] [Google Scholar]
- 24.Hornik CP, Fort P, Clark RH, et al. Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Hum Dev. 2012;88 Suppl 2(Suppl 2):69. doi: 10.1016/S0378-3782(12)70019-1 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoll BJ, Hansen NI, Adams-Chapman I, et al. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA. 2004;292(19):2357–2365. doi: 292/19/2357 [pii]. [DOI] [PubMed] [Google Scholar]
- 26.Chiu CH, Michelow IC, Cronin J, Ringer SA, Ferris TG, Puopolo KM. Effectiveness of a guideline to reduce vancomycin use in the neonatal intensive care unit. Pediatr Infect Dis J. 2011;30(4):273–278. doi: 10.1097/INF.0b013e3182011d12 [doi]. [DOI] [PubMed] [Google Scholar]
- 27.Hamdy RF, Bhattarai S, Basu SK, et al. Reducing vancomycin use in a level IV NICU. Pediatrics. 2020;146(2):e20192963. doi: 10.1542/peds.2019-2963. Epub 2020 Jul 1. doi: e20192963 [pii]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


