Abstract
We screened the genome of Saccharomyces cerevisiae for the genes responsive to oxidative stress by using the lacZ transposon-insertion library. As a result, we found that expression of the DOG2 gene coding for 2-deoxyglucose-6-phosphate phosphatase was induced by oxidative stress. The expression of DOG2 was also induced by osmotic stress. We found a putative cis element (STRE, a stress response element) in the DOG2 promoter adjacent to a consensus sequence to which the Mig1p repressor is known to bind. The basal levels of DOG2 gene expression were increased in a mig1Δ mutant, while the derepression of DOG2 was not observed in a snf1Δ mutant under glucose-deprived conditions. Induction of the DOG2 gene expression by osmotic stress was observed in any of the three disruptants pbs2Δ, hog1Δ, and snf1Δ. However, the osmotic induction was completely abolished in both the snf1Δ pbs2Δ mutant and the snf1Δ hog1Δ mutant. Additionally, these single mutants as well as double mutants failed to induce DOG2 expression by oxidative stress. These results suggest that Snf1p kinase and the high-osmolarity glycerol–mitogen-activated protein kinase cascade are likely to be involved in the signaling pathway of oxidative stress and osmotic stress in regulation of DOG2.
In all aerobic cells, oxygen respiration plays a critical role to acquire the energy efficiently; however, it leads to the formation of harmful reactive oxygen species which can damage many cellular components, such as DNA, proteins, and lipid membranes (4). Because reactive oxygen species are commonplace in aerobic cells, they have diverse defense systems (2, 20). Some antioxidant systems are known to be regulated at the transcriptional step (13, 23). Yap1p belongs to a Jun family of transcriptional activators in Saccharomyces cerevisiae (24). Yap1p regulates the expression of several genes encoding antioxidant enzymes, such as TRX2 (thioredoxin) (14), GSH1 (γ-glutamylcysteine synthetase) (40), and GPX2 (glutathione peroxidase) (12). In addition to Yap1p, several transcription factors, such as Msn2p, Msn4p, Skn7p, Mac1p, Ace1p, and Hap1p, are also known to mediate oxidative stress in S. cerevisiae (13, 23). For example, expression of the CTT1 gene encoding cytosolic catalase is induced by several environmental stimuli, and such signals are thought to be transmitted by Msn2p and Msn4p to the stress response element (STRE; consensus sequence, 5′-AGGGG-3′ or 5′-CCCCT-3′) in its promoter (31). Msn2p and Msn4p can bind to the STRE under several environmental stress conditions (7, 21, 33).
Changes in the intracellular and/or extracellular environment, which includes temperature, limitation of nutrients, osmotic pressure, redox status, and so on, enhance the synthesis of a number of stress proteins to adapt to environmental stress in both prokaryotic and eukaryotic cells. Environmental stress response of the budding yeast S. cerevisiae has been the focus of attention, because the yeast is known to have similar regulatory mechanisms of gene expression and signal transduction system to those of higher eukaryotes. Additionally, the yeast S. cerevisiae has a great advantage of gene analysis owing to the utility of the complete genome sequence. For example, expression databases of whole open reading frames of this microorganism in various genetic backgrounds as well as different growth conditions have being established by the microarray analysis.
In this study, we used the lacZ transposon-insertion library (3) to search the genes that are responsive to oxidative stress from the yeast genome. tert-Butyl hydroperoxide (t-BHP) was used as a stressor, because we have been studying the oxidative stress response of yeast caused by lipid hydroperoxide (9, 10, 12, 34). As a result, the DOG2 gene encoding 2-deoxyglucose-6-phosphate phosphatase was found among the genes whose expression was responsive to oxidative stress. The expression of DOG2 was also induced by osmotic stress and glucose starvation. To investigate the regulatory mechanism of DOG2 gene expression under these stress conditions, we applied two approaches. One is disruption of genes coding for transcription factors which could be predicted to be involved in the regulation of DOG2. The other is breakage of signal transduction systems which can transfer the signal to such transcription factors.
MATERIALS AND METHODS
Strains.
Yeast strains used in this study are listed in Table 1. The diploid strain (YPH274), which was obtained from the Yeast Genetic Stock Center, University of California at Berkeley, was used to generate the lacZ-insertion library. The plasmid library was provided by M. Snyder. Construction of a yeast insertion library was done as described by Burns et al. (3).
TABLE 1.
Strains used in this study
| Straina | Relevant genotype |
|---|---|
| YPH274 | MATa/α trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ801/lys2-Δ801 ade2-101/ade2-101 ura3-52/ura3-52 |
| YPH252 | MATα trp1-Δ1 his3-Δ200 leu2-Δ1 lys2-Δ801 ade2-101 ura3-52 |
| SET8 | Isogenic of YPH274, except for DOG2/dog2::lacZ-LEU2 |
| SET8-1-C | MATα trp1-Δ1 his3-Δ200 leu2-Δ1 lys2-Δ801 ade2-101 ura3-52 dog2::lacZ-LEU2 |
| SCP2 | Isogenic of SET8-1-C, except for pbs2-Δ1::URA3 |
| SCH1 | Isogenic of SET8-1-C, except for hog1Δ::URA3 |
| SCM2 | Isogenic of SET8-1-C, except for msn2-Δ3::HIS3 |
| SCM4 | Isogenic of SET8-1-C, except for msn4Δ::URA3 |
| SCM24 | Isogenic of SET8-1-C, except for msn2-Δ3::HIS3 msn4Δ::URA3 |
| SCS1 | Isogenic of SET8-1-C, except for snf1Δ::HIS3 |
| SCMG1 | Isogenic of SET8-1-C, except for mig1Δ::HIS3 |
| SCSP12 | Isogenic of SET8-1-C, except for snf1Δ::HIS3 pbs2-Δ1::URA3 |
| SCSH11 | Isogenic of SET8-1-C, except for snf1Δ::HIS3 hog1Δ::URA3 |
| SCDG1 | Isogenic of SET8-1-C, except for dog1Δ::HIS3 |
| YPB2 | Isogenic of YPH252, except for pbs2-Δ1::URA3 |
| YHG1 | Isogenic of YPH252, except for hog1Δ::URA3 |
All strains except for YPH274 and YPH252, which were obtained from the Yeast Genetic Stock Center, University of California at Berkeley, were constructed in this study.
Construction of mutants.
Disruption of each gene in the SET8-1-C background (Table 1) was done by the one-step gene replacement method. Each of the pbs2Δ::URA3, hog1Δ::URA3, msn2Δ::HIS3, and msn4Δ::URA3 strains was constructed by the use of the plasmids pJB4D (1), pUHOGΔUra3 (11), pt32-DXB::HIS3 (6), and pUCmsn4ΔUra3 (11), respectively. The SNF1 gene was cloned by PCR using primers 5′-GCGCAAGAAACGGCAGAACAGAAGCTGCTC-3′ and 5′-TCCCGATAACGCTCTGGAATTCAGTGTTGG-3′. The PCR fragment (3,376 bp) was digested with EcoRI and cloned into the EcoRI site of pUC19. The resultant plasmid (pUCSNF1) was digested with AflII and MluI, and the 816-bp fragment was replaced with the HIS3 gene. The resultant plasmid (pUSNF1ΔHis3) was digested with EcoRI, and the DNA fragment containing the snf1Δ::HIS3 cassette was introduced to S. cerevisiae. The MIG1 gene was obtained by PCR using primers 5′-GCATATCAACGCATGCGTTACACAAGATAT-3′ and 5′-GGGATTATGTCGACCTGAAGATTAACCCAC-3′, which were designed to contain recognition sites for SphI and SalI, respectively (underlined). The PCR product was cloned between the SphI and SalI sites of pUC19 to yield pUCMIG1. The region between the XhoI and StyI sites was replaced with the HIS3 gene to construct pUMIG1ΔHis3. To obtain the mig1Δ::URA3 cassette, pUMIG1ΔHis3 was digested with ClaI and PvuII. To disrupt the DOG1 gene, a plasmid which was rescued by YIp5 to determine the SET8 (DOG2) locus was used because this plasmid contained the DOG1 locus. These two genes are linked on chromosome VIII (Fig. 1). The rescued plasmid was digested with EcoRI and PstI, and the fragment containing the DOG1 gene was inserted into the EcoRI and PstI sites of pUC19. The resultant plasmid (pUDOG1) was digested with NruI and SacI, and then the HIS3 gene was inserted. The resultant plasmid was digested with EcoRI and PstI, and the dog1Δ::HIS3 cassette was used to disrupt the DOG1 gene of SET8 clone. The resultant transformant was sporulated, and the dog1 dog2 double mutant was isolated by tetrad analysis. Disruption of each gene was verified by PCR, Southern blot analysis, and the corresponding mutant's phenotype. Tetrad analysis was done by a standard method (30).
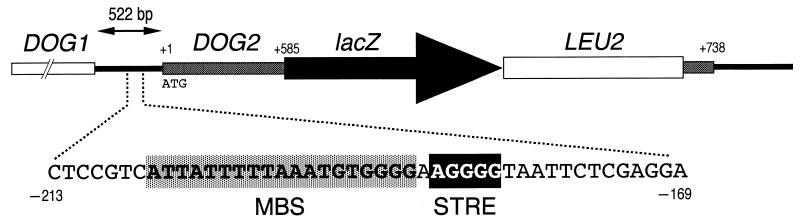
FIG. 1.
Diagram of lacZ-LEU2 insertion in the DOG2 locus. Adenine (A) of the translational initiation codon (ATG) of the DOG2 gene was taken as +1. MBS, Mig1p-binding site; STRE, stress response element.
Screening of oxidative stress-responsive genes.
To screen the oxidative stress-responsive genes, approximately 3,000 Leu+ clones (mTn-lacZ/LEU2 insertion into the genome of strain YPH274) were replica plated onto nylon membranes (Hybond-N; Amersham) on SD (2% glucose, 0.67% yeast nitrogen base without amino acids supplemented with appropriate amino acids and bases; pH 5.5) agar plates containing 0.8 mM t-BHP and were cultured at 28°C for 1 day. At this concentration, all transformants could form colonies. Nylon membranes with yeast colonies were peeled off from the plates, dipped in the liquid nitrogen for 10 s, and then put onto filter papers previously soaked in the Z-buffer (16.1 g of Na2HPO4 · 7H2O/liter, 5.5 g of NaH2PO4 · H2O/liter, 0.75 g of KCl/liter, 0.246 g of MgSO4 · 7H2O/liter) containing 2.7 ml of β-mercaptoethanol/liter and 330 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per liter. Nylon membranes were incubated at room temperature for 12 to 24 h, and then colonies that turned blue were selected as first candidates. In the first screening, we obtained 320 candidates by this screening method. Because the objective of this study was to isolate the oxidative stress-responsive genes, we subjected these clones to the second screening to select the genes whose expression is predominantly induced under oxidative conditions. For the second screening, first candidates on the master plates were replica plated in triplicate on the nylon membranes, and each membrane was put onto the SD agar plates with 0, 0.08, or 0.8 mM t-BHP. Cells were incubated at 28°C for 1 day. After colonies appeared, nylon membranes were treated as described above. Sixty-two clones that mostly turned blue in the presence of a higher concentration of t-BHP were obtained. The plasmid YIp5 was used to rescue the lacZ-LEU2 cassette from the genome DNA of a candidate clone (3), and the nucleotide sequence of a 5′ region of the lacZ-LEU2 cassette-inserted locus of the candidate was sequenced by using a primer (5′-CGTTGTAAAACGACGGGATCCCCCT-3′) as described by Burns et al. (3).
β-Galactosidase assay.
Cells were cultured in a 200-ml flask containing 50 ml of YPD (2% glucose, 1% yeast extract, 2% peptone; pH 5.5) medium at 28°C. When the optical density at 610 nm (OD610) reached 0.8 to 1.0, 0.6 mM t-BHP or solid NaCl was added and the cells were cultured for another 1 h at 28°C. Cell extracts were prepared as described previously (11). β-Galactosidase activity was measured by the method of Miller (22), and 1 U was expressed as the amount of enzyme increasing A420 by a factor of 1,000 per min at 30°C. Protein was determined by the method of Lowry et al. (16).
RESULTS AND DISCUSSION
Expression of DOG2 is induced by oxidative stress.
We obtained 62 clones whose lacZ-reporter gene expression was enhanced by oxidative stress caused by t-BHP. We chose one of them arbitrarily, named SET8, and it was used for further analysis. The remaining clones will be described elsewhere. The SET8 locus was found to be the DOG2 gene encoding 2-deoxyglucose-6-phosphate phosphatase (32), and the lacZ-LEU2 cassette was inserted 585 bp downstream of the ATG codon of DOG2 (Fig. 1). Because the host cell (YPH274) for construction of the lacZ-insertion library is a diploid strain, the DOG2-lacZ insertion of the SET8 clone was heterozygous. The SET8 clone was then sporulated, and tetrads were dissected. All spores from 17 asci were able to germinate; thus, the DOG2 gene was confirmed to be not essential (29). The LEU2 marker and β-galactosidase activity were completely linked and segregated in a 2:2 ratio. A haploid strain obtained by tetrad analysis (SET8-1-C; dog2::lacZ) was used for further investigation.
DOG2 is repressed by Snf1p-Mig1p pathway.
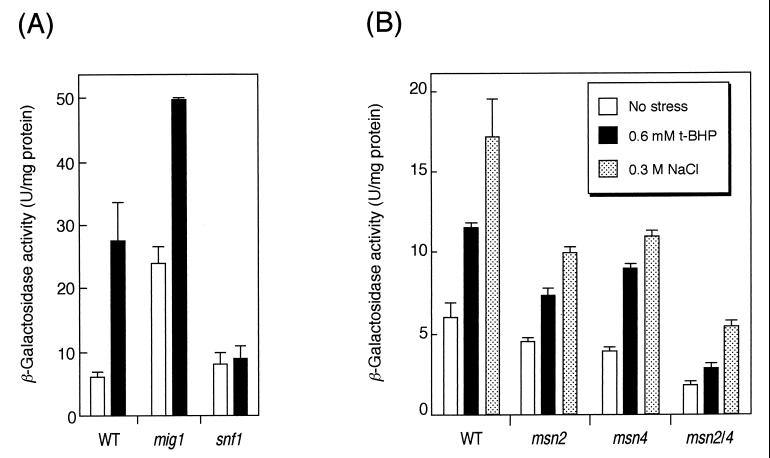
Randez-Gil et al. (29) reported that the DOG2 promoter has a putative Mig1p binding site (MBS; consensus sequence, AT-rich plus GGGG [17]) (Fig. 1), and Lutfiyya et al. reported that expression of DOG2 was regulated by Mig1p (19). Mig1p is a repressor for the glucose-repressed genes (25, 35, 37). The MIG1 gene was then disrupted to confirm whether the DOG2-lacZ reporter construct in SET8-1-C was repressed by Mig1p in the presence of glucose. As shown in Fig. 2A, the basal expression levels of DOG2-lacZ were increased in a mig1Δ mutant compared with those of the wild-type strain. Glucose repression by Mig1p is known to be derepressed by Snf1p, a Ser/Thr protein kinase, if the cells are cultured in a glucose-deprived medium (15, 36). To verify whether the same mechanism was working on this DOG2-lacZ reporter construct, the SNF1 gene was disrupted. As shown in Fig. 2A, derepression of DOG2-lacZ under glucose-deprived conditions was not observed for a snf1Δ mutant. These results indicate that the DOG2-lacZ gene is subject to glucose repression via the Snf1p-Mig1p pathway. We then used this DOG2-lacZ reporter gene to monitor the expression profile of DOG2 under several stress conditions. Induction of DOG2 gene expression under glucose-deprived conditions was still observed in the mig1Δ mutant. According to Lutfiyya et al., Mig2p is not inactivated by Snf1p under glucose-deprived conditions but DOG2 is still glucose regulated, even in a mig1Δ mig2Δ double mutant (19). Since the induction of DOG2 expression under glucose-starved conditions was completely abolished for the snf1Δ mutant (Fig. 2A), this suggests that a third regulator downstream of Snf1p may be involved in regulation of DOG2.
FIG. 2.
(A) Effect of glucose starvation on derepression of DOG2-lacZ gene. Cells were cultured in YPD medium until the OD610 reached approximately 0.8 to 1.0. After cultivation, the cells were collected by centrifugation, resuspended in fresh YPD medium (white bars) or fresh YEP medium without glucose (1% yeast extract, 2% peptone [pH 5.5]) (black bars), and cultured for another 1 h. Strains used were as follows: wild type (WT), SET8-1-C, mig1 mutant, SCMG1; snf1 mutant, SCS1. Data are a summary of three independent experiments (mean ± standard deviation). (B) Regulation of DOG2-lacZ expression by Msn2p and Msn4p. Cells were cultured in YPD medium until the OD610 reached approximately 0.8 to 1.0, and 0.6 mM t-BHP (black bars) or solid NaCl (final concentration, 0.3 M) was added. Cells were cultured for another 1 h, and cell extracts were prepared to measure β-galactosidase activity. Strains used are as follows: wild type (WT), SET8-1-C; msn2 mutant, SCM2; msn4 mutant, SCM4; msn2/4 mutant, SCM24. Data are a summary of three independent experiments (mean ± standard deviation).
Regulation of DOG2 expression by Msn2p and Msn4p.
As shown in Fig. 1, we found a consensus sequence to the STRE (5′-AGGGG-3′) just adjacent to the MBS in the DOG2 promoter. According to Ruis and Schuller (31), several environmental stress signals, including oxidative stress, are targeted to the STRE. We then speculated that the oxidative stress response of DOG2 caused by t-BHP was mediated by Msn2p and Msn4p, which can bind to the STRE under stressful conditions (7, 21, 33). Since the basal expression levels of DOG2 were decreased by simultaneous disruption of MSN2 and MSN4 (Fig. 2B), these two transcription factors may be involved in the expression of DOG2 under normal conditions. Induction of DOG2 gene expression by oxidative stress was reduced in an msn2Δ msn4Δ double mutant (Fig. 2B).
Next, we examined whether Msn2p and Msn4p are involved in the osmotic stress response of DOG2, because expression of many genes carrying the STRE in their promoters has been known to be positively regulated by these C2H2 zinc-finger proteins under hyperosmotic conditions (11, 23, 33). Expression of DOG2 was induced by osmotic stress, as we expected. We confirmed that induction occurred not only by NaCl (Fig. 2B) but also by KCl and sorbitol (data not shown). As shown in Fig. 2B, the single mutant of msn2Δ or msn4Δ as well as the msn2Δ msn4Δ double mutant could still respond to 0.3 M NaCl stress. Although deletion of both MSN2 and MSN4 reduced the basal expression levels of DOG2, the fold increase in induction by osmotic stress in the msn2Δ msn4Δ mutant (3-fold) was the same as that of the wild type (2.9-fold). In contrast to the case of oxidative stress, Msn2p and Msn4p were not likely to function as transcriptional activators for DOG2 under osmotic stress conditions.
Regulation of DOG2 expression by Snf1p-Mig1p pathway and HOG-MAP kinase cascade.
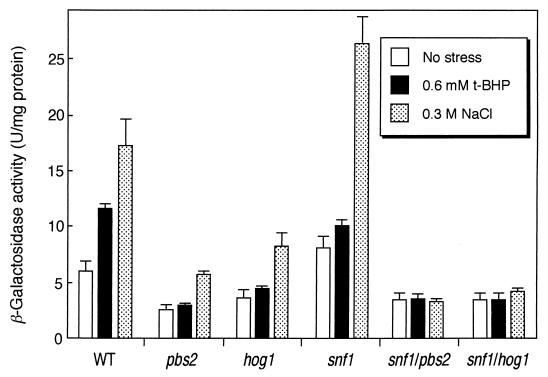
Induction of DOG2 gene expression was observed with between 0.1 and 0.7 M NaCl, with the maximum at 0.3 M, but not at 1.4 M (data not shown). Hog1p, one of the mitogen-activated protein (MAP) kinases in S. cerevisiae, is phosphorylated by Pbs2p (MAP kinase kinase) during the osmotic stress response, and the maximum phosphorylation has been reported to occur at 0.3 M NaCl stress (1). To assess whether the osmotic stress response of DOG2 is dependent upon the HOG (high-osmolarity glycerol)–MAP kinase cascade, we disrupted the genes involved in this signaling pathway, PBS2 and HOG1. The basal expression levels of DOG2 were decreased in both the pbs2Δ mutant and the hog1Δ mutant; however, induction of the DOG2 gene expression by 0.3 M NaCl stress was not repressed in these mutants (Fig. 3). Thus far, we have demonstrated that expression of DOG2 was regulated by Snf1p protein kinase, so we investigated the roles of the Snf1p-Mig1p pathway in the osmotic induction of DOG2. As shown in Fig. 3, the induction of DOG2 expression by osmotic stress was observed for the snf1Δ mutant; however, interestingly, the induction was completely repressed in the snf1Δ pbs2Δ and in the snf1Δ hog1Δ double mutants. These results suggest that the HOG-MAP kinase cascade and Snf1p protein kinase cooperatively transmit the osmotic stress signal to the DOG2 promoter. Since the msn2Δ msn4Δ mutant still responded to hyperosmotic stress (Fig. 2B), the osmotic stress signal from both Snf1p and Hog1p protein kinases might be targeted to an unknown factor(s) other than Msn2p and Msn4p.
FIG. 3.
Regulation of DOG2-lacZ expression by Snf1p kinase and HOG-MAP kinase cascade. Cells were cultured in YPD medium until the OD610 reached approximately 0.8 to 1.0, and 0.6 mM t-BHP or solid NaCl (final concentration, 0.3 M) was added. Cells were cultured for another 1 h, and β-galactosidase activity was measured. Strains used are as follows: wild type (WT), SET8-1-C; pbs2 mutant, SCP2; hog1 mutant, SCH1; snf1 mutant, SCS1; snf1/pbs2 mutant, SCSP12; snf1/hog1 mutant, SCSH11. Data are a summary of three independent experiments (mean ± standard deviation).
The ENA1 gene encodes a cation extrusion P-type ATPase, and its expression is induced by osmotic stress (39). Osmotic regulation of the ENA1 gene is subject to derepression by the Ssn6p-Tup1p complex. The Ssn6p-Tup1p complex itself does not have the ability to bind to DNA directly, but it is recruited to the DNA through interaction with different DNA binding proteins, such as Rox1p, α2/Mcm1, α2/a1, and Mig1p (5, 26, 27, 35, 38). In the case of the ENA1 gene, the SKO1 gene product, a b-Zip DNA binding protein, makes a repressor complex with Ssn6p and Tup1p to bind the cAMP response element (CRE)-like sequence. The Sko1p-Ssn6p-Tup1p complex is dissociated from the ENA1 promoter under highly osmotic conditions which are under the control of the HOG-MAP kinase signaling pathway (28). Deactivation of the Sko1p-Ssn6p-Tup1p repressor complex by the HOG-MAP kinase cascade led us to suspect a possibility that Hog1p and/or Snf1p might also interact with Mig1p-Ssn6p-Tup1p to alleviate the repression of DOG2 by this complex in response to osmotic stress. However, osmotic induction of DOG2 was observed in a mig1Δ mutant (fold increase in induction: wild type, 2.9-fold; mig1Δ, 2.3-fold). This suggests that induction of DOG2 expression by osmotic stress is not caused by deactivation of the Mig1p-Ssn6p-Tup1p repressor complex and that other osmotic stress-responsive transcription factor(s) may function on the DOG2 promoter.
To investigate the contribution of the HOG-MAP kinase cascade and the Snf1p-Mig1p pathway to induction of DOG2 gene expression under oxidative stress conditions, we treated the mutants defective in these pathways with 0.6 mM t-BHP. As shown in Fig. 3, induction of DOG2 expression was not observed for the pbs2Δ and hog1Δ mutants as well as for the snf1Δ mutant. Consequently, the induction was abolished by simultaneous disruption of SNF1 and PBS2 or SNF1 and HOG1 (Fig. 3). These results suggest that Snf1p protein kinase and the HOG-MAP kinase cascade transfer the oxidative stress signal to the DOG2 promoter. It has been reported that AMP-activated protein kinase in mammals is activated by various types of stress (8). The Snf1p protein kinase is a yeast homolog of the mammalian AMP-activated protein kinase. Therefore, Snf1p might be activated by oxidative stress and osmotic stress in addition to glucose starvation.
Role of Dog2p in stress resistance.
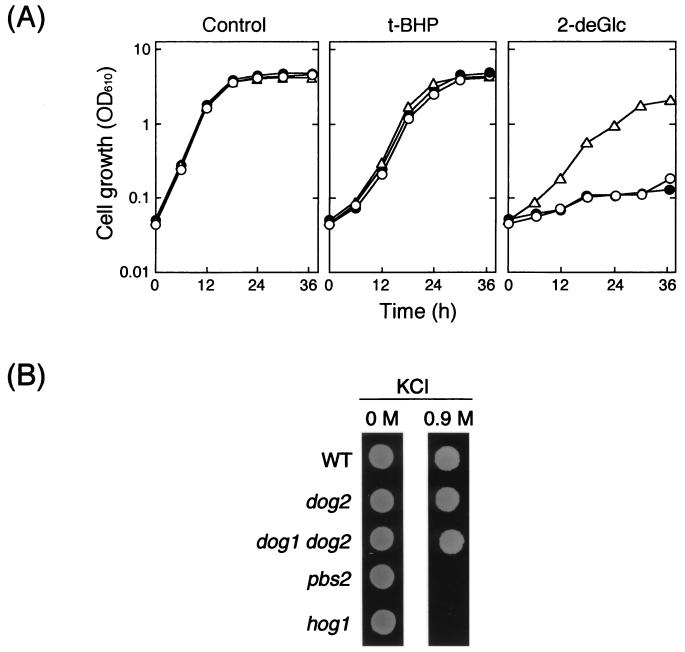
Because the expression of DOG2 was enhanced by t-BHP and osmotic stress, we examined whether a dog2 mutant became hypersensitive to these stresses. No distinct difference was observed in susceptibility to t-BHP between the wild type and a dog2 mutant, although the mutant was sensitive to 2-deoxyglucose (Fig. 4A). Similarly, the dog2 mutant did not exhibit susceptibility to osmotic stress (Fig. 4B). The DOG2 gene was originally cloned as a multicopy suppressor of 2-deoxyglucose toxicity, and the gene product has 2-deoxyglucose-6-phosphate phosphatase activity (32). It is known that twin genes DOG1 and DOG2, which share 92% identity at the amino acid level, are able to confer resistance to 2-deoxyglucose when overexpressed (29). We then disrupted the DOG1 gene in the dog2 background, and susceptibility to oxidative stress and osmotic stress of the resultant double mutant (dog1 dog2) was investigated. The dog1 dog2 mutant did not show an increased susceptibility to these stresses compared with its isogenic wild-type strain (Fig. 4). 2-Deoxyglucose is not a natural substance, and an in vivo substrate for Dog2p as well as Dog1p has not yet been identified. However, since the expression of DOG2 was induced under several stressful conditions (oxidative stress, osmotic stress, and glucose starvation), it must have physiological significance to be induced under such conditions.
FIG. 4.
Effects of disruption of DOG2 and DOG1 on susceptibility to various stresses. (A) Cells were grown in fructose medium (2% fructose, 0.67% yeast nitrogen base without amino acids supplemented with appropriate amino acids and bases; pH 5.5) at 28°C with shaking for 16 h. A small portion of the culture was transferred to fresh fructose medium containing 0.4 mM t-BHP or 0.1% 2-deoxyglucose (2-deGlc) and cultured at 28°C to monitor the OD610. Strains used are as follows: open triangles, YPH252 (wild type); open circles, SET8-1-C (dog2); closed circles, SCDG1 (dog1 dog2). (B) Cells (105 cells/ml) were spotted (5 μl) on YPD agar plates with or without 0.9 M KCl and incubated at 28°C for 2 days. Strains used are as follows: wild type (WT), YPH252; dog2, SET8-1-C; dog1 dog2, SCDG1; pbs2, YPB2; hog1, YHG1.
As far as we know, this is the first report proposing a possibility that the HOG-MAP kinase cascade is likely to mediate oxidative stress signal, as well as proposing that Snf1p protein kinase seems to mediate the signals for osmotic stress and oxidative stress through analyses of the expression pattern of DOG2. Our observations are expected to add a new aspect to the stress response of S. cerevisiae.
ACKNOWLEDGMENTS
We thank M. Snyder for the plasmid for lacZ-insertion library. We are grateful to C. Schuller and F. Estruch for the plasmids pJB4D and pt32-DXB::HIS3, respectively, and to K. Fukuda for assistance in tetrad analysis. We also thank G. Storz, A. Dancis, and M. C. Merlotti.
REFERENCES
- 1.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 2.Bunn H F, Poyton R O. Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 3.Burns N, Grimwade B, Ross-Macdonald P B, Choi E, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 4.Cadenas E. Biochemistry of oxygen toxicity. Annu Rev Biochem. 1989;58:79–110. doi: 10.1146/annurev.bi.58.070189.000455. [DOI] [PubMed] [Google Scholar]
- 5.Deckert J, Rodriguez Torres A M, Simon J T, Zitomer R S. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6109–6117. doi: 10.1128/mcb.15.11.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardie D G. Roles of the AMP-activated/SNF1 protein kinase family in the response to cellular stress. Biochem Soc Symp. 1999;64:13–27. [PubMed] [Google Scholar]
- 9.Inoue Y, Ichiryu T, Yoshikawa K, Tran L-T, Murata K, Kimura A. Induction of glutathione peroxidase by linoleic acid hydroperoxide in Hansenula mrakii. Agric Biol Chem. 1990;54:3289–3293. [Google Scholar]
- 10.Inoue Y, Tran L-T, Kamakura M, Izawa S, Miki T, Tsujimoto Y, Kimura A. Oxidative stress response in yeast: glutathione peroxidase of Hansenula mrakii is bound to the membrane of both mitochondria and cytoplasm. Biochim Biophys Acta. 1995;1245:325–330. doi: 10.1016/0304-4165(95)00117-4. [DOI] [PubMed] [Google Scholar]
- 11.Inoue Y, Tsujimoto Y, Kimura A. Expression of the glyoxalase I gene of Saccharomyces cerevisiae is regulated by high osmolarity glycerol mitogen-activated protein kinase pathway in osmotic stress response. J Biol Chem. 1998;273:2977–2983. doi: 10.1074/jbc.273.5.2977. [DOI] [PubMed] [Google Scholar]
- 12.Inoue Y, Matsuda T, Sugiyama K, Izawa S, Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J Biol Chem. 1999;274:27002–27009. doi: 10.1074/jbc.274.38.27002. [DOI] [PubMed] [Google Scholar]
- 13.Jamieson D J, Storz G. Transcriptional regulators of oxidative stress responses. In: Scandalios J G, editor. Oxidative stress and the molecular biology of antioxidant. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 91–115. [Google Scholar]
- 14.Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator, a new role for SNF1 in the glucose response. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 17.Lundin M, Nehlin J O, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutfiyya L L, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutfiyya L L, Iyer V R, DeRisi J, DeVit M J, Brown P O, Johnston M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–1391. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mager W H, de Kruijff A J J. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Moradas-Ferreira P, Costa V, Piper P, Mager W H. The molecular defences against reactive oxygen species in yeast. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- 24.Moye-Rowley W S, Harshman K D, Parker C S. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989;3:283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- 25.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumor finger proteins. EMBO J. 1990;9:2891–2895. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostling J, Ronne H. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 27.Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proft M, Serrano R. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Randez-Gil F, Blasco A, Prieto J A, Sanz P. DOGR1 and DOGR2, two genes from Saccharomyces cerevisiae that confer 2-deoxyglucose resistance when overexpressed. Yeast. 1995;11:1233–1240. doi: 10.1002/yea.320111303. [DOI] [PubMed] [Google Scholar]
- 30.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 31.Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 32.Sanz P, Randez-Gil F, Prieto J A. Molecular characterization of a gene that confers 2-deoxyglucose resistance in yeast. Yeast. 1994;10:1195–1202. doi: 10.1002/yea.320100907. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran L-T, Inoue Y, Kimura A. Oxidative stress response in yeast: purification and some properties of a membrane-bound glutathione peroxidase from Hansenula mrakii. Biochim Biophys Acta. 1993;1164:166–172. doi: 10.1016/0167-4838(93)90244-l. [DOI] [PubMed] [Google Scholar]
- 35.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treitel M A, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trumbly R J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 38.Wahi M, Johnson A D. Identification of genes required for α2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wieland J, Nitsche A M, Strayle J, Steiner H, Rudolph H K. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu A-L, Moye-Rowley W S. GSH1, which encodes γ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol. 1994;14:5822–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]