Abstract
Serratia entomophila and Serratia proteamaculans cause amber disease in the grass grub Costelytra zealandica (Coleoptera: Scarabaeidae), an important pasture pest in New Zealand. Larval disease symptoms include cessation of feeding, clearance of the gut, amber coloration, and eventual death. A 115-kb plasmid, pADAP, identified in S. entomophila is required for disease causation and, when introduced into Escherichia coli, enables that organism to cause amber disease. A 23-kb fragment of pADAP that conferred disease-causing ability on E. coli and a pADAP-cured strain of S. entomophila was isolated. Using insertion mutagenesis, the pathogenicity determinants were mapped to a 17-kb region of the clone. Sequence analysis of the 17-kb region showed that the predicted products of three of the open reading frames (sepA, sepB, and sepC) showed significant sequence similarity to components of the insecticidal toxin produced by the bacterium Photorhabdus luminescens. Transposon insertions in sepA, sepB, or sepC completely abolished both gut clearance and cessation of feeding on the 23-kb clone; when recombined back into pADAP, they abolished gut clearance but not cessation of feeding. These results suggest that SepA, SepB, and SepC together are sufficient for amber disease causation by S. entomophila and that another locus also able to exert a cessation-of-feeding effect is encoded elsewhere on pADAP.
Amber disease of the New Zealand grass grub Costelytra zealandica (Coleoptera: Scarabaeidae) is caused by some strains of Serratia entomophila and Serratia proteamaculans (Enterobacteriaceae). The disease was first described by Trought et al. (40), and an isolate of S. entomophila was subsequently developed into a commercially available biological control agent for C. zealandica in New Zealand (26). The disease is highly host specific, affecting only a single species of scarab that is indigenous to New Zealand (24). The disease is chronic, with a prolonged infection phase before bacteria invade the hemocoelic cavity, causing death (25). Amber disease has a distinct phenotypic progression, with infected hosts ceasing feeding within 2 to 4 days of ingesting pathogenic bacteria. At this time, levels of the major gut digestive enzymes decrease sharply (23) and the normally black-gray gut clears (25), resulting in a characteristic amber color of the infected insects. The infected larva may remain in this state for a prolonged period (1 to 3 months) before the infecting bacteria eventually invade the hemocoel, causing death.
The disease determinants of S. entomophila are encoded on a 115-kb plasmid designated pADAP, for amber disease-associated plasmid (17). pADAP has been transferred by conjugation to Enterobacter agglomerans, Escherichia coli, a Klebsiella sp., and the Serratia species S. proteamaculans, S. marcescens, and S. liquefaciens. Acquisition of pADAP by these species confers pathogenicity towards grass grub larvae (18).
To identify pathogenicity determinants on pADAP, Grkovic et al. (20) mutated a number of cloned HindIII fragments from pADAP with the mini-Tn10 derivative 103 and recombined the insertion mutations back into pADAP to form pADK derivatives. Bioassays of S. entomophila strains containing the various pADK derivatives against grass grub larvae showed that 21 of the mutations had no detectable effect on pathogenicity toward the grass grub. However, in contrast to larvae infected with wild-type S. entomophila(pADAP), where there is a cessation of feeding and clearance of the larval gut, seven mutants induced a phenotype of nonfeeding with no gut clearance. The mutations in these strains were all located throughout a single 11-kb HindIII fragment, but one insertion (pADK-35) in the central region of the fragment had no effect on the disease process. Complementation analysis of the pADAP recombinants that contained insertions on either side of the pADK-35 insertion (pADK-10 and pADK-13) showed that only the pADK-13 region was complemented by the subcloned 11-kb fragment. The subclone itself did not enable a pADAP-cured strain of S. entomophila to induce any disease symptoms, indicating that it did not contain all of the essential pathogenicity determinants of pADAP.
In this report, we describe the identification, cloning, mutagenesis, and nucleotide sequence analysis of a region of pADAP that is sufficient to confer pathogenicity toward grass grub on both E. coli and pADAP-cured S. entomophila bacteria.
MATERIALS AND METHODS
Bacterial strains and methods of culture.
Table 1 lists bacterial strains and plasmids used in this study. Bacteria were grown in Luria-Bertani (LB) broth or on LB agar (37) at 37°C for E. coli and 30°C for S. entomophila. For Serratia, the antibiotics kanamycin, chloramphenicol, and tetracycline were used at 100, 90, and 30 μg/ml, respectively; for E. coli, kanamycin, chloramphenicol, tetracycline, and ampicillin were used at 50, 30, 15, and 100 μg/ml, respectively.
TABLE 1.
Bacterial strains, plasmids, and bacteriophage used in this study
| Strain, plasmid, or phage | Description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH5α | F− φ80d lacZΔM15 ΔlacZYA-argFU169 recA1 endA1 supE44 | 22 |
| DH10B | F−mcrA Δmrr-hsdRMS-mcrBCφ80d lacZΔM15 ΔlacX74 endA1 recA1 deoRΔara leu 7697 araD139 galU galK nupG rpsL λ− | 31 |
| DF1 | γδ transposase tnpA | Gibco BRL |
| MC1061 | sup0hsdR mcrB araD139 ΔaraABC-leu7679 ΔlacX74 galU galK rpsL thi | 7 |
| MC4100 | araD139 ΔlacZYA-argFU169 rpsL150 StrrelA1 flbB5301 deoC1 ptsF25 rbsR | 38 |
| XL1-BlueMRA | ΔmcrA183 ΔmcrCB-hsdSMR-mrr-173 endA1 supE44 thi-1 reA1 gyrA96 relA1 | Stratagene |
| S. entomophila | ||
| A1MO2 | Apr, pADAP, pathogenic | 19 |
| 5.6 | Heat-cured pADAP-minus derivative of A1MO2 | 17 |
| 5.6RC | CmrrecA pADAP-minus strain | 20 |
| 5.6RK | KnrrecA pADAP-minus strain | This study |
| Plasmids | ||
| pACYC184 | Cmr Tcr | 8 |
| pADAP | Amber disease-associated plasmid | 17 |
| pBR322 | Apr Tcr | 3 |
| pBM32 | 23-kb BamHI fragment from pMH32 cloned in pBR322 | This study |
| pBM32-1-40 | pBM32 containing mini-Tn10 insertions | This study |
| pDELTA1 | Apr Smr Knr, sucroser | Gibco BRL |
| pLAFR3 | Tcr pRK290 with λcos, lacZα, and multicloning site from pUC8 | 39 |
| pGLA20 | 11-kb HindIII pADAP fragment cloned in pLAFR3 | 20 |
| pADK-13 | pADAP::mini-Tn10 insertion in 10.6-kb HindIII fragment, Knr, nonpathogenic | 20 |
| pADK-35 | pADAP::mini-Tn10 insertion in 10.6-kb HindIII fragment, Knr, pathogenic | 20 |
| pMH32 | 23-kb BamHI fragment of pADAP cloned into pLAFR3 | This study |
| pMH41 | 33-kb BamHI fragment of pADAP cloned into pLAFR3 | This study |
| pBM32 | 23-kb BamHI fragment of pMH32 cloned into pBR322 | This study |
| pUC19 | Apr, lacZα, multicloning site | 42 |
| Bacteriophage | ||
| λNK1316 | Mini-Tn10 derivative 103 donor λb522 cI857 Pam80 nin5 | 27 |
DNA isolation and manipulation.
pADAP DNA was isolated from a 50-ml overnight culture of bacteria using a Qiagen (Hilden, Germany) plasmid maxi kit according to the manufacturer's instructions. Standard DNA techniques were carried out as described by Sambrook et al. (37). Radioactive probes were made using the Amersham (Buckinghamshire, United Kingdom) Megaprime DNA labeling system. Southern blot and colony hybridizations were performed as described by Sambrook et al. (37).
Introduction of plasmid DNA into E. coli and S. entomophila.
pLAFR3- and pBR322-based plasmids were electroporated into E. coli and S. entomophila strains using a Bio-Rad Gene Pulser (25 μF, 2.5 kV, and 200 ohms) (10).
Mutagenesis.
Transposon insertions were generated in recombinant plasmids using the mini-Tn10 derivative 103 (kanamycin resistant) carried on λNK1316, as described by Kleckner et al. (27). Insertions were recombined into pADAP by transforming strain A1MO2 (Table 1) with the desired pLAFR3-based construct. After 5 days of growth in nonselective medium, bacteria were selected for resistance to kanamycin and screened for loss of the pLAFR3 tetracycline resistance marker (approximately 34% of the kanamycin-resistant colonies were tetracycline sensitive).
Bioassay against C. zealandica larvae.
Infection of C. zealandica larvae was determined by a standard bioassay (20) where healthy larvae, collected from the field, were individually fed cubes of carrot (3 mm3) which had been rolled in colonies of bacteria grown overnight on solid medium, resulting in approximately 107 cells per carrot cube. Twelve second- or third-instar larvae were used for each treatment. Inoculated larvae were maintained at 15°C in ice cube trays. Larvae were fed treated carrot at day 1; at days 3 and 6, they were transferred to fresh trays containing untreated carrot (3 mm3). The occurrence of gut clearance and cessation of feeding were monitored at days 3, 6, and 12. Strains were tested for loss of disease-causing ability by comparing numbers of diseased larvae in treated with known pathogenic and nonpathogenic bacterial controls after 12 days using a one-tailed paired t test.
Recovery of bacteria from larvae.
To isolate bacteria from inoculated grubs, larvae were first surface sterilized by submersion in 70% methanol for 30 s. The larvae were then shaken in sterile distilled water, removed, and individually macerated in 1.5-ml microcentrifuge tubes. Each macerate was serially diluted and plated on LB medium containing antibiotics selective for the host S. entomophila strain. To assess the maintenance of the plasmid in the bioassayed strain, colonies were patched onto a plate containing antibiotics selective for the recombinant plasmid. Identity of plasmids in the recovered strains was checked by restriction enzyme profiling.
Nucleotide sequencing.
A 9-kb BamHI-EcoRI fragment derived from pBM32-8 (Fig. 1C) and the 8-kb HindIII fragment of pBM32 were separately cloned into the appropriate sites of the deletion factory plasmid pDELTA1 (Gibco BRL, Rockville, Md.). Deletions were generated using the deletion factory system as outlined in the manufacturer's instructions.
FIG. 1.
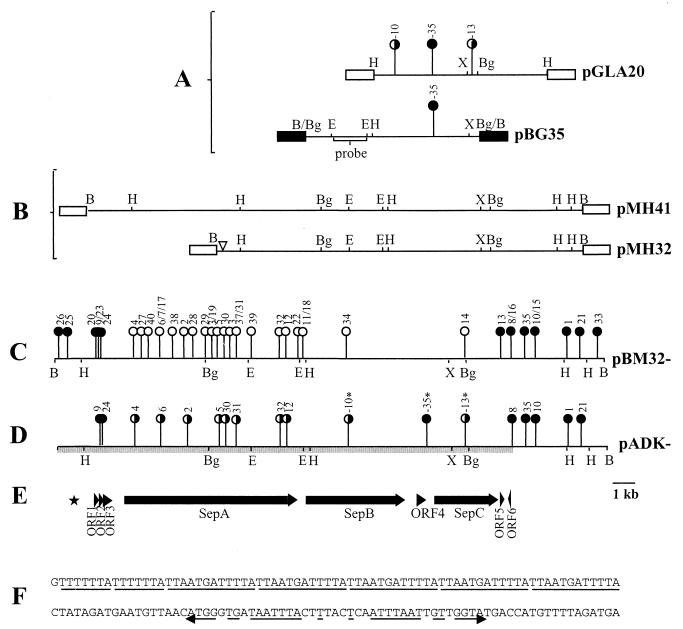
(A) The HindIII fragment from pADAP cloned into pLAFR3 to form pGLA20, showing locations of the mini-Tn10 103 insertion mutations at positions -10, -13, and -35 (18). Results of bioassay of mutants against the grass grub are shown. The map of pBG35 shows the relative position of pGLA20-35 mutation and the location of the 2.2-kb EcoRI fragment used as a probe to screen the pADAP BamHI library. (B) Restriction enzyme maps of the pathogenic clones pMH32 and pMH41. (C) Locations and phenotypes of mini-Tn10 insertions in pBM32. (D) Bioassay results of the pADK recombinants. (E) Schematic diagram of the sequenced region. (F) Nucleotide sequence of the 7-bp repeat, five-copy 12-bp repeat, and the downstream degenerate 34-bp inverted repeat. ∗, pADK mutations isolated by Grkovic et al. (20); filled circles, mutations that resulted in an unaltered pathogenic phenotype (clear gut, nonfeeding); open circles, mutations that resulted in the abolition of pathogenicity; half-filled circles, mutations that induced a nonfeeding pathotype without clearance of the gut; ▿, site of internal deletion; ■, pBR322 vector DNA; □, pLAFR3; ★, location of nucleotide repeats. Arrows indicate ORFs and their orientation. Abbreviations for restriction enzymes: B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; X, XbaI.
To identify the precise locations of mini-Tn10 insertions, the peripheral BamHI sites located within the ends of mini-Tn10 were used in conjunction with the BamHI sites of the cloned region to subclone the regions flanking the mini-Tn10 insertion into either pACYC184 or pUC19. Sequences were generated using the mini-Tn10-specific primer 5′ ATGACAAGATGTGTATCCACC 3′ (27).
Plasmid templates for sequencing were prepared using Wizard (Promega, Madison, Wis.) or Quantum-Prep (Bio-Rad) miniprep kits. Sequences were determined on both strands using combinations of subcloned fragments, custom primers, and deletion products derived from the deletion factory system. The DNA was sequenced either by using [33P]dCTP and the Thermosequenase cycle sequencing kit (Amersham) or by automated sequencing using an Applied Biosystems 373A or 377 autosequencer. Sequence data were assembled using SEQMAN (DNASTAR Inc., Madison, Wis.). Databases at the National Center for Biotechnology Information were searched using BLASTN and BLASTX (1). Searches for open reading frames (ORFs), DNA repeats, and inverted repeats were undertaken using DNAMAN (Lynnon BioSoft, Quebec, Quebec, Canada). Searches for protein motifs were carried out using Blocks (http://www.blocks.fhcrc.org/), ExPASy (http://www.expasy.ch/), and Gene Quiz (http://columba.ebi.ac.uk:8765/gqsrv/submit).
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in GenBank under accession number AF135182.
RESULTS
Cloning the major virulence determinants from pADAP.
Complementation analysis of the pADK-10 and -13 mutants with the 11-kb HindIII fragment cloned in pLAFR3 to give pGLA20 showed that only the pADK-13 insertion was complemented, suggesting that the locus inactivated by the pADK-10 insertion was not fully contained within the fragment (20). To define the region required to complement the pADK-10 mutation, a 13-kb BglII fragment from pADK-35 that included the mini-Tn10 insertion and encompassed the sites of both the pADK-10 and pADK-35 mutations was cloned into the BamHI site of pBR322 to form pBG35. pBG35 was placed separately in trans with pADK-10 and pGLA20 in the pADAP-cured S. entomophila strain 5.6RC, and the resultant strains were bioassayed against grass grub larvae. Results showed that pBG35 complemented strain 5.6RC(pADK-10) but did not confer the ability to induce any of the disease symptoms on strain 5.6RC(pGLA20), suggesting that there must be a region of pADAP in addition to that encoded by the pGLA20 and pBG35 fragments needed to induce amber disease.
Restriction enzyme mapping of pGLA20 and pBG35 showed that neither contained a BamHI site, indicating that the cloned DNA from both plasmids was contained within a large (>15-kb) BamHI fragment of pADAP. A BamHI library of pADAP was made and screened using a 2.2-kb EcoRI fragment derived from pBG35 (Fig. 1A) as the probe. Several probe-positive clones were isolated, and all had similar restriction enzyme profiles. However, one (designated pMH32) was smaller, with an inserted BamHI fragment of only 23 kb, compared with the 33-kb insert of the other clones (e.g., pMH41 [Fig. 1B]). The difference between pMH32 and pMH41 was found to be a 10-kb truncation at one end of pMH32 that included one HindIII site (Fig. 1B). Recent sequence data have shown that the site of truncation is at bp 170 of the sequence generated in this study (M. R. H. Hurst, unpublished data).
When bioassayed against grass grub larvae, E. coli strains containing pMH32 or pMH41 induced the full symptoms of amber disease (i.e., gut clearance and cessation of feeding activity). However, about 10 days after infection, approximately 25% of the grass grub larvae fed the E. coli strains recovered from a diseased to a healthy phenotype. This may reflect either poor persistence of the E. coli strains in the grass grub or poor expression of the cloned genes. Therefore, all further studies of the cloned loci were done in S. entomophila backgrounds.
Plasmids pMH32 and pMH41 were subsequently introduced into an S. entomophila strain cured of pADAP (5.6RC), and the strains were bioassayed against grass grub larvae. The strains gave the same disease progression as the wild type, and no larvae were recovered (Table 2).
TABLE 2.
Disease-causing ability of the cloned virulence-encoding region, its mutated derivatives, and pADAP recombined mutationsa
| Positive (A1MO2), 91–100%b
|
Negative (untreated carrot), 5.3–22.8%b
|
||||||
|---|---|---|---|---|---|---|---|
| Construct | Mean % showing disease symptoms | One-tailed t testc
|
Construct | Mean % showing disease symptoms | One-tailed t test
|
||
| df | P | df | P | ||||
| Virulence-encoding clones | |||||||
| pBM32 | 97.8 | 3 | NV | pMH32 | 97.0 | 2 | 0.211 |
| pMH41 | 95.5 | 1 | 0.250 | ||||
| pBM32 derivativesd | |||||||
| pBM32-1 | 97.0 | 2 | 0.500 | pBM32-21 | 88.7 | 2 | 0.211 |
| pBM32-2 | 16.5 | 1 | 0.052 | pBM32-22 | 8.3 | 2 | 0.005* |
| pBM32-3 | 4.0 | 1 | 0.015* | pBM32-23 | 91.5 | 3 | 0.196 |
| pBM32-4 | 12.50 | 1 | 0.050 | pBM32-24 | 94.0 | 2 | 0.500 |
| pBM32-5 | 25.0 | — | pBM32-25 | 94.0 | 2 | 0.211 | |
| pBM32-6 | 4.0 | 1 | 0.029* | pBM32-26 | 93.3 | 3 | NV |
| pBM32-7 | 8.0 | — | pBM32-27 | 10.5 | 1 | 0.041* | |
| pBM32-8 | 94.0 | 2 | 0.211 | pBM32-28 | 94 | 2 | 0.001* |
| pBM32-9 | 93.3 | 3 | 0.091 | pBM32-29 | 8.0 | 1 | NV |
| pBM32-10 | 95.8 | 5 | 0.298 | pBM32-30 | 8.0 | 1 | NV |
| pBM32-11 | 2.9 | 3 | 0.029* | pBM32-31 | 10.5 | 1 | 0.056 |
| pBM32-12 | 18.5 | 1 | 0.011* | pBM32-32 | 10.7 | 2 | 0.002* |
| pBM32-13 | 94.0 | 2 | 0.500 | pBM32-33 | 91.0 | 4 | 0.035* |
| pBM32-14 | 8.0 | 1 | 0.031* | pBM32-34 | 0.0 | 2 | 0.000* |
| pBM32-15 | 95.5 | 1 | NV | pBM32-35 | 87.5 | 3 | 0.187 |
| pBM32-16 | 100.0 | — | pBM32-36 | 97.0 | 2 | 0.211 | |
| pBM32-17 | 21.0 | — | pBM32-37 | 2.7 | 2 | 0.000* | |
| pBM32-18 | 91.0 | 1 | 0.250 | pBM32-38 | 4.0 | 1 | 0.015* |
| pBM32-19 | 8.0 | — | pBM32-39 | 4.0 | 1 | 0.029* | |
| pBM32-20 | 93.0 | 4 | 0.102 | pBM32-40 | 0.0 | 1 | NV |
| pADAP pADK derivativese | |||||||
| pADK-1 | 97.0 | 2 | 0.211 | pADK-21 | 94.0 | 2 | 0.092 |
| pADK-4 | 8.3 (NF) | 2 | 0.004* | pADK-22 | 12.0 (NF) | 1 | 0.002* |
| pADK-5 | 12.0 (NF) | 1 | 0.014* | pADK-23 | 93.3 | 3 | 0.091 |
| pADK-6 | 4.0 (NF) | 1 | 0.013* | pADK-24 | 91.0 | — | |
| pADK-8 | 8.0 (NF) | — | pADK-30 | 13.7 (NF) | 2 | 0.006* | |
| pADK-9 | 97.0 | 2 | 0.211 | pADK-31 | 4.0 (NF) | 1 | 0.013* |
| pADK-10 | 88.7 | 2 | 0.131 | pADK-32 | 19.0 (NF) | 2 | 0.004* |
| pADK-11 | 12.0 (NF) | 1 | 0.014* | pADK-34 | 13.3 (NF) | 2 | 0.000* |
| pADK-12 | 95.5 | 1 | 0.250 | pADK-35 | 100.0 | 3 | NV |
| pADK-13 | 97.8 | 3 | 0.196 | pADK-39 | 8.0 (NF) | 1 | 0.028* |
NF, larvae were healthy in appearance but unable to feed.
Range of controls over 80 batches.
One-tailed paired t test over batches of the percentage of C. zealandica larvae showing full disease symptoms (amber coloration and inhibition of feeding) after ingestion of Serratia entomophila at day 12. NV, no variation; ∗, significant difference (P < 0.05); —, only one batch.
See Fig. 1C for locations of pBM32 mutations.
See Fig. 1D for locations of pADK mutations.
Effects of mini-Tn10 insertions in pBM32 on disease-causing ability.
To facilitate mutagenesis, the 23-kb BamHI fragment from pMH32 was cloned into the medium-copy-number plasmid pBR322 to give pBM32. Bioassays of the strains containing pBM32 showed that it conferred the ability to induce amber disease in an S. entomophila (5.6RC) background (Table 2). Plasmid pBM32 was mutated with the mini-Tn10 transposon derivative 103, and the sites of the insertions were mapped (Fig. 1C). Bioassays of S. entomophila strain 5.6RC derivatives containing the resultant mutated plasmids showed that the disease determinants were confined to a central 17-kb region of the BamHI fragment. Each strain either had no effect or caused full disease symptoms (cessation of feeding and gut clearance) (Table 2).
Effects of mini-Tn10 insertions in pADAP on disease-causing ability.
Grkovic et al. (20) recombined the pGLA20-based mutations −10 and −13 into pADAP by homologous recombination (Fig. 1A and D). When bioassayed, S. entomophila strains containing either of these mutant pADAP plasmids caused a partial disease condition, inducing cessation of feeding but not gut clearance and amber coloration. This was in contrast to the complete abolition of disease observed with pADAP-cured S. entomophila strains containing mutant pBM32 plasmids with similar insertions (Table 2). To determine the phenotype of the pBM32-based insertions in a pADAP background, DNA fragments containing the pBM32 insertions at positions −1, −2, −4, −5, −6, −8, −9, −10, −12, −21, −24, −30, −31, −32, and −35 and flanking DNA were cloned separately into pLAFR3, and the inserted transposon was introduced into pADAP by homologous recombination (Fig. 1D). The resultant recombinant S. entomophila strains were checked by Southern analysis to confirm that recombination had occurred as expected and that no pLAFR3 vector sequences were present (data not shown). The strains were then assayed against grass grub larvae (Table 2). Mutations that did not affect the disease process in pBM32 also had no effect on disease when recombined into pADAP. However, strains with the pADAP mutations that totally abolished the disease process when in pBM32 caused cessation of feeding but not gut clearance of the grubs (Fig. 1C and D).
Assessment of the stability of pBM32 and its mutated derivatives during the course of the bioassays showed that greater than 90% of the recovered Serratia strains contained the plasmid of interest.
Sequence analysis of the disease-encoding region.
The BamHI fragment (18,937 bp) derived from pBM32-8 was sequenced on both strands using a combination of constructed deletions, plasmid subclones, and custom-made primers. Structural analysis of the DNA sequence using DNAMAN showed that there was a 7-bp direct repeat at bp 671 to 684, followed by a 12-bp sequence repeated five times between positions 685 and 744. Downstream of the repeat region was a degenerate 39-bp inverted repeat (bp 763 to 801) (Fig. 1E and F). These repeat motifs are in a region of DNA that is AT rich and lacks any potential ORFs.
Translation of the nucleotide sequence revealed the presence of nine ORFs of more than 90 codons. Eight of the ORFs were oriented in the same direction, and the other was oriented in the opposite direction (Fig. 1E). Sequence similarity searches showed that the deduced products of seven of these ORFs showed similarity with known proteins (Table 3). ORF1 (100 amino acid residues) and ORF2 (91 amino acid residues) had no similarity to any proteins in the current databases. Products of three of the ORFs showed similarity to different protein components of insecticidal toxins of Photorhabdus luminescens (5). These ORFs were designated sep (sepA, sepB, and sepC), for Serratia entomophila pathogenicity.
TABLE 3.
Similarities of products of putative ORFs to protein sequences in the database detected using BlastP
| ORF, size (amino acids) | Protein homologue, size (amino acids) | Degree of similaritya | Function of similar protein | Organism | Accession no. |
|---|---|---|---|---|---|
| SepA, 2,376 | TcbA, 2,504 | 34/50, 41–1628; 57/72, 1630–2374 | Insecticidal toxin complex protein | P. luminescens | AF047457 |
| TcdA, 2,516 | 38/55, 33–1289; 40/54, 1499–1625; 58/71, 1630–2374 | Insecticidal toxin complex protein | P. luminescens | AF188483 | |
| TcaB, 1,189 | 29/50, 936–1198; 38/54, 1625–2374 | Insecticidal toxin complex protein | P. luminescens | AF046867 | |
| TccB, 1,565 | 31/51, 930–1204; 36/51, 1575–2373 | Insecticidal toxin complex protein | P. luminescens | AF047028 | |
| TcaA, 1,095 | 36/56, 94–183; 18/39, 435–928 | Insecticidal toxin complex protein | P. luminescens | AF046867 | |
| TccA, 965 | 27/45, 115–280 | Insecticidal toxin complex protein | P. luminescens | AF047028 | |
| SepB, 1,428 | TcaC, 1,485 | 49/63, 1–1263; 64/78, 1270–1421 | Insecticidal toxin complex protein | P. luminescens | AF046867 |
| SpvB, 591 | 40/52, 9–365 | Salmonella virulence protein | Salmonella serovar Typhimurium | S22664 | |
| SepC, 938 | TccC, 1,043 | 53/66, 3–782 | Insecticidal toxin complex protein | P. luminescens | AF047028 |
| Gene sc2h4.02, 2,183 | 23/34, 68–677 | Hypothetical wall-associated protein | S. coelicolor A3(2) | CAA20596.1 | |
| WapA, 2,334 | 20/36, 48–625; 22/34, 255–677 | Wall-associated protein precursor | B. subtilis | Q07833 | |
| Orf 774, 334 | 21/34, 181–684 | Hypothetical wall-associated protein | C. burnetii | CAA75841 | |
| Rhs core, 1,420 | 21/36, 35–300; 21/35, 237–677 | Rhs core protein | E. coli | ACC32471 | |
| ORF3, 144 | Gene 15, 263 | 45/62, 1–139 | Morphogenesis protein of bacteriophage B103 | B. subtilis | CAA67646.1 |
| Gene 19, 146 | 46/61, 1–143 | Phage P22 lysozyme EC3.2.1.17 | Salmonella | Q37896 | |
| ORF4, 191 | Gp55, 181 | 28/42, 1–184 | Bacteriophage N15 protein | E. coli | AAC19092.1 |
| ORF5, 236 | Tnp1294, 312 | 50/69, 18651–18391; 55/74, 18934–18752 | Transposase | E. coli | S49612 |
| ORF6, 310 | IS91 | 39/58, 18675–18394; 39/56, 18934–18725; 30/48, 18391–18164 | IS91 transposase | E. coli | S23782 |
Amino acid similarity (percent identity/percent similarity over the indicated range of amino acid residues) in relation to sequence generated in this study. Percent identities and similarities were calculated in relation to the deduced gene products of the sequenced ORF. Similarities were considered potentially significant if the BlastP score exceeded e−5.
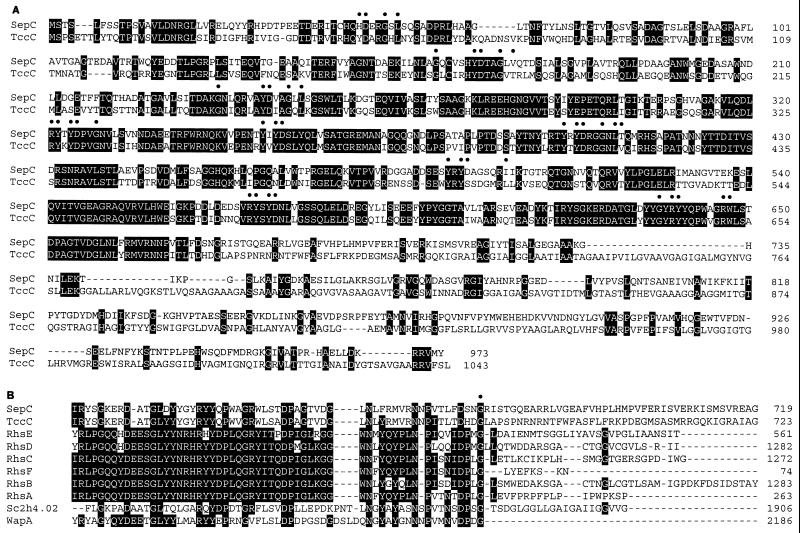
The predicted protein product of sepA had high similarity throughout its entire length to the P. luminescens insecticidal toxin complex proteins TcbA, TcdA, TcaB, and TccB, with greatest similarity at the carboxyl terminus (Table 3; Fig. 2). Analysis for protein motifs showed that the tripeptide cell-binding motif Arg-Gly-Asp (RGD), implicated in the binding of various adhesion proteins produced by parasites and viruses to eukaryotic cells (29), is present in SepA and the P. luminescens TcdA, TcbA, and TcaB proteins (Fig. 2).
FIG. 2.
Alignment of amino acid sequences of the SepA and P. luminescens toxin components TcbA, TcdA, TcaB, and TccB. ■, RGD motif.
SepB and the P. luminescens insecticidal toxin complex protein TcaC showed similarity throughout their length, and both SepB and TcaC showed high amino-terminal similarity to the Salmonella enterica serovar Typhimurium virulence protein SpvB (21) (Fig. 3). The similarity of SepB and TcaC to SpvB diminished after SpvB amino acid residue 356.
FIG. 3.
Alignment of amino acid sequences of the SepB, P. luminescens toxin component TcaC, and SpvB.
SepC showed strong similarity to the amino-terminal region of the insecticidal toxin complex protein TccC, up to amino acid residue 732 of SepC (Fig. 4A). A number of putative bacterial cell wall-associated proteins also showed similarity to SepC, including the wall-associated precursor protein of Bacillus subtilis (WapA) (15), members of the E. coli rhs (recombination hot spot) elements (41), and hypothetical wall-associated proteins from Streptomyces coelicolor A3(2) and Coxiella burnetii (Table 3; Fig. 4B). Comparison of SepC to its homologues showed that all showed amino acid similarity to the carboxyl end of a so-called Rhs core region (41) (Fig. 4B) and diverged from each other in amino acid composition at a conserved glycine residue (SepC amino acid residue 666 [Fig. 4B and 5]). Further comparison of SepC with members of the Rhs family showed that it contained nine partial or complete matches to the Rhs core protein peptide motif GxxRYxYDxxGRL(I/T) (12, 41) (Fig. 4A).
FIG. 4.
(A) Alignment of amino acid sequences of the SepC and P. luminescens toxin component TccC. Conserved positions of the repeat motif GxxRYxYDxxGRL(I/T) (●) are marked. (B) Alignment of amino acid sequences of SepC to the P. luminescens toxin component TccC, the Rhs elements (RshE, P24211; RshD, P16919; RshC, P16918; RshF, I69801; RshB, P16917; RshA, P16916), the hypothetical protein SC2H4.02 from S. coelicolor A3(2), and the wall-associated protein of B. subtilus (WapA). ●, position of the conserved glycine residue which characterizes the junction between the conserved carboxyl end of the Rhs core and the variable carboxyl terminus.
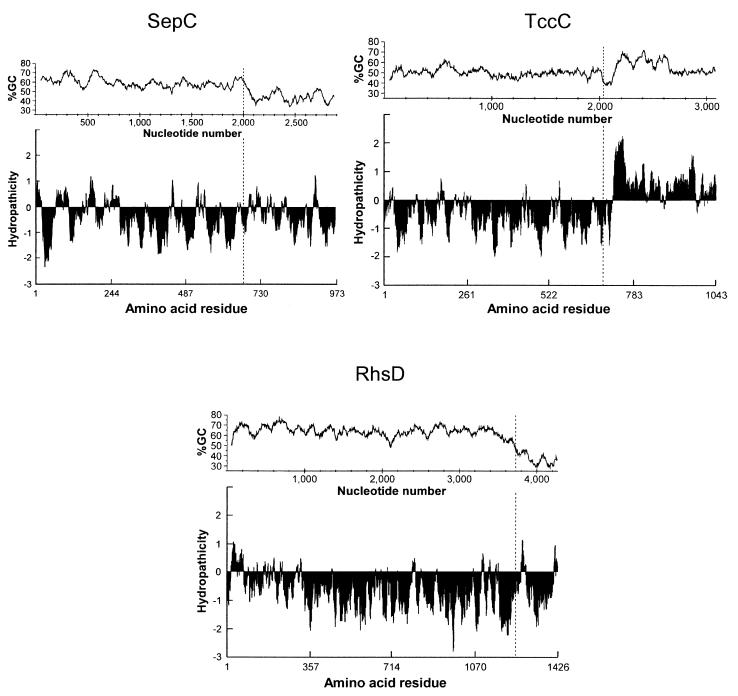
FIG. 5.
GC content (window size, 100; window position shift, 3) and hydropathicity plots of SepC, TccC, and RhsD (scanning window of 17 amino acid residues). Each vertical dashed bar denotes the position of the conserved glycine residue which characterizes the junction between the conserved carboxyl end of the Rhs core and the variable carboxyl terminus.
We found that the sepA and sepB genes were more GC rich (54 and 58% G+C) than their P. luminescens counterparts (43 and 44% [tcbA and tcdA] and 51% [tcaC] G+C), while sepC and tccC had similar GC contents (55 and 54% G+C). Similar to rhs elements, sepC is relatively GC rich (sepC bp 1 to 2031, 60% G+C) preceding the juxtaposition of the core and variable region but decreases in GC content thereafter (sepC bp 2032 to 2922, 44% G+C) (Fig. 5). The GC content of the sep genes was similar to that of the S. entomophila genome, which is 58% G+C (19). tccC also shows a strong reduction in GC content at the junction of the core and variable regions, but thereafter its 3′ region is GC rich. This may reflect the highly hydrophobic nature of the carboxyl terminus of TccC together with its high content of glycine and alanine residues, which together comprise 40% of the amino acids of the region and are encoded by GC-rich codons.
The hydropathicity profile of each of the Sep proteins was examined using the Kyte and Doolittle algorithm (28) and compared to the profiles of relevant P. luminescens homologues. None of the Sep or Tc proteins contained a characteristic signal sequence (16) or regions capable of spanning the cell membrane, except for the P. luminescens protein TccC, which has a highly hydrophobic carboxyl terminus from amino acid residue 719 onward (Fig. 5).
ORF3 precedes sepA (Fig. 1E) and has high similarity to the morphogenesis protein encoded by gene 15 of the B. subtilis bacteriophage B103 (33) and the product of gene 19 from the Salmonella bacteriophage P2, a protein essential for the lysis of the bacterial cell wall (35). Hydropathicity analysis showed that the translated product of ORF3 contains a hydrophobic amino terminus capable of spanning the lipid bilayer.
Located between sepB and sepC is ORF4, the translated product of which has similarity to the E. coli bacteriophage N15 gp55 protein, a protein of unknown function (Table 3), and contains an amino terminus capable of spanning the lipid bilayer.
Identification of mini-Tn10 location by sequence analysis.
Analysis of the insertion points of the mini-Tn10 insertions (Fig. 1C) within the putative ORFs (Table 4) revealed that ORF3 and ORF4 were interrupted by the insertions that had no effect on the disease process. However, the pADAP-35 mutation was at the 3′ end of ORF4, resulting in a truncation of the final 11 amino acid residues of ORF4, which may not have affected protein function. Further mutagenesis of ORF4 is therefore required to confirm that it has no role in disease. The mutations that caused loss of disease-causing ability all resided within sepA, sepB, or sepC. No mutation mapped to ORF1, ORF2, or ORF5.
TABLE 4.
Positions of mini-Tn10 insertions
| Mini-Tn10 insertion | ORF | Position (bp) downstream of initiation codon |
|---|---|---|
| 9/23 | ORF3 | 120 |
| 24 | ORF3 | 345 |
| 4 | sepA | 747 |
| 27 | sepA | 1037 |
| 40 | sepA | 1097 |
| 6 | sepA | 1727 |
| 38 | sepA | 2887 |
| 2 | sepA | 3197 |
| 5 | sepA | 3737 |
| 3 | sepA | 3697 |
| 30 | sepA | 4467 |
| 31 | sepA | 4627 |
| 12 | sepB | 182 |
| 22 | sepB | 172 |
| 11 | sepB | 362 |
| 10 | sepB | 2162 |
| 35 | ORF4 | 557 |
| 13 | sepC | 2525 |
| 8 | 18937 |
Complementation analysis.
The sequence data indicated that pBG35 and pGLA20 contain sepB and sepC, respectively. The complementation data obtained with these plasmids indicate that both genes are essential for virulence. Attempts to complement sepA mutants with the 8.45-kb HindIII fragment encompassing the sepA gene cloned into pLAFR3 were unsuccessful. In these bioassays, 80% of the grubs remained healthy but nonfeeding. However, over 90% of S. entomophila bacteria isolated from macerates of healthy nonfeeding grubs had lost the complementing plasmid, whereas 80% isolated from diseased grubs retained the plasmid.
DISCUSSION
The large conjugative plasmid pADAP is present in all S. entomophila and S. proteamaculans strains capable of causing amber disease of the New Zealand grass grub C. zealandica; it encodes the genes responsible for the symptoms of amber disease, including cessation of feeding and the gut clearance that results in amber coloration of the grub (17). We have defined a 16.9-kb region of pADAP that is sufficient to confer disease-causing ability to C. zealandica on pADAP-cured strains of S. entomophila and on strains of E. coli. Mutagenesis and sequence analyses of the region indicated that it encodes three genes, sepA, sepB, and sepC, that are required for pathogenicity. The products of these genes show similarity to components of the insecticidal toxin complexes of the entomopathogen P. luminescens.
The cloned pathogenicity region conferred the ability to initiate all symptoms of amber disease on pADAP-cured S. entomophila strains, and insertion mutations in any of sepA, sepB, or sepC abolished disease. Complementation studies confirmed that sepB and sepC were both required for disease, but attempts to complement sepA mutants were unsuccessful. This was attributed to instability of the plasmid clone encoding SepA, suggesting that overexpression of the SepA protein may be detrimental to the growth of the host bacterium. Nevertheless, the fact that sepB mutants were complemented by a clone lacking sepA strongly suggests that the sepA mutant phenotype was not due to a downstream effect of the transposon insertion on sepB or sepC expression. Hence, it is likely that sepA, sepB, and sepC together comprise the entire complement of essential virulence genes on pADAP. However, when the sep insertion mutations were transferred to pADAP, the resultant strains showed a partial disease phenotype, inducing cessation of feeding but not gut clearance. This result suggests that another locus able to exert a cessation-of-feeding effect may be present elsewhere on pADAP. The findings that pADAP-cured strains of S. entomophila containing the cloned sep genes cause cessation of feeding and that higher doses of sepB(pADK-10) or sepC(pADK-13) mutants are required to induce cessation of feeding compared to the wild-type strain, as shown in dose-response assays (20), indicate that the sep gene products are likely to play a key role in induction of the cessation-of-feeding response.
Another locus, amb2, that is required for induction of both symptoms of amber disease has already been described for S. entomophila (32). The cloned amb2 locus confers a cessation-of-feeding effect on E. coli strains harboring it, and amb2 mutants of S. entomophila are nonpathogenic. However, amb2 is different from the loci described in this work, as it maps to the chromosome of S. entomophila and does not show sequence similarity to the sep genes. Further work is required to determine the relationships or interactions, if any, between the amb2 and sep loci.
Comparison of the predicted translated products of the sep genes showed they have similarity to the proteins that are components of the insecticidal toxin complexes of the enterobacterium P. luminescens (a symbiont of entomopathogenic nematodes of the family Heterorhabditidae). Bowen et al. (5) found that several P. luminescens strains secrete high-molecular-weight toxins that have strong insecticidal activity toward a large number of insects, including species of Coleoptera, Dictyoptera, Hymenoptera, and Lepidoptera. The physiological effects exerted by these toxins on susceptible insects are very similar to the effects exerted by δ-endotoxins of Bacillus thuringiensis and include cessation of feeding, loss of gut peristalsis, paralysis of the insect, and death (2, 5). Four P. luminescens toxin complexes were resolved on a native gel and termed toxin complexes Tca, Tcb, Tcc, and Tcd (5, 11). The Tcb and Tcd complexes are encoded by single-gene loci, but Tca and Tcc could be further resolved into a number of different polypeptides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The loci encoding Tca and Tcc comprise four genes, three of which (tcaABC or tccABC) are oriented in the same direction, while the fourth, tcaZ or tccZ, is located downstream and oriented in the opposing direction.
The sep gene products show similarity to members of each of the P. luminescens loci, tca (SepA to TcaB; SepB to TcaC), tcb (SepA to TcbA), tcc (SepA to TccB; SepC to TccC), and tcd (SepA to TcdA). SepA and its P. luminescens homologues share an RGD motif. The RGD motif is present in cell surface adhesins produced by the human pathogen Bordetella pertussis, namely, the filamentous hemagglutinin (34) and the outer membrane protein pertactin (29), and has been implicated in enhancing the binding of B. pertussis to eukaryotic cells. The RGD motif found in SepA falls in a region of high similarity between SepA and its P. luminescens counterparts, and it seems possible that it may play a role in mediating the attachment of the proteins and/or the bacteria to the insect cell membrane.
SepB shows strong similarity to P. luminescens TcaC throughout its length, and both proteins show strong amino-terminal similarity to the amino terminus of the Salmonella virulence gene product SpvB (21). The region of similarity in relation to SpvB terminates 10 amino acid residues upstream of the proline-rich region postulated to divide SpvB into separate domains (36). This may indicate a vital role for the amino termini of the three proteins in interacting with an evolutionarily conserved eukaryotic protein. SpvB is believed to enhance the survival of virulent Salmonella in macrophages, but its mechanism of action is unknown (30). Based on its similarity to SpvB, it was suggested that TcaC may act by attacking insect hemocytes (5). However, hemocytes reside within the insect hemocoel, and S. entomophila does not invade the hemocoel until late in the infection process (25), suggesting that SepB may act in some other way.
The strong similarity of SepC to TccC is confined to the first 680 amino acids of the ∼1,000-amino-acid proteins. The region common to SepC and TccC also shows similarity to the B. subtilis wall-associated protein WapA (a prototype of a family of hypothetical cell wall-associated bacterial proteins) and to members of the E. coli rhs element family. The Rhs family of elements has an unusual structure, with a GC-rich (62%) core of about 3.7 kb common to all members, followed by an AT-rich (60%) extension region of 400 to 600 bp that is unique to each member of the family. A single ORF runs through the GC-rich core and terminates in the extension region, which also encodes a second small ORF (12, 41). Though smaller than the typical Rhs elements, sepC encodes a hydrophilic protein core that contains nine variants of the Rhs peptide motif repeat GxxRYxYDxxGRL(IT) (12, 41) (Fig. 4A). There is also high similarity between SepC, TccC, WapA, and SC2H4.02 from S. coelicolor A3(2) to the carboxyl end of the Rhs core, which ends at a conserved glycine residue (Fig. 4B) (41). After the glycine residue the similarity between each of the proteins diminishes, resulting in different carboxy termini, as also occurs with the Rhs elements. The function of Rhs proteins is yet to be established, but they have been proposed to be cell surface-associated ligand-binding proteins on the basis of their similarity to WapA (15). The variable carboxyl termini may be the result of acquisition of new protein domains by modular evolution.
The similarity between the sep and tc gene products suggests that they are members of a new family of insecticidal toxins. The lack of DNA similarity as opposed to protein similarity between sep and P. luminescens tc genes, together with the difference in GC content of the sepA and sepB genes compared to their tc homologues, suggests that these genes were present in a common enterobacterial ancestor of P. luminescens and S. entomophila and were not acquired by a more recent horizontal transfer event.
The involvement of similar disease determinants suggests that the histopathology of the diseases induced by P. luminescens and S. entomophila might be similar, despite the fact that amber disease is chronic whereas P. luminescens causes acute infections. Blackburn et al. (2) examined histopathology of the midgut of Manduca sexta larvae after treatment with purified Tca (TcaA, TcaB/SepA, TcaC/SepB, and TcaZ) through feeding on a diet cube and intracoelomic injection. Ingestion of Tca protein resulted in cessation of feeding, swelling of the midgut columnar epithelial cells, extrusion of vesicles into the gut lumen, and complete destruction of the midgut epithelium within 12 h. Injection of protein also resulted in distortion of the midgut cells. In contrast, S. entomophila infection has no observed histological effect on the midgut epithelial cells of C. zealandica but did show a reduction in the number of fat cells to almost undetectable levels and an emptying of the larval gut (25). Studies with purified Sep proteins are required to determine whether these differences reflect a different mode of action of the proteins or a toxin dose effect. Such studies will also indicate whether the remarkable host specificity of amber disease is a property of the Sep proteins or some other aspect of the disease process.
In summary, we have identified three S. entomophila genes, sepA, sepB, and sepC, that encode proteins with strong similarity to P. luminescens insecticidal toxins and are required for the causation of amber disease in the scarab C. zealandica. The similarity between S. entomophila and P. luminescens toxins suggests that they are members of a new family of insecticidal toxins. To further understand the disease process, purification of the Sep proteins and analysis of their expression and mode of action are being undertaken.
ACKNOWLEDGMENTS
We thank Richard Townsend for collecting grass grub larvae and Alison Inwood for help with bioassays.
This study was supported by a contract from the New Zealand Foundation for Research, Science and Technology.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn M, Golubeva E, Bowen D, Ffrench-Constant R H. A novel insecticidal toxin from Photorhabdus luminescens, toxin complex a (Tca), and its histopathological effects on the midgut of Manduca sexta. Appl Environ Microbiol. 1998;64:3036–3041. doi: 10.1128/aem.64.8.3036-3041.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 4.Bowen D, Ensign J C. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl Environ Microbiol. 1998;64:3029–3035. doi: 10.1128/aem.64.8.3029-3035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen D, Rocheleau T A, Blackburn M, Andreev O, Golubeva E, Bhartia R, Ffrench-Constant R H. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;280:2129–2132. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 6.Bowen D, Blackburn M, Rocheleau T, Andreev O, Golubeva E, Ffrench-Constant R H. Insecticidal toxins from the bacterium Photorhabdus luminescens: gene cloning and toxin histopathology. Pestic Sci. 1999;55:633–675. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ensign J C, Bowen D J, Petell J, Fatig R, Schoonover S, Ffrench-Constant R H, Rocheleau T A, Blackburn M B, Hey T D, Merlo D J, Orr G L, Roberts J L, Strickland J A, Guo L, Ciche T A, Sukhapinda K. Insecticidal protein toxins from Photorhabdus. Patent WO 97/17432. Geneva, Switzerland: World Intellectual Property Organisation; 1997. [Google Scholar]
- 12.Feulner G, Gray J A, Kirschman J A, Lehner A F, Sadosky A B, Vlazny D A, Zhang J, Zhao S, Hill C W. Structure of the rhsA locus from Escherichia coli K-12 and comparison of rhsA with other members of the rhs multigene family. J Bacteriol. 1990;172:446–456. doi: 10.1128/jb.172.1.446-456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finnegan J, Sherratt D. Plasmid ColE1 conjugal mobility: the nature of bom, a region required in cis for transfer. Mol Gen Genet. 1982;185:344–351. doi: 10.1007/BF00330810. [DOI] [PubMed] [Google Scholar]
- 15.Foster S J. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol Microbiol. 1993;8:299–310. doi: 10.1111/j.1365-2958.1993.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 16.Gierasch L M. Signal sequences. Biochemistry. 1989;28:923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- 17.Glare T R, Corbett G E, Sadler A J. Association of a large plasmid with amber disease of the New Zealand grass grub, Costelytra zealandica, caused by Serratia entomophila and Serratia proteamaculans. J Invertebr Pathol. 1993;62:165–170. [Google Scholar]
- 18.Glare T R, Hurst M R H, Grkovic S. Plasmid transfer among several members of the family Enterobacteriaceae increases the number of species capable of causing experimental amber disease in grass grub. FEMS Microbiol Lett. 1996;139:117–120. [Google Scholar]
- 19.Grimont P A D, Jackson T A, Ageron E, Noonan M J. Serratia entomophila sp. nov. associated with amber disease in the New Zealand grass grub, Costelytra zealandica. Int J Syst Bacteriol. 1988;38:1–6. [Google Scholar]
- 20.Grkovic S, Glare T R, Jackson T A, Corbett G E. Genes essential for amber disease in grass grub are located on the large plasmid found in Serratia entomophila and Serratia proteamaculans. Appl Environ Microbiol. 1996;61:2218–2223. doi: 10.1128/aem.61.6.2218-2223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulig P A, Caldwell A L, Chiodo V A. Identification, genetic analysis and DNA sequence of a 7.8-kb virulence region of the Salmonella typhimurium virulence plasmid. Mol Microbiol. 1992;6:1395–1411. doi: 10.1111/j.1365-2958.1992.tb00860.x. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Jackson T A. Amber disease reduces trypsin activity in midgut of Costelytra zealandica Coleoptera; Scarabaeidae larvae. J Invertebr Pathol. 1995;65:68–69. [Google Scholar]
- 24.Jackson T A, Glare T R, O'Callaghan M. Pathotypic boundaries for Serratia spp. causing amber disease in the New Zealand grass grub, Costelytra zealandica. In: Smits P H, editor. Proceedings of the 3rd European Meeting of Microbial Control of Pests. IOBC/WPRS bulletin XIV/7. 1991. pp. 148–152. Wageningen, The Netherlands. [Google Scholar]
- 25.Jackson T A, Huger A M, Glare T R. Pathology of amber disease in the New Zealand grass grub, Costelytra zealandica Coleoptera: Scarabaeidae. J Invertebr Pathol. 1993;61:123–130. [Google Scholar]
- 26.Jackson T A, Pearson J F, O'Callaghan M, Mahanty H K, Willocks M. Pathogen to product—development of Serratia entomophila Enterobacteriaceae as a commercial biological control agent for the New Zealand grass grub Costelytra zealandica. In: Jackson T A, Glare T R, editors. Use of pathogens in scarab pest management. Andover, Mass: Intercept Ltd.; 1992. pp. 191–198. [Google Scholar]
- 27.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–179. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Leininger E, Roberts M, Kenimer J G, Charles I G, Fairweather N, Novotny P, Brennan M J. Pertactin, an Arg-Gly-Asp-containing Bordetella pertussis surface protein that promotes adherence to mammalian cells. Proc Natl Acad Sci USA. 1991;88:345–349. doi: 10.1073/pnas.88.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libby S J, Adams L G, Ficht T A, Allen C, Whitford H A, Buchmeier N A, Bossie S, Guiney D G. The spv genes on the Salmonella dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect Immun. 1997;65:1786–1792. doi: 10.1128/iai.65.5.1786-1792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorow D, Jessee J. Max efficiency DH10BTM: a host for cloning methylated DNA. Focus. 1990;12:19. [Google Scholar]
- 32.Nunez-Valdez M E, Mahanty H K. The amb2 locus from Serratia entomophila confers a cessation of feeding effect on larvae of Costelytra zealandica Coleoptera: Scarabaeidae. Gene. 1996;172:75–79. doi: 10.1016/0378-1119(96)00055-8. [DOI] [PubMed] [Google Scholar]
- 33.Pecenkova T, Benes V, Paces J, Vlcek C, Paces V. Bacteriophage B103: complete DNA sequence of its genome and relationship to other Bacillus phages. Gene. 1996;199:157–163. doi: 10.1016/s0378-1119(97)00363-6. [DOI] [PubMed] [Google Scholar]
- 34.Relman D A, Domenighini M, Tuomanen E, Rappuoli R, Falkow S. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc Natl Acad Sci USA. 1989;86:2637–2641. doi: 10.1073/pnas.86.8.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rennell D, Poteete A R. Phage P22 lysis genes: nucleotide sequences and functional relationships with T4 and lambda genes. Virology. 1985;143:280–289. doi: 10.1016/0042-6822(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 36.Roudier C, Fierer J, Guiney D G. Characterization of translation termination mutations in the spv operon of the Salmonella virulence plasmid pSDL2. J Bacteriol. 1992;174:6418–6423. doi: 10.1128/jb.174.20.6418-6423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 39.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trought T E T, Jackson T A, French R A. Incidence and transmission of a disease of grass grub Costelytra zealandica in Canterbury. N Z J Exp Agric. 1982;10:79–82. [Google Scholar]
- 41.Wang Y D, Zhao S, Hill C H. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J Bacteriol. 1998;180:4102–4110. doi: 10.1128/jb.180.16.4102-4110.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]