Abstract
FtsZ and ZipA are essential components of the septal ring apparatus, which mediates cell division in Escherichia coli. FtsZ is a cytoplasmic tubulin-like GTPase that forms protofilament-like homopolymers in vitro. In the cell, the protein assembles into a ring structure at the prospective division site early in the division cycle, and this marks the first recognized event in the assembly of the septal ring. ZipA is an inner membrane protein which is recruited to the nascent septal ring at a very early stage through a direct interaction with FtsZ. Using affinity blotting and protein localization techniques, we have determined which domain on each protein is both sufficient and required for the interaction between the two proteins in vitro as well as in vivo. The results show that ZipA binds to residues confined to the 20 C-terminal amino acids of FtsZ. The FtsZ binding (FZB) domain of ZipA is significantly larger and encompasses the C-terminal 143 residues of ZipA. Significantly, we find that the FZB domain of ZipA is also required and sufficient to induce dramatic bundling of FtsZ protofilaments in vitro. Consistent with the notion that the ability to bind and bundle FtsZ polymers is essential to the function of ZipA, we find that ZipA derivatives lacking an intact FZB domain fail to support cell division in cells depleted for the native protein. Interestingly, ZipA derivatives which do contain an intact FZB domain but which lack the N-terminal membrane anchor or in which this anchor is replaced with the heterologous anchor of the DjlA protein also fail to rescue ZipA− cells. Thus, in addition to the C-terminal FZB domain, the N-terminal domain of ZipA is required for ZipA function. Furthermore, the essential properties of the N domain may be more specific than merely acting as a membrane anchor.
The first recognized event in bacterial cell division is assembly of the key division protein FtsZ into a ring (the Z-ring) which spans the circumference of the cell and resides just underneath the cytoplasmic membrane. Other division proteins are then recruited to the Z-ring, forming the septal ring organelle, which mediates cell wall invagination (for recent reviews, see references 19 and 32). The biochemical properties and tertiary structure (15, 25, 26) of FtsZ reveal that the protein shares many features with eukaryotic tubulins. Like tubulin, FtsZ is a GTPase (5, 21, 31, 37) which forms dynamic polymer filaments upon binding GTP (9, 16–18, 22–24, 39). Furthermore, hydrolysis of the nucleotide substrate is required for depolymerization and thus contributes to the dynamic nature of the filaments (22, 23). Unlike tubulin, however, FtsZ filaments are homopolymers of a single peptide species, and FtsZ readily binds nucleotide (5, 21, 31) as well as polymerizes (17, 22) in the absence of divalent metal.
We initially identified the ZipA protein of Escherichia coli based on its ability to directly bind FtsZ in vitro and showed it to be an essential cytoplasmic membrane protein which associates with the Z-ring in vivo (10). Depletion of ZipA leads to the formation of nonseptate filaments. FtsZ rings can still assemble in these filaments, although the number of rings per cell mass is significantly smaller than expected (11, 13). These observations suggest that ZipA is required for constriction of the septal ring and that the protein may also play a role in stabilizing the ring structure. Support for a stabilizing role of ZipA has also come from the finding that moderate overexpression of the protein suppresses the instability of the Z-ring in strains with the temperature-sensitive ftsZ84 allele (30).
To better understand the interaction between FtsZ and ZipA, we have tested the ability of deletion derivatives of these proteins to interact both in vitro and in vivo. We show that a small domain at the extreme C terminus of FtsZ and a significantly larger C-terminal domain of ZipA are both required and sufficient for the specific interaction between the two proteins. In agreement with a recent report (30), we also found that addition of purified ZipA protein to FtsZ polymers causes the arrangement of polymers into extensive networks of thick bundles in which individual polymers are present in a staggered side-by-side fashion. Using deletion derivatives of ZipA, we show that the interaction between the minimal FtsZ binding domain of ZipA and FtsZ polymers is also sufficient to induce bundling of the polymers.
The existence of some homology between a highly charged domain in ZipA and a domain in MAP-Tau proteins involved in binding microtubules has led to the assertion that ZipA may be considered a MAP-Tau homolog (30). Our results, however, show that this domain of ZipA is dispensable for both binding to FtsZ and mediating the bundling of FtsZ polymers.
We also tested the ability of various deletion derivatives of ZipA to substitute for the native protein in supporting cell division in vivo. Interestingly, ZipA derivatives which lacked the small N-terminal membrane anchor or in which the membrane anchor was replaced with that of another type Ib protein of E. coli (DjlA), were incapable of substituting for the native protein. Thus, whereas the C-terminal portion of ZipA is sufficient for binding and bundling of FtsZ polymers, it is clearly not sufficient to support cell division. In particular, our results suggest that the membrane anchor domain of ZipA is not merely required to anchor the protein to the membrane but may have more specific properties which are essential to proper functioning of ZipA in the division process.
MATERIALS AND METHODS
Strains.
Strains PB103 (dadR trpE trpA tna), PB143/pDB346 [PB103 ftsZ0 recA::Tn10/cI857(Ts) PλR::ftsZ+], CH5/pCH32 [PB103 zipA::aph recA::Tn10/repA(Ts) ftsZ+ zipA+], and CH5/pDB361 (zipA::aph recA::Tn10/cI857 PλR::zipA+) have been described previously (11). Strains BL21(λDE3)/plysS [ompT rB− mB−(PlacUV5::T7gene1)/T7lysS+)] and HMS174(λDE3) [r m+ recA Rifr (PlacUV5::T7gene1)] were purchased from Novagen. Unless stated otherwise, cells were grown at 37°C in Luria-Bertani (LB) medium supplemented, where appropriate, with antibiotics at 50 μg/ml (ampicillin, kanamycin, and spectinomycin), 25 μg/ml (chloramphenicol), and 12.5 μg/ml (tetracycline).
Plasmid construction.
Plasmids pMLB1113 (6), pDB319, pDB324, pDB326, and pDR10 (10), pDR112 (28), pDR107a, pDR107b, and pDR107c (29), pAR(ΔRI)59/60 (1), pGFPS65T (12), and pPSG961-31 (3) have been described previously. Plasmids pET16b, pET21a, pET21b, pET21c, and plysS were purchased from Novagen, and pUC4-KIXX was purchased from Pharmacia.
Plasmids for this study were created by a combination of PCR and standard subcloning techniques. The PCRs were performed using Vent DNA polymerase as recommended by the supplier (New England Biolabs) and, unless stated otherwise, using genomic DNA of strain PB103 as template. To place transcription under control of the lac or T7 promoter, fragments were inserted into the vector pMLB1113 (or derivatives) or into one of the pET vectors (or derivatives), respectively. PCR-derived portions of inserts were sequenced using an ABI PRISM automated sequencer to ensure the absence of undesired mutations.
(i) FtsZ plasmids.
To construct plasmid pDB312, we performed PCR using primers 5′-GGAGGATCCCATATGTTTGAACCAATGGAAC-3′ and 5′-TTCCGGTCGACTCTTAATCAGCTTGCTTACG-3′ introducing BamHI, NdeI, and SalI sites (underlined) flanking the ftsZ open reading frame (ORF). Digestion with NdeI and SalI resulted in an 1,156-bp fragment, which was ligated to similarly treated pET21a, yielding pDB312.
For pDR118, the above PCR product was treated with BamHI and SalI, yielding an 1,163-bp fragment, which was ligated to BamHI- and SalI-digested pDR107a. The resulting plasmid encodes a 68.6-kDa Gfp-T-FtsZ fusion protein, in which the GFPmut2 peptide is fused to the N terminus of the complete FtsZ peptide with the linker peptide ASMTGGQQMGRGSH, which includes the Novagen T7.tag peptide (T, underlined).
For pCH94, we used primers 5′-GGAGGATCCCATATGTTTGAACCAATGGAAC-3′ and 5′-GGTACTCGAGATAATCCGGCTCTTTCGC-3′, designed to amplify the first 1,113 bp of ftsZ. The PCR product was treated with EcoRI, which cuts within ftsZ, and XhoI (underlined), generating a 1,002-bp fragment which was ligated to EcoRI- and XhoI-digested pDB312. Plasmid pCH94 encodes a 40-kDa FtsZ-H fusion protein, in which the His tag peptide E(H)6 is fused to the C terminus of FtsZ at amino acid 372.
Plasmid pCH95 was constructed in a similar fashion using primers 5′-GGAGGATCCCATATGTTTGAACCAATGGAAC-3′ and 5′-TCCACTCGAGACCTGTCGCAACAACGG-3′ to amplify the first 942 bp of ftsZ. The plasmid encodes a 33.6-kDa FtsZ-H fusion protein, in which the His tag peptide LE(H)6 is fused to the C terminus of FtsZ at amino acid 314.
To construct plasmid pCH105, we used primers 5′-GTCTGGATCCAACGCGACTGTGGTTATCGG-3′ and 5′-TTCCGGTCGACTCTTAATCAGCTTGCTTACG-3′, designed to amplify a fragment consisting of the last 285 bp of the ftsZ ORF flanked by BamHI and SalI sites (underlined). Digestion with these enzymes yielded a 296-bp fragment, which was ligated to BamHI- and SalI-digested pDR107a, yielding pCH105. This plasmid encodes a 38.6-kDa Gfp-T-FtsZ fusion protein, in which GFPmut2 is fused to the N terminus of FtsZ at amino acid 289 by the linker peptide ASMTGGQQMGRGS. Plasmid pCH109 was then obtained by ligating a 150-bp Sau3AI-HindIII fragment from pCH105 to BamHI- and HindIII-digested pDR107b. The resulting plasmid encodes a 33.3-kDa Gfp-T-FtsZ fusion protein, in which GFPmut2 is fused to the N terminus of FtsZ at amino acid 337 with the linker peptide ASMTGGQQMGR.
For pCH122, the 1,363-bp EcoRV-PstI fragment of pCH109 was ligated to pDR107c which had been treated sequentially with BamHI, Klenow enzyme plus deoxynucleoside triphosphate (dNTP), and PstI. Plasmid pCH122 codes for a 29.2-kDa Gfp-T-FtsZ fusion protein, in which GFPmut2 is fused to the N terminus of FtsZ at amino acid 374 by the linker peptide ASMTGGQQMGRI.
To construct pCH127, pCH109 DNA was used as template in a PCR with primers 5′-ACACTGGATCCCAAACTGCGAAAGAGCCGG-3′ and 5′-CTGAAGCTTACCAATGCTTAATCAGTGAGGC-3′, which were designed to anneal, respectively, upstream of the last 60 bp of the ftsZ ORF and downstream of the bla gene in pCH109. Digestion with BamHI and PstI yielded a 1,398-bp fragment which was ligated to BamHI- and PstI-digested pDR107a, resulting in plasmid pCH127. The plasmid codes for a 30.4-kDa Gfp-T-FtsZ fusion protein, in which the GFPmut2 peptide is fused to the N terminus of FtsZ at amino acid 364 by the linker ASMTGGQQMGRGS.
For pCH130, we used pDB312 DNA as template in a PCR with primers 5′-GGAGGATCCCATATGTTTGAACCAATGGAAC-3′ and 5′-CGTCTACTCGAGATCAGCTTGCTTACGCAGG-3′, introducing NdeI and XhoI sites (underlined) upstream and downstream, respectively, of the ftsZ ORF. The 1,155-bp fragment that resulted from digestion with NdeI and XhoI was ligated to similarly treated pET21b. Plasmid pCH130 encodes a 41.4-kDa FtsZ-H fusion protein, in which the peptide tag LE(H)6 is fused to the C terminus of the complete FtsZ protein.
(ii) ZipA plasmids.
For pDB322, zipA was amplified by PCR with primers 5′-ACAGAGATCCATATGATGCAGGATTTGCGTCTG-3′ and 5′-TTAACCAAGCTTAAGTGTATCAGGCGTTGG-3′, designed to introduce a NdeI site (underlined) at the translation start codon of zipA. The PCR product was treated with NdeI and HindIII, and the 994-bp fragment was ligated to NdeI- and HindIII-digested pET21a, resulting in pDB317. The 676-bp AgeI-HindIII fragment of pDB317 was then replaced with the 1,015-bp AgeI-HindIII fragment of pDB315, yielding pDB318. To obtain pDB319, pDB318 was treated successively with AflII plus HindIII, Klenow enzyme, and ligase, thereby removing all lig sequences and retaining a HindIII site. Plasmid pDB319 encodes the complete ZipA protein under the control of the T7 promoter. To place zipA expression under the control of the lac promoter, the 1,089-bp BglII-HindIII fragment of pDB319 was ligated to BamHI- and HindIII-digested pMLB1113, yielding pDB322.
For pCH35, the 1,733-bp chromosomal BamHI-HindIII fragment containing the entire zipA ORF, as well as a portion of the upstream cysZ and downstream lig ORFs, was cloned into M13mp19 (10) and a nested set of deletions was obtained as described previously (6). The 1,136-bp EcoRI-HindIII fragment of one of these deletions was ligated to similarly treated pET21c, yielding pCH35. Plasmid pCH35 encodes a 30.5-kDa protein in which the peptide tag MASMTGGQQMGRIRIPPPPP is fused to the N terminus of ZipA at amino acid 70.
To construct pCH38, we performed a PCR with primers 5′-GGATCTAGACATATGATGCAGGATTTG-3′ and 5′-AAGTCTCGAGGGCGTTGGCGTCTTTGAC-3′ so as to introduce NdeI and XhoI sites (underlined) upstream and downstream, respectively, of the zipA ORF. The 986-bp fragment that resulted from an NdeI-XhoI digestion was ligated to similarly treated pET21b. Plasmid pCH38 encodes a 37.5-kDa ZipA-H fusion protein in which the peptide tag LE(H)6 is fused to the C terminus of the complete ZipA protein.
Plasmid pCH49 was obtained in multiple steps. The 1,265-bp BglII-HindIII fragment of pCH35 was inserted into BamHI- and HindIII-digested pMLB1113, yielding pCH39. Next, pCH38 was treated sequentially with Styl, Klenow enzyme plus dNTP, and XbaI, and the 1,112-bp fragment was ligated to pDB326 that had been treated with AccI, Klenow enzyme plus dNTP, and XbaI, yielding pCH41. Finally, the 1,218-bp XbaI-HindIII fragment of pCH39 was replaced with the 1,148-bp XbaI-HindIII fragment of pCH41. Plasmid pCH49 encodes the same fusion as pCH38, but transcription is under control of the lac promotor.
For pCH50, zipA was amplified by a PCR with primers 5′-ACAGAGATCCATATGATGCAGGATTTGCGTCTG-3′ and 5′-AAGTCTCGAGGGCGTTGGCGTCTTTGAC-3′, designed to introduce a NdeI site (underlined) at the translation start codon of zipA and to replace the translation stop codon with an XhoI site (underlined). The product was treated with NdeI and XhoI, and the 984-bp fragment was ligated to NdeI- and XhoI-digested pET21b, resulting in pCH38. Plasmid pGFPS65T contains gfpS65T on a 729-bp BamHI fragment in the vector pRSETB (12). This fragment was ligated to BamHI-digested pET16b, yielding plasmid pDB338, which contains an XhoI site immediately upstream of the gfp coding sequence. The small ApaI-XhoI fragment of pDB338 was next replaced by that of pCH38, resulting in pDB341. This plasmid encodes a 64.2-kDa ZipA-GfpS65T fusion protein which includes the complete ZipA and Gfp proteins, fused by the linker peptide LEDPPAEF. To place expression of this fusion under control of the lac promoter, the 2,126-bp BglII-HindIII fragment of pDB341 was next ligated to BamHI- and HindIII-digested pMLB1113, yielding pCH50.
Plasmid pCH77 was also constructed in several steps. First, plasmid pDB324 was digested with BamHI and HindIII, generating a 1,214-bp fragment, which was ligated to similarly treated pET21c. The resulting plasmid, pCH14, was digested with BglII and HindIII, yielding a 1,354-bp fragment that was ligated to BamHI- and HindIII-digested pMLB1113. This new plasmid, pCH56, was treated with AflII and HindIII, filled in with Klenow, and then religated, removing the 328-bp AflII-HindIII fragment at the end of the zipA ORF, regenerating a HindIII site, and yielding pCH57. Lastly, the 687-bp AgeI-HindIII fragment of pCH57 was replaced with the 1,710-bp AgeI-HindIII fragment of pCH50, resulting in pCH77. This plasmid encodes a 61.2-kDa T-ZipA-Gfp fusion protein, in which the peptide MASMTGGQQMGRI is fused to the N terminus at amino acid 39 and in which the complete GfpS65T peptide is fused to the C-terminal residue (at position 328) of ZipA with the linker sequence LEDPPAEF.
To construct pCH78, pCH79, and pCH80, we used primers 5′-GCTTTACATATGCATGGTTTCTGGACCAGC-3′ and 5′-TTAACCAAGCTTAAGTGTATCAGGCGTTGG-3′, introducing NdeI and HindIII sites (underlined), respectively, upstream of bp 67 and downstream of the translational stop signal in the zipA ORF. Treatment with NdeI and HindIII resulted in a 933-bp fragment, which was ligated to similarly treated pET21a. The resulting plasmid, pCH78, encodes a 34.2-kD ZipA peptide, beginning at amino acid 23 and continuing to its natural stop. Plasmid pCH79 was then constructed by ligating the 1,039-bp BglII-HindIII fragment of pCH78 to BamHI- and HindIII-digested pMLB1113. Finally, for pCH80, the 1,021-bp ApaI-AgeI fragment of pCH79 was substituted for the 1,084-bp ApaI-AgeI fragment of pCH50. Plasmid pCH80 encodes a 62.0-kDa ZipA-Gfp fusion in which GfpS65T is fused to the C-terminal residue of ZipA(23–328) with the linker sequence LEDPPAEF.
Plasmid pCH93 was obtained with primers 5′-CGGTGGATCCCTTCTTAACAGCATTCAAC-3′ and 5′-TTAACCAAGCTTAAGTGTATCAGGCGTTGG-3′, introducing BamHI and HindIII sites (underlined) upstream and downstream, respectively, of the last 348 bp of the zipA ORF. Digestion with BamHI and HindIII yielded a 360-bp fragment, which was ligated to BamHI- and HindIII-digested pDR107a. The resulting plasmid, pCH92, encodes a 41.2-kDa Gfp-T-ZipA fusion protein, in which the complete GFPmut2 peptide is fused to the N terminus of the ZipA peptide at amino acid 212 by the linker peptide MTGGQQMGRGS. To place the expression of this fusion under the control of Plac, the 1,150-bp BglII-HindIII fragment of pCH92 was ligated to BamHI- and HindIII-digested pMLB1113, giving rise to pCH93.
For pCH103, we used primers 5′-ACAGAGATCCATATGATGCAGGATTTGCGTCTG-3′ and 5′-ATCGCTCGAGGTCAAGCACGACACCGC-3′, designed to amplify the first 906 bp of the zipA ORF. The resulting product was treated with AgeI, which cuts within the zipA gene, and XhoI (underlined), generating a 598-bp fragment, which was then ligated to similarly treated pCH38. Plasmid pCH103 encodes a 34.3-kDa ZipA-H fusion protein, in which the peptide tag LE(H)6 is fused to the C terminus of the ZipA peptide at amino acid 302.
Plasmid pCH106 was constructed in several steps. First, plasmid pCH14 was digested with ApaI and NdeI and the small fragment was replaced with the 1,093-bp ApaI-NdeI fragment of pET16b. The resulting plasmid, pCH15, encodes a fusion protein in which the first 38 amino acids of ZipA have been replaced with a His-T7.tag (H-T) peptide. Then, the 1,710-bp AgeI-HindIII fragment from pCH50 was ligated to the large AgeI-HindIII fragment of pCH15, yielding pCH106. The plasmid encodes a 63.7-kDa H-T-ZipA-Gfp fusion protein, in which GfpS65T is fused to the C-terminal amino acid of ZipA with the linker sequence LEDPPAEF and in which the H-T peptide MG(H)10SSGHIEGRHMASMTGGQQMGRI is fused to the N terminus at amino acid 39.
For pCH121, PCR primers 5′-GCTTTACATATGCATGGTTTCTGGACCAGC-3′ and 5′-TTGGAAGCTTACAGCTCGTCACCGTAAGAC-3′ were used to introduce NdeI and HindIII sites (underlined) upstream and downstream, respectively, of bp 67 and 837 in the zipA ORF. The 786-bp fragment that resulted from an NdeI-HindIII digestion was ligated to similarly treated pET21a, generating pCH121. The plasmid encodes a 28.5-kDa peptide, consisting of residues 23 to 279 of ZipA.
To construct plasmids pCH131 and pCH132, we performed PCR with primers 5′-ACCAGGATCCGATAAACCGAAGCGCAAAGAAGCGG-3′ and 5′-AAGTCTCGAGGGCGTTGGCGTCTTTGAC-3′, introducing BamHI and XhoI sites (underlined) upstream and downstream, respectively, of the last 426 bp of the zipA ORF. Digestion with BamHI and XhoI yielded a 431-bp fragment, which was ligated to either BamHI- and XhoI-digested pET21a, resulting in plasmid pCH131, or similarly treated pDR107a, to yield pCH132. Plasmid pCH131 encodes a 18.4-kDa T-ZipA-H fusion protein, in which the T7 tag MASMTGGQQMGRGS is fused to the N terminus of ZipA at amino acid 186, while pCH132 encodes a 45.1-kDa Gfp-T-ZipA-H fusion protein in which GFPmut2 is fused to the N terminus of ZipA at amino acid 186 by the linker ASMTGGQQMGRGS. In addition, both fusions carry the peptide tag LE(H)6 fused to their C-terminal ends.
For pCH136, we used primers 5′-CGGTGGATCCCTTCTTAACAGCATTCAAC-3′ and 5′-AAGTCTCGAGGGCGTTGGCGTCTTTGAC-3′, introducing BamHI and XhoI sites (underlined) upstream and downstream, respectively, of the last 348 bp of the zipA ORF. Digestion with BamHI and XhoI yielded a 353-bp fragment, which was ligated to BamHI- and XhoI-digested pDR107a. The resulting plasmid, pCH136, encodes a 42.3-kDa Gfp-T-ZipA-H fusion protein, in which GFPmut2 is fused to the N terminus of the ZipA peptide at amino acid 212 by the linker peptide ASMTGGQQMGRGS and in which the peptide tag LE(H)6 is fused at the C-terminal end.
Plasmid pCH138 was obtained by replacing the 962-bp XbaI-KpnI fragment of pCH93 with the 1,040-bp XbaI-KpnI fragment of pCH132. Plasmid pCH138 encodes a 44.0-kDa Gfp-T-ZipA fusion protein which is similar to that encoded by pCH132 except that it ends at the natural stop codon of zipA and thus lacks the C-terminal LE(H)6 tag.
Plasmid pCH148 was constructed by first replacing the 1,026-bp XbaI-XhoI fragment of pCH50 with the 948-bp XbaI-XhoI fragment of pCH103, generating plasmid pCH139. Then the 2,052-bp ApaI-NcoI fragment of pDR112 was replaced with the 948-bp ApaI-NcoI fragment of pCH139. Plasmid pCH148 encodes a 61.0-kDa ZipA-Gfp fusion protein in which the complete GFPmut2 peptide is fused to the C terminus of the ZipA peptide at amino acid 302 by the linker sequence LEDPPAEF.
For pCH172, plasmid pCH166 was first constructed by ligating the 936-bp NdeI-HindIII fragment, carrying the coding sequence for ZipA(23–328), of pCH78 to similarly treated pPSG961-31. The djlA-zipA fusion was then put under control of Plac by ligating the 1,068-bp EcoRI-HindIII fragment from pCH166 to EcoRI- and HindIII-digested pMLB1113, yielding pCH168. Loss of a base during the generation of pPSG961-31 inadvertently resulted in a translational fusion between the 5′ end of lacZ, present in the pMLB1113 vector, and the djlA-zipA ORF. To place the fusion out of frame with lacZ sequences, pCH168 was digested with EcoRI, treated with Klenow, and religated. The resulting plasmid, pCH172, encodes a 37.4-kDa DjlA-ZipA fusion protein in which the transmembrane domain of DjlA (residues 1 to 32) is fused to the N terminus of ZipA at amino acid 23.
Plasmid pCH174 was constructed by replacing the 1,040-bp ApaI-AgeI fragment of pCH50 with the 1,043-bp ApaI-AgeI fragment of pCH172. Plasmid pCH174 encodes a 65.2-kDa DjlA-ZipA-Gfp fusion protein in which GfpS65T is fused to the C-terminal amino acid of DjlA-ZipA with the linker sequence LEDPPAEF.
For pCH182, the 1,026-bp XbaI-XhoI fragment of pCH49 was replaced with the 948-bp XbaI-XhoI fragment of pCH103, thereby placing the expression of the ZipA(1–302)-H fusion under the control of Plac.
For pAB10, we made use of plasmid pAR(ΔRI)59/60 (1), which contains a 54-bp NdeI fragment encoding the peptide MDYKDDDDKARRASVEF. This peptide, denoted as FK, is a fusion of the Flag peptide (IBI) with a heart muscle kinase substrate peptide (underlined). To ease the manipulation of the NdeI cassette, a ∼1,800-bp EcoRI fragment, isolated from pUC4-KIXX and carrying the Tn5 aph(neo) gene, was inserted in the EcoRI site of pAR(ΔRI)59/60, which is flanked by the NdeI sites. The ∼1,850-bp NdeI fragment of the resulting plasmid, pTD1, was treated with Klenow and ligated to XhoI-digested and Klenow-treated pCH38 in the desired orientation to yield pAB9.5. To remove the ∼1,800-bp stuffer fragment, the latter was treated with EcoRI and recircularized, generating pAB10. This plasmid encodes a 39.9-kDa ZipA-FKH fusion protein in which the peptide LDMDYKDDDDKARRASVEFHIE(H)6 is fused to the C terminus of the complete ZipA protein.
Protein purification.
BL21(λDE3)/plysS cells carrying the appropriate pET-derived plasmid were grown overnight in LB medium with 50 μg of ampicillin per ml, 25 μg of chloramphenicol per ml, and 0.1% glucose. Cultures were diluted 1:200 in 500 ml of LB with 50 μg of ampicillin per ml and 0.04% glucose and grown at 30 or 37°C to an optical density at 600 nm (OD600) of 0.4 to 0.5. Isopropyl-β-d-thiogalactoside (IPTG) was added to 0.84 mM, and growth continued for another 2 h. Cells were harvested by centrifugation, washed once in 0.9% saline, and resuspended in cell breakage buffer (specified below) to a final volume of 5 ml. Cell suspensions were subjected to three rapid freeze-thaw cycles using a dry ice-acetone bath and a 37°C water bath. Cell lysis was monitored by phase-contract microscopy. The cell lysates were briefly sonicated to reduce viscosity and fractionated into pellet (P200) and supernatant (S200) fractions by centrifugation at 200,000 × g for 3 h at 5°C. After purification, the proteins were rapidly frozen in a dry ice-acetone bath and stored at −80°C.
ZipA(23–328), ZipA(23–279), and T-ZipA(70–328) were expressed from plasmids pCH78, pCH121, and pCH35, respectively. Cells were broken in 20 mM Tris-Cl (pH 8.0)–25 mM NaCl–5 mM EDTA, and the bulk of these proteins fractionated with the supernatant as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses. Proteins in the S200 fractions were precipitated by addition of ammonium sulfate to 30% saturation and centrifugation at 8,000 × g for 20 min at 4°C. The pellets were resuspended in a minimal volume (0.5 to 1.0 ml) of 20 mM Tris-Cl (pH 8.0)–25 mM NaCl–5 mM EDTA and dialyzed extensively against the same solution. The dialysates were further fractionated by fast protein liquid chromatography on an Uno Q column (Bio-Rad) with a linear 25 to 500 mM NaCl gradient in the same buffer. ZipA(23–328) and ZipA(23–279) both eluted in a sharp peak at 210 mM NaCl, while ZipA(70–328) eluted at 100 mM NaCl. Peak fractions were dialyzed extensively against 20 mM Tris-Cl (pH 8.0)–25 mM NaCl–2 mM EDTA before storage.
H-T-ZipA(39–328)-Gfp, T-ZipA(186–328)-H, Gfp-T-ZipA(186–328)-H, and Gfp-T-ZipA(212–328)-H were expressed from plasmids pCH106, pCH131, pCH132, and pCH136, respectively. Cells were broken in 20 mM Tris-Cl (pH 7.9)–70 mM NaCl–50 mM imidazole [NiB(70/50)]. Almost all of the desired proteins were present in the corresponding S200 fractions. These were passed through a 1-ml fast-flow chelating Sepharose column (Pharmacia) that had been charged with NiCl2 and equilibrated with NiB(70/50) buffer. The columns were washed with NiB(500/50) (as above but containing 500 mM NaCl and 50 mM imidazole), and bound protein was eluted in NiB(500/500) (500 mM NaCl and 500 mM imidazole). EDTA was added to 5 mM, and peak fractions were dialyzed extensively against 20 mM Tris-Cl (pH 8.0)–25 mM NaCl–2 mM EDTA.
ZipA(1–328)-H, ZipA(1–328)-FKH, and ZipA(1–302)-H were expressed from plasmids pCH38, pAB10, and pCH103, respectively, and cells were broken in NiB(70/50) as above. Very little of these proteins was recovered in the S200 fractions. Rather, ZipA(1–328)-H and ZipA(1–328)-FKH were almost equally distributed in the pellet fraction and in an opaque interphase zone which was easily visible just above the solid pellet material. The interphase material was collected, and Triton X-100 was added to 0.1%. For ZipA(1–302)-H, no interphase zone was visible and the protein was present exclusively in the pellet fraction. This fraction was resuspended in NiB(70/50) containing 0.5% Triton X-100. After a second centrifugation step, the bulk of all three proteins remained in the supernatant fraction, indicating that they were effectively solubilized by the detergent, as is also the case with native ZipA (10). The proteins were further purified on chelating Sepharose as described above, except that 0.02% [ZipA(1–328)-H and ZipA(1–328)-FKH], or 0.05% [ZipA(1–302)-H] Triton X-100 was included in all the buffers. ZipA(1–328)-FKH and ZipA(1–302)-H were used after this step. The ZipA(1–328)-H was further purified by fast protein liquid chromatography on a Mono-Q column (Pharmacia) with a linear 25 to 500 mM NaCl gradient in 20 mM Tris.Cl (pH 8.0)–2 mM EDTA–0.02% Triton X-100. The protein eluted in a sharp peak at 380 mM NaCl. Peak fractions were dialyzed against 20 mM Tris-Cl (pH 8.0)–25 mM NaCl–2 mM EDTA–0.02% Triton X-100, and the protein was concentrated with a Centricon-10 device (Amicon) before storage.
Native FtsZ(1–383) was expressed from plasmid pDB312, and cells were lysed in 50 mM Tris.Cl (pH 7.9)–50 mM KCl–1 mM EDTA–10% glycerol. The protein was subsequently purified as described previously (23).
FtsZ(1–383)-H, FtsZ(1–372)-H, and FtsZ(1–314)-H were expressed from plasmids pCH130, pCH94, and pCH95, respectively. The proteins were purified from S200 fractions on chelating Sepharose essentially as described above for the soluble His-tagged ZipA derivatives, except that KCl was substituted for NaCl and glycerol was present at 10% in all buffers. Peak fractions were dialyzed into 50 mM HEPES hydroxide (pH 7.2)–0.1 mM EDTA–10% glycerol for storage. HFKT-FtsZ(1–383) was obtained as described previously (10).
FtsZ polymerization.
FtsZ was added to polymerization buffer (50 mM morpholineethanesulfonic acid [MES], 50 mM KCl, 10 mM MgCl2, 1 mM GTP [pH 5.8]) and incubated at 30°C for 5 min. A ZipA derivative (or ZipA storage buffer for ZipA− controls) was added, and incubation was continued for another 10 min. The final reaction volume was 50 μl, and the final concentration of each protein was 5 μM. A 10-μl volume of the reaction mixture was spotted on a glow-discharged, carbon-coated copper grid (300 mesh). After 20 s, the grid was wicked dry with filter paper, stained with 1% uranyl acetate for 45 s, and wicked dry again. For some reactions, GDP was used instead of GTP, and/or MgCl2 was omitted and EDTA was added to 2 mM. Polymerization of FtsZ(1–314)-H was carried out in the same way, except that the pH of the buffer was 6.0. The grids were viewed and photographed on a JEOL 100CX or JEOL 1200CX transmission electron microscope at 80 kV. Negatives were scanned using a Dimage Scan Multi (Minolta), and images were manipulated using Adobe Photoshop.
For fluorescence-based assays, polymerization reactions were carried out as above, except that FtsZ was used at 6.0 μM and Gfp-tagged ZipA derivatives were used at either 6.0 or 0.6 μM. Samples were applied to a microscope slide and viewed with a plan-NEOFLUAR (100×; numerical aperture [NA] = 1.3) objective on a Zeiss Axioplan-2 fluorescence microscope equipped with a cooled charge-coupled device camera (Hamamatsu), using a 495-nm dichroic mirror, a 450- to 490-nm excitation filter, and a 500- to 550-nm barrier filter. Images were captured using QED software and further manipulated using Adobe Photoshop.
Affinity blotting.
Purified HFKT-FtsZ and ZipA-FKH were phosphorylated as described previously (10) to specific activities of 1.0 × 107 and 4.3 × 106 cpm/μg, respectively. Subsequent procedures were performed as described previously (10).
Protease accessibility.
The preparation of spheroplasts and determination of protease accessibility were done essentially as described previously (2). Briefly, an overnight culture of strain PB103 was diluted 100-fold in 20.0 ml of LB broth and grown 37°C to an OD600 of 0.5. The cells were harvested by centrifugation and resuspended in 0.15 ml of 30 mM Tris-Cl (pH 8.0) containing 25% sucrose. The resuspended cells were mixed with an equal volume of a solution containing 10 mM EDTA and 0.4 mg of lysozyme per ml in water, and the mixture was incubated at room temperature (RT) for 3.0 min. As judged by phase-contrast microscopy, virtually all the cells had been converted to spheroplasts at this point. Aliquots of 50 μl were either left untreated or treated with protease and/or detergent by addition of proteinase K to 150 μg/ml (1.6 μl of a 5-mg/ml stock) and/or Triton-X100 to 0.1% (1.0 μl of a 5% stock), respectively. After 5 min at RT, 5.0 μl of a 50 mM solution of phenylmethylsulfonyl fluoride in ethanol was added to each sample, and after an additional 5 min at RT, each sample was mixed with 55 μl of 2× SDS-PAGE sample buffer. Samples were incubated at 100°C for 10 min, aliquots (20 μl per lane) were used to prepare three identical SDS-PAGE gels, and proteins were blotted to nitrocellulose filters. One filter was incubated with monoclonal antibody 4H4 to detect TonB (27). FtsZ was detected with a polyclonal antiserum (11), and ZipA was detected by incubation of the remaining filter with radiolabeled HFKT-FtsZ (10).
Quantitative immunoblotting.
To determine the cellular levels of plasmid-encoded ZipA and derivatives (see Table 2), cells were harvested by centrifugation, resuspended in SDS-PAGE electrophoresis sample-lysis buffer (7) to the equivalent of 13.3 OD600 units per ml, and incubated at 100°C for 5 min. The resulting lysates were further diluted in sample buffer as needed. Samples were separated by SDS-PAGE and transferred to nitrocellulose filters. The filters were incubated with affinity-purified ZipA antibodies and subsequently with enhanced chemiluminescence reagents using an Amersham-Pharmacia ECL kit as recommended by the manufacturer. Chemiluminescent signals were visualized and quantified using a Fluor-S MAX Multiimager system and Quantity One software (Bio-Rad). Each filter contained a reference lane loaded with 10.0 × 10−3 OD600 equivalent (corresponding to 8.0 μg of total protein) of a PB103/pMLB1113ΔH extract to establish the level of chromosomally encoded native ZipA in wild-type cells. Adjacent lanes were loaded with 2.0 × 10−3 and 1.0 × 10−3 OD600 equivalents (corresponding to 1.6 and 0.8 μg of total protein, respectively) of each relevant extract to determine the level of plasmid-encoded antigen. Signals showed optimal linearity with the amount of antigen under these conditions, as established in pilot experiments. Since the ZipA derivatives migrated close to the position of native ZipA in the gels, the combined signal was measured. Based on the signal in the reference lane, and accounting for dilution factors, the contribution of chromosomally encoded native ZipA was calculated and subtracted from the total signal to obtain the contribution of plasmid encoded ZipA.
TABLE 2.
Effect of zipA plasmids on division of wild-type cellsa
| Plasmid | Inducible protein | IPTG concnb (μM) | Relative protein concnc | Division blockd |
|---|---|---|---|---|
| pMLB1113ΔHe | None | >500 | − | − |
| pDB322 | ZipA | 25 | 7 | + |
| pCH49 | ZipA-H | 100 | 6 | + |
| pCH182 | ZipA(1–302)-H | >500 | 6 | − |
| pCH79 | ZipA(23–328) | 100 | 11 | + |
| pCH172 | D-ZipA(23–328) | 500 | 6 | + |
Overnight cultures of zipA+ strain PB103, containing the indicated plasmids, were diluted 100-fold in fresh LB medium containing ampicillin and 0, 5, 10, 25, 50, 100, 250, or 500 μM IPTG. The cultures were incubated at 37°C to an OD600 of 0.6 to 0.8. The cells were examined by phase-contrast microscopy to determine the concentrations of IPTG at which expression of ZipA (or derivative) caused the formation of nonseptate filaments. In parallel, a portion of each culture was used to prepare a whole-cell lysate, and relative protein levels were determined by quantitative immunoblotting as described in Materials and Methods.
Minimum IPTG concentration at which a clear division block was observed.
Level of the plasmid-encoded ZipA protein (or derivative) in cells that were grown at this minimum IPTG concentration. For ZipA(1–302)-H, the level in cells grown with 500 μM inducer is given. Levels are expressed as multiples of the level of chromosomally encoded native ZipA in PB103/pMLB1113ΔH control cells that had been grown in the presence of 500 μM IPTG.
Division phenotype at the indicated concentration of IPTG; +, division block (i.e., the majority of the cells were nonseptate filaments over 20 μm in length); −, no division block at any of the IPTG concentrations tested.
Plasmid is a lacZ derivative of vector pMLB1113.
RESULTS
A small C-terminal domain of FtsZ is required and sufficient for interaction with ZipA.
We previously detected the ZipA protein by an affinity-blotting procedure in which a radiolabeled derivative of the FtsZ protein was incubated with Western blots containing ZipA on nitrocellulose filters (10). To delineate the portion of FtsZ involved in the interaction with ZipA, we used a converse approach in which radiolabeled ZipA was incubated with immobilized FtsZ. As probe we used a derivative of ZipA (ZipA-FKH) consisting of the complete ZipA peptide carrying a C-terminal tag peptide (FKH) which includes a substrate site for heart muscle kinase (K) and a stretch of six histidine residues (H). Purified ZipA-FKH was radiolabeled with 32P and incubated with filters containing native FtsZ as well as a variety of derivatives in which portions of FtsZ were fused to Gfp and/or polyhistidine tags (Fig. 1).
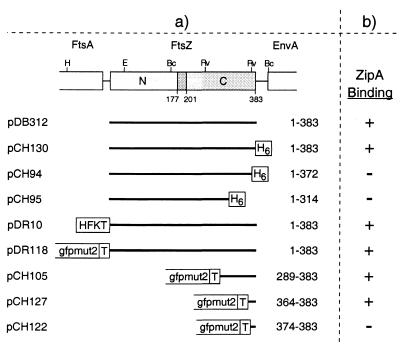
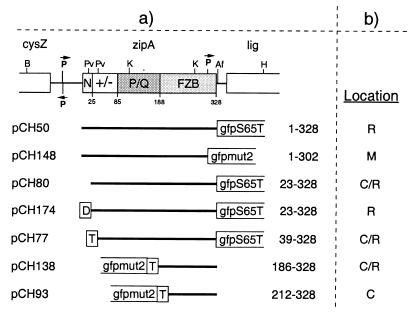
FIG. 1.
FtsZ plasmids used to define the ZipA-binding domain. (a) The physical map of ftsZ and portions of flanking genes in E. coli are shown at the top of the panel. The positions of BclI (Bc), EcoRI (E), EcoRV (Rv), and HindIII (H) restriction sites are indicated. The positions in the E. coli FtsZ polypeptide of two domains (N and C) and a connecting core helix (residues ca. 177 to 201) were inferred from the crystal structure of FtsZ from Methanococcus jannaschii (14, 15). Inserts of plasmids are presented below the map, and the FtsZ residues they encode are given at the right of each insert. All plasmids were derivatives of pET21, such that transcription of inserts was under control of the T7lac promoter (Novagen). Plasmid pDB312 encodes native FtsZ. All others encode either full-length or portions of FtsZ fused to various tags, as indicated. H6, stretch of six histidine residues; HFKT, combination tag peptide which includes a stretch of 10 histidine residues and a substrate site for heart muscle kinase (10); T, T7.tag peptide. (b) The ability of these FtsZ derivatives to bind radiolabeled ZipA-FKH was determined by affinity blotting. +, protein binds ZipA; −, protein does not bind ZipA.
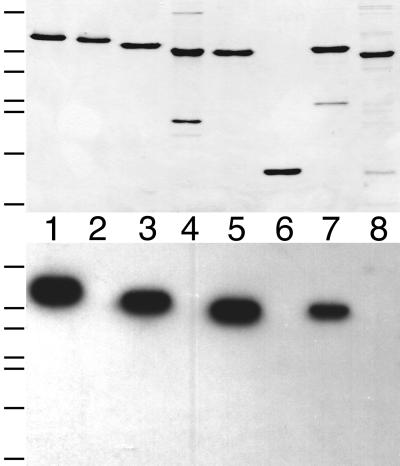
As shown in Fig. 2, the ZipA probe readily recognized purified untagged FtsZ (383 amino acids [aa]) on Western blots (lanes 1). ZipA-FKH also bound well to a version containing a His6 tag at the C terminus of the complete FtsZ protein [FtsZ(1–383)-H] (lanes 2), indicating that addition of the tag did not substantially interfere with the interaction between the two proteins. In contrast, deletion derivatives containing residues 1 to 314 or 1 to 372 failed to bind the ZipA probe (lanes 4 and 3, respectively), indicating that removal of as little as 10 residues from the C terminus of FtsZ abolished its interaction with ZipA-FKH.
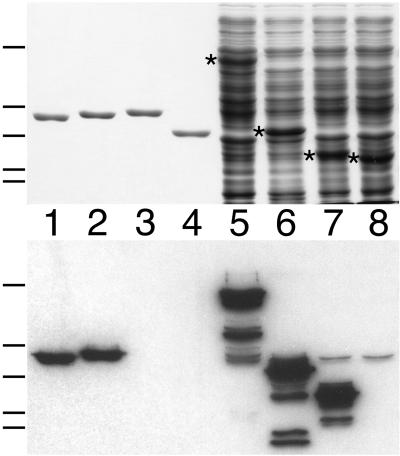
FIG. 2.
Binding of radiolabeled ZipA to a C-terminal domain of FtsZ. Purified proteins (lanes 1 to 4) and whole-cell extracts (lanes 5 to 8) were separated on two identical SDS-PAGE gels. One gel was stained with Coomassie brilliant blue to visualize protein bands (top), and proteins in the other gel were blotted to a nitrocellulose filter which was subsequently incubated with radiolabeled ZipA-FKH (bottom). Lanes 1 to 4 contain 50 pmol of native FtsZ(1–383) (lane 1), FtsZ(1–383)-H (lane 2), FtsZ(1–372)-H (lane 3), or FtsZ(1–314)-H (lane 4). Lanes 5 to 8 contain 10 μl of extract of cells overexpressing Gfp-T-FtsZ(1–383) (lane 5), Gfp-T-FtsZ(289–383) (lane 6), Gfp-T-FtsZ(364–383) (lane 7), or Gfp-T-FtsZ(374–383) (lane 8). Extracts were prepared from cells of strain BL21(λDE3)/plysS containing the appropriate plasmid (Fig. 1) after growth in the presence of IPTG and by resuspension of cells in SDS-PAGE lysis buffer to the equivalent of 20.0 OD600 units. Bands corresponding to the overexpressed proteins are indicated by an asterisk in the upper panel. The positions of molecular mass standards (66, 45, 36, 29, and 24 kDa [top to bottom]) are indicated on the left of the panels.
To delineate the ZipA binding domain of FtsZ further, we next tested the binding of ZipA to a set of derivatives in which Gfp was fused to various portions of the C terminus of FtsZ. The fusions were all overexpressed in strain BL21(λDE3)/plysS, and whole-cell lysates were used for affinity blotting with radiolabeled ZipA-FKH. As shown in Fig. 2, a fusion containing the last 20 residues of FtsZ (lanes 7) bound the probe as well as did fusions containing larger portions of the protein (lanes 5 and 6), whereas a fusion containing only the last 10 residues completely failed to bind (lanes 8). We conclude that the C-terminal 20 residues of FtsZ include elements that are both required and sufficient for the specific interaction of the protein with ZipA.
FtsZ interacts with the cytoplasmic C-terminal domain of ZipA in vitro.
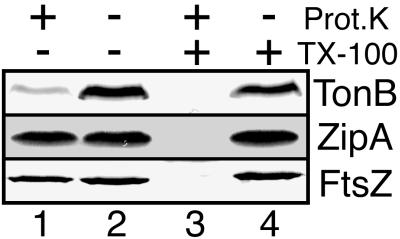
Membrane fractionation studies, in combination with the deduced primary sequence of ZipA, indicated that the protein is a bitopic integral inner membrane species of type lb, with the N terminus anchored in the membrane and the rest of the protein in the cytoplasm (10). To further validate this assignment, spheroplasts derived from wild-type strain PB103 were incubated with proteinase K in the presence or absence of the detergent Triton X-100, and the fate of ZipA was monitored by affinity blotting with radiolabeled HFKT-FtsZ. As controls, we performed immunoblot analyses on the same samples to determine the fates of FtsZ, a cytoplasmic protein, and of TonB, a type II transmembrane protein of which the bulk is present in the periplasm (27). Addition of proteinase K to detergent-permeabilized spheroplasts led to the complete degradation of all three proteins (Fig. 3, lanes 3), confirming that all three are good substrates for the protease. In contrast, treatment of intact spheroplasts with protease led to the specific degradation of TonB whereas the bulk of both FtsZ and ZipA remained unaffected (lanes 1). These results support a largely cytoplasmic localization of ZipA and indicate that if any part of ZipA is exposed to the periplasm (such as the extreme N-terminal portion), it must be very small.
FIG. 3.
Protease inaccessibility of ZipA in intact spheroplasts. Spheroplasts of PB103 cells were either left untreated (lanes 2) or treated with protease (lanes 1 and 3) and/or detergent (lanes 3 and 4). Samples were used to prepare three identical Western blots. TonB (top) and FtsZ (bottom) were detected with specific antibodies; ZipA (middle) was detected by incubation of the blot with radiolabeled HFKT-FtsZ.
The proposed membrane anchor domain of ZipA (aa 1 to 24) is followed by a highly charged domain (aa 25 to 84), a proline- and glutamine-rich domain (aa 85 to 187), and a C-terminal domain (aa 188 to 328) (10). To delineate the portion of ZipA required for FtsZ binding, we tested the ability of purified derivatives of ZipA, lacking various portions of the native protein, to bind radiolabeled HFKT-FtsZ in affinity blot assays (Fig. 4 and 5). During purification of these derivatives it became evident that each species that still contained the proposed membrane anchor domain of native ZipA fractionated with the insoluble material of broken cells. As with native ZipA, however, these species could be readily solubilized by extraction of pellet fractions with nonionic detergent. In contrast, each ZipA derivative lacking the first 22 (or more) residues behaved as soluble species, even in the absence of detergent. These observations are fully consistent with the proposal that the N terminus of ZipA functions as the sole membrane anchor.
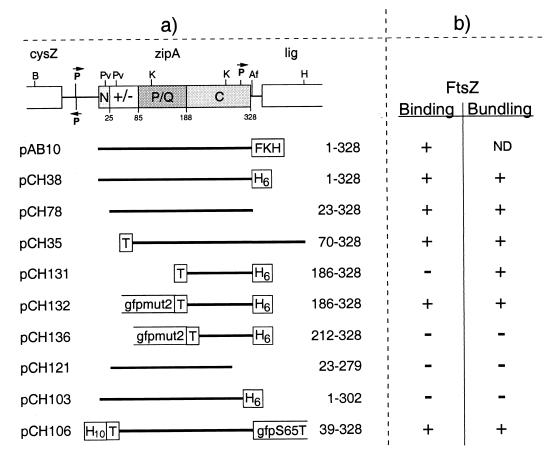
FIG. 4.
ZipA plasmids used to define the FtsZ-binding domain. (a) The physical map of zipA and portions of flanking genes in E. coli are shown at the top of the panel. The positions of AflII (Af), BamHI (B), HindIII (H), KpnI (K), and PvuII (Pv) restriction sites are indicated. The four domains of the ZipA polypeptide we previously proposed are denoted N (N-terminal membrane anchor), +/− (highly charged domain), P/Q (proline- and glutamine-rich domain), and C (C-terminal domain). The +/− domain includes the MAP-Tau repeat-like sequence proposed by RayChaudhuri to mediate binding to and bundling of FtsZ polymers (30). Inserts of plasmids are presented below the map, and the ZipA residues they encode are given at the right of each insert. All plasmids were derivatives of pET21, such that transcription of inserts was under the control of the T7lac promoter (Novagen). As indicated, inserts encode either full-length or portions of ZipA fused to various tags. H10, stretch of 10 histidine residues; FKH, combination tag which includes a stretch of six histidine residues and a substrate site for heart muscle kinase. See also the legend to Fig. 1. (b) The ability of these ZipA derivatives to bind radiolabeled HFKT-FtsZ was determined by affinity blotting. +, protein binds FtsZ; −, protein does not bind FtsZ. The ability of proteins to bundle FtsZ polymers was assessed by electron microscopy in all cases. The results obtained with the three Gfp-tagged derivatives were confirmed by fluorescence microscopy. +, numerous bundles and bundle networks observed. −, no bundles observed.
FIG. 5.
Binding of radiolabeled FtsZ to a C-terminal domain of ZipA. Purified proteins (50 pmol/lane) were separated on two identical SDS-PAGE gels. One gel was stained with Coomassie brilliant blue to visualize protein bands (top), and proteins in the other gel were blotted to a nitrocellulose filter which was subsequently incubated with radiolabeled HFKT-FtsZ (bottom). Lanes contained ZipA(1–328)-H (lane 1), ZipA(1–302)-H (lane 2), ZipA(23–328) (lane 3), ZipA(23–279) (lane 4), T-ZipA(70–328) (lane 5), T-ZipA(186–328)-H (lane 6), Gfp-T-ZipA(186–328)-H (lane 7), or Gfp-T-ZipA(212–328)-H (lane 8). The positions of molecular mass standards (66, 45, 36, 29, 24, 20, and 14 kDa [top to bottom]) are indicated on the left of the panels.
As shown in Fig. 5, radiolabeled HFKT-FtsZ readily recognized ZipA(23–328) on affinity blots, demonstrating that the membrane anchor of ZipA is not required for binding FtsZ (lanes 3). Moreover, the probe bound equally well to ZipA(70–328), a derivative which lacks both the membrane anchor and 25 of the 29 charged residues of the highly charged domain, showing that the charged domain is also not required for binding to FtsZ in vitro (lanes 5). Evidence that the PQ-rich domain is also dispensable came from the finding that the probe also bound to Gfp-T-ZipA(186–328)-H (lanes 7). The latter is a protein fusion in which the N-terminal three domains of ZipA were replaced with a 251-aa peptide, which consists of Gfpmut2 (Gfp) and the T7.tag peptide (T) and which carries a His6 tag fused to the C-terminal residue of ZipA. Binding of HFKT-FtsZ to this fusion was specific to the ZipA portion since the Gfp-T peptide or the H tag by themselves showed no affinity for the probe (data not shown) and since a similar fusion, from which additional ZipA residues were removed [Gfp-T-ZipA(212–328)-H] was not recognized by the probe (lanes 8). These results demonstrated that FtsZ binds the C-terminal domain (C-domain, aa 186 to 328) of ZipA and, moreover, that residues at the N terminus of this domain (aa 186 to 211) are required for binding. Residues at the C-terminal end of this domain are also important for this property, since deletion of as little as 26 residues from the C terminus of ZipA [ZipA(1–302)-H (lanes 2)] also completely abolished its ability to bind HFKT-FtsZ.
We conclude that the FtsZ binding domain (FZB domain) of ZipA is confined to the C-terminal portion (aa 186 to 328) of the protein. The finding that small deletions from either end of this domain preclude it from binding the probe suggests that the integrity of this complete domain is important for binding FtsZ in this assay. In this regard, it is noteworthy that T-ZipA(186–328)-H, which contains the complete FZB domain but lacks the Gfp tag, failed to bind probe in the affinity-blotting assay (lanes 6). The simplest interpretation of this result is that direct binding of the untagged FZB domain to the nitrocellulose support precludes a subsequent interaction with FtsZ because the FtsZ binding site is no longer accessible, or because binding to the support prevents it from adopting a proper configuration. We envision that the Gfp peptide in the Gfp-T-ZipA(186–328)-H fusion allows molecules of the fusion to adhere to the support through the Gfp portion while allowing the FZB domain to adopt or retain its proper conformation for FtsZ binding.
The FZB domain directs ZipA to the septal ring in vivo.
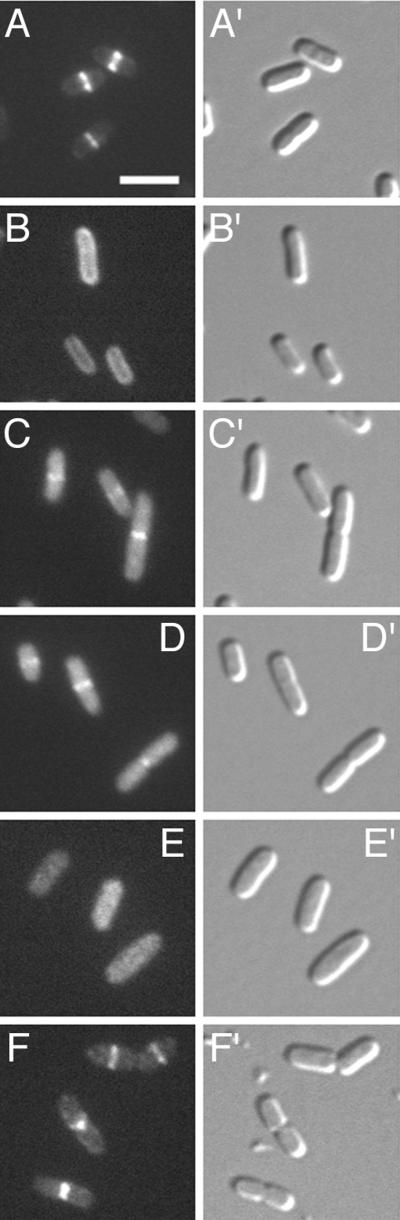
We previously showed that ZipA localizes to the septal ring organelle in a FtsZ-dependent manner (10, 11). As shown above, the FZB domain of ZipA is required and sufficient for binding to FtsZ in vitro. To test whether this domain is also sufficient to direct the ZipA protein to the FtsZ ring in vivo, we examined the cellular localization of derivatives containing various portions of ZipA fused to Gfp (Fig. 6 and 7). As observed previously (10, 11), virtually all fluorescence was seen associated with the septal ring structure in the vast majority of cells of strain PB103/pCH50 [wt/Plac::zipA(1–328)-gfp], which express a fusion of the full-length ZipA peptide with Gfp (Fig. 7A). In contrast, in PB103/pCH148 [wt/Plac::zipA(1–302)-gfp] cells, which express a derivative lacking the C-terminal 26 aa of ZipA, fluorescence failed to accumulate at the septal ring. Rather, this fusion appeared to be evenly distributed along the entire periphery of the cell (Fig. 7B). Combined with the observations that ZipA(1–302)-H fractionates with the insoluble fraction of cells and that the protein fails to bind FtsZ in vitro, this distribution indicates that whereas the fusion is still anchored to the cytoplasmic membrane by the ZipA N-terminal membrane domain, it no longer recognizes FtsZ that has accumulated within the septal ring.
FIG. 6.
Plasmids used to sublocalize ZipA derivatives. (a) The C-domain of ZipA is renamed FZB (FtsZ-binding domain) to indicate that this domain coincides with the portion of ZipA found here to be required and sufficient for binding FtsZ (see the text). Inserts were cloned into the vector pMLB1113 such that expression of the fusion proteins is under control of the lac promoter and lacIq. D, transmembrane domain corresponding to residues 1 to 32 of DjlA. See also the legend to Fig. 1. (b) Cellular location of fusion proteins in strain PB103. R, virtually all fluorescence associated with the septal ring; M, fluorescence evenly distributed along the entire cell membrane; C, fluorescence evenly distributed throughout the cytoplasm; C/R, a significant portion of total fluorescence throughout the cytoplasm and the rest associated with the septal ring.
FIG. 7.
Localization of Gfp-tagged ZipA derivatives in wild-type cells. Cells were chemically fixed and observed under fluorescence (A to F) and differential interference contrast (A′ to F′) optics. Images faithfully reflected the distributions of fluorescence seen prior to fixation. Panels show cells of strain PB103 (wild type) expressing ZipA derivatives from plasmids pCH50 [Plac::zipA(1–328)- gfp] (A), pCH148 [Plac::zipA(1–302)-gfp] (B), pCH80 [Plac::zipA(23–328)-gfp] (C), pCH138 [Plac::gfp-t-zipA(186–328)] (D), pCH93 [Plac::gfp-t-zipA(212–328)] (E), or pCH174 [Plac::djlA(1–32)-zipA(23–328)-gfp] (F). Cells were grown in the presence of 5 μM (A), 10 μM (B), 25 μM (C), or 100 μM (D to F) IPTG. None of the plasmids interfered noticeably with the normal division phenotype under these conditions. Bar, 3 μm.
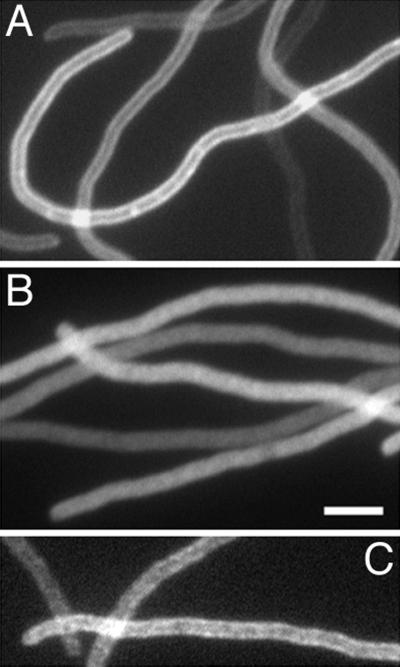
Fusions containing an intact FZB domain but lacking the membrane domain showed a different distribution. Thus, cells expressing ZipA(23–328)-Gfp (PB103/pCH80) showed a high signal throughout the cell body in addition to a distinct accumulation at the middle of cells (Fig. 7C). Consistent with the binding studies above, this pattern indicates that the absence of the transmembrane domain of ZipA does not preclude the protein from interacting with the FtsZ ring in vivo. To ensure that the apparent accumulation at the septal ring we observed with this fusion was in fact dependent on FtsZ, we also determined the distribution of this fusion in cells from which FtsZ had been depleted (Fig. 8). For this experiment, we made use of the previously described strain PB143/pDB346 [ftsZ0/cI857 PλR::ftsZ], which allows for a cold-induced depletion of FtsZ (11). This strain, containing either pCH50 or pCH80, was grown in the presence of IPTG and shifted from 42 to 30°C to repress the expression of ftsZ. Fluorescent rings were absent in the resulting filaments of both PB143/pDB346/pCH50 and PB143/pDB346/pCH80. The bulk of ZipA-Gfp in PB143/pDB346/pCH50 filaments appeared evenly distributed along the membrane (Fig. 8A). In a small fraction of filaments, some periodic accumulations of fluorescence, which might represent ZipA-Gfp binding to minor assemblies of the remaining FtsZ, could still be discerned. In contrast, ZipA(23–328)-Gfp appeared dispersed throughout the cytoplasm in the FtsZ-depleted filaments of strain PB143/pDB346/pCH80 (Fig. 8B). In combination with the fractionation and binding studies above, these results show that removal of the membrane anchor of ZipA yields a soluble species capable of binding FtsZ both in vitro and in vivo.
FIG. 8.
Localization of Gfp-tagged ZipA derivatives in FtsZ− cells. Filaments of the FtsZ CID strain PB143/pDB346 (ftsZ0/cI857 PλR::ftsZ) carrying pCH50 [Plac::zipA(1–328)-gfp] (A), pCH80 [Plac::zipA(23–328)-gfp] (B), or pCH174 [Plac::djlA(1–32)-zipA(23–328)-gfp] (C) are shown. Cells were grown at 30°C (leading to depletion of FtsZ) in the presence of 5 μM (A and B) or 100 μM (C) IPTG. The resulting filaments were chemically fixed and observed under fluorescence optics. Images faithfully reflected the distributions of fluorescence seen prior to fixation. Bar, 3 μm.
A fusion containing just the FZB domain [Gfp-ZipA(186–328)] of ZipA showed a distribution (Fig. 7D) indistinguishable from that of ZipA(23–328)-Gfp (Fig. 7C). In contrast, Gfp-ZipA(212–328) did not accumulate at septal rings but was evenly distributed throughout the cytoplasm (Fig. 7E). In summary, the results of these localization studies directly paralleled those of our binding studies, and we conclude that the FZB domain of ZipA is not only required and sufficient for binding to FtsZ in vitro but also required and sufficient for recruitment to the FtsZ ring in vivo.
ZipA-induced bundling of FtsZ protein filaments in vitro.
Using an optimized purification protocol for FtsZ, Mukherjee and Lutkenhaus (23) recently showed that the protein can readily form homopolymeric protein filaments in a strictly GTP-dependent fashion and in the absence of additional promoting agents such as DEAE-dextran, cationic phospholipids, or high concentrations of Ca2+, which had been used in earlier studies (9, 24, 35, 39). In addition, it was shown that whereas FtsZ can bind nucleotide and form filaments in the absence of divalent metals (5, 21, 23, 31), Mg2+ is required for hydrolysis of GTP (5, 21, 31), which, in turn, is correlated with the ability of filaments to depolymerize (22).
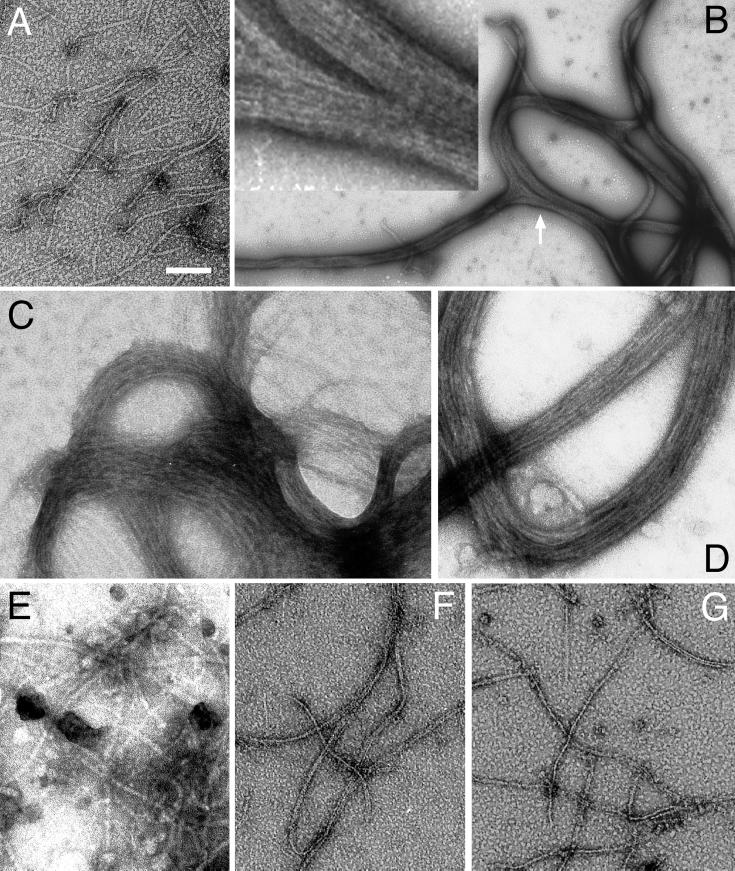
To assess the effect of ZipA on FtsZ polymers, we purified FtsZ by the method of Mukherjee and Lutkenhaus (23), and incubated the protein with GTP and Mg2+ for 5 min at 30°C to allow polymers to form. We next added buffer or buffer containing various purified ZipA derivatives, and after an additional 10 min of incubation, samples were prepared for analysis by negative-stain electron microscopy (EM). Similar to what was described before (23), buffer control samples showed the presence of numerous polymers of variable length and a diameter of approximately 5 to 7 nm (Fig. 9A). The vast majority of structures appeared as individual filaments, and bundles in which more than a few polymers were closely apposed were rare. Furthermore, consistent with the previous reports (22, 23), polymers formed readily when Mg2+ was omitted but were completely absent when GDP was substituted for GTP (see below) (data not shown).
FIG. 9.
ZipA-induced bundling of FtsZ polymer filaments. Purified native FtsZ (A to E) and FtsZ(1–314)-H (F and G) was incubated in the presence of 1.0 mM GTP and in the presence of either 10 mM Mg2+ (A, B, and D to G) or 2 mM EDTA (C). After 5 min, buffer (A and F), ZipA(23–328) (B, C, and G), Gfp-T-ZipA(186–328)-H (D), or Gfp-T-ZipA(212–328)-H (E) was added. After an additional 10 min, samples were applied to a microscope grid, stained with uranyl acetate, and examined under an electron microscope. Each protein in the reactions was present at 5 μM. The inset in panel B represents a portion (arrow) of the bundle network in more detail, emphasizing the side-by-side arrangement of polymers within the bundles. Polymers or polymer bundles were completely absent in control reactions in which GTP or FtsZ were omitted (data not shown). Bar, 76 nm (C), 100 nm (A, E to G, and inset in B), 125 nm (D), or 600 nm (B).
Interestingly, addition of purified ZipA(23–328) induced the formation of large protein structures in which numerous FtsZ polymers were arranged in a side-by-side fashion, resulting in bundles which themselves were most often arranged in extensive networks (Fig. 9B). Whereas the bulk of individual filaments were 100 to 500 nm long, bundles were frequently a few micrometers in length and their width typically ranged from 70 to 500 nm. Identical structures were seen in a similar reaction in which FtsZ was incubated with ZipA(1–328)-H in the presence of 0.1% Triton X-100, showing that the presence of the membrane anchor, a C-terminal His tag, and detergent did not affect this bundling activity of ZipA (Fig. 4b). Very few solitary FtsZ polymers were observed in these preparations, indicating that the bulk of protofilaments were present in the bundles. Formation of bundles was dependent on both FtsZ and GTP, since individual filaments, as well as bundles, were completely absent in control samples in which FtsZ or nucleotide was omitted or in which GTP was substituted with GDP (data not shown). In contrast, bundles readily formed in reaction mixtures containing EDTA instead of Mg2+, demonstrating that the interaction of ZipA with FtsZ polymers did not require the presence of free divalent metals (Fig. 9C). Bundles also formed readily when FtsZ and ZipA(23–328) were mixed before polymerization was induced by addition of GTP, indicating that binding of ZipA to FtsZ does not prevent the latter from polymerizing (data not shown). Most of our polymerization and bundling assays were performed under slightly acidic conditions (pH 5.8 to 6.0), which favor the formation of relatively long and stable FtsZ polymers (22, 23). ZipA also induced bundling at higher pH (6.5 and 7.0), but both the number and size of the bundle networks we observed tended to be smaller (data not shown). Whether this is solely due to a less efficient formation of FtsZ protofilaments or whether ZipA-induced bundling itself is pH sensitive is currently not clear.
Domains required for bundling.
RayChaudhuri reported on the bundling activity of ZipA during the course of this study and proposed that ZipA is a MAP-Tau homolog, based on some homology between residues 45 to 70 of ZipA and a MAP-Tau repeat motif involved in the binding of microtubule-associated proteins (MAPs) to microtubules (30). To simultaneously test this proposal and delineate the protein domains required for FtsZ polymer bundling, we next assayed the ability of various other purified ZipA derivatives to induce bundling (Fig. 4B and 9).
Interestingly, the FZB domain proved to be not only required but also sufficient for mediating the bundling of FtsZ polymers. Addition of purified T-ZipA(70–328), Gfp-T-ZipA(186–328)-H, or T-ZipA(186–328)-H to FtsZ filaments in each case led to the formation of extensive bundle networks very similar to those described above (Fig. 4b and 9D). In contrast, ZipA(23–279), ZipA(1–302)-H, and Gfp-T-ZipA(212–328)-H all failed to induce bundling and also did not appear to otherwise affect FtsZ protofilaments (Fig. 4b and 9E).
Since we defined the extreme C terminus of FtsZ as the domain which interacts with the FZB domain of ZipA (see above), the latter should be unable to bundle polymers of FtsZ derivatives that do not contain the C terminus. To test this prediction, we used FtsZ(1–314)-H, in which aa 315 to 383 of native FtsZ were replaced with a His6 tag. Upon incubation with GTP, this protein formed numerous protein filaments, demonstrating that the C-terminal 69 residues of FtsZ are not required for polymerization (Fig. 9F). Compared to filaments of the native protein, the filaments of FtsZ(1–314)-H showed an increased tendency to align, and most were present as thin bundles composed of two to five filaments. Similar observations were made before with the somewhat larger peptide FtsZ(1–320) (36). As shown in Fig. 9G, addition of ZipA(23–328) had no effect on the FtsZ(1–314)-H polymers, showing that the C terminus of FtsZ is indeed required for ZipA-mediated bundling of FtsZ filaments.
We conclude that the same domain we found to be required and sufficient for binding of ZipA to FtsZ is also required and sufficient for ZipA-mediated bundling of FtsZ polymers. It follows that residues 1 to 185, which include the MAP-Tau repeat-like motif (30), are dispensable for both the FtsZ binding and bundling activities of ZipA.
Detection of FtsZ polymer bundles by fluorescence microscopy.
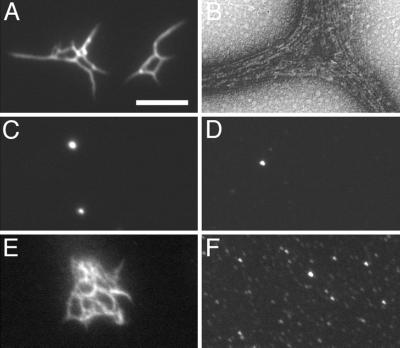
Since the size of the bundle networks we observed by EM was significant, we reasoned that they should also be visible by light microscopy. We first tested this by using the Gfp-tagged ZipA derivative HT-ZipA(39–328)-Gfp. Consistent with the results above, this protein readily bound FtsZ on affinity blots (Fig. 4b) and partially accumulated at the septal ring in cells of strain PB103/pCH77 [wt/Plac::t-zipA(39–328)-gfp] (Fig. 6b). As shown in Fig. 10A, addition of purified HT-ZipA(39–328)-Gfp to preformed FtsZ polymers led to the formation of large protein structures which could indeed be easily visualized by fluorescence microscopy. Fluorescence was concentrated in structures of variable shapes ranging from single thick rods to very extensive networks of thinner filaments. When using FtsZ and HT-ZipA(39–328)-Gfp at equimolar concentration (6.0 μM), we observed very large and highly fluorescent networks which were difficult to image in a single plane of focus. When we reduced the concentration of the fluorescent protein to 0.6 μM, as was done for the experiments in Fig. 10, numerous structures were still observed, but they tended to be smaller and were subsequently easier to image in some detail. Analyses of these structures by EM showed thick bundles of FtsZ polymers, identical in appearance to the ones described above (Fig. 10B). Formation of these structures, furthermore, was clearly dependent on the presence of FtsZ polymers. In control experiments in which FtsZ and/or GTP was omitted or in which GDP was substituted for GTP, the fluorescence signal was largely homogenous. In addition, some fluorescence was concentrated in what appeared as small vesicular structures (Fig. 10C and D). Analyses by EM confirmed the absence of FtsZ polymers and polymer bundles in these samples (data not shown).
FIG. 10.
Detection of polymer bundles by fluorescence microscopy. Purified native FtsZ (6.0 μM) was incubated in the presence of GTP (1 mM) and Mg2+ (10 mM) at 30°C. After 5 min, purified H-T-ZipA(39–328)-Gfp was added to 0.6 μM. After an additional 10 min, one sample was applied to a microscope slide and observed immediately by fluorescence microscopy (A) and another was used for observation by EM (B). The fluorescent samples shown in panels C to F were prepared identically, except that GTP was replaced with GDP (C), FtsZ was replaced with buffer (D), or H-T-ZipA(39–328)-Gfp was replaced with either Gfp-T-ZipA(186–328)-H (E) or Gfp-T-ZipA(212–328)-H (F). EM grids prepared from the reactions shown in panels C to F showed extensive bundle networks as in panel A (E), no bundles but many individual FtsZ protofilaments (F), or no individual protofilaments or polymer bundles (C and D) (data not shown). Bar, represents 0.1 μm (B) or 3.4 μm (A and C to F).
Addition of Gfp-T-ZipA(186–328), which essentially constitutes a Gfp-tagged version of the minimal FZB domain, to FtsZ polymers produced structures similar to those obtained with HT-ZipA(39–328)-Gfp (Fig. 10E). As expected, furthermore, Gfp-T-ZipA(212–328) failed to induce FtsZ polymers to form visible networks (Fig. 10F). The results obtained by fluorescence microscopy were thus fully consistent with those obtained by EM (Fig. 9), indicating that analysis of ZipA-mediated bundling of FtsZ filaments by fluorescence microscopy provides a valid and convenient qualitative assay for FtsZ polymerization.
The membrane anchor of ZipA is required for function.
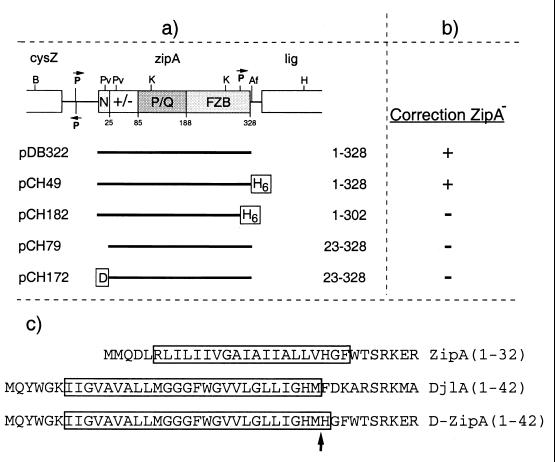
The results above showed that the FZB domain of ZipA is sufficient and required to bind and bundle FtsZ polymers in vitro and to associate with the FtsZ ring in vivo. To test whether the FZB domain is also sufficient to support cell division, we assessed the ability of various deletion derivatives of ZipA to complement the division defect of cells carrying a chromosomal zipA null allele. To this end, we used the ZipA HID (heat-induced depletion) strain CH5/pCH32 [zipA::aph recA::Tn10/repA(Ts) ftsZ+ zipA+] in which the chromosomal copy of zipA is destroyed and ZipA is expressed from plasmid pCH32. This plasmid is temperature sensitive for replication, such that growth of these cells at 42°C results in depletion of ZipA and a subsequent block in septation (11). CH5/pCH32 was transformed with plasmids expressing various portions of ZipA under the control of the lac promoter (Fig. 11), and the ability of transformants to form colonies at 42°C in the presence of IPTG (at a wide range of concentrations) was determined (Table 1).
FIG. 11.
Plasmids used to test correction of ZipA− by ZipA derivatives. (a) Inserts were cloned into the vector pMLB1113 such that expression of the proteins is under control of the lac promoter and lacIq. See also the legends to Fig. 1 and 6. (b) The ability of the plasmids to correct a ZipA− phenotype was determined as described in the text. +, correction; −, no correction. (c) Shown are the N-terminal domains of ZipA, DjlA, and DjlA(1–32)-ZipA(23–328). Transmembrane segments as predicted by the Dense Alignment Surface method (4) (http://www.biokemi.su.se/∼server/DAS/) are boxed. Residue numbers are indicated in parentheses. The arrow marks the junction between DjlA and ZipA residues in the DjlA-ZipA fusion.
TABLE 1.
Ability of zipA plasmids to correct a chromosomal zipA null allelea
| Plasmid | Inducible protein | CFU on plate at:
|
Efficiency (%)c | ||
|---|---|---|---|---|---|
| 30°C, no IPTG | 42°C, no IPTG | 42°C, plus IPTGb | |||
| pMLB1113 | LacZ | 246 | 0 | 0 | <0.4 |
| pDB322 | ZipA | 358 | 0 | 238 | 66 |
| pCH49 | ZipA-H | 516 | 0 | 350 | 68 |
| pCH182 | ZipA(1–302)-H | 454 | 0 | 0 | <0.2 |
| pCH79 | ZipA(23–328) | 278 | 0 | 0 | <0.4 |
| pCH172 | D-ZipA(23–328) | 234 | 0 | 0 | <0.4 |
Overnight cultures of strain CH5/pCH32 [recA::Tn10 zipA0/repA(Ts) zipA+ftsZ+] containing the indicated plasmids were diluted in LB broth to an OD600 of 2 × 10−5, and 0.1-ml aliquots were spread on nine LB agar plates containing 100 μg of ampicillin per ml. One plate was supplemented with 0.2% glucose and was incubated at 30°C. The other plates were supplemented with 0, 5, 10, 25, 50, 100, 250, or 500 μM IPTG and were incubated at 42°C. For each plate, the number of CFU was determined.
The value represents the number of colonies on the plate with the optimal concentration of IPTG, which was 5 μM for pDB322 and 50 μM for pCH49. The highest concentration(s) of IPTG allowing colony formation at 42°C were 50 and 100 μM for pDB322 and pCH49, respectively. Overexpression of ZipA blocks cell division, explaining the inability of these plasmids to support colony formation at higher IPTG concentrations (10). Cells containing any of the other plasmids failed to form any colonies on any of the plates at 42°C.
Plating efficiency at the optimal IPTG concentration at 42°C relative to that at 30°C.
Both pDB322 [Plac::zipA(1–328)], encoding native ZipA, and pCH49 [Plac::zipA(1–328)-h], encoding full-length ZipA fused to a C-terminal His tag, allowed colony formation at 42°C with high efficiency (Table 1) and at low concentrations of inducer (5 to 50 μM). At high concentrations of inducer, pDB322 and pCH49 failed to support colony formation. This was not unexpected, since we previously showed that overexpression of native ZipA leads to a division block which can be relieved by co-overexpression of FtsZ (10). Accordingly, when pDB322 or pCH49 was introduced into wild-type strain PB103, cells formed nonseptate filaments when grown in the presence of IPTG at concentrations equal to or higher than 25 and 100 μM, respectively. Quantitative immunoblotting, furthermore, showed that an increase of ca. sixfold in the cellular concentration of either ZipA or ZipA-H is sufficient to induce this division block (Table 2). In contrast, overexpression of the ZipA(1–302)-H protein to this level had no effect on the division phenotype of PB103/pCH182 cells, consistent with a role for the FZB domain in the division block caused by overexpression of full-length ZipA (Table 2). As shown in Table 1, plasmid pCH182 also failed to support colony formation of ZipA− cells at any of the inducer concentrations (0 to 500 μM), underscoring the notion that the interaction between FZB and FtsZ is essential to ZipA function (Table 1).
Although less toxic than native ZipA, overexpression of the ZipA(23–328) protein in PB103/pCH79 cells to ∼11-fold the normal level of ZipA still led to the formation of nonseptate filaments, showing that the membrane anchor is not required for the division block (Table 2). Interestingly, however, plasmid pCH79 [Plac::zipA(23–328)] completely failed to correct the ZipA− phenotype of CH5/pCH32 cells at 42°C under any conditions, including those (<100 μM IPTG) under which the levels of ZipA(23–328) (over)expression are too low to induce a division block in wild-type cells (Table 1). Thus, although ZipA(23–328) has an intact FZB domain which allows a significant fraction of the protein to accumulate at the septal ring (Fig. 7C), the protein does not support cell constriction.
This result implied that in addition to a functional FZB domain, the membrane anchor is required for ZipA function. A comparison of the distributions in PB103 cells of ZipA(23–328)-Gfp and Gfp-T-ZipA(186–328) with that of ZipA(1–328)-Gfp (Fig. 7C, D, and A, respectively) indicated that the presence of the membrane anchor in the latter allows a more efficient localization to the septal ring, which is not unexpected since the anchor causes the protein to become concentrated at the membrane. [Note that the relatively high level of cytoplasmic fluorescence observed in cells expressing ZipA(23–328)-Gfp and Gfp-T-ZipA(186–328) was not caused by cleavage of the fusions, since we detected no significant breakdown products in Western analyses using both ZipA- and Gfp-specific antibodies (data not shown). It seemed unlikely, however, that a more efficient localization alone could explain the need for the anchor, because pCH79 [Plac::zipA(23–328)] failed to support cell division in CH5/pCH32 cells even when significantly overexpressed (Table 1). Other possibilities are that a precise orientation of ZipA relative to the membrane and the FtsZ ring is required for full function of the protein and the anchor is required to constrain this orientation, or that the membrane anchor serves a more specific essential function such as mediating interactions with other septal-ring components.
To study these possibilities, we constructed plasmids pCH172 [Plac::djlA(1–32)-zipA(23–328)] and pCH174 [Plac::djlA(1–32)-zipA(23–328)-gfp] (Fig. 6 and 11). The former encodes a fusion protein in which the ZipA membrane anchor is replaced with that of DjlA (MucZ), and the latter encodes a Gfp-tagged version of this fusion. DjlA is an inner membrane-associated DnaJ-like protein which, besides ZipA, is the only other E. coli protein shown to insert in the membrane in the unusual type lb (also designated as type III) topology, with a short N-terminal domain that traverses the bilayer once (residues 1 to 32) and the rest of the protein resident in the cytoplasm (3). The predicted transmembrane domains of ZipA, DjlA, and the DjlA-ZipA fusions are indicated in Fig. 11c. Expression of the DjlA-ZipA fusions from pCH172 and pCH174 was relatively inefficient compared to expression of native ZipA from pDB322. Since we detected no significant breakdown products in Western analyses (data not shown), we suspect that this is due to the poor translation initiation signal of the djlA gene, which was retained in the djlA-zipA fusion constructs. Even so, cell division appeared equally sensitive to overexpression of DjlA-ZipA and native ZipA. Thus, when PB103/pCH172 cells were grown in the presence of 500 μM IPTG, the cellular level of DjlA(1–23)-ZipA(23–328) was equal to ca. sixfold the normal level of native ZipA, and cells formed nonseptate filaments (Table 2).
As shown in Fig. 7, fusion of the DjlA membrane anchor to ZipA(23–328)-Gfp appeared to restore the ability of the protein to efficiently associate with the septal ring. In fact, the distribution of fluorescence in PB103/pCH174 cells (Fig. 7F) appeared identical to that in cells expressing ZipA(1–328)-Gfp (Fig. 7A), with virtually all fluorescence being associated with the septal ring and very little being present in the cytoplasm. This suggested that the DjlA portion of the fusion indeed acted as a functional membrane anchor. Consistent with this notion, we found that the bulk (>80%) of DjlA(1–32)-ZipA(23–328) fractionated with insoluble material of broken cells (data not shown) and that the bulk of fluorescent DjlA(1–32)-ZipA(23–328)-Gfp in FtsZ-depleted filaments of strain PB143/pDB346/pCH174 was distributed along the cell periphery (Fig. 8C). Nevertheless, plasmid pCH172 [Plac::djlA(1–32)-zipA(23–328)] completely failed to support growth of CH5/pCH32 cells at 42°C at any level of expression (Table 1).
To explore the possibility that ZipA(23–328) and/or DjlA(1–32)-ZipA(23–328) might be capable of rescuing ZipA− cells at a low temperature, we also introduced pDB322, pCH79, and pCH172 into the ZipA cold-induced depletion strain CH5/pDB361 (zipA::aph recA::Tn10/cI857 PλR::zipA), in which expression of native ZipA is shut down at 30°C (11). At 30°C, CH5/pDB361/pDB322 formed colonies in an IPTG-dependent manner with a maximum efficiency close to 100% (at 5 μM IPTG). In contrast, neither pCH79 nor pCH172 was capable of rescuing the lethal division block of CH5/pDB361 cells at any of a wide range of IPTG concentrations (data not shown). We conclude that the N-terminal domain of ZipA is essential for its function. In addition, our data suggest that the essential function(s) of this domain extends beyond merely anchoring the protein to the membrane.
DISCUSSION
In this study we have used a variety of techniques to identify protein domains that are both required and sufficient for the specific interaction between ZipA and FtsZ in vitro as well as in vivo. The results allow us to narrow down the domains involved to the 20 C-terminal residues (aa 364 to 383) of FtsZ and the 143 C-terminal residues (aa 186 to 328) of ZipA. Our findings support and extend the recent reports of Liu et al., who detected an interaction between ZipA(176–328) and FtsZ(1–383) but not FtsZ(1–320) in two-hybrid assays (13), and of Ma and Margolin, who found that a ZipA-Gfp fusion failed to colocalize with FtsZ(1–371) in filaments that had been depleted of native FtsZ (20).
Structure determinations indicate that both tubulin and FtsZ roughly consist of two structural domains which are separated by a long helix corresponding to residues 177 to 201 in the E. coli peptide. The N-terminal two-thirds of both tubulin and FtsZ include all residues which contact the nucleotide and also show the highest degree of primary sequence conservation among peptides from different organisms (14, 15, 26). Alignments of E. coli FtsZ with peptides from various organisms show that the region with a high degree of conservation extends from the N terminus up to approximately residue 314. It was shown previously that FtsZ(1–320) (36) and FtsZ(1–316) (20) contain all elements required for nucleotide-dependent polymer formation, and we found here that the same is true for FtsZ(1–314). This region is followed by a region which is variable in length and sequence in diverse organisms and which extends approximately to residue 369 in E. coli. This variable region is followed, in turn, by a 10-residue peptide which, once again, shows a high degree of conservation and which was named the C-terminal core domain by Ma and Margolin (20). Although FtsZ derivatives lacking this domain can polymerize, they fail to support cell division but instead display a prominent trans-dominant negative phenotype (references 8, 20, and 36 and data not shown), demonstrating the domain is essential for FtsZ function. In E. coli this small domain (DYLDIPAFLR) corresponds to residues 370 to 379. Deletion of the C-terminal 10 residues of FtsZ removes most of this core domain and, as shown here, also abolishes the interaction with ZipA, implicating residues within the core domain in providing a binding interface for ZipA.
Interestingly, the core domain peptide has also been implicated in the interaction between FtsZ and FtsA in Caulobacter crescentus, Bacillus subtilis, and E. coli. Thus, mutation or deletion of the C-terminal ends of FtsZ from each of these species was found to abolish an interaction with FtsA, as detected by two-hybrid analyses (8, 20, 36) and colocalization studies (20). We show here that the C-terminal 20 residues of FtsZ include all elements that are not only required but also sufficient for binding to ZipA. This may not be the case for binding of FtsZ to FtsA, however, since deletion of N-terminal portions of FtsZ also abolished interaction with FtsA in two-hybrid assays (36), suggesting that FtsA may require multiple and/or larger binding surfaces on FtsZ for stable engagement. ZipA and FtsA were shown to be recruited to the FtsZ ring in a mutually independent fashion (11, 13). The finding that the C-core is required for recruitment of both proteins raises the issue of whether ZipA and FtsA bind in a mutually exclusive fashion or whether both can be engaged at the same time. The latter seems unlikely, given the small size of the C-terminal peptide, but formation of a tripartite complex cannot yet be excluded. Assuming that ZipA and FtsA do potentially compete for binding FtsZ, it is presently unclear whether such competition would be physiologically relevant, given the large number of FtsZ molecules estimated to make up the FtsZ ring (18).
RayChaudhuri first reported that ZipA induces extensive bundling of FtsZ polymers in vitro (30). We have confirmed this by both EM and fluorescence microscopy and have found that bundling also occurs in the absence of free Mg2+. Furthermore, our results show that the FZB domain of ZipA is both required and sufficient for this bundling activity. This domain does not include the MAP-Tau repeat motif, which is embedded in the highly charged domain near the N terminus of ZipA and which was proposed to mediate FtsZ binding and bundling (30). Although it is possible that this motif plays some role in ZipA function, it is clearly not required or sufficient for binding and bundling FtsZ polymers. Therefore, although ZipA and MAPs have some interesting functional analogies (30), they should not be considered homologs.
It is not clear how binding of the FZB domain of ZipA stimulates lateral associations between FtsZ protofilaments. The assembly of FtsZ protofilaments into bundles or sheets also occurs in the presence of DEAE-dextran (9, 24), at millimolar (>7 mM) concentrations of Ca2+ (16, 22, 39), or on a surface of cationic lipids (9). Since these treatments all involve cationic substances and since FtsZ is an acidic protein, it is likely that bundling of FtsZ polymers in these cases is stimulated by a reduction of electrostatic repulsion between individual protofilaments due to charge compensation by the cationic counterions. These counterions are thought to exist in a condensed state, where they are free to diffuse along the polymer axis but are inhibited from diffusing away (22, 33, 34, 38). Because the ZipA protein (as well as its FZB domain) is actually slightly acidic and because the protein binds only to a specific site on the polymer subunit, such nonspecific charge compensation cannot easily explain ZipA mediated bundling. It is possible, however, that ZipA shields repulsive charges on some specific residues and/or that binding of ZipA provokes conformational changes within polymer subunits, causing repulsive groups to become buried or attractive groups to become exposed. Alternatively, bundling might involve noncovalent cross-linking of adjacent polymers through FZB-FZB interactions. Further work is needed to explore these possibilities.
Previous evidence suggests that one role of ZipA is to provide stability to the FtsZ ring (11, 13, 30). The in vitro bundling activity of ZipA provides an attractive mechanism of how the protein might contribute to Z-ring stability in vivo. A strengthening of the lateral association between FtsZ protofilaments would increase the structural integrity and stress-bearing capacity of the whole septal ring structure, which is likely to be important both before and during septal invagination. However, our results suggest that binding and bundling of FtsZ is not the only relevant property of ZipA. In particular, our findings that removal of the membrane anchor domain of ZipA or its replacement with the equivalent domain of DjlA is not tolerated indicate that the N-terminal domain of ZipA may be involved in an essential activity additional to its role as a membrane tether. One possibility is that this domain mediates an essential interaction with another membrane component. Other transmembrane septal ring proteins would be logical candidates for such a component. If so, however, such an interaction must be either weak or dependent on the binding of the FZB domain with the FtsZ ring, since the ZipA(1–302)-Gfp fusion, which lacks just a small portion of the FZB domain, showed no obvious preference for the septal ring (Fig. 6b and 7B).
The FZB domain and the membrane anchor are the most highly conserved regions among ZipA peptides of different species (10), which is consistent with the notion that both domains play essential roles. The charged and PQ-rich domains are not as well conserved, but whether or not these domains are essential for function still needs to be addressed.
ACKNOWLEDGMENTS
We thank David Raskin and Anita Boyapati for help with plasmid construction, Rui-Zhen Wang and Joe Polak for help with electron microscopy, and Annick Jacq, Barry Holland, Michael Maguire, and Katleen Postle for generous gifts of plasmids and antibodies.
This work was supported by NIH grant GM-57059 and by a donation from Wyeth-Ayerst Research (to P.D.B.).
Footnotes
This study is dedicated to the memory of Joe Polak, whose expertise and cheerfulness are sorely missed.
REFERENCES
- 1.Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 2.Broome-Smith J K, Spratt B G. A vector for the construction of translational fusions to TEM β-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 3.Clarke D J, Jacq A, Holland I B. A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol Microbiol. 1996;20:1273–1286. doi: 10.1111/j.1365-2958.1996.tb02646.x. [DOI] [PubMed] [Google Scholar]
- 4.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 5.de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 6.de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 7.de Boer P A J, Crossley R E, Rothfield L I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Din N, Quardokus E M, Sackett M J, Brun Y V. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 9.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 11.Hale C A, de Boer P A J. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ, and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- 14.Löwe J. Crystal structure determination of FtsZ from Methanococcus jannaschii. J Struct Biol. 1998;124:235–243. doi: 10.1006/jsbi.1998.4041. [DOI] [PubMed] [Google Scholar]
- 15.Löwe J, Amos L A. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 16.Löwe J, Amos L A. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 1999;18:2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C, Reedy M, Erickson H P. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C, Stricker J, Erickson H P. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima-quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 19.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee A, Lutkenhaus J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol. 1999;181:823–832. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Lutkenhaus L. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nogales E, Downing K H, Amos L A, Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 26.Nogales E, Wolf S G, Downing K H. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 27.Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomembr. 1993;25:591–601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- 28.Raskin D M, de Boer P A J. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 29.Raskin D M, de Boer P A J. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.RayChaudhuri D, Park J T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 32.Rothfield L, Justice S, García-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 33.Tang J X, Ito T, Tao T, Traub P, Janmey P A. Opposite effects of electrostatics and steric exclusion on bundle formation by F-actin and other filamentous polyelectrolytes. Biochemistry. 1997;36:12600–12607. doi: 10.1021/bi9711386. [DOI] [PubMed] [Google Scholar]
- 34.Tang J X, Janmey P A. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J Biol Chem. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- 35.Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 38.Wolff J, Knipling L, Sackett D L. Charge-shielding and the “paradoxical” stimulation of tubulin polymerization by guanidine hydrochloride. Biochemistry. 1996;35:5910–5920. doi: 10.1021/bi9527395. [DOI] [PubMed] [Google Scholar]
- 39.Yu X-C, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]