Abstract
Characterization in Thermotoga neapolitana of a catabolic gene cluster encoding two glycosyl hydrolases, 1,4-β-d-glucan glucohydrolase (GghA) and cellobiose phosphorylase (CbpA), and the apparent absence of a cellobiohydrolase (Cbh) suggest a nonconventional pathway for glucan utilization in Thermotogales. GghA purified from T. neapolitana is a 52.5-kDa family 1 glycosyl hydrolase with optimal activity at pH 6.5 and 95°C. GghA releases glucose from soluble glucooligomers, with a preference for longer oligomers: kcat/Km values are 155.2, 76.0, and 9.9 mM−1 s−1 for cellotetraose, cellotriose, and cellobiose, respectively. GghA has broad substrate specificity, with specific activities of 236 U/mg towards cellobiose and 251 U/mg towards lactose. With p-nitrophenyl-β-glucoside as the substrate, GghA exhibits biphasic kinetic behavior, involving both substrate- and end product-directed activation. Its capacity for transglycosylation is a factor in this activation. Cloning of gghA revealed a contiguous upstream gene (cbpA) encoding a 93.5-kDa cellobiose phosphorylase. Recombinant CbpA has optimal activity at pH 5.0 and 85°C. It has specific activity of 11.8 U/mg and a Km of 1.42 mM for cellobiose, but shows no activity towards other disaccharides or cellotriose. With its single substrate specificity and low Km for cellobiose (compared to GghA's Km of 28.6 mM), CbpA may be the primary enzyme for attacking cellobiose in Thermotoga spp. By phosphorolysis of cellobiose, CbpA releases one activated glucosyl molecule while conserving one ATP molecule per disaccharide. CbpA is the first hyperthermophilic cellobiose phosphorylase to be characterized.
To utilize polysaccharides such as glucans to meet carbon and energy requirements, heterotrophic organisms depend on a catabolic pathway involving the interaction of multiple hydrolytic enzymes, transporter complexes, and regulatory systems coordinating gene expression of pathway-specific proteins (41). During hydrolysis of the homopolymer cellulose, for example, the consortium of catalytic enzymes consists of endoglucanases (1,4-β-d-glucan 4-glucohydrolase [EC 3.2.1.4]), which randomly hydrolyze internal β-1,4 glycosidic bonds; cellobiohydrolases (β-d-glucan cellobiohydrolase [EC 3.2.1.91]), also referred to as exoglucanases, which remove cellobiose from either the nonreducing or reducing ends of cellooligomers; and β-glucosidases (β-d-glucoside glucohydrolase [EC 3.2.1.21]), which convert cellobiose to glucose. These classes of enzymes have been documented in a number of fungal and bacterial systems (4, 5, 39), while other glucan-catabolyzing enzymes are less well understood.
Thermotoga neapolitana, a marine hyperthermophile isolated from geothermally heated biotopes, belongs to the order Thermotogales. T. neapolitana shares with other Thermotogales, specifically Thermotoga maritima, both the capacity to catabolize a wide variety of α- and β-linked glucans and a fermentative metabolism. While Thermotogales elaborate hydrolases such as amylases, cellulases, glucosidases, galactosidases, mannanases, and xylanases (6, 7, 10, 11, 23, 37), neither T. neapolitana (D. Y. Yernool, G. Swiatek, and J.-D. Bok, unpublished data) nor T. maritima (27) has been shown to have a gene for a cellobiohydrolase, a key enzyme in classical cellulolytic systems.
In this paper, we report the characterization of a catabolic gene cluster in T. neapolitana encoding two enzymes: a cellobiose phosphorylase (CbpA) and a β-glucan glucohydrolase (GghA). Cellobiose phosphorylases (EC 2.4.1.20) cleave cellobiose by phosphorolysis, yielding glucose-1-phosphate as one of the products, while β-glucan glucohydrolases or exoglucohydrolases (1,4-β-d-glucan glucohydrolase [EC 3.2.1.74]) preferentially act on cellooligomers, releasing glucose (17, 31, 37, 43). In addition to the functional and molecular characterization of GghA and CbpA, we propose a nonconventional, ATP-conserving, catabolic pathway for glucan utilization consisting of endoglucanases CelA and CelB (7), a β-glucan glucohydrolase (GghA), and a cellobiose phosphorylase (CbpA).
MATERIALS AND METHODS
Bacterial strains and culture media.
T. neapolitana NS-E (kindly provided by K. O. Stetter and R. Huber, University of Regensburg, Regensburg, Germany) was grown anaerobically in MMS medium (9) at 77°C with cellobiose or glucose (1%, wt/vol) as a carbon source in static culture for 24 h. A cosmid DNA library in vector pLAFR3 (36) of T. neapolitana strain NS-E in Escherichia coli DH5α (gift from Kenneth Knoll, University of Connecticut, Storrs, Conn.) was used. Commercially available E. coli strains were grown in Luria-Bertani (LB) medium or super broth (32) containing either ampicillin (50 μg/ml) or tetracycline (25 μg/ml). Some of the abbreviations used in this report are as follows: ORF, open reading frame; oNP, o-nitrophenol; PC, sodium phosphate-sodium citrate; pNPA, pNP-α-d-arabinoside; pNPC, pNP-β-d-cellobioside; pNPG, pNP-β-d-glucoside; pNPL, pNP-β-d-lactoside; pNP, p-nitrophenol; pNPX, pNP-β-d-xyloside; pNPαG, pNP-α-d-glucoside; oNPG, oNP-β-d-glucoside; oNPGal, oNP-β-d-galactoside.
Purification of glucan glucohydrolase.
T. neapolitana was grown on cellobiose, collected by centrifugation, washed twice in Tris-HCl (0.1 M [pH 7.5] at 4°C), resuspended in the same buffer, and lysed by sonication. Cell debris was removed, the proteins in the supernatant were precipitated using ammonium sulfate (final concentration, 80%), collected, and dissolved in 50 mM Tris-HCl (pH 7.5) (20 ml). The ammonium sulfate precipitation was repeated twice, and the resultant cell extract was dissolved in 20 mM piperazine-HCl buffer (pH 5.1) (20 ml) and desalted by ultrafiltration using a PM 10 membrane (Amicon, Cambridge, Mass.). The pH of this cell-free extract was adjusted to 4.3 by addition of sodium citrate (0.15 M, pH 4.1); precipitated proteins were removed by centrifugation. The pH of the supernatant containing aryl β-glucosidase activity was raised to 5.1 using 0.2 M sodium citrate. Purification of the aryl-glycosidase was achieved by anion-exchange chromatography using an FFQ-Sepharose column (XK26; 60-ml bed volume; Pharmacia, Piscataway, N.J.) equilibrated with 20 mM piperazine buffer (pH 5.1). Bound proteins were eluted with a linear gradient of 0 to 0.3 M NaCl. Fractions from a single peak with activity towards aryl-glycosides were pooled, desalted by ultrafiltration, and subjected to gel filtration (BioGel P-60 column, 1.5 by 97 cm; 50 mM MOPS [pH 7.0]). Active fractions were pooled, the concentration of ammonium sulfate was adjusted to 0.2 M, and the proteins were then fractionated by hydrophobic-interaction chromatography (Phenyl-Sepharose FF; XK26 column; 20-ml bed volume; Pharmacia). Proteins bound to the column were eluted using a reverse gradient (0.2 M ammonium sulfate in 20 mM Tris-HCl [pH 7.5] to distilled water). Active fractions were pooled, desalted, and further purified using a galactose-agarose column (p-aminobenzyl-1-thio-β-d-galactopyranoside; Sigma, St. Louis, Mo.) equilibrated with binding buffer (0.1 M NaPO4 [pH 7.0], 0.15 M NaCl). Bound proteins were eluted with elution buffer (binding buffer with 0.3 M glucose). After buffer exchange to 20 mM BisTris (pH 6.5), pooled active fractions were applied to a Mono-Q column (HR 10/10; Pharmacia) and eluted with a linear gradient of 0.05 to 0.5 M NaCl. Final purification was achieved by discontinuous preparative polyacrylamide gel electrophoresis (PAGE) using a PrepCell (Bio-Rad, Richmond, Calif.) following the manufacturer's instructions.
Purification of recombinant cellobiose phosphorylase.
E. coli clone 13CBPFM was grown in 5 liters of LB medium containing ampicillin (50 μg/ml) to an optical density at 600 nm of 0.5. Expression of the cbpA gene was induced by addition of arabinose (final concentration, 0.015%). Four hours postinduction, cells were collected by centrifugation (4,000 × g, 15 min, 4°C). Cell pellets were resuspended in buffer (50 mM Tris-HCl buffer [pH 7.9], 50 mM dextrose, 1 mM EDTA) containing lysozyme (4 mg/ml) (Sigma) and sonicated. Heat-labile E. coli proteins were removed from the cell extract by heat treatment (75°C for 15 min) and precipitated by centrifugation (15,000 × g, 30 min, 4°C). Supernatant was equilibrated with ammonium sulfate (final concentration, 1.0 M) and loaded onto a phenyl-Sepharose column (XK26; Pharmacia; 100-ml bed volume). Bound proteins were eluted with a reverse gradient, ammonium sulfate (1.0 M) in Tris-HCl buffer (20 mM, pH 7.8) to distilled H2O and distilled H2O to 50% acetonitrile. Active fractions from a single peak were pooled, desalted, and further purified using immobilized metal-ion chromatography (Ni-nitrilotriacetic acid resin; Qiagen, Valencia, Calif.; 10-ml bed volume) with a linear gradient of 0.01 to 0.3 M imidazole, following the manufacturer's instructions. Active fractions were pooled, buffer exchanged by ultrafiltration (Amicon YM10; Millipore, Bedford, Mass.), and further purified by anion-exchange chromatography (Mono-Q HR 5/5; Pharmacia). The pool was equilibrated with loading buffer (20 mM piperazine [pH 6.0], 10 mM NaCl), and bound protein was eluted with a linear gradient of 0.01 to 0.5 M NaCl. Active fractions from the single peak were pooled and further purified by ultrafiltration (Microcon YM50; Millipore).
Glucan glucohydrolase assays.
Aryl-glycosidase activity was determined using either pNP- or oNP-glycoconjugates at 5 mM final concentration in a 15-min assay (42). The glycoconjugates used included pNPA, pNPC, pNPG, pNPL, pNPX, pNPαG, oNPG, and oNPGal. Released pNP and oNP were measured at 405 and 420 nm, respectively; activity was calculated using a standard curve developed under assay conditions. Hydrolysis of cellobiose and lactose was analyzed using the glucose hexokinase kit following the manufacturer's instructions (Sigma). One unit of enzyme activity corresponds to release of 1 μmol of pNP, oNP, or glucose/min (for cellobiose as the substrate, one unit corresponds to the release of 2 μmol/min). All assays were carried out in 100 mM (final concentration) PC buffer (pH 6.4) at 85°C unless otherwise cited.
Cellobiose phosphorylase assay.
Cellobiose phosphorylase activity was determined by measuring the amount of glucose 1-phosphate formed from phosphorolysis of cellobiose (Sigma). The enzyme was incubated at 85°C for 15 min with 10 mM cellobiose in PC buffer (pH 5.5). The reaction was stopped by boiling for 10 min, and the amount of glucose 1-phosphate produced was determined by a coupled enzyme assay measuring the appearance of NADPH at 340 nm (30). The reaction mixture contained phosphoglucomutase (4 U ml−1), glucose-6-phosphate dehydrogenase (2.0 U ml−1) (Boehringer Mannheim, Indianapolis, Ind.), 3 mM NADP, and 5 μM glucose 1,6-bisphosphate (Sigma) in 80 mM triethanolamine buffer (with 4.4 mM MgCl2 [pH 7.5]). One unit of activity corresponds to the release of 1 μmol of glucose 1-phosphate/min.
Analytical methods and enzymatic characterization.
The molecular mass of glucan glucohydrolase and cellobiose phosphorylase was determined by sodium dodecyl sulfate (SDS)-PAGE according to Laemmli (24) and by gel filtration using an analytical Suparose 12 HR column (Pharmacia). Protein bands in SDS-PAGE gels were visualized with either silver stain for GghA or Coomassie blue for CbpA. To determine the N-terminal sequence, the protein band was transferred to a polyvinylidene difluoride membrane, and the sequence was determined using an Applied Biosystems 475A gas phase sequenator (Applied Biosystems, Foster City, Calif.). Protein concentrations were estimated by the Bradford dye-binding method (Bio-Rad) with bovine serum albumin as the standard and using A280 and the molar extinction coefficient (12). Isoelectric focusing was used to determine the isoelectric point (pI) (Phast System; Pharmacia). pH optima were determined using PC buffer (0.1 M, pH 3.6 to 7.0) and phosphate buffer (pH 6.0 to 7.5). To determine the optimum temperature for activity for GghA, a mixture of GghA in PC buffer (0.1 M, pH 6.4) and pNPG (5 mM) were sealed in 2-ml gas chromatography vials (Wheaton, Vineland, N.J.). Activity was measured after heating in an oil bath (70 to 120°C) for 20 min. Released pNP was quantified as described for the enzyme assays. The temperature optimum for CbpA was determined by the cellobiose phosphorylase assay run for 15 min at various temperatures prior to the coupled enzyme assay as described above. The glucan glucohydrolase's thermal stability was evaluated by heating the enzyme (0.19 μg) in sealed vials in an oil bath at 90 and 95°C for up to 9 h in MOPS buffer (20 mM, pH 7.0). After cooling on ice, residual activity was estimated using pNPG as the substrate. The effect of end product on hydrolytic activity was assessed by measuring the rate of release of pNP from pNPG in the presence of increasing concentrations of glucose (0 to 1.0 M).
Purified glucan glucohydrolase (37 to 185 ng) was mixed with each substrate (2 to 40 mM) in a total volume of 120 μl and incubated at 75°C in 5 mM HEPES buffer (pH 7.2). The cellooligosaccharide substrates used included cellobiose, cellotriose, and cellotetraose (V-Labs, Covington, La.). The products of hydrolysis of cellooligosaccharides and transglycosylation were fractionated on an Aminex HPX42A column (Bio-Rad) attached to a high-pressure liquid chromatograph (HPLC; model LC600; Shimadzu, Kyoto, Japan) and detected using a refractometer (Waters, Milford, Mass.) linked to a Hewlett-Packard 3390A integrating recorder. The released sugars were eluted isocratically with distilled water (0.6 ml/min). The data were compared to commercially available standards analyzed under identical conditions. One unit is defined as the amount of enzyme required to hydrolyze 1 μmol of cellobiose to glucose (2 μmol) or to release 1 μmol of glucose from cellotriose and cellotetraose in 1 min under assay conditions.
Determination of anomeric carbon configuration of the hydrolysis products.
The method described by Baker and Himmel (3) was used with minor modifications. GghA (0.9 μg) was mixed with oNPG (10 mM final concentration) in 5 mM HEPES buffer (pH 7.2), incubated at 22°C for 10 min, and immediately placed on ice. A 20-μl aliquot of the reaction mixture was fractionated on an Aminex HPX-87C column run at 25°C at 0.5 ml/min. The retention times and the extent of mutarotation were compared to those of freshly prepared controls (α-glucose and β-glucose). Under these conditions, mutarotation was negligible (β/α = 4.26; at equilibrium, β/α = 1.6).
Cloning, sequencing, and analysis of gghA and cbpA from T. neapolitana.
All DNA manipulations were carried out using standard methods (32). A genomic DNA library of T. neapolitana was constructed and screened for thermostable glycosidases as described earlier (7). The DNA sequence (partial) of the inserts contained in selected clones was determined using fluorescent dye terminator sequencing chemistry with an ABI 373 automated sequencer (Applied Biosystems). The sequence was assembled and analyzed using BLAST (2). Initially, clone p46B1 containing part of the sequence of the gghA gene was identified. The sequence at the other end of the p46B1 insert revealed similarities to known cellobiose phosphorylases. Based on this sequence, two PCR primers, OT305PA1 (5′TCCAGTC CGCCCTTCCTATGAG3′) and OT317PA1 (5′AAGGCATT TCACAGGAGAG GTC 3′), were designed. A PCR screen was conducted using these primers on a T. neapolitana cosmid library. By analyzing PCR amplicons by agarose gel electrophoresis, two cosmid clones, RUC 270 and RUC 278, containing the gghA and cbpA genes were identified; both gghA and cbpA genes were cloned, and the DNA sequence was determined.
Comparison of amino acid sequences of cellobiose phosphorylases and β-(1-2) glucan synthetases.
Percent similarity was calculated using the GAP program of GCG version 9.1 (Genetics Computer Group, Madison, Wis.). Complete amino acid sequences were used except for domains from Clostridium thermocellum Cbp (amino acids 564 to 975), Rhizobium meliloti NdvB (amino acids 2090 to 2843), and Brucella abortus Cgs (amino acids 2084 to 2817).
Expression of the cellobiose phosphorylase cbpA gene in E. coli.
The cbpA gene was amplified using primers CBP.509 (5′AGGATCCATAGCATGAAGTTCGGCTACTT 3′) and CBP.R510 (5′CAAGCTTTCGCTCTTTGCTGTAGTTTG 3′), designed with BamHI and HindIII restriction sites, respectively (underlined sequence). The amplicon was cloned into the pBAD-TOPO-TA vector (Invitrogen, Carlsbad, Calif.) under the control of an inducible arabinose promoter, resulting in the addition of a C-terminal (His)6 tag.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to GenBank and EMBL Data Bank under accession number AF039487.
RESULTS
Purification of glucan glucohydrolase (GghA).
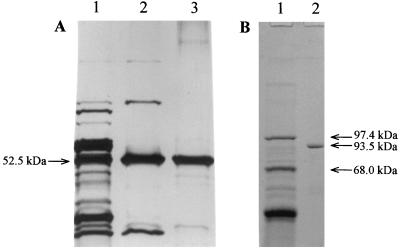
A seven-step protocol was used to purify GghA, giving 184-fold purification and a total recovery of 32% (Table 1). The specific activity of the purified enzyme is 807 U/mg toward oNPG. SDS-PAGE analysis showed that the purified enzyme had an apparent molecular mass of 52.5 kDa (Fig. 1). A few faint bands were visible on the silver-stained gel as purified sample (610 ng) was loaded in excess (sensitivity of silver staining is 2 to 10 ng of protein/band [13]). The N-terminal sequence of GghA was MKKFPEGFLWGVATASYQIEG, which agrees with the DNA sequence data (see below).
TABLE 1.
Purification of glucan glucohydrolase from T. neapolitana
| Purification step | Total protein (mg) | Total activitya (U) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Crude extract | 2,425 | 4,994 | 2.06 | —b | —b |
| Acidification to pH 4.3 (supernatant) | 628 | 2,747 | 4.37 | 1 | 100 |
| Anion-exchange chromatography (Q-Sepharose) | 152 | 2,200 | 14.47 | 3 | 80 |
| Gel filtration (Biogel P-60) | 82.8 | 2,025 | 24.45 | 6 | 74 |
| Hydrophobic-interaction chromatography (phenyl-Sepharose) | 24.4 | 1,735 | 71.1 | 16 | 63 |
| Affinity chromatography (galactose-agarose) | 3 | 1,460.9 | 374.3 | 86 | 53 |
| Anion-exchange chromatography (Mono-Q) | 3.2 | 1,413 | 441.5 | 97 | 51 |
| Preparative PAGE (native) | 1.1 | 888 | 807.27 | 184 | 32 |
The substrate used in enzyme assays was oNPG.
Purification and recovery were not calculated because the crude extract contains at least two enzymes with aryl-glycosidase activity.
FIG. 1.
SDS-PAGE analysis of purified native glucan glucohydrolase (GghA) and recombinant cellobiose phosphorylase (CbpA). (A) Glucan glucohydrolase. Lane 1, fraction after hydrophobic interaction chromatography (2 μg); lane 2, fraction after affinity chromatography (1 μg); lane 3, purified GghA after preparative PAGE (0.61 μg). (B) Cellobiose phosphorylase. Lane 1, high-range protein molecular size markers; lane 2, purified CbpA after Mono-Q chromatography (6.7 μg).
Characterization of GghA.
GghA exhibited optimal activity over the pH range 6.4 to 7.0 and at 95°C. The enzyme was highly thermostable, retaining 85% activity after incubation for 9 h at 90°C and 88% activity after 1 h at 95°C. Active GghA was a monomer (determined by gel filtration) with an apparent molecular mass of 58.9 kDa. GghA showed broad-spectrum glycosidase activity, releasing oNP from both oNPG and oNPGal with high specific activities, 807 and 778 U/mg, respectively (Table 2). GghA showed higher activity towards aryl-glucosides in which the nitrophenol group was attached to the ortho rather than the para position. In addition, GghA hydrolyzed pNPC and pNPL, and the lower rate of pNP release compared to the release of reducing sugars suggests that the attack on the substrate takes place from the nonreducing end of the glycoside. To determine the true substrate specificity of the enzyme, GghA was tested for its ability to hydrolyze naturally occurring disaccharides, cellobiose and lactose. The specific activity towards lactose (251 U/mg) was marginally higher than that towards cellobiose (236 U/mg). In view of the organism's ability to grow on glucans, and to establish the precise role for GghA in the glucan catabolic pathway, the kinetic constants for hydrolysis of glucooligosaccharides were determined. Comparable kcat values of 345.7 and 333.7 s−1 were found for cellotriose and cellotetraose, respectively (Table 3). The Km for cellotetraose (2.15 mM), however, is significantly lower than that for cellotriose (4.55 mM), resulting in a twofold increase in the kcat/Km value with cellotetraose as the substrate. Interestingly, the Km for cellobiose is 13-fold higher than the Km for cellotetraose, indicating a weak interaction between the enzyme and cellobiose (Table 3).
TABLE 2.
Substrate specificity of T. neapolitana glucan glucohydrolase towards aryl-glycosides
| Substrate | Sp acta (U/mg) | Relative activity (%) |
|---|---|---|
| oNPG | 807 | 100 |
| oNPGal | 778 | 96.4 |
| pNPG | 664 | 82.3 |
| pNPL | 260 | 32.2 |
| pNPC | 225 | 27.9 |
| pNPX | 117 | 14.5 |
| pNPA | 86 | 10.6 |
| pNPαG | 0 | 0 |
Determined with 5 mM (final concentration) of aryl-glycosides as the substrate.
TABLE 3.
Kinetic parameters of T. neapolitana GghA towards glucooligosaccharides and pNPG
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Sugarsa | |||
| Cellobiose | 28.6 | 285.04 | 9.96 |
| Cellotriose | 4.55 | 345.71 | 75.98 |
| Cellotetraose | 2.15 | 333.68 | 155.2 |
| pNPb | |||
| Hydrolysis (low [S] = 0 to 2 mM) | 0.28 | 512.55 | 1,830.54 |
| Transglycosylation (high [S] = 2 to 40 mM) | 1.93 | 883.9 | 457.97 |
Activity determined by analysis of products of hydrolysis using HPLC (see text).
Activity determined by measuring the rate of release of pNP.
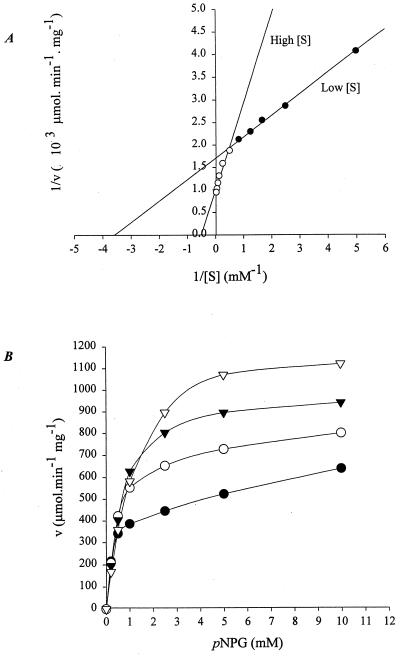
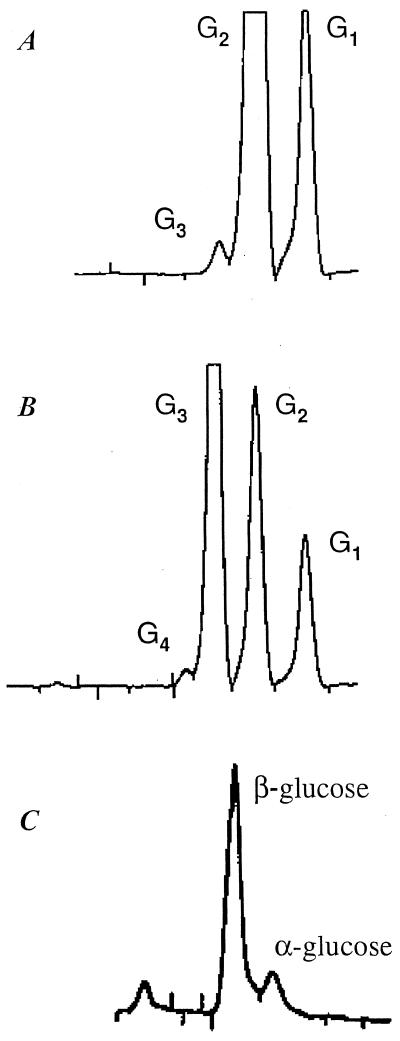
The kinetics for the hydrolysis of pNPG by GghA is described in Fig. 2A. The double reciprocal plot of v versus [S] shows a biphasic pattern. The inflection point on the curve, at a substrate concentration of 2 mM, suggests the binding of an additional substrate molecule. The Michaelis-Menten equation was used to calculate the kinetic constants for both low (0 to 2 mM) and high (2 to 40 mM) substrate concentrations by fitting the data at the extremes of v and [S]. At lower substrate concentrations (0.2 to 2.0 mM pNPG), the Km and kcat are 0.28 mM and 512.55 s−1, respectively, while at higher concentrations of the substrate (2 to 40 mM), both Km and kcat increase, to 1.93 mM and 883.9 s−1, respectively (Table 3). The biochemical basis for such biphasic kinetic behavior was investigated by analyzing the effect of the end product (glucose, 0 to 200 mM) on GghA's hydrolytic activity towards pNPG. Surprisingly, glucose acts as an activator even up to a concentration of 200 mM, and there was a direct relationship between glucose concentration and the rate of activation (Fig. 2B). End product inhibition was observed only at glucose concentrations of >200 mM. The activation of GghA both at higher substrate concentrations (2 to 40 mM pNPG) and in the presence of end product glucose (0 to 200 mM) may be due to transglycosylation activity of GghA, where the substrate or end product can act as the preferential glycosyl acceptor compared to water. To test the possibility of simultaneous occurrence of hydrolysis and transglycosylation, GghA was reacted with 40 mM cellobiose and 16 mM cellotriose, and the products were fractionated by HPLC. Figure 3A and B show the production of cellotriose from cellobiose and cellotetraose from cellotriose, respectively. Moreover, cellopentaose was produced when GghA was reacted with 16 mM cellotetraose (data not shown). The stereochemistry of the newly formed chiral center was assayed using purified GghA enzyme under conditions in which the mutarotation is negligible (see Materials and Methods). The results show that GghA retains the anomeric configuration at the C-1 atom of the glucoside (Fig. 3C).
FIG. 2.
Effect of substrate (pNPG) and end product (glucose) on activity of GghA. (A) Lineweaver-Burke plot of rate of pNP release as a function of pNPG concentration. (B) Effect of end product (glucose) on rate of pNP release from pNPG. ●, v(U/mg)@[I] = 0; ○, v(U/mg)@[I] = 50; ▾, v(U/mg)@[I] = 100; ▿, v(U/mg)@[I] = 200. v(U/mg)@[I] = specific activity at various concentrations of inhibitor.
FIG. 3.
HPLC analysis of products of hydrolysis and transglycosylation of GghA. (A) Production of cellotriose (G3) and glucose (G1) from cellobiose (G2). (B) Production of cellotetraose (G4), cellobiose (G2), and glucose (G1) from cellotriose (G3). (C) Determination of the configuration at the newly formed anomeric center.
Purification of recombinant cellobiose phosphorylase (CbpA).
Recombinant CbpA was purified by a three-step protocol, resulting in 17-fold purification. The specific activity of the purified enzyme is 11.8 U/mg towards cellobiose. SDS-PAGE analysis showed a homogeneous enzyme preparation with an apparent molecular mass of 93 kDa (Fig. 1B), which is in agreement with sequence-derived data.
Characterization of CbpA.
CbpA showed optimal activity at 85°C over a pH range of 4.5 to 6.0. Its pI was 4.8. The thermostability of CbpA was affected by the presence or absence of substrate: when incubated with substrate at 85°C, CbpA remained active for 2 h; when incubated without substrate, there was little activity after 15 min. Without phosphate in the reaction mixture, CbpA failed to hydrolyze the substrate. CbpA shows activity only towards cellobiose; it was not active towards cellotriose or the disaccharides lactose, chitobiose, and xylobiose. The kinetic constants were determined for the phosphorolysis of cellobiose by fitting data to the Michaelis-Menten equation using linear regression: the Km for cellobiose was 1.42 mM, the kcat was 26.3 s−1, and kcat/Km was 18.3 s−1 mM−1.
Cloning, sequencing, and analysis of the glucan catabolic cluster.
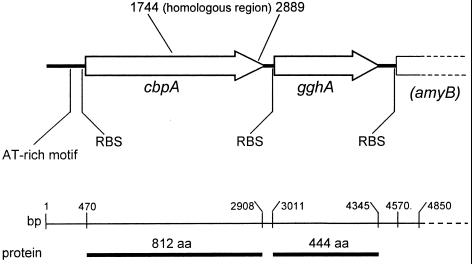
Preliminary DNA sequencing and analysis were conducted on the inserts from eight clones identified in a prior screening protocol (7). Of these clones, p46B1 contained the partial sequence of the gghA gene and an incomplete upstream ORF with similarities to cellobiose phosphorylases. Together, the two genes constitute a cluster involved, in all likelihood, in the catabolism of glucooligosaccharides. To define this cluster, two cosmid clones, RUC 270 and RUC 278, with similar restriction maps were identified by screening the T. neapolitana cosmid library. An insert within clone RUC 270 was subcloned to obtain the full-length gghA gene, and primer walking was used to completely sequence the cellobiose phosphorylase (cbpA) gene. cbpA and gghA, encoding 812 and 444 amino acid proteins, respectively, are separated by a 103-bp intergenic region (Fig. 4). The structural organization of the cluster consisting of cbpA and gghA was confirmed by Southern blotting. Further analysis of the sequence 3′ to the gghA gene revealed the presence of a partial ORF encoding a putative α-amylase (amyB); and 5′ to the cbpA gene, several short AT-rich sequence motifs are present, one of which has 95% identity over its 23-bp length (Fig. 4) to motifs present 5′ of endoglucanase clusters from T. maritima and T. neapolitana (10, 25). Such regions may be involved in the regulation of glucan catabolic gene clusters.
FIG. 4.
Molecular organization of oligosaccharide catabolic cluster. Arrows indicate the direction of gene transcription. cbpA, cellobiose phosphorylase; gghA, glucan glucohydrolase; amyB, partial ORF of putative α-amylase; AT-rich region, conserved-sequence motif present upstream of other endoglucanase genes in Thermotoga spp.; RBS, ribosome-binding site; bases 1744 to 2889, domain common to C. thermocellum cellodextrin phosphorylase (1, 38); aa, amino acids.
Based on amino acid sequence comparisons, cellobiose and cellodextrin phosphorylases form a homologous group of proteins. Interestingly, CbpA is 84.7, 82.5, and 76.9% similar to the cellobiose phosphorylases of the cellulolytic organisms C. thermocellum (GenBank accession no. AB013109), Clostridium stercorarium (30), and Cellvibrio gilvus (26), respectively. It is also noted that the cellobiose phosphorylases, including T. neapolitana CbpA, have >50% similarity to the C-terminal domains of R. meliloti NdvB (21) and B. abortus Cgs proteins, which are responsible for β-(1,2) glucan synthetase activity (22). The similarity in primary sequence between glucan synthetic enzymes and phosphorolytic enzymes with significant transglycosylation activity indicates common biochemical mechanisms in these functionally diverse groups of enzymes.
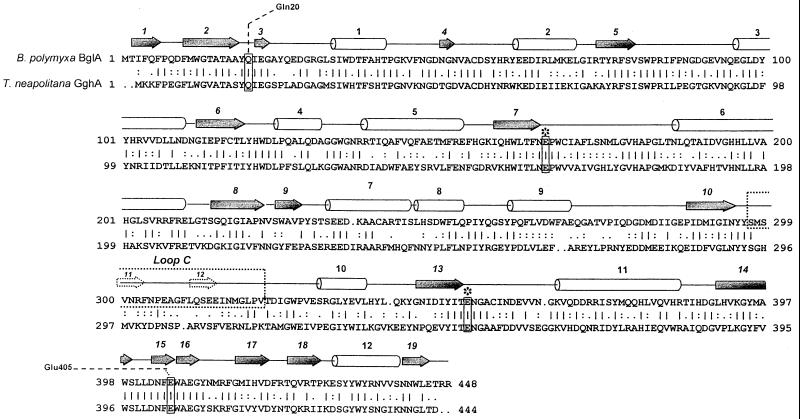
The GghA protein belongs to family 1 of glycosyl hydrolases, containing mainly β-glucosidases, phospho-β-glucosidases, and phospho-β-galactosidases (16). Sequence alignment of T. neapolitana gghA and the Bacillus polymyxa β-glucosidase gene (bglA) (33), shown in Fig. 5, reveals 68% sequence similarity and 46% identity over their entire length. The crystal structure of B. polymyxa BglA has been determined, and those structural data have been used to delineate its secondary structural elements. Conserved active-site glutamates, E166 and E342, are located at the ends of β-sheets 7 and 13, respectively. Of particular interest are Gln-20, conserved in all members of family 1, and Glu-405, conserved among glycosidases but not phosphoglycosidases, both of which are capable of forming bidentate hydrogen bonds and also loop C (amino acids 296 to 327), which is involved in substrate binding, including glucooligomers above G2. The sequences of B. polymyxa BglA loop C and the homologous region in T. neapolitana GghA exhibit 52% similarity.
FIG. 5.
Comparison of primary sequence of GghA with that of β-glucosidase from B. polymyxa. The secondary-structure elements were defined using the crystal structure of B. polymyxa β-glucosidase (33). Barrel, helix; arrow, β-strand; dashed arrow, secondary structure within loop C; ∗, conserved active-site amino acids; Gln20 and Glu405, amino acids capable of bidentate hydrogen bonding.
DISCUSSION
We report the purification, characterization, and cloning of a cellobiose phosphorylase and a β-glucan glucohydrolase cluster from T. neapolitana. Few cellobiose phosphorylases or glucan glucohydrolases have been defined at the biochemical and molecular level; this may be due in part to the lack of methods for direct screening and selection of these enzymes. For instance, characterization of an enzyme as a glucan glucohydrolase requires HPLC analysis of the products of hydrolysis of glucooligomers. Also, no direct colorimetric or fluorometric methods are available for analysis of cellobiose phosphorylase activity.
GghA was purified from T. neapolitana extracts using a multistep purification protocol, as Thermotoga spp. have the potential to produce a number of glycosyl hydrolases (7, 8, 10, 11, 23, 37). Purified GghA has broad substrate specificity, with similar specific activities towards the disaccharides lactose and cellobiose, reflecting the polyspecific nature of family 1 glycosyl hydrolases to which it belongs (15). GghA, like other family 1 enzymes, hydrolyzes various aryl-β-glycosides, but it also exhibits biphasic kinetic behavior towards pNPG: its activity increases at higher substrate concentrations (Fig. 2A). This apparent activation may be due to higher concentrations of substrate or the de novo end product or both. Our data show the involvement of both substrate-directed and end product-directed activation, suggesting that GghA's greater kinetic activity at higher substrate or end product concentrations is due to its transglycosylating capability. For example, the production of oligomers of N + 1 length by GghA (N = length of substrate) is indicative of both substrate-induced and end product-activated (glucose concentrations up to 200 mM) transglycosylation (Fig. 3A and B). This effect is similar to that reported for a cytosolic β-glucosidase from guinea pig liver (14). Although similar end product activation of glucoside hydrolases has been reported for a β-glucosidase from a Streptomyces sp. (29), and a family 1 glycosyl hydrolase from Microbispora bispora (44), the biochemical basis was not defined. While transglycosylation clearly plays a role in activation of GghA, our data do not rule out other mechanisms of activation operating in concert with transglycosylation.
T. neapolitana GghA was identified initially as an aryl-β-glycosidase based on its activity towards pNPG, an activity it shares with other glucan glucohydrolases (17, 18, 28, 31). The basis for designating T. neapolitana GghA a glucan glucohydrolase (EC 3.2.1.74) instead of a β-glucosidase (EC 3.2.1.21) is its greater catalytic efficiency towards cellotetraose: the kcat/Km for GghA's hydrolysis of cellotetraose is 16-fold greater than that for cellobiose, while its overall substrate preference is cellotetraose > cellotriose > cellobiose. GghA's kcat values for cellotetraose (333.6 s−1) and cellotriose (345.7 s−1) are comparable, indicating that the rate of formation of the covalent glucosyl-enzyme intermediate and subsequent release of glucose from the complex are essentially the same for both cellooligomers. The Km for cellotetraose, however, is half that of cellotriose, implying a better fit for the enzyme with the longer substrate. Indeed, it can be suggested that GghA has at least four binding subsites, all of which, in the case of cellotetraose, interact with the substrate and result in the strongest enzyme-substrate interaction.
Such tandem multisugar binding sites and a tendency towards greater catalytic efficiency (kcat/Km) with increasing degrees of substrate polymerization have been reported for a family 1 glucan glucohydrolase (βII) from barley (19), an exoglucosidase from the archaeon Sulfolobus solfataricus (28), and a β-glucosidase (BglA) from B. polymyxa (33). Loop C in B. polymyxa BglA (amino acids 297 to 328) (Fig. 5), for example, has been specifically identified both with substrate binding and with providing additional subsites for the binding of larger glucooligomers. Given the high level of primary sequence conservation between T. neapolitana GghA and B. polymyxa BglA (68% sequence similarity and 46% identity over the entire length), it is not surprising that GghA shows greater catalytic efficiency towards glucooligomers longer than cellobiose. In light of GghA's activity towards cellobiose and lactose, it is interesting that conserved amino acids Gln-20 and Glu-405, which enable the enzyme to form bidentate hydrogen bonds with oxygen atoms of glycosides, may account for the dual glycosidase and galactosidase activity of family 1 members, including GghA from T. neapolitana (33, 40).
From a bioenergetic perspective, cellobiose phosphorylases are interesting enzymes, for the energy of the β-1,4 glycosidic bond is conserved during phosphorolysis in the form of glucose 1-phosphate. One ATP molecule is needed to activate the other glycosyl product of the reaction. Glucose 1-phosphate is then converted to glucose 6-phosphate by phosphoglucomutase, a non-energy-consuming isomerization. Conversely, when a β-glucosidase hydrolyzes cellobiose, glycosidic bond energy is lost, and activation of the two glycosyl products requires two ATP molecules rather than one. To date, only cellobiose phosphorylases from C. gilvus (34), C. thermocellum (1, 38), and C. stercorarium (30) have been purified and characterized. The CbpA from T. neapolitana is the first report of a cellobiose phosphorylase from any hyperthermophile.
T. neapolitana and T. maritima are closely related species with minor differences in their physiological characteristics (20). A comparison of their glycosyl hydrolases, made possible by recent publication of the T. maritima genome (27), reveals that both gene sequences and the arrangement of catabolic gene clusters are highly conserved. Notably, both species apparently lack a gene encoding a cellobiohydrolase (27; D. Y. Yernool, G. Swiatek, and J.-D. Bok, unpublished data). The absence of the enzyme may explain the inability of Thermotoga spp. to hydrolyze crystalline substrates such as Avicel despite their capacity for hydrolysis of noncrystalline substrates such as acid-swollen cellulose and barley β-glucan. As cellobiohydrolases play a key role in classical cellulolysis and as sugar catabolism has been studied in detail in several hyperthermophilic genera, including Thermotoga (35), we propose a previously unreported catabolic pathway for glucan utilization. This nonconventional, ATP-conserving pathway consists of endoglucanases (CelA and CelB) (7), a β-glucan glucohydrolase (GghA), and a cellobiose phosphorylase (CbpA) (Fig. 6). Other potential components of this catabolic pathway might include a laminarinase (LamA) and a laminaribiase/β-glucosidase (BglB), which act on both β-1,3 and β-1,4 linkages (45, 46).
FIG. 6.
Proposed glucan catabolic pathway in T. neapolitana. A schematic conversion of cellooligomers to activated glucosyl molecules. Enzymes indicated are CelA and CelB, endoglucanases (7) active in or on the toga; GghA, glucan glucohydrolase, and CbpA, cellobiose phosphorylase, intracellular enzymes (this work); and LamA and BglB, laminarinase and β-glucosidase, respectively (45, 46).
Hydrolysis of internal glycosidic linkages of polymeric glucans by endoglucanases CelA and CelB begins the attack and results in the production of soluble glucooligomers. As CelB has an N-terminal signal peptide sequence, this initial processing probably occurs in or on the toga, the proteinaceous outer sack covering all Thermotogales. That neither CelB nor CelA is secreted but is extracted from the pellet when purified from T. neapolitana grown on cellobiose (6) further suggests that initial activity involves the toga. The subcellular location of CelA, however, is not clear, as it lacks a secretory signal peptide sequence. The soluble products of the initial hydrolysis are then acted on by GghA, presumably an intracellular enzyme, which preferentially attacks the longer glucooligomers cellotriose to cellohexaose (the longer the glucooligomers, the lower their Kms), releasing glucose from the nonreducing end and eventually producing the disaccharide intermediate cellobiose. At low concentrations of cellobiose (CbpA's Km for cellobiose is 1.42 mM, compared to 28.6 mM for GghA), and given the enzyme's preference for a single substrate, CbpA, also presumably intracellular, then attacks cellobiose, producing the activated molecule glucose 1-phosphate and glucose.
Some cellobiose might be processed by other resident glycosyl hydrolases; however, the Km of BglB (46) for cellobiose (50 mM) is 35-fold greater than that of CbpA, a strong indication that CbpA may be the primary enzyme for processing cellobiose in T. neapolitana. Further support for inclusion of CbpA in this pathway is the enzyme's energy-conserving mode of action: when cellobiose is converted to its glycosyl substituents by CbpA, one fewer ATP molecule is expended for every cellobiose molecule processed. For an organism with fermentative metabolism such as T. neapolitana, this represents a significant metabolic advantage. It is interesting that if cellobiohydrolase were present, the energy conserved would be greater, as there would be higher yields of cellobiose per molecule of glucooligosaccharide processed. Other important questions that bear on this issue include the type of glucans and polysaccharides present in the organism's ecological niche and what role, if any, other microbes play in complementing the degradative capability of Thermotogales. Finally, a number of genes encoding putative glycosyl hydrolases have been described in T. maritima (27). Functional analyses of these gene products and studies on regulation of their expression will undoubtedly further our understanding of the process of acquisition of carbon and energy by Thermotogales.
ACKNOWLEDGMENTS
D. A. Yernool and J. K. McCarthy contributed equally to this work.
This research was supported by U.S.D.A. National Research Initiative Competitive Grants Program grant 97-35503-4557, NJ Marine Sciences Consortium grant B/T-12, and McIntire-Stennis grant 0181520. J.K.M. was supported by the National Institutes of Health Biotechnology Training Program.
Footnotes
New Jersey Agricultural Experiment Station publication no. D-01111-01-00.
REFERENCES
- 1.Alexander J K. Purification and specificity of cellobiose phosphorylase from Clostridium thermocellum. J Biol Chem. 1968;243:2899–2904. [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Baker J O, Himmel M E. Separation of sugar anomers by aqueous chromatography on calcium and lead-form ion-exchange columns—application to anomeric analysis of enzyme reaction products. J Chromatogr. 1986;357:161–181. [Google Scholar]
- 4.Barr B K, Hsieh Y L, Ganem B, Wilson D B. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35:586–592. doi: 10.1021/bi9520388. [DOI] [PubMed] [Google Scholar]
- 5.Beguin P, Aubert J P. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 6.Bibel M, Brettl C, Gosslar U, Kriegshauser G, Liebl W. Isolation and analysis of genes for amylolytic enzymes of the hyperthermophilic bacterium Thermotoga maritima. FEMS Microbiol Lett. 1998;158:9–15. doi: 10.1111/j.1574-6968.1998.tb12793.x. [DOI] [PubMed] [Google Scholar]
- 7.Bok J D, Yernool D A, Eveleigh D E. Purification, characterization, and molecular analysis of thermostable cellulases CelA and CelB from Thermotoga neapolitana. Appl Environ Microbiol. 1998;64:4774–4781. doi: 10.1128/aem.64.12.4774-4781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronnenmeier K, Kern A, Liebl W, Staudenbauer W L. Purification of Thermotoga maritima enzymes for the degradation of cellulosic materials. Appl Environ Microbiol. 1995;61:1399–1407. doi: 10.1128/aem.61.4.1399-1407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Childers S E, Noll K M. Characterization and regulation of sulfur reductase activity in Thermotoga neapolitana. Appl Environ Microbiol. 1994;60:2622–2626. doi: 10.1128/aem.60.7.2622-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dakhova O N, Kurepina N E, Zverlov V V, Svetlichnyi V A, Velikodvorskaya G A. Cloning and expression in Escherichia coli of Thermotoga neapolitana genes coding for enzymes of carbohydrate substrate degradation. Biochem Biophys Res Commun. 1993;194:1359–1364. doi: 10.1006/bbrc.1993.1974. [DOI] [PubMed] [Google Scholar]
- 11.Duffaud G D, McCutchen C M, Leduc P, Parker K N, Kelly R M. Purification and characterization of extremely thermostable beta-mannanase, beta-mannosidase, and alpha-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl Environ Microbiol. 1997;63:169–177. doi: 10.1128/aem.63.1.169-177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill S C, von Hippel P H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 13.Giulian G G, Moss R L, Greaser M. Improved methodology for analysis and quantitation of proteins on one-dimensional silver-stained slab gels. Anal Biochem. 1983;129:277–287. doi: 10.1016/0003-2697(83)90551-1. [DOI] [PubMed] [Google Scholar]
- 14.Hays W S, VanderJagt D J, Bose B, Serianni A S, Glew R H. Catalytic mechanism and specificity for hydrolysis and transglycosylation reactions of cytosolic beta-glucosidase from guinea pig liver. J Biol Chem. 1998;273:34941–34948. doi: 10.1074/jbc.273.52.34941. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280(Pt. 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrissat B. Glycosidase families. Biochem Soc Trans. 1998;26:153–156. doi: 10.1042/bst0260153. [DOI] [PubMed] [Google Scholar]
- 17.Himmel M E, Tucker M P, Lastick S M, Oh K K, Fox J W, Spindler D D, Grohmann K. Isolation and characterization of a 1,4-beta-d-glucan glucohydrolase from the yeast Torulopsis wickerhamii. J Biol Chem. 1986;261:12948–12955. [PubMed] [Google Scholar]
- 18.Hrmova M, Harvey J, Wang J, Shirley N J, Jones G P, Stone B A, Hoj P B, Fincher G B. Barley beta-d-glucan exohydrolases with beta-d-glucosidase activity—purification, characterization, and determination of primary structure from a cDNA clone. J Biol Chem. 1996;271:5277–5286. doi: 10.1074/jbc.271.9.5277. [DOI] [PubMed] [Google Scholar]
- 19.Hrmova M, Macgregor E A, Biely P, Stewart R J, Fincher G B. Substrate binding and catalytic mechanism of a barley beta-d-glucosidase/(1,4)-beta-d-glucan exohydrolase. J Biol Chem. 1998;273:11134–11143. doi: 10.1074/jbc.273.18.11134. [DOI] [PubMed] [Google Scholar]
- 20.Huber R, Stetter K, editors. The prokaryotes. 2nd ed. IV. New York, N.Y: Springer-Verlag; 1992. [Google Scholar]
- 21.Ielpi L, Dylan T, Ditta G S, Helinski D R, Stanfield S W. The ndvB locus of Rhizobium meliloti encodes a 319-kDa protein involved in the production of beta-(1----2)-glucan. J Biol Chem. 1990;265:2843–2851. [PubMed] [Google Scholar]
- 22.Inon de Iannino N, Briones G, Tolmasky M, Ugalde R A. Molecular cloning and characterization of cgs, the Brucella abortus cyclic beta (1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J Bacteriol. 1998;180:4392–4400. doi: 10.1128/jb.180.17.4392-4400.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King M R, Yernool D A, Eveleigh D E, Chassy B M. Thermostable alpha-galactosidase from Thermotoga neapolitana—cloning, sequencing and expression. FEMS Microbiol Lett. 1998;163:37–42. doi: 10.1111/j.1574-6968.1998.tb13023.x. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Liebl W, Ruile P, Bronnenmeier K, Riedel K, Lottspeich F, Greif I. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology. 1996;142:2533–2542. doi: 10.1099/00221287-142-9-2533. [DOI] [PubMed] [Google Scholar]
- 26.Liu A M, Tomita H, Li H B, Miyaki H, Aoyagi C, Kaneko S, Hayashi K. Cloning, sequencing and expression of the cellobiose phosphorylase gene of Cellvibrio gilvus. J Ferment Bioeng. 1998;85:511–513. [Google Scholar]
- 27.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson L D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, White O, Fraser C M, et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 28.Nucci R, Moracci M, Vaccaro C, Vespa N, Rossi M. Exo-glucosidase activity and substrate specificity of the beta-glycosidase isolated from the extreme thermophile Sulfolobus solfataricus. Biotechnol Appl Biochem. 1993;17:239–250. [PubMed] [Google Scholar]
- 29.Perez-Pons J A, Rebordosa X, Querol E. Properties of a novel glucose-enhanced beta-glucosidase purified from Streptomyces sp. (ATCC 11238) Biochim Biophys Acta. 1995;1251:145–153. doi: 10.1016/0167-4838(95)00074-5. [DOI] [PubMed] [Google Scholar]
- 30.Reichenbecher M, Lottspeich F, Bronnenmeier K. Purification and properties of a cellobiose phosphorylase (CepA) and a cellodextrin phosphorylase (CepB) from the cellulolytic thermophile Clostridium stercorarium. Eur J Biochem. 1997;247:262–267. doi: 10.1111/j.1432-1033.1997.00262.x. [DOI] [PubMed] [Google Scholar]
- 31.Rixon J E, Ferreira L M, Durrant A J, Laurie J I, Hazlewood G P, Gilbert H J. Characterization of the gene celD and its encoded product 1,4-beta-d-glucan glucohydrolase d from Pseudomonas fluorescens subsp. cellulosa. Biochem J. 1992;285:947–955. doi: 10.1042/bj2850947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanz-Aparicio J, Hermoso J A, Martinez-Ripoll M, Gonzalez B, Lopez-Camacho C, Polaina J. Structural basis of increased resistance to thermal denaturation induced by single amino acid substitution in the sequence of beta-glucosidase A from Bacillus polymyxa. Proteins. 1998;33:567–576. doi: 10.1002/(sici)1097-0134(19981201)33:4<567::aid-prot9>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki T, Tanaka T, Nakagawa S, Kainuma K. Purification and properties of Cellvibrio gilvus cellobiose phosphorylase. Biochem J. 1983;209:803–807. doi: 10.1042/bj2090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schonheit P, Schafer T. Metabolism of hyperthermophiles. World J Microbiol Biotechnol. 1995;11:26–57. doi: 10.1007/BF00339135. [DOI] [PubMed] [Google Scholar]
- 36.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sunna A, Moracci M, Rossi M, Antranikian G. Glycosyl hydrolases from hyperthermophiles. Extremophiles. 1997;1:2–13. doi: 10.1007/s007920050009. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka K, Kawaguchi T, Imada Y, Ooi T, Arai M. Purification and properties of cellobiose phosphorylase from Clostridium thermocellum. J Ferment Bioeng. 1995;79:212–216. [Google Scholar]
- 39.Teeri T T. Crystalline cellulose degradation—new insight into the function of cellobiohydrolases. Trends Biotechnol. 1997;15:160–167. [Google Scholar]
- 40.Vyas N K. Atomic features of protein carbohydrate interactions. Curr Opin Struct Biol. 1991;1:732–740. [Google Scholar]
- 41.Warren R A. Microbial hydrolysis of polysaccharides. Annu Rev Microbiol. 1996;50:183–212. doi: 10.1146/annurev.micro.50.1.183. [DOI] [PubMed] [Google Scholar]
- 42.Wood T M, Bhat K M. Methods for measuring cellulase activities. Methods Enzymol. 1988;160:87–112. [Google Scholar]
- 43.Wood T M, McCrae S I. Purification and some properties of a (1-4)β-d-glucan glucohydrolase associated with the cellulase from the fungus Penicillium funiculosum. Carbohydr Res. 1982;110:291–303. [Google Scholar]
- 44.Wright R M, Yablonsky M D, Shalita Z P, Goyal A K, Eveleigh D E. Cloning, characterization, and nucleotide sequence of a gene encoding Microbispora bispora BglB, a thermostable beta-glucosidase expressed in Escherichia coli. Appl Environ Microbiol. 1992;58:3455–3465. doi: 10.1128/aem.58.11.3455-3465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zverlov V V, Volkov I Y, Velikodvorskaya T V, Schwarz W H. Highly thermostable endo-1,3-beta-glucanase (laminarinase) LamA from Thermotoga neapolitana—nucleotide sequence of the gene and characterization of the recombinant gene product. Microbiology. 1997;143:1701–1708. doi: 10.1099/00221287-143-5-1701. [DOI] [PubMed] [Google Scholar]
- 46.Zverlov V V, Volkov I Y, Velikodvorskaya T V, Schwarz W H. Thermotoga neapolitana bglB gene, upstream of lamA, encodes a highly thermostable beta-glucosidase that is a laminaribiase. Microbiology. 1997;143:3537–3542. doi: 10.1099/00221287-143-11-3537. [DOI] [PubMed] [Google Scholar]