Abstract
Thiamine pyrophosphate is an essential cofactor that is synthesized de novo in Salmonella enterica serovar Typhimurium and other bacteria. In addition to genes encoding enzymes in the biosynthetic pathway, mutations in other metabolic loci have been shown to prevent thiamine synthesis. The latter loci identify the integration of the thiamine biosynthetic pathway with other metabolic processes and can be uncovered when thiamine biosynthesis is challenged. Mutations in gshA, encoding γ-l-glutamyl-l-cysteine synthetase, prevent the synthesis of glutathione, the major free thiol in the cell, and are shown here to result in a thiamine auxotrophy in some of the strains tested, including S. enterica LT2. Phenotypic characterization of the gshA mutants indicated they were similar enough to apbC and apbE mutants to warrant the definition of a class of mutants unified by (i) a requirement for both the hydroxymethyl pyrimidine (HMP) and thiazole (THZ) moiety of thiamine, (ii) the ability of l-tryosine to satisfy the THZ requirement, (iii) suppression of the thiamine requirement by anaerobic growth, and (iv) suppression by a second-site mutation at a single locus. Genetic data indicated that a defective ThiH generates the THZ requirement in these strains, and we suggest this defect is due to a reduced ability to repair a critical [Fe-S] cluster.
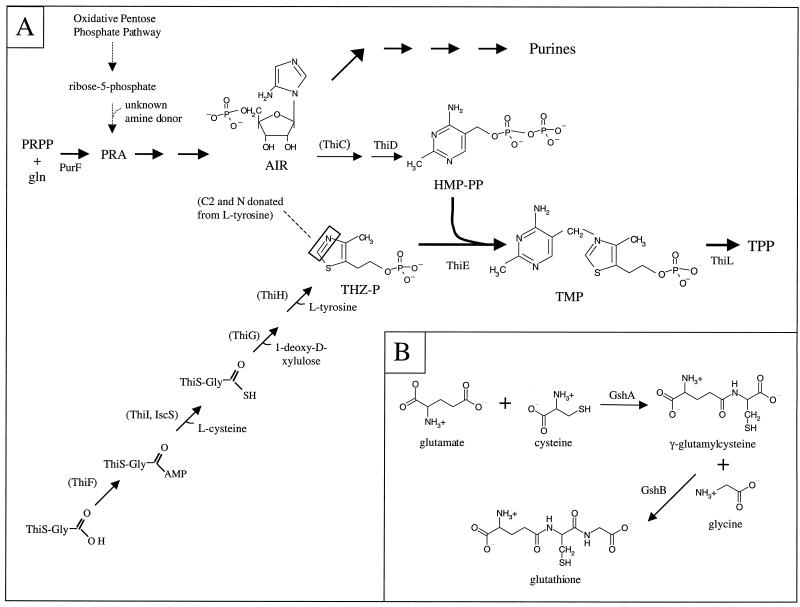
Thiamine pyrophosphate (TPP) is an essential cofactor for several enzymes, including pyruvate dehydrogenase, transketolase, and acetolactate synthase, for which it functions as a carbon unit carrier and electron sink. TPP is composed of two independently synthesized moieties, 4-methyl-5-β-hydroxyethyl-thiazole monophosphate (THZ-P) and 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate (HMP-PP). Although several recent studies have increased our understanding of the synthesis of TPP in Escherichia coli and Salmonella enterica (5, 31, 39, 46, 51, 52, 53), many central issues have yet to be resolved. The current understanding of the substrates and enzymes in TPP synthesis are outlined in Fig. 1A (6, 15, 20–23, 43, 54, 55). Significantly, l-Tyr donates a single carbon and the nitrogen atom to the thiazole (THZ) ring.
FIG. 1.
Biosynthetic pathways for thiamine and GSH. (A) Our current understanding of thiamine biosynthesis in S. enterica. Enzymes for each of the reactions are indicated above the relevant arrows. Enzymes that have not been demonstrated but are predicted to function at certain steps in thiamine biosynthesis are in parentheses. The nitrogen and carbon atoms donated by tyrosine are boxed in the THZ structure. The position of ThiH with respect to ThiG, -F, and -S is based partially on work reported here. (B) The GSH biosynthetic pathway. Abbreviations: PRPP, phosphoribosylpyrophosphate; Gln, glutamine; AIR, 5-aminoimidazole ribotide; THZ-P, 4-methyl-5-β-hydroxyethyl-thiazole monophosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate; TMP, thiamine monophosphate; TPP, thiamine pyrophosphate.
Of the five genes implicated in the synthesis of THZ-P in S. enterica and E. coli (5, 49, 51), the products of thiFSGI have been assigned functions based on in vitro activity assays (31, 46, 47). However, the order of the steps and the specific metabolites in this biosynthesis have not been rigorously defined. The precursor for the pyrimidine (HMP) moiety was shown to be the purine biosynthetic intermediate, aminoimidazole ribotide (AIR) (22). The steps involved in the conversion of AIR to HMP are unknown, and to date, mutations in only one gene, thiC, have resulted in an unconditional requirement for HMP or thiamine.
Past work showed that mutations in several loci distinct from the thi genes could result in a thiamine auxotrophy. Characterization of several mutations that indirectly affected thiamine synthesis has increased our understanding of thiamine synthesis and its integration with other metabolic processes (12, 13, 17, 18, 24, 25, 40, 42; J. Zilles, J. Kappock, J. Stubbe, and D. M. Downs, unpublished data). Mutations that affected thiamine synthesis often resulted in phenotypes dependent on growth conditions and strain backgrounds. Functional characterization of two of these loci in particular, apbE and apbC, has been difficult since each contains a novel open reading frame (ORF) and no significant functional information has been uncovered by sequence analyses. ApbE is a periplasmic lipoprotein (3, 4), and apbC (mrp) encodes a protein of unknown function (38) that has been implicated in a variety of metabolic processes (J. Escalante-Semerena, J. R. Roth, and R. LaRossa, personal communication). Both apbE and apbC were required for thiamine synthesis under some conditions, and lesions in either of these genes resulted in a requirement for both the HMP and THZ moieties of thiamine for growth (4).
Here we show that organisms of a laboratory strain of S. enterica with mutations in the gshA gene, encoding γ-l-glutamyl-l-cysteine synthetase (EC 6.3.2.2), are thiamine auxotrophs. The gshA mutants displayed phenotypes similar to apbC and apbE mutants, including (i) a requirement for both HMP and THZ, (ii) the ability of l-Tyr to satisfy the THZ requirement, (iii) the suppression of the thiamine requirement by anaerobic growth, and (iv) the suppression by second-site mutations at a single locus. GshA is one of two gene products that are required for the synthesis of glutathione (GSH), the major free thiol in the cell (Fig. 1B). The extensive literature on GshA and the role of GSH in metabolism, the demonstration that mutant strains defective in [Fe-S] cluster formation or repair have similar THZ phenotypes (i to iv above) (42), and the work described here led to a model to explain the THZ requirement in these strains (27, 37). Results from genetic experiments here are consistent with the THZ requirement in this group of strains (gshA, apbE, and apbC) being caused by inhibition of ThiH activity. We suggest that ThiH contains an [Fe-S] cluster, and in these mutants, synthesis and/or repair of this cluster is defective. This defect results in a loss of ThiH activity and compromises the ability of the cell to synthesize THZ. The specific defect resulting in a requirement for HMP in these strains has not yet been addressed.
MATERIALS AND METHODS
Strains, media, and chemicals.
[35S]methionine (specific activity, 1,000 Ci/mmol) was purchased from New England Nuclear (Boston, Mass.). All other chemicals were purchased from Sigma (St. Louis, Mo.). Unless otherwise indicated, all strains used in this study are derivatives of S. enterica LT2 and are described in Table 1. MudJ refers to the Mud1734 insertion element (11), and Tn10d(Tc) refers to the transposition-defective mini-Tn10 described by Way et al. (50). NCE medium supplemented with MgSO4 (1 mM) (14) and glucose (11 mM) was used as minimal medium. Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) added was used as rich medium. Difco BiTek agar was added to a final concentration of 1.5% for solid medium. The final concentrations of antibiotics in rich medium (in micrograms/milliliter) were as follows: tetracycline (Tc), 20; kanamycin (Km), 50; ampicillin (Ap), 30; and chloramphenicol (Cm), 20.
TABLE 1.
Strains
| Strain | Genotype |
|---|---|
| LT2 | Wild type |
| LT7 | Wild type |
| Q1 | Wild type |
| DM271 | apbE42::Tn10d(Tc) |
| DM460 | thiH910::MudJ |
| DM3012 | thiL953::MudJ |
| DM4104 | zii-8039::Tn10d thiH1105 |
| DM4106 | zii-8039::Tn10d thiH1106 |
| DM4108 | zii-8039::Tn10d thiH1107 |
| DM4331 | gshA101::Tn10d(Tc) |
| DM4447 | BL21λDE3 pThiH-7(−) |
| DM4448 | BL21λDE3 pThiH-7(+) |
| DM4496 | gshB103::MudJa |
| DM4620 | gshA102::MudJ |
| DM4797 | zii-8039::Tn10d(Tc) thiH1106 thiG1113 |
| DM4801 | zii-8039::Tn10d(Tc) thiH1106 thiG1116 |
| DM4804 | zii-8039::Tn10d(Tc) thiH1106 thiFS1115 |
| DM5490 | apbE42::Tn10d(Tc) thiL953::MudJ |
| DM5537 | thiL953::MudJ thi1120(ΔthiCEFSGH) |
| DM5655 | (LT7) gshA101::Tn10d(Tc) |
| DM5669 | (LT7) apbE42::Tn10d(Tc) |
| DM5670 | (LT7) apbC55::Tn10d(Tc) |
| DM5747 | (Q1) apbC55::Tn10d(Tc) |
| DM5748 | (Q1) apbE42::Tn10d(Tc) |
| DM5749 | (Q1) gshA101::Tn10d(Tc) |
gshB103::MudJ was the gift of J. C. Escalante-Semerena.
Genetic methods. (i) Transduction methods.
The high-frequency generalized transducing mutant of bacteriophage P22 (HT105/1, int-201) (41) was used in all transductional crosses. The method for transduction and subsequent purification of transductants has been previously described (16).
(ii) Mutant isolation.
Mutations causing a thiamine auxotrophy in a purF mutant (DM1936) were isolated by insertion mutagenesis using one of two defective transposons, Tn10d(Tc) or MudJ, as has been described elsewhere (40). After reintroduction of the insertions into DM1936 (purF) and confirmation of an appropriate phenotype, the sequence adjacent to the insertion was determined (10). In subsequent experiments, the insertion mutations were transduced into other strains by selecting the antibiotic resistance associated with the insertion.
Mutations in the thi operon were isolated by local mutagenesis (29) with a strain carrying a Tn10d(Tc) transposon (DM851) 80% linked to the thiCEFSGH operon at minute 90 as the donor. Tetracycline-resistant transductants were scored for the appropriate thiamine phenotype. Strains that required THZ and were satisfied by l-Tyr were designated Thz* mutants. Temperature-sensitive mutants with lesions in the thiCEFSGH operon were isolated by transducing a strain carrying a polar insertion in thiC (DM95) to prototrophy at 30°C using a mutagenized P22 phage lysate. Prototrophic transductants were scored for a THZ requirement at 42°C. Double mutants were generated by transducing the temperature-sensitive mutants to tetracycline resistance with a phage lysate grown on the respective Thz* mutant. The double mutant strains were identified by their requirement for THZ or l-Tyr at 30°C and a requirement for THZ or thiamine at 42°C.
Molecular biology techniques.
Enzymes for use in molecular biology were purchased from Promega (Madison, Wis.) and used per the manufacturer's instructions. Sequencing was carried out by the University of Wisconsin-Madison Biotechnology Sequencing Center, and sequence alignments of thiH were performed using the SeqEd program (PE Biosystems, Foster City, Calif.).
Phenotypic analysis.
Procedures for testing nutritional requirements on solid medium by use of soft agar overlays and by use of growth curves have been described (12, 40).
Enzyme assay for aconitase.
Aconitase activity in crude cell extracts was assayed by the protocol of Gruer and Guest (28), as modified by Skovran and Downs (42). Specific activity was described in units/milligram of protein, where a unit was the change in absorbance at 240 nm per minute. Protein concentration was determined by the method of Bradford (8).
TMP synthesis in resting cells.
Cultures were grown overnight in a 50-ml volume of minimal medium supplemented with TPP (20 nM). Cells were pelleted and washed twice with cold, sterile double-distilled water. The final cell pellet was resuspended in 5 ml of minimal media. A sample (200 μl) of this cell suspension was used to inoculate tubes containing 5 ml of minimal media resulting in cultures with an A650 of 0.3 to 0.4. Following a 1-h incubation at 37°C in a shaking water bath, THZ and/or HMP were added to the cultures at the indicated concentrations. At various times, samples were removed, and cells were pelleted, resuspended in 300-μl of double-distilled water, and frozen in a dry ice-ethanol bath for measurement of thiamine monophosphate (TMP) and TPP pools.
TMP and TPP were extracted and assayed by a CNBr thiochrome derivatization procedure as has been described (19, 32, 36).
ThiH expression.
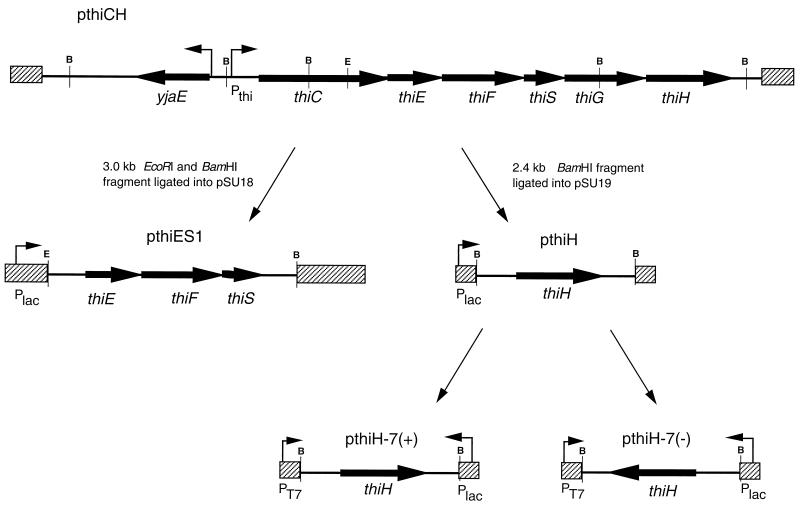
T7-RNAP-specific protein expression and [35S]methionine labeling of ThiH were performed as has been described (39, 44) with the plasmids pthiH-7(+) and pthiH-7(−) diagrammed in Fig. 6.
FIG. 6.
Plasmid construction. Plasmid pthiCH was isolated as described in the text. BamHI (B) digestion allowed individual subcloning of thiH on a 2.4-kb fragment, generating plasmid pthiH. The insert of pthiH was ligated into BamHI-cut pGEM7 (Promega), generating plasmids pthiH-7(+) and pthiH-7(−), with thiH in the correct orientation for expression from the T7 promoter and from the lac promoter, respectively. Plasmid pthiES1 was generated via BamHI and EcoRI (E) digestion of pthiCH and ligation into pSU18. Black arrows represent complete ORFs, and boxes represent vector DNA.
Nucleotide sequence accession number.
The sequence of the insert from plasmid pthiH was submitted to the GenBank database under accession no. AF154064.
RESULTS AND DISCUSSION
Mutants defective in gshA are thiamine auxotrophs.
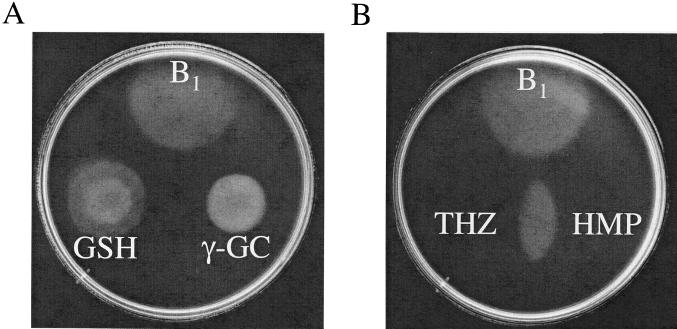
Screens to isolate thiamine auxotrophs in purF mutants identified insertion mutations in gshA, encoding γ-l-glutamyl-l-cysteine (γ-GC) synthetase (Fig. 1B). In each case, the causative lesions were sequenced, the mutants were reconstructed, and the phenotype of the strain was confirmed. Unlike a number of previously described mutations that were obtained in similar screens (i.e., nuo, gnd, and zwf), mutations in gshA also generated a thiamine auxotrophy in otherwise wild-type strains. As shown in Fig. 2A, thiamine-independent growth of gshA mutants was restored by the addition of GSH to the medium. This result suggested that the product of the GshA reaction, and not an additional activity of the protein, was involved in thiamine synthesis.
FIG. 2.
Growth requirements of gshA mutations in S. enterica. Soft agar containing strain DM4620 (gshA) was overlaid on minimal glucose medium. Compounds were spotted in 2-μl aliquots on the solidified agar as indicated, at the following concentrations: thiamine (B1), HMP, and THZ, 100 μM; GSH and γ-GC, 10 mM.
Glutathione or γ-glutamylcysteine is needed for thiamine synthesis.
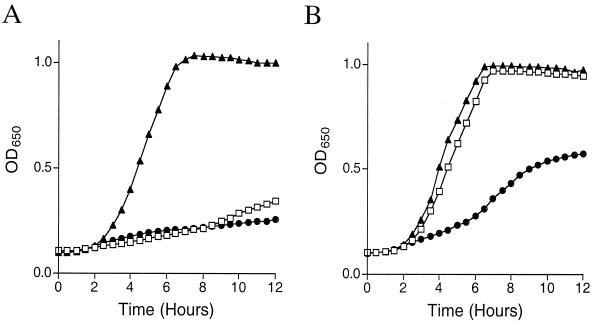
Although synthesis of GSH requires two enzymes, GshA and GshB (Fig. 1B), mutations in gshB did not generate a thiamine requirement in S. enterica strain LT2. Further, gshB mutants were shown to accumulate γ-GC (26), and the addition of this metabolite also restored thiamine-independent growth of a gshA mutant (Fig. 2A). Taken together, these results suggested that the thiamine requirement of gshA mutants was due to the loss of a prevalent cellular free thiol species, not a specific requirement for GSH. Consistently, gshA mutants had additional phenotypes not caused by gshB mutations. In E. coli, gshA mutants had reduced aconitase activity, which was suggested to result from an inability to repair oxidatively damaged [Fe-S] clusters in the absence of GSH (27). Cell extracts of wild-type, gshA mutant, and gshB mutant strains of S. enterica LT2 had aconitase activities of 4.2 ± 0.31 (mean ± standard deviation), 1.7 ± 0.17, and 4.19 ± 0.25 U/mg of protein, respectively, suggesting that γ-GC was also sufficient to facilitate [Fe-S] cluster repair. Furthermore, as shown in Fig. 3, gshA mutants displayed an increased sensitivity to the superoxide-generating compound paraquat, yet gshB mutants were more resistant than the wild type. This result suggested that γ-GC was proficient at protecting the cell against superoxide damage. Thus, in each of the above phenotypes, the requirement for gshA reflected the need for a thiol, and either GSH or γ-GC would suffice.
FIG. 3.
Mutations in gshB and gshA have opposite effects on sensitivity of the strain to paraquat. Cultures were grown as described in Materials and Methods. (A) Cultures were grown in Luria broth supplemented with 4 μM paraquat (methyl viologen). (B) Strains were grown in Luria broth supplemented with 0.4 μM paraquat. In each case, the strains were LT2 (wild type, □), DM4496 (gshB mutant, ▴), and DM4620 (gshA mutant, ●).
Definition of a class of thiamine auxotrophs.
More detailed phenotypic analyses of gshA mutants identified enough similarity to previously described mutants defective in apbC or apbE (4, 38) to group these three mutants as a class of thiamine auxotrophs. In all three mutants, the growth requirement for thiamine represented a need for both the HMP and THZ moieties of thiamine. As shown in Fig. 2B, neither HMP nor THZ alone were sufficient to allow growth of DM4620 (gshA). Additionally, exogenous l-Tyr satisfied the THZ requirement of DM4620 (gshA), as it did for mutants described previously to require both HMP and THZ (3; data not shown). Other phenotypes that were conserved among the mutants in this class were (i) elimination of the thiamine requirement by anaerobic growth conditions (data not shown), (ii) background specificity (see below), and (iii) suppression by a frequently arising second-site mutation at a locus near 66 min (J. Gralnick and D. M. Downs, unpublished data).
We showed that gshA mutants of S. enterica were thiamine auxotrophs, yet no nutritional requirement was reported for gshA mutants of E. coli (2). To confirm this apparent difference, a gshA mutant of E. coli was obtained from the E. coli Genetic Stock Center (JTG10) and the gshA insertion was transduced by phage P1 into E. coli K12. As expected, the resulting strain showed no growth defect on minimal medium. When a gshA mutation was introduced into two other wild-type strains of S. enterica, LT7 and Q1 (7), the LT7 derivative (DM5655) was prototrophic while the Q1 derivative (DM5749) required thiamine. Insertion mutations in apbE and apbC also resulted in a thiamine requirement in LT2 and Q1, but not the LT7 background (i.e., the same pattern as the gshA mutation). From these results, we concluded that there was a significant, but undefined, metabolic difference between strains that impacted the thiamine phenotype caused by these three mutations.
An HMP or THZ requirement can be caused by defective step(s) prior to condensation of HMP-PP and THZ-P.
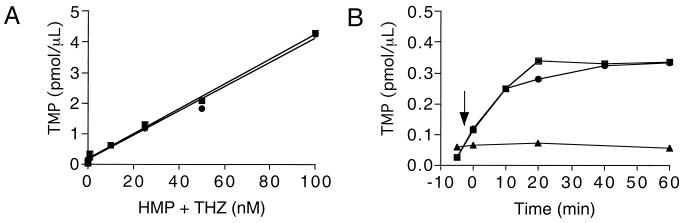
The GshA protein has a defined enzymatic function, and GSH has been implicated in various metabolic processes (37). We sought to use this knowledge to dissect the thiamine defect in this group of mutants. It was formally possible that the dual requirement for the independently synthesized HMP and THZ moieties of thiamine that characterized this class of mutants was due to inhibition of ThiE activity that is responsible for the condensation of HMP-PP and THZ-P (Fig. 1A). In considering ThiE as a target, we were influenced by results of an experiment addressing this possibility in an apbE mutant. ThiE activity in resting cells was monitored by the formation of TMP from added THZ and HMP. All strains in these experiments carried an insertion mutation in thiL to prevent conversion of the generated TMP to TPP (Fig. 1). The data in Fig. 4A show that in either an apbE mutant or a wild-type strain, the amount of TMP synthesized was proportional to the concentration of substrate (HMP and THZ) added. Figure 4B showed that there was no difference between the apbE+ and apbE-negative strains when TMP formation was monitored over time. A mutant completely lacking the thiE gene served as a control strain that was unable to convert HMP and THZ to detectable levels of TMP.
FIG. 4.
TMP synthesis in resting cells. (A) Titration of TMP synthesis in DM3012 (thiL mutant, ■) and DM5490 (thiL apbE mutant, ●). Equal amounts of cells were resuspended in minimal media supplemented with HMP and THZ as described in Materials and Methods. (B) Measurement of TMP formation over time in resting cells from strains DM3012 (thiL mutant, ■), DM5490 (thiL apbE mutant, ●), and DM5537 (thiL thiE mutant, ▴). Equal amounts of cells were resuspended in minimal media, and samples were removed for TMP determination. The time of addition of HMP plus THZ (25 nM each) is indicated by the arrow.
Identification of mutants requiring THZ or l-Tyr.
The results above were consistent with the double nutritional requirement of the apbE, apbC, and gshA mutants being generated by two distinct defects in thiamine biosynthesis, one affecting the THZ pathway and one affecting HMP synthesis. The ability of l-Tyr to satisfy the THZ requirement in these strains suggested a means to identify the defective step in the THZ pathway, and the HMP requirement was not addressed further. We reasoned that if an inhibited enzyme in the THZ biosynthetic pathway resulted in the THZ or l-Tyr requirement, point mutations generating a similar requirement would identify the relevant gene.
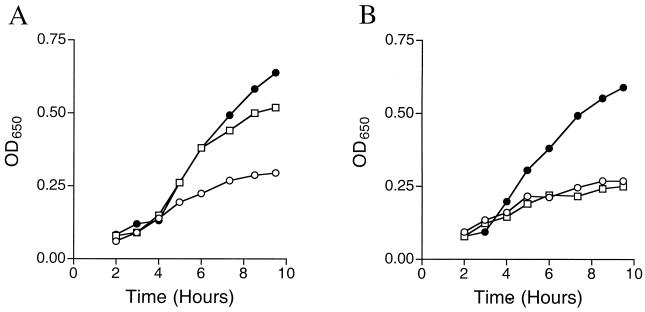
Twenty-five independent point mutants that had a nutritional requirement for THZ were identified with lesions linked to the thiCEFSGH operon. Phenotypic analysis determined that in 4 of the 25 strains, l-Tyr or THZ was able to satisfy the nutritional requirement (denoted Thz* mutants). The ability of l-Tyr to satisfy the thiamine requirement of one such mutant strain (DM4106) is shown in the growth data in Fig. 5. Two points are illustrated by these data. First, addition of l-Tyr or thiamine restored similar growth to the Thz* mutants, although significantly more l-Tyr (100 μM) than thiamine (1 μM) was required to do so. Titration experiments determined that concentrations of l-Tyr less than 100 μM failed to restore wild-type growth rates. Secondly, strains carrying null mutations in thiH (Fig. 5), and similarly, thiF, -S, -G, or -I (data not shown), failed to respond to exogenous l-Tyr. The latter result eliminated the formal possibility that l-Tyr could substitute for a THZ requirement in a general way.
FIG. 5.
Representative growth curves of Thz* and thiH null mutants. (A) Representative growth of Thz* mutant DM4106 (thiH1106). (B) Growth response of a thiH null mutant, DM460. Strains were grown in minimal medium (○), minimal medium supplemented with 100 μM l-tyrosine (□), or 1 μM thiamine (●).
Thz* mutants are defective in thiH.
The Thz* mutants were further characterized to determine the affected gene(s). A plasmid (pthiCH) carrying the complete operon complemented the growth defect of each of the four Thz* mutants. Plasmid pthiCH was subcloned, and the relevant plasmids generated through this process are described in Fig. 6.
Plasmid pthiH allowed each of the four Thz* mutants to grow on minimal medium (both solid and liquid), while pthiES1 failed to complement the growth defect of these strains. From this result, it was concluded that each of the four Thz* mutants were defective in thiH. Additional complementation analyses determined that 9 of the 21 remaining mutants auxotrophic for THZ (but not correctable by l-Tyr) were also defective in thiH.
Physical and molecular characterization of thiH.
The insert from plasmid pthiH was sequenced entirely on both strands (accession no. AF154064). Database analyses using basic local alignment search tool (BLAST) programs (1) confirmed that the insert contained a single complete ORF of 1,134 bp that was 89% identical to the ThiH protein in E. coli and was predicted to encode a 43-kDa protein with the potential to carry an [Fe-S] cluster. T7-RNA polymerase-specific protein expression and [35S]methionine labeling (44) confirmed that thiH encoded a protein of the expected size. Specifically labeled proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized via a phosphorimager showed that a ∼44-kDa protein was present in strain DM4448 [pthiH-7(+)] and not in the control strain, DM4447 [pthiH-7(−)].
The thiH gene was amplified via PCR from three Thz* backgrounds [DM4104 (zii-8039::Tn10d thiH1105), DM4106 (zii-8039::Tn10d thiH1106), and DM4108 (zii-8039::Tn10d thiH1107) and sequenced. Sequence analysis determined that the three mutants contained different missense mutations in thiH: H124Y (thiH1105), P347S (thiH1106), and V257A A351T (thiH1108).
Role and position of ThiH in THZ synthesis.
While other formal possibilities existed, identification of mutations causing a requirement for either THZ or l-Tyr in only the thiH gene was most consistent with the class of thiamine mutants described here affecting ThiH activity in vivo. Distinguishing the possible mechanisms for this inhibition demanded a better understanding of the role and position of the ThiH protein in THZ synthesis. It was not feasible to assay ThiH, partly because the order of the biochemical steps and in vivo intermediates in THZ biosynthesis have not been rigorously determined. If the order of biosynthetic reactions were known, development of an activity assay for ThiH would be facilitated by potential substrates. Previous attempts to order the steps by cross-feeding experiments have been unsuccessful. The identification of l-Tyr-correctable mutants provided a genetic means to address the position of ThiH in the THZ biosynthetic pathway.
To determine the position of ThiH activity with respect to other THZ biosynthetic enzymes, a rationale based on the method of Jarvik and Botstein (30) was used; double mutant strains defective in thiH and a distinct gene involved in THZ biosynthesis were constructed, with each mutation resulting in a distinct conditional requirement for THZ that could be manipulated by growth conditions. Thiamine-independent growth was measured after condition shifts designed to chase accumulated substrate to product. While a similar approach was used to dissect the cell cycle pathway in yeast (34), to our knowledge, this approach has not been utilized to order steps in a biosynthetic pathway.
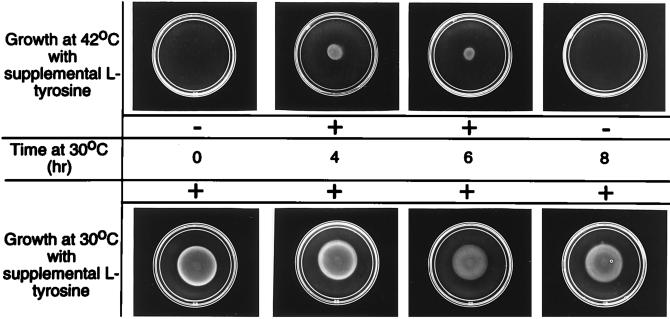
A Thz* mutation (required THZ in the absence of l-Tyr) and three independent temperature-sensitive mutations in other THZ biosynthetic genes were combined. Three such double mutant strains were constructed as described in Materials and Methods. Each double mutant contained a Thz* mutation (thiH1106) and a temperature-sensitive Thz− mutation (thiG1113, thiG1115, or thiFS1116; strains DM4797, -4801, and -4804, respectively). In each case, the double mutant was auxotrophic for THZ or l-Tyr at 30°C but required THZ or thiamine at 42°C. By incubating the double mutants under conditions permissive for one lesion (i.e., 30°C), nonpermissive for the other (i.e., no l-Tyr), and then shifting to the reverse (i.e., adding l-Tyr, 42°C), an order for the respective steps could be inferred.
Results from these experiments (some of which are presented in Fig. 7) supported the conclusion that the activities of ThiFS and ThiG were required prior to the activity of ThiH in THZ synthesis. The three relevant double mutant strains were incubated on minimal medium at 30°C for the indicated time, after which they were shifted to 42°C (or kept at 30°C) with the addition of l-Tyr spotted in the center of the plate. From these experiments, two significant points were noted. First, only when the double mutants had been preincubated at 30°C did addition of l-Tyr allow growth at 42°C. This result demonstrated that l-Tyr was not itself able to satisfy the requirement of the double mutants but rather required prior to function of the gene products that were active at 30°C. Thus, this result was consistent with addition of l-Tyr chasing a metabolite formed at 30°C to THZ. As expected, incubation of the double mutant strain at 30°C was not required for thiamine or THZ to allow growth at 42°C (data not shown).
FIG. 7.
Activity of ThiH follows that of ThiG-, -F, and -S in THZ synthesis. Cells of strain DM4797 (thiH1106 thiG1113) were overlaid in soft agar on a minimal glucose plate and incubated at 30°C for the indicated time. At the appropriate times, 2 μmol of l-tyrosine in 20 μl was spotted in the middle of duplicate plates. One of the plates was then returned to 30°C and one was incubated at 42°C. Growth was assessed after 18 h. Pluses and minuses represent our interpretation of growth on the relevant plates.
Although these experiments were indirect, a simple interpretation of the above results was that ThiH enzyme activity followed the activity of both ThiG and ThiFS in the synthesis of THZ. The definitive control to validate this interpretation of these results (i.e., a condition shift in the reverse order) was not technically feasible, and thus these results were only suggestive of the reaction order. Nonetheless, these results were consistent with a working model that ThiH catalyzes the last step in THZ-P, resulting in the incorporation of atoms from l-Tyr and the oxidative closure of the THZ ring. Biochemical assays to address this prediction are being pursued.
Interestingly, while the other four genes involved in THZ synthesis (thiFGS and thiI) have homologues in Bacillus subtilus, no homologue of thiH is found in this organism. Consistent with the finding is that in B. subtilus, glycine, not l-Tyr, provides the respective carbon and nitrogen atoms for the THZ moiety (48). Similarly, an oxidase (ThiO) that has been implicated in THZ synthesis in Rhizobium etli (33) has a homologue in B. subtilus and no homologue to this enzyme exists in E. coli.
Conclusions.
The work described here contributes to the understanding of thiamine biosynthesis and its integration with metabolism. The demonstration that gshA mutants belong to a larger phenotypic class of thiamine auxotrophs provided a perspective with which to consider the mechanism of the indirect effect(s) on thiamine synthesis in this class of mutants.
Based primarily on the work presented here, we suggest that ThiH is the site of the THZ biosynthetic defect caused by mutations in gshA, apbC, and apbE. It was recently shown that mutations in the isc gene cluster result in phenotypically similar defects in THZ synthesis (42). The involvement of the Isc proteins in the formation and/or repair of [Fe-S] clusters (35, 42, 45, 56) suggested an attractive model for the general mechanism of ThiH inhibition in this class of mutants. The sequence of ThiH is consistent with the presence of an [Fe-S] cluster as judged by a CXXXCXXCXnC motif. Our working model suggests that this putative [Fe-S] center is essential for efficient function of ThiH, either because it is involved in catalysis or because it increases stability of the protein. In isc and gshA mutants, there is evidence that [Fe-S] cluster formation and/or repair is defective (27, 42), and we suggest that this process might also be compromised in apbC and apbE mutants. In this scenario, the role of l-Tyr would be to stabilize the protein or provide enough substrate to increase turnover of the limited number of active protein molecules. The case of aconitase provides precedent for stabilization of an [Fe-S] protein by its substrate (9). By either scenario, l-Tyr could eliminate the thiamine requirement of the strains. Such a general model would explain why the thiamine requirement of the relevant mutants is suppressed anaerobically, since damage to [Fe-S] centers would be more prevalent during aerobic metabolism. This model is also consistent with recent work that shows the second-site mutation suppressing the thiamine requirement in these strains restores aconitase activity in a gshA mutant (Gralnick and Downs, unpublished data).
ACKNOWLEDGMENTS
Some of the point mutations in the thi operon were identified by Brad Paris as an undergraduate.
This work was supported by competitive grant MCB9723830 from the National Science Foundation, GM47296 from the National Institutes of Health, and a Shaw Scientists Award from the Milwaukee Foundation.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Apontoweil P, Berends W. Mapping of gshA, a gene for the biosynthesis of glutathione in Escherichia coli K12. Mol Gen Genet. 1975;141:91–95. doi: 10.1007/BF00267676. [DOI] [PubMed] [Google Scholar]
- 3.Beck B, Downs D M. A periplasmic location is essential for the role of the ApbE lipoprotein in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1999;181:7285–7290. doi: 10.1128/jb.181.23.7285-7290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck B J, Downs D M. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1998;180:885–891. doi: 10.1128/jb.180.4.885-891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley T P, Downs D M, Ealick S, McLafferty F, van Loon D, Taylor S, Chiu H, Kinsland C, Reddick J, Xi J, Campobasso N. Thiamin synthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 6.Bellion E, Kirkley D H. The origin of the sulfur atom in thiamine. Biochim Biophys Acta. 1977;497:323–328. doi: 10.1016/0304-4165(77)90166-0. [DOI] [PubMed] [Google Scholar]
- 7.Boyd J S K, Bidwell D E. The Q1(A) strans of Salmonella typhimurium identification by standardized cross immunity test. J Gen Microbiol. 1959;21:635–651. doi: 10.1099/00221287-16-1-217. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Buchanan J M, Anfinsen C B. Partial purification of aconitase. J Biol Chem. 1949;180:47–54. [PubMed] [Google Scholar]
- 10.Caetano-Annoles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–92. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 11.Castilho B A, Olfson P, Casadaban M J. Plasmid insertion mutagenesis and lac gene fusion with mini Mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian T, Downs D M. Defects in pyruvate kinase cause a conditional increase of thiamine synthesis in Salmonella typhimurium. Can J Microbiol. 1999;45:565–572. [PubMed] [Google Scholar]
- 13.Claas K, Weber S, Downs D M. Lesions in the nuo operon, encoding NADH dehydrogenase complex I, prevent PurF-independent thiamine synthesis and reduce flux through the oxidative pentose phosphate pathway in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:228–232. doi: 10.1128/jb.182.1.228-232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 15.DeMoll E, Shive W. Determination of the metabolic origin of the sulfur atom in thiamine of Escherichia coli by mass spectrometry. Biochem Biophys Res Commun. 1985;132:217–222. doi: 10.1016/0006-291x(85)91010-1. [DOI] [PubMed] [Google Scholar]
- 16.Downs D M. Evidence for a new, oxygen-regulated biosynthetic pathway for the pyrimidine moiety of thiamine in Salmonella typhimurium. J Bacteriol. 1992;174:1515–1521. doi: 10.1128/jb.174.5.1515-1521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enos-Berlage J, Downs D M. Biosynthesis of the pyrimidine moiety of thiamine independent of the PurF enzyme (phophoribosylpyrophosphate amidotransferase) in Salmonella typhimurium: incorporation of stable isotope-labeled glycine and formate. J Bacteriol. 1999;181:841–848. doi: 10.1128/jb.181.3.841-848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enos-Berlage J L, Downs D M. Involvement of the oxidative pentose phosphate pathway in thiamine biosynthesis in Salmonella typhimurium. J Bacteriol. 1996;178:1476–1479. doi: 10.1128/jb.178.5.1476-1479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enos-Berlage J L, Langendorf M J, Downs D M. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J Bacteriol. 1998;180:6519–6528. doi: 10.1128/jb.180.24.6519-6528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estramareix B. Biosynthesis of the pyrimidine moiety of thiamine: origin of carbon-6 in Salmonella typhimurium. Biochim Biophys Acta. 1970;208:170–171. [PubMed] [Google Scholar]
- 21.Estramareix B, Lesieur M. Biosynthesis of the pyrimidine moiety of thiamine: origin of carbons in positions 2 and 4 in Salmonella typhimurium. Biochim Biophys Acta. 1969;192:375–377. [PubMed] [Google Scholar]
- 22.Estramareix B, Therisod M. Biosynthesis of thiamine: 5-aminoimidazole ribotide as the precursor of all the carbon atoms of the pyrimidine moiety. J Am Chem Soc. 1984;106:3857–3860. [Google Scholar]
- 23.Estramareix B, Therisod M. Tyrosine as a factor in biosynthesis of the thiazole moiety of thiamine in Escherichia coli. Biochim Biophys Acta. 1972;273:275–282. [PubMed] [Google Scholar]
- 24.Frodyma M, Downs D M. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180:4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frodyma M, Rubio A, Downs D M. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:236–240. doi: 10.1128/jb.182.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs J A, Warner H R. Isolation of an Escherichia coli mutant deficient in glutathione synthesis. J Bacteriol. 1975;124:140–148. doi: 10.1128/jb.124.1.140-148.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner P R, Fridovich I. Effect of glutathione on aconitase in Escherichia coli. Arch Biochem Biophys. 1993;301:98–102. doi: 10.1006/abbi.1993.1120. [DOI] [PubMed] [Google Scholar]
- 28.Gruer M J, Guest J R. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994;140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 29.Hong J S, Ames B N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci USA. 1971;68:3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarvik M, Botstein D. A genetic method for determining the order of events in a biological pathway. Proc Natl Acad Sci USA. 1973;72:2738–2742. doi: 10.1073/pnas.70.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kambampati R, Lauhon C T. IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry. 1999;38:16561–16568. doi: 10.1021/bi991119r. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki T. Determination of thiamin and its phosphate esters by high-performance liquid chromatography. In: Chytil F, McCormick D B, editors. Vitamins and coenzymes, part G. Vol. 122. New York, N.Y: Academic Press, Inc.; 1986. pp. 15–29. [DOI] [PubMed] [Google Scholar]
- 33.Miranda-Rios J, Morera C, Taboada H, Dávalos A, Encarnación S, Mora J, Soberón M. Expression of thiamin biosynthetic genes (thiCOGE) and production of symbiotic terminal oxidase cbb3 in Rhizobium etli. J Bacteriol. 1997;179:6887–6893. doi: 10.1128/jb.179.22.6887-6893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moir D, Botstein D. Determination of the order of gene function in the yeast nuclear division pathway using cs and ts mutants. Genetics. 1982;100:565–577. doi: 10.1093/genetics/100.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura M, Saeki K, Takahashi Y. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J Biochem (Tokyo) 1999;126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 36.Nosaka K, Kaneko Y, Nishimura H, Iwashima A. Isolation and characterization of a thiamin pyrophosphokinase gene, TH180, from Saccharomyces cerevisiae. J Biol Chem. 1993;268:17440–17447. [PubMed] [Google Scholar]
- 37.Penninckx M J, Elskens M T. Metabolism and functions of glutathione in micro-organisms. Adv Microb Physiol. 1993;34:239–301. doi: 10.1016/s0065-2911(08)60031-4. [DOI] [PubMed] [Google Scholar]
- 38.Petersen L, Downs D M. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J Bacteriol. 1996;178:5676–5682. doi: 10.1128/jb.178.19.5676-5682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen L A, Downs D M. Identification and characterization of an operon in Salmonella typhimurium involved in thiamine biosynthesis. J Bacteriol. 1997;179:4894–4900. doi: 10.1128/jb.179.15.4894-4900.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen L A, Enos-Berlage J E, Downs D M. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143:37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 42.Skovran E, Downs D M. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. JBacteriol. 2000;182:3896–3903. doi: 10.1128/jb.182.14.3896-3903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprenger G A, Schorken U, Wiegert T, Grolle S, DeGraaf A A, Taylor S V, Begley T P, Bringer-Meyer S, Sahm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausube P A, Brent R, Kingston R E, Moore D D, Seidman J C, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 45.Takahashi Y, Nakamura M. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J Biochem (Tokyo) 1999;126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 46.Taylor S V, Kelleher N L, Kinsland C, Chiu H-J, Costello C A, Backstrom A D, McLafferty F W, Begley T P. Thiamin biosynthesis in Escherichia coli: identification of ThiS thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem. 1998;273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 47.Taylor S V, Vu L D, Begley T P, Sprenger G A, Gringer-Meyer S, Sahm H. Chemical and enzymatic synthesis of 1-deoxy-D-xylulose-5-phosphate. J Org Chem. 1998;63:2375–2377. [Google Scholar]
- 48.Tazuya K, Yamada K, Nakamura K, Kumaoka H. The origin of the sulfur atom of thiamine. Biochim Biophys Acta. 1987;924:210–215. doi: 10.1016/0304-4165(87)90089-4. [DOI] [PubMed] [Google Scholar]
- 49.Vander Horn P B, Backstrom A D, Stewart V, Begley T P. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J Bacteriol. 1993;175:982–992. doi: 10.1128/jb.175.4.982-992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 51.Webb E, Claas K, Downs D. Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J Bacteriol. 1997;179:4399–4402. doi: 10.1128/jb.179.13.4399-4402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webb E, Claas K, Downs D. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem. 1998;273:8946–8950. doi: 10.1074/jbc.273.15.8946. [DOI] [PubMed] [Google Scholar]
- 53.Webb E, Downs D M. Characterization of thiL, encoding thiamine monophosphate kinase, in Salmonella typhimurium. J Biol Chem. 1997;272:15702–15707. doi: 10.1074/jbc.272.25.15702. [DOI] [PubMed] [Google Scholar]
- 54.White R H. Stable isotope studies on the biosynthesis of the thiazole moiety of thiamine in Escherichia coli. Biochemistry. 1978;17:3833–3840. doi: 10.1021/bi00611a024. [DOI] [PubMed] [Google Scholar]
- 55.White R H, Rudolph F B. The origin of the nitrogen atom in the thiazole ring of thiamine in Escherichia coli. Biochim Biophys Acta. 1978;542:340–347. doi: 10.1016/0304-4165(78)90029-6. [DOI] [PubMed] [Google Scholar]
- 56.Yuvaniyama P, Agar J N, Cash V L, Johnson M K, Dean D R. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci USA. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]