Abstract
The nos (nitrous oxide reductase) operon of Paracoccus denitrificans contains a nosX gene homologous to those found in the nos operons of other denitrifiers. NosX is also homologous to NirX, which is so far unique to P. denitrificans. Single mutations of these genes did not result in any apparent phenotype, but a double nosX nirX mutant was unable to reduce nitrous oxide. Promoter-lacZ assays and immunoblotting against nitrous oxide reductase showed that the defect was not due to failure of expression of nosZ, the structural gene for nitrous oxide reductase. Electron paramagnetic resonance spectroscopy showed that nitrous oxide reductase in cells of the double mutant lacked the CuA center. A twin-arginine motif in both NosX and NirX suggests that the NosX proteins are exported to the periplasm via the TAT translocon.
Denitrification is an anaerobic respiratory process, found in many genera of bacteria, whereby nitrate is reduced sequentially to nitrite, nitric oxide, nitrous oxide, and ultimately dinitrogen gas by a set of corresponding oxidoreductases (5). The last reduction in the pathway, that of nitrous oxide to nitrogen, is performed by nitrous oxide reductase (NosZ), a soluble dimeric enzyme located in the periplasm. NosZ contains two spectroscopically distinct copper centers, CuA, at which the enzyme is thought to receive electrons from small redox proteins and CuZ, believed to be the catalytic center (13). Recently, two lines of evidence which suggest that the structural complexity of nitrous oxide reductase is reflected in its biosynthetic pathway have emerged. First, the enzyme contains an unusually long periplasmic targeting sequence containing a so-called twin-arginine motif (3), indicating that the enzyme belongs to a family of metalloproteins in which targeting and metal insertion are coupled by a Sec-independent secretory mechanism (28). Second, the structural gene for nitrous oxide reductase (nosZ) is part of a cluster of genes whose products are involved in the maturation of the enzyme. In the organism Pseudomonas stutzeri, nosZ is followed by the genes nosD, -F, -Y, and -L, and it has been suggested that NosD, -F, and -Y form a multisubunit ABC-type transporter which catalyzes the transport and insertion of copper to the CuZ center (36). NosL is predicted to be an outer membrane lipoprotein. The NosL protein from P. stutzeri contains sequence motifs for a disulfide isomerase protein (12), but these are absent in NosL from Sinorhizobium meliloti and mutations of nosL yielded no apparent phenotype in either organism (7, 12). Similar nos gene clusters have been identified in the organisms Bradyrhizobium japonicum (GenBank accession number AJ002531), Achromobacter cycloclastes (20), and Sinorhizobium meliloti (17), and there is evidence from partial sequence data that this operon structure is conserved in Paracoccus denitrificans (16) and Pseudomonas aeruginosa (35). Each of these clusters also contain the nosR gene immediately upstream of nosZ. NosR is predicted to be an integral membrane protein with six transmembrane helices and a C-terminal cytoplasmic domain containing sequence motifs for two [4Fe-4S] clusters (9). Insertion mutagenesis of nosR in P. stutzeri resulted in failure to transcribe the nosZ gene.
In B. japonicum, A. cycloclastes, and S. meliloti, an additional nos gene, nosX, has been identified downstream of nosZDFYL. Tn5 mutagenesis of the S. meliloti nosX gene resulted in a Nos− phenotype (7), but the molecular basis for the defect is unknown. NosX is predicted to be a soluble periplasmic protein, but to date there has been no detailed characterization of NosX or of the phenotype exhibited by the nosX mutant. A homologue of nosX designated nirX has been identified in the P. denitrificans nir (nitrite reductase) gene cluster (29). Inactivation of this gene yielded no apparent phenotype, suggesting that another protein, possibly a counterpart of nosX, takes over the role of NirX. The aim of the present study was to test this hypothesis and to determine the function of the NosX and NirX proteins during denitrification.
MATERIALS AND METHODS
Bacterial strains and growth media.
Escherichia coli TG1 (27) was used routinely for cloning procedures. E. coli HB101(pRK2020) was used as the helper strain in the conjugative transfer of plasmids via triparental mating. E. coli strains were grown aerobically in Luria-Bertani medium. P. denitrificans Pd1222 (10) was grown aerobically in brain heart infusion broth (Oxoid) or anaerobically in minimal succinate medium containing 25 mM succinate and 100 mM KNO3 (29). Antibiotics were used at different concentrations, as follows: ampicillin, 100 μg ml−1; kanamycin, 25 μg ml−1 (E. coli) or 100 μg ml−1 (P. denitrificans); rifampin, 100 μg ml−1; and streptomycin, 25 μg ml−1 (E. coli) or 100 μg ml−1 (P. denitrificans).
DNA manipulations.
Routine cloning procedures were performed according to standard protocols (1). Fusions of chromosomal promoter-lacZ and the nosZ gene were carried out using the suicide plasmid pBK11 as described previously (31). The construction of marked and unmarked mutations in the nirX and nosX genes is described in Results and was performed as described previously (32). Identification and characterization of the P. denitrificans nirX gene has been described previously (29). The nosX gene was identified by subcloning and sequencing of plasmid pFUH21 and cosmid pJLA601 (gifts from S. J. Ferguson, University of Oxford), as described in Results. Generation of subclones in M13mp18 or M13mp19 and automated sequencing of single-stranded DNA were performed as described previously (14).
Protein techniques and enzyme assays.
Total soluble cell extracts were prepared by French press and ultracentrifugation as previously described (29). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out according to the method of Laemmli (19) using a Bio-Rad Mini Protean II apparatus. Immunoblotting of nitrous oxide reductase was performed as described previously (25), using a rabbit polyclonal antiserum against Paracoccus pantotrophus nitrous oxide reductase for the primary antibody (a gift from B. Berks, University of East Anglia) (4) and goat anti-rabbit immunoglobulin G alkaline phosphatase conjugate (Sigma). The β-galactosidase activity of promoter-lacZ fusion strains was measured as described previously (21). Nitrous oxide reductase activity in whole cells using succinate as the electron donor was measured by monitoring the steady-state redox level of cytochrome c as described previously (6). Nitrite was assayed colorimetrically according to a method described elsewhere (22).
Analysis of nitrous oxide.
Gas samples (150 μl) were withdrawn from the headspace of denitrifying cultures and injected in a gas chromatograph (type 8500; Perkin-Elmer) equipped with a J&W Scientific column (type GS-Q). Helium was used as carrier gas. Concentrations of nitrous oxide in each sample were calculated from standards of pure nitrous oxide.
EPR and optical spectroscopy.
Electron paramagnetic resonance (EPR) spectra were recorded on a Varian 109 EPR spectrometer equipped with a home-built He flow system. Optical spectra were recorded on an Aminco DW2000 spectrophotometer.
Nucleotide sequence accession number.
The nucleotide sequence of the nosX gene has been deposited in the GenBank database under accession number AJ010260.
RESULTS
Isolation and sequence analysis of the nosX gene.
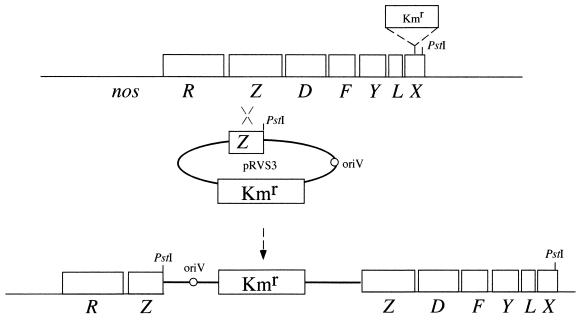
As the nosX gene is located in the downstream part of the nos operon in other organisms, we decided to determine whether P. denitrificans also contained a nosX gene in this region. A 1.3-kbp HindIII-SalI fragment of the plasmid pFUH21 carrying part of the nosZ gene from P. denitrificans was cloned into the suicide vector pRVS3 and transferred to P. denitrificans via conjugation. Strains that had integrated the construct via homologous recombination were isolated and shown to contain two truncated versions of nosZ separated by pRVS3 (not shown). These strains were used to recover the plasmid as a recircularized chromosomal PstI fragment containing the nosZ gene and approximately 4.6 kbp of downstream DNA. A map of the nos locus along with the recombination event are presented in Fig. 1. Sequencing at the 3′ end of the fragment revealed a partial open reading frame with substantial identity to both nirX of P. denitrificans and the previously identified nosX genes of other denitrifying bacteria. The P. denitrificans nosX gene sequence was completed using subclones from the cosmid pJLA601. It encoded a protein of 304 amino acids, with a molecular mass of 32.1 kDa. The NosX and NirX protein sequences from P. denitrificans showed 37% identity and 49% similarity.
FIG. 1.
Maps of the nos gene cluster in P. denitrificans wild-type strain (top) and a strain that had integrated suicide vector pRVS3 with a copy of the nosZ gene via homologous recombination (bottom). The PstI sites used for recovery of the plasmid along with adjacent chromosomal DNA are indicated. The position of a kanamycin resistance cassette, a 1.2-kbp PstI fragment from the plasmid pUC4K (Kmr) in the nosX mutant, is also indicated.
Analysis of the NirX and NosX proteins from P. denitrificans using the program Prosite (2) revealed no known motifs. However, the program SignalP (24) suggested that both proteins are periplasmic with a cleavable N-terminal signal sequence predicted to be at residue 21 for NirX (ARA-AT) and residue 24 for NosX (LRA-QP). The BLAST results using NirX and NosX were used to retrieve all known NosX sequences from the GenBank database, and these were aligned using the program ClustalX (30) (Fig. 2). Included also in this alignment is a NosX protein retrieved from the Rhodobacter capsulatus genome sequencing project (University of Chicago [http://rhodol.uchicago.edu/capsulapedia/capsulapedia.shtml]). Overall, the NosX family is highly conserved. The NosX proteins from B. japonicum and S. meliloti are somewhat longer than the rest, at 366 and 334 residues, respectively. Of particular note is the conserved motif R-R-R at the N terminus of each sequence. This motif is absent from the GenBank sequence of NosX from S. meliloti. However, translation of the reading frame starting upstream of the chosen start codon and continuing to the next start codon revealed an R-R-R pattern in that protein as well. The correctness of the start codon assignment for this protein awaits further studies. Analysis of the most conserved regions within NosX using the Blocks Server (15) or the Swiss Institute for Experimental Cancer Research PatternFind server (Bioinformatics Group, Swiss Institute for Experimental Cancer Research [http://www.isrec.isb-sib.ch/software/PATFND_form.html]) failed to identify any specific groups of proteins that might be related to NosX.
FIG. 2.
Multiple alignment of the NosX protein family. The alignment was constructed using the programs ClustalX and GeneDoc (23). The twin-arginine motif is indicated by asterisks. The consensus sequence shown on top includes identical amino acid residues found in all sequences (uppercase) and those found uniquely in the NosX sequences (lowercase). PdnirX, P. denitrificans Pd1222; PdnosX, P. denitrificans Pd1222; SmnosX, S. meliloti; AcnosX, A. cycloclastes; RcnosX, R. capsulatus; BjnosX, B. japonicum. –, amino acid residue is absent.
Construction and analysis of nosX, nirX, and double-mutant strains.
To investigate the phenotype of nosX and nirX mutants, mutations were introduced into each gene both separately and in combination. A marked nirX mutant strain, Pd76.21, was already available (29). In the case of nosX, a PstI site was first introduced in the middle of that gene at nucleotide position 469 relative to its start codon using a PCR approach with primers that contained a PstI site at the corresponding position. This site was used for the introduction of a kanamycin cassette as a 1.2-kbp PstI fragment from the plasmid pUC4K. The nosX mutant strain was named Pd101.21. To construct a double nosX nirX mutant, an unmarked deletion was first created in nirX by exchanging the chromosomally integrated kanamycin resistance cassette with a plasmid-borne nirX copy from which a 0.2-kbp NruI-HincII fragment had been removed. The unmarked ΔnirX mutant strain, Pd76.31, was then mutated by the insertion of the kan gene into nosX as described previously, resulting in the ΔnirX nosX::Kmr double mutant, Pd92.36. All mutant strains were analyzed using Southern blotting to confirm that the correct recombinational events had taken place.
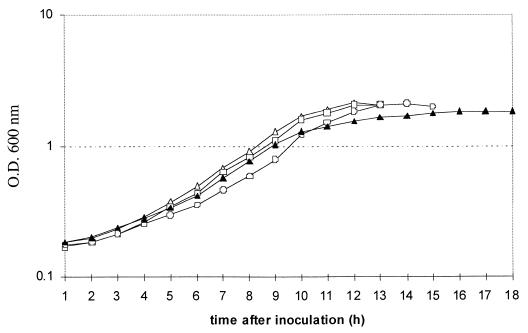
The wild-type strain and three mutant strains were cultured anaerobically, and growth curves for the four strains were obtained (Fig. 3). In the single-mutant strains, growth was virtually unaffected, with both strains growing at a rate similar to that of the wild type and reaching a turbidity at 600 nm of around 2.2. The nirX nosX double mutant reached a final turbidity at 600 nm of 1.8. In this mutant, growth was biphasic, entering a linear phase approximately 10 h after inoculation. The double mutant also failed to produce visible gas bubbles in the culture medium, in contrast to the wild-type and single-mutant strains.
FIG. 3.
Growth curve of P. denitrificans Pd1222 (open triangles), compared with those of the mutant strains Pd76.21 (open squares), Pd101.21 (open circles), and Pd92.36 (closed triangles), during denitrifying growth.
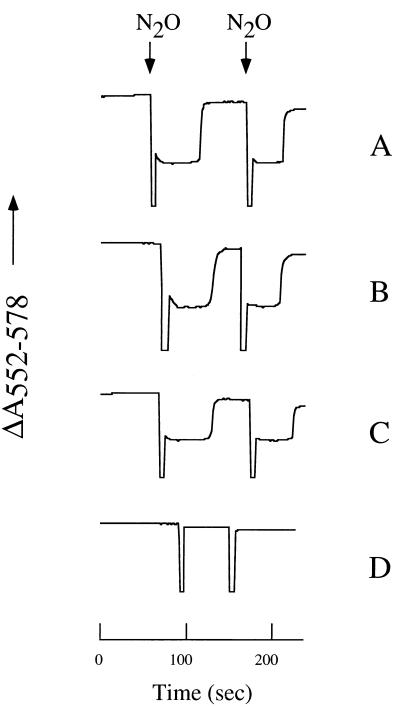
The absence of gas bubbles in denitrifying cultures of Pd92.36 indicated that the nirX nosX double mutant was unable to produce dinitrogen gas, which has a relatively low solubility in solution (100 μM at 30°C). In order to test whether the double mutant had accumulated nitrous oxide, which has a relatively high solubility in solution (20 mM at 30°C), samples were withdrawn from the headspace of the denitrifying cultures and analyzed for the presence of nitrous oxide using a gas chromatograph. These experiments revealed that the concentration of nitrous oxide in the gas phase of the wild-type culture was below the detection level, while it was 20% in that of the mutant culture. Apparently, the nirX nosX mutant was deficient in nitrous oxide reductase activity. To test this hypothesis, whole cells of each strain were assayed by monitoring the kinetics of oxidation and reduction of c-type cytochromes in the cell suspensions when pulses of N2O were added, using succinate as the electron donor (Fig. 4). In the single-mutant strains, NosZ activity was comparable to that in the wild-type. However, nitrous oxide reductase activity was completely absent in the double mutant, as shown by the failure of cytochromes to become oxidized transiently upon addition of N2O. Activity was also absent when ascorbate plus tetramethyl-p-phenylenediamine (TMPD) was used as electron donor to NosZ (data not shown), indicating that the defect did not lie in the pathway of electron transfer to the reductase. Nitrate and nitrite reductases in the nirX nosX double mutant were both active as judged by the same assay (data not shown).
FIG. 4.
Nitrous oxide reductase activity in whole cells of Pd1222 (A) compared to that of Pd76.21 (B) Pd101.21 (C), and Pd92.36 (D). Cell suspensions (with a turbidity at 660 nm of 50) of anaerobically grown cultures were incubated with succinate under a stream of nitrogen gas to allow full reduction of c-type cytochromes. Changes in the absorption of light at 552 nm, which is diagnostic for the redox state of c-type cytochromes, were recorded in time using an Aminco spectrophotometer. At the points indicated, aliquots of buffer saturated with nitrous oxide were added to the suspensions.
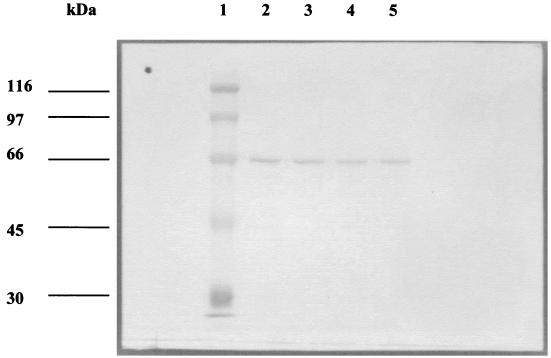
To determine whether nitrous oxide reductase was induced in the mutant strains, nosZ promoter activity was measured using a construct in which a 2.0-kbp EcoRI fragment containing the 5′ end of the nosZ gene and approximately 2.8 kbp of upstream DNA was cloned in front of a promoterless lacZ gene in the suicide plasmid pBK11. This construct was transferred to Pd1222 and to the mutant strains, and the β-galactosidase activity was assayed in denitrifying cultures. The activity was similar (about 1,500 Miller units) in each case, showing that mutations of nirX and/or nosX did not affect transcription of the nosZ gene. In addition, proteins in total soluble extracts from the wild-type and the three mutants were separated using SDS-PAGE, transferred to nitrocellulose, and immunoblotted using a polyclonal antiserum against nitrous oxide reductase from P. pantotrophus, an organism closely related to P. denitrificans (25) (Fig. 5). A single band of around 66 kDa was detected in extracts from all strains, showing that the NosZ polypeptide was synthesized at wild-type levels in each case. Furthermore, the position of the band was identical in each lane, indicating that in each case the periplasmic targeting sequence of about 5 kDa was recognized and cleaved.
FIG. 5.
Lane 1, prestained protein molecular weight standards, the sizes of which are shown to the left of the figure; lanes 2, 3, 4, and 5, immunodetection of nitrous oxide reductase in total soluble extracts from Pd1222, Pd76.21, Pd101.21, and Pd92.36, respectively.
EPR spectroscopy of nirX nosX double mutant.
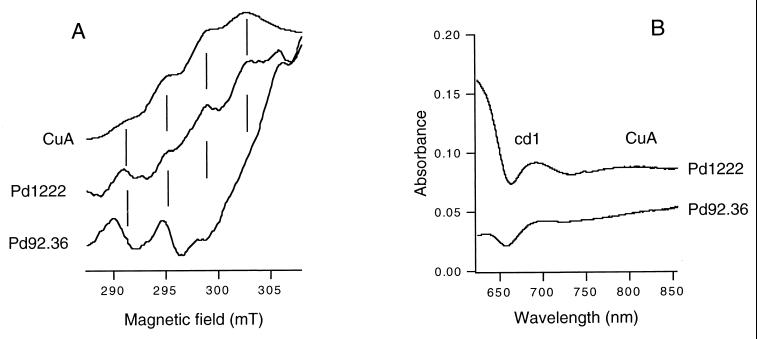
The EPR spectrum of total soluble extract from wild-type P. denitrificans cells in the oxidized state showed four lines separated by 3.83 mT with a gz value of 2.17, characteristic for CuA of NosZ (18) or cytochrome c oxidase (34) (Fig. 6A). The relative intensity of the lines is consistent with a 1:2:3:4 ratio, as expected for the first four of the seven spectral lines arising from a dinuclear mixed-valence copper center. The g- perpendicular region of the spectrum is obscured by resonances of unknown origin and has been omitted from the figure. The features mentioned above are absent from the spectrum of the soluble fraction obtained from Pd92.36. Therefore, we conclude that the CuA center is absent from NosZ in the nirX nosX mutant strain. A similar conclusion can be drawn from the optical spectra (Fig. 6B). The reduced minus oxidized spectrum of the wild-type strain shows a broad (weak) absorbance peak around 800 nm, which is diagnostic for CuA. This band is lacking from the spectrum of the nirX nosX mutant.
FIG. 6.
(A) EPR spectra of total soluble cell extracts from wild-type (Pd1222) and nirX nosX mutant (Pd92.36) strains, oxidized with solid potassium ferricyanide. CuA is the water-soluble fragment of cytochrome caa3 from Bacillus subtilis, shown here for comparison. The vertical lines are spaced by 3.83 mT, the hyperfine coupling value for both the CuA center of nitrous oxide reductase and CuA in the B. subtilis fragment. Spectra are corrected for differences in protein concentrations. The origin of the peaks around 290 and 295 mT in the mutant is unknown. Experimental conditions: frequency, 9.235 GHz; modulation amplitude, 1.0 mT; microwave power, 2.0 mW; temperature, 39K. (B) Optical reduced minus oxidized spectra of total soluble cellular extracts from the wild-type strain (Pd1222) and the nirX nosX mutant strain (Pd92.36). When isolated, the soluble extracts were found to be completely reduced. For oxidation a small amount of solid potassium ferricyanide was added. The absorbance maximum around 800 nm in the wild-type strain is ascribed to CuA and that at 650 to 700 nm to cytochrome cd1. The spectra were corrected for differences in protein concentrations.
Cultures of Pd92.36 that were grown aerobically contained a redox-active cytochrome aa3-type oxidase (5) which became reduced following the addition of succinate to the cells, as judged by the appearance of a visible absorbance peak at 605 nm. This demonstrated that the CuA center in the oxidase was properly assembled and indicated that the absence of the CuA center from NosZ in Pd92.36 was a specific defect that did not result from a general defect in copper transport or processing. In an attempt to recover the CuA center in NosZ, Pd92.36 was cultured under denitrifying growth conditions until mid-exponential phase and then switched to aerobic growth for a further 4 h. The rationale for this experiment was to investigate whether the mechanism by which CuA is inserted into the aa3-type oxidase could also reconstitute NosZ. However, whereas wild-type cells still exhibited nitrous oxide reductase activity 4 h after the switch to aerobiosis, no activity was observed in cell suspensions of Pd92.36. Furthermore, the addition of increasing concentrations of copper ions to the assay buffer had no apparent effect. These data strengthen the case for a specific role for the NosX and NirX proteins in assembly of the CuA center of NosZ during denitrification.
DISCUSSION
P. denitrificans is the first organism reported to date that contains two homologues of the NosX protein. Thus far P. denitrificans is also unique in containing a second homologue (named nirI) of the gene nosR within the nir gene cluster (29). Insertion mutagenesis in either of the two genes nirX or nosX results in no observable phenotype. This is in contrast to the observation that Tn5 mutagenesis of nosX from S. meliloti gives a Nos-deficient phenotype and suggests that there may be no second homologue of nosX in the latter organism. Despite its position in the P. denitrificans nir gene cluster, nirX appears not to play an essential role in expression of functional nitrite reductase, nor is nosX essential with respect to nitrous oxide reductase activity. However, mutagenesis of both nirX and nosX eliminates nitrous oxide reductase activity in P. denitrificans, implying that the two proteins are functional homologues and that each can take over the other's role.
Our analyses of the nirX nosX double mutant of P. denitrificans showed that the deficiency in nitrous oxide reduction was the consequence of a failure to activate rather than to express nitrous oxide reductase. The analyses of the promoter-reporter gene fusions as well as the Western analyses have shown that nosZ transcription and translation levels in the double mutant were comparable to those in the wild-type strain. NosZ produced by the double mutant, however, was inactive in nitrous oxide reduction regardless of whether endogenous substrates or ascorbate was used as the electron donor. EPR spectral analyses showed that the signals that are diagnostic for CuA were completely absent from the soluble fraction of the double mutant cells grown anaerobically with nitrate, whereas they were present in that of the wild-type cells. The most likely explanation for the observation that NosZ was inactive in the double mutant is therefore that its CuA center was not properly assembled. It is then tempting to speculate that the NosX and NirX have their task in the transport of copper to NosZ or in catalyzing its insertion into the metal binding site. However, the absence of the CuA center might also be the result of a secondary effect of the mutations rather than the primary cause or it may result from a concomitant failure to assemble the CuZ center in NosZ.
An alignment of all known NosX protein sequences revealed several highly conserved blocks, but these did not indicate a possible function of NosX when used to search for similar regions in other protein sequences. However, the N-terminal region of the NosX proteins contains the sequence (S/T/N)-R-R-R-(F/A/L/M)-(I/L), corresponding closely to the twin-arginine motif (S/T)-R-R-X-F-L-K. This conserved motif has been identified in a large number of periplasmic metalloproteins that contain complex cofactors and are exported to the periplasm via the Sec-independent TAT translocon (3). Therefore, we hypothesize that NosX is exported via this pathway in a partially folded state.
One intriguing aspect of NosX is its absence from the nos gene cluster of P. stutzeri. A BLAST search of the emerging sequence from the P. aeruginosa and Neisseria gonorrhoeae genomes, both of which contain functional nos operons, also failed to reveal a NosX-like sequence. It may be that a NosX homologue remains to be discovered in these organisms or that the mechanism of CuA insertion differs between bacteria. However, there is evidence that CuA insertion into P. stutzeri nitrous oxide reductase is an enzyme-catalyzed event. Expression of the nosZ gene from P. stutzeri in E. coli yielded an apoprotein lacking all copper centers, into which copper could be reconstituted in vitro (33). This form of the enzyme was catalytically inactive and contained only CuA, but the CuA center was spectroscopically distinct from the native protein. A similar result was achieved by reconstitution of purified nitrous oxide reductase from which the copper centers had been chemically removed (8). In contrast, the characteristic electronic spectrum of CuA could be obtained when a translocation-incompetent mutant of nitrous oxide reductase in which the twin-arginine motif was mutated (R20D) was reconstituted with copper, but the insertion process was slow and resulted in a weak retention of copper (11). These data led to the suggestion that the twin-arginine motif of NosZ may confer both translocational competence and delivery to a specific system involved with copper insertion.
ACKNOWLEDGMENTS
This work was supported by the Netherlands Foundation for Chemical Research (SON), with financial aid from the Netherlands Organization for Scientific Research (NWO). This work was partly financed by the European Commission under contract ERB-FMB-ICT972594.
We thank S. J. Ferguson for the gift of plasmid plasmid pFUH21 and cosmid pJLA601, B. Berks for antibodies to nitrous oxide reductase, B. Beaumont for gas chromatographic analyses, and M. D. Page for careful reading of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. [Google Scholar]
- 2.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 4.Berks B C, Baratta D, Richardson D J, Ferguson S J. Purification and characterization of a nitrous oxide reductase from Thiosphaera pantotropha. Implications for the mechanism of aerobic nitrous oxide reduction. Eur J Biochem. 1993;212:467–476. doi: 10.1111/j.1432-1033.1993.tb17683.x. [DOI] [PubMed] [Google Scholar]
- 5.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 6.Boogerd F C, van Verseveld H W, Stouthamer A H. Electron transport to nitrous oxide in Paracoccus denitrificans. FEBS Lett. 1980;113:279–284. doi: 10.1016/0014-5793(80)80609-0. [DOI] [PubMed] [Google Scholar]
- 7.Chan Y, McCormick W, Watson R. A new nos gene downstream from nosDFY is essential for dissimilatory reduction of nitrous oxide by Rhizobium (Sinorhizobium) meliloti. Microbiology. 1997;143:2817–2824. doi: 10.1099/00221287-143-8-2817. [DOI] [PubMed] [Google Scholar]
- 8.Coyle C L, Zumft W G, Kroneck P M H, Körner H, Jakob W. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina. Purification and properties of a novel multicopper enzyme. Eur J Biochem. 1985;153:459–467. doi: 10.1111/j.1432-1033.1985.tb09324.x. [DOI] [PubMed] [Google Scholar]
- 9.Cuypers H, Viebrock-Sambale A, Zumft W G. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J Bacteriol. 1992;174:5332–5339. doi: 10.1128/jb.174.16.5332-5339.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vries G E, Harms N, Hoogendijk J, Stouthamer A H. Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a n(GATC)n DNA-modifying property. Arch Microbiol. 1989;152:52–57. [Google Scholar]
- 11.Dreusch A, Burgisser D M, Heizmann C W, Zumft W G. Lack of copper insertion into unprocessed cytoplasmic nitrous oxide reductase generated by an R20D substitution in the arginine consensus motif of the signal peptide. Biochim Biophys Acta. 1997;1319:311–318. doi: 10.1016/s0005-2728(96)00174-0. [DOI] [PubMed] [Google Scholar]
- 12.Dreusch A, Riester J, Kroneck P M H, Zumft W G. Mutation of the conserved Cys165 outside of the Cu-A domain destabilizes nitrous oxide reductase but maintains its catalytic activity—evidence for disulfide bridges and a putative protein disulfide isomerase gene. Eur J Biochem. 1996;237:447–453. doi: 10.1111/j.1432-1033.1996.0447k.x. [DOI] [PubMed] [Google Scholar]
- 13.Farrar J A, Thomson A J, Cheesman M R, Dooley D M, Zumft W G. A model of the copper centres of nitrous oxide reductase (Pseudomonas stutzeri)—evidence from optical, EPR and MCD spectroscopy. FEBS Lett. 1991;294:11–15. doi: 10.1016/0014-5793(91)81331-2. [DOI] [PubMed] [Google Scholar]
- 14.Harms N, Reijnders W N M, Anazawa H, Van der Palen C J N M, Van Spanning R J M, Oltmann L F, Stouthamer A H. Identification of a 2-component regulatory system controlling methanol dehydrogenase synthesis in Paracoccus denitrificans. Mol Microbiol. 1993;8:457–470. doi: 10.1111/j.1365-2958.1993.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff S, Henikoff J G. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 16.Hoeren F U, Berks B C, Ferguson S J, McCarthy J E G. Sequence and expression of the gene encoding the respiratory nitrous-oxide reductase from Paracoccus denitrificans. New and conserved structural and regulatory motifs. Eur J Biochem. 1993;218:49–57. doi: 10.1111/j.1432-1033.1993.tb18350.x. [DOI] [PubMed] [Google Scholar]
- 17.Holloway P, McCormick W, Watson R J, Chan Y K. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J Bacteriol. 1996;178:1505–1514. doi: 10.1128/jb.178.6.1505-1514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroneck P M, Antholine W A, Riester J, Zumft W G. The cupric site in nitrous oxide reductase contains a mixed-valence [Cu(II),Cu(I)] binuclear center: a multifrequency electron paramagnetic resonance investigation. FEBS Lett. 1988;242:70–74. doi: 10.1016/0014-5793(88)80987-6. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.McGuirl M A, Nelson L K, Bollinger J A, Chan Y K, Dooley D M. The nos (nitrous oxide reductase) gene cluster from the soil bacterium Achromobacter cycloclastes: cloning, sequence analysis, and expression. J Inorg Biochem. 1998;70:155–169. doi: 10.1016/s0162-0134(98)10001-6. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Nicholas D J D, Nason A. Determination of nitrate and nitrite. Methods Enzymol. 1957;3:981–984. [Google Scholar]
- 23.Nicholas K B, Nicholas H B, Jr, Deerfield D W., II EMBNEW NEWS. 1997;4:14. [Google Scholar]
- 24.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Page M D, Ferguson S J. Apo forms of cytochrome c550 and cytochrome cd1 are translocated to the periplasm of Paracoccus denitrificans in the absence of haem incorporation caused either by mutation or inhibition of haem synthesis. Mol Microbiol. 1990;4:1181–1192. doi: 10.1111/j.1365-2958.1990.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 26.Rainey F A, Kelly D P, Stackebrandt E, Burghardt J, Hiraishi A, Katayama Y, Wood A P. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int J Syst Bact. 1999;49:645–51. doi: 10.1099/00207713-49-2-645. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd. ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders N F W, Houben E, Koefoed S, De Weert S, Reijnders W N M, Westerhoff H V, de Boer A P N, van Spanning R J M. Transcription regulation of the nir gene cluster encoding nitrite reductase of Paracoccus denitrificans involves NNR and NirI, a novel type of membrane protein. Mol Microbiol. 1999;34:24–36. doi: 10.1046/j.1365-2958.1999.01563.x. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Spanning R J M, Houben E, Reijnders W N, Spiro S, Westerhoff H V, Saunders N. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J Bacteriol. 1999;181:4129–4132. doi: 10.1128/jb.181.13.4129-4132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Spanning R J M, Wansell C W, Reijnders W N M, Harms N, Ras J, Oltmann L F, Stouthamer A H. A method for introduction of unmarked mutations in the genome of Paracoccus denitrificans: construction of strains with multiple mutations in the genes encoding periplasmic cytochromes c550, c551i, and c553i. J Bacteriol. 1991;173:6962–6970. doi: 10.1128/jb.173.21.6962-6970.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viebrock A, Zumft W G. Molecular cloning, heterologous expression and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J Bacteriol. 1988;170:4658–4668. doi: 10.1128/jb.170.10.4658-4668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Von Wachenfeldt C, De Vries S, Van Der Oost J. The CuA site of the caa3-type oxidase of Bacillus subtilis is a mixed-valence binuclear copper centre. FEBS Lett. 1994;340:109–113. doi: 10.1016/0014-5793(94)80182-7. [DOI] [PubMed] [Google Scholar]
- 35.Zumft W G, Dreusch A, Lochelt S, Cuypers H, Friedrich B, Schneider B. Derived amino acid sequences of the nosZ gene from Alcaligenes eutrophus, Pseudomonas aeruginosa and Pseudomonas stutzeri reveal potential copper-binding residues. Eur J Biochem. 1992;208:31–40. doi: 10.1111/j.1432-1033.1992.tb17156.x. [DOI] [PubMed] [Google Scholar]
- 36.Zumft W G, Viebrock-Sambale A, Braun C. Nitrous oxide reductase from denitrifying Pseudomonas stutzeri. Genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. Eur J Biochem. 1990;192:591–599. doi: 10.1111/j.1432-1033.1990.tb19265.x. [DOI] [PubMed] [Google Scholar]