Abstract
Macrophages are armed with multiple oxygen-dependent and -independent bactericidal properties. However, the respiratory burst, generating reactive oxygen species, is believed to be a major cause of bacterial killing. We exploited the susceptibility of Escherichia coli in macrophages to characterize the effects of the respiratory burst on intracellular bacteria. We show that E. coli strains recovered from J774 macrophages exhibit high rates of mutations. We report that the DNA damage generated inside macrophages includes DNA strand breaks and the modification 8-oxo-2′-deoxyguanosine, which are typical oxidative lesions. Interestingly, we found that under these conditions, early in the infection the majority of E. coli cells are viable but gene expression is inhibited. Our findings demonstrate that macrophages can cause severe DNA damage to intracellular bacteria. Our results also suggest that protection against the macrophage-induced DNA damage is an important component of the bacterial defense mechanism within macrophages.
Reactive oxygen intermediates can damage proteins, nucleic acids, and cell membranes and have been implicated in cancer, aging, and numerous degenerative diseases. To counter oxidative stress, both prokaryotic and eukaryotic cells maintain inducible defense systems to detoxify the oxidants and to repair the damage. The antioxidant defense systems have been best characterized in Escherichia coli and Salmonella enterica serovar Typhimurium, in which the OxyR and SoxR transcription factors activate genes to protect against H2O2 and O2·−, respectively (reviewed in reference 28).
While cells of aerobic organisms generate deleterious reactive oxygen metabolites under normal physiological conditions, stimulated macrophages generate reactive oxygen and nitrogen species as a defense mechanism during infection. Macrophages are armed with multiple oxygen-dependent and -independent bactericidal properties (9). However, the respiratory burst, which generates reactive oxygen and nitrogen species, is believed to be a major cause of bacterial killing. In chronic granulomatous disease, the neutrophils are incapable of producing the respiratory burst, and individuals with chronic granulomatous disease have high rates of mortality due to bacterial infections (20, 23). The deleterious effects of reactive oxygen and nitrogen species have been demonstrated with bacterial cells in culture in numerous studies (28). However, the toxic effects of these reactive species have not been characterized in phagocytosed bacteria. Given that S. enterica serovar Typhimurium recombination-deficient (recA and recBC) mutants showed attenuated virulence in mice and increased sensitivity in macrophages, DNA damage might be an important consequence of the activities inside macrophages (4, 26). In addition, it was shown that human peripheral phagocytes have some mutagenic activity against S. enterica serovar Typhimurium while phagocytes from patients with chronic granulomatous disease do not (32). Therefore, we examined the macrophage-induced lesions in phagocytosed, intracellular bacteria.
Although S. enterica serovar Typhimurium and E. coli are genetically related and share many functions to counter oxidative stress, E. coli is sensitive to the intracellular macrophage environment while S. enterica serovar Typhimurium cells multiply within macrophages (3). We exploited the difference in the abilities of E. coli and S. enterica serovar Typhimurium to survive within macrophages to characterize the effects of the respiratory burst on intracellular bacteria and to examine the intracellularly induced lesions. We present direct evidence that macrophages can cause severe DNA damage to intracellular bacteria.
MATERIALS AND METHODS
Plasmid construction.
To construct PoxyS-gfp (pSA8), 135 nucleotides of 5′ promoter sequences of the oxyS gene, including the OxyR binding region, were PCR amplified from S. enterica serovar Typhimurium LT2 and E. coli MC4100 chromosomal DNA using primers (5′-TCAGGATCCCGAATATTCATTATTCATC and 5′-CGGTCGACGTCTTCAAGGGTTAAACG). The LT2 fragment was then digested with BamHI and SalI and cloned into the corresponding sites of pKK177-3 (2). The fluorescence-enhanced green fluorescent protein (GFP) gene (gfp) of Aquorea victorea (SmaI and HindIII) from plasmid pGFPmut3 (6) was then cloned downstream the oxyS promoter into SalI (filled in) and HindIII. To construct PoxySEc-gfp (pSA9), the MC4100 fragment (filled in) was cloned into the BamHI sites (filled in) of pSA8. To construct Ptac-gfp-lacI (pSA11), the lacI gene (SalI filled in) from plasmid pDMI was cloned into the unique HindIII site (filled in) of pKK177-3, generating pSA10. The gfp gene (EcoRI and PstI) from pGFPmut3 was then cloned into the corresponding sites of pSA10.
Bacterial strains and media.
The bacterial strains used in this study were MC4100 [F−araD139 Δ(arg-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR] (27); CC102 [ara Δ (lac-proB)XIII thi F′-lacI378 lacZ461 proA+ B+) (7); CC104, which is identical to CC102 except for the lacZ mutation; SL1344 (hisG46) (13); LT2 (wild type) (18); and LT2 (galE zbi-812::Tn10) (24). Strains were routinely grown in Luria-Bertani (LB) medium. CC102 and CC104 were grown in Vogel-Bonner minimal medium (18) supplemented with biotin (5 mg/liter) and dextrose (0.5%) to maintain the episome.
Macrophage-induced mutagenesis.
J774 macrophages (5 × 105 to 10 × 105/well) were seeded in 24-well microtiter plates containing F-12 medium supplemented with 10% fetal calf serum. The next day the cells were activated with phorbol 12-myristate 13-acetate (PMA) (6 μg/ml) and infected (multiplicity of infection [MOI], 10:1) with E. coli cultures grown to mid- to late-log phase or with S. enterica serovar Typhimurium cultures grown without agitation to late log phase (17). At 30 min after infection, the macrophages were washed and incubated for an additional 30 or 60 min in medium containing gentamicin at 50 μg/ml. Thereafter, the cells were washed with phosphate-buffered saline and lysed in 1% Triton X-100 for 10 min. To determine the frequencies of mutagenesis, the recovered intracellular bacteria were diluted in fresh medium (1:5) and grown for 18 h as previously described (1, 7). The overnight cultures were plated on LB plates for viable cells, on MacConkey lactose plates for Lac+ revertants, or on LB plates containing 100 μg of rifampin per ml for Rifr mutants. The numbers of Rifr mutants and of Lac+ revertants were normalized to the numbers of viable cells in the overnight cultures. As a control, overnight cultures after mock infection were plated as above. Where indicated, we monitored the rates of mutagenesis of bacteria infecting epithelial HeLa cells as a control for the macrophage experiments. Infection of HeLa cells (3 × 105 to 5 × 105/well) with bacterial strains (MOI, 25:1) and analysis of mutagenesis were done as described above. To achieve invasion of HeLa cells by E. coli, the bacteria were transformed with plasmid pBF1001 expressing the inv gene of Yersinia pseudotuberculosis, previously shown to enable noninvasive bacteria to invade HeLa cells (15).
Measurement of oxo8dG.
J774 macrophages (4 × 107) were activated (with PMA at 6 μg/ml) and infected for 30 min with E. coli bearing pKK177-3. Thereafter, the cells were washed and further incubated for 15 min in medium containing gentamicin (50 μg/ml). Plasmid DNA was extracted from intracellular bacteria by the alkali lysis procedure. The samples were treated with RNase A (0.1 mg/ml) for 60 min at 37°C. The RNase A was then removed by phenol-chloroform treatment, and the DNA was precipitated with sodium acetate and ethanol. The recovered DNA was hydrolyzed with 0.5 U of nuclease P1 (Sigma) in 20 mM sodium acetate (pH 4.8) for 30 min at 37°C and then incubated in 0.1 M Tris-HCl (pH 7.4)–0.02 U of E. coli alkaline phosphatase (Sigma) for 60 min at 37°C (25). The hydrolyzed samples were analyzed by high-performance liquid chromatography with electrochemical detection (Kontron) using a C18 reversed-phase analytical column (LichroCart-5μ; 250 by 4 mm [Merck]). 8-Oxo-2′-deoxyguanosine (oxo8dG) was detected using an LC4A amperometric electrochemical detector (BAS, West Lafayette, Ind.) with an applied potential of +0.6 V. The intact nucleosides including 2′-deoxyguanosine (dG) were analyzed online with a UV detector (at 260 nm) (Spectra series UV100 [Thermo Separation Products]). The eluent was 10% methanol–50 mM potassium phosphate (pH 5.5), and the flow rate was 0.7 ml/min. External standards (Sigma) were used to calculate the amounts of oxo8dG and dG. The number of molecules of oxo8dG obtained was 92 (control, before infection) and 1,789 (postinfection) per 104 dG. Plasmid pKK177-3 (2,901 bp) contains 717 dG. A total of 104 dG is equivalent to 14 plasmids. The amount of oxo8dG formed per plasmid molecule is given in Results.
Analysis of DNA topology within macrophages.
J774 macrophages plated in 10-cm-diameter dishes (107 macrophages) were activated and infected with E. coli (MC4100) or S. enterica serovar Typhimurium (SL1344) bearing pKK177-3. Plasmid DNA was extracted by alkali lysis both before infection and from intracellular bacteria 30 min after infection. Intracellular bacteria were recovered as described above. The DNA was quantified by the dot blot assay using γ-32P-end-labeled primer 374 (5′-CCTGTGTGAAATTCTTATCC) corresponding to pKK177-3. Equal amounts of DNA were separated on a 1.4% agarose gel (Bethesda Research Laboratories) containing 10 μg of chloroquine (Sigma) per ml and transferred to a nylon membrane. The membrane was probed with labeled primer 374.
Survival in macrophages.
J774 macrophages (∼106) seeded in 24-well microtiter plates were activated and infected with bacterial cultures as above (MOI, 10:1). At 20 min after infection, the macrophages were washed and lysed with 1% Triton X-100, and aliquots were plated to determine the number of viable intracellular bacteria or incubated for an additional 20, 40, or 70 min in medium containing gentamicin (50 μg/ml) and then plated.
Analysis of GFP expression.
Cultures of E. coli (MC4100) and S. enterica serovar Typhimurium (SL1344) carrying the plasmid PoxyS-gfp (pSA8) or Ptac-gfp-lacI (pSA11) were grown in LB medium to an absorbance at 600 nm of 0.25. The cultures were split; half of each culture was treated with 0.2 mM H2O2 or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the other half remained untreated. At the indicated time points, samples (0.7 ml) were fixed with 0.7 ml of 3.2% paraformaldehyde, washed, and resuspended in 0.5 ml of filtered phosphate-buffered saline. GFP expression was analyzed with a FACScan cytometer (Becton Dickinson).
Immunostaining and confocal microscopy.
PMA-activated J774 (∼106) cells seeded on coverslips in 24-well microtiter plates were infected (MOI, 10:1) with E. coli/PoxyS-gfp and S. enterica serovar Typhimurium/PoxyS-gfp cultures as before. At 30 min of infection, the macrophages were washed with PBS, fixed in 2% PFA for 10 min, and extensively washed with PBS. After fixation, the macrophages were permeabilized with 0.1% Triton X-100 for 5 min, washed, incubated with 1 ml of antibodies raised against S. enterica serovar Typhimurium (1:50) or E. coli (1:100) for 60 min, and then stained with 2 ml of Cy5 (Jackson Immuno Research Inc.)-conjugated secondary antibodies (1:400) for an additional 60 min. The coverslips were placed on 2 μl of mounting solution (50% glycerol, 0.1% sodium azide, and 3% 1,4-diazabicyclo[2.2.2]octane [Sigma] in PBS), sealed with UHU glue, and analyzed by confocal microscopy. For analysis of Ptac-gfp-lacI expression within macrophages, J774 were infected for 30 min as above, washed, and further incubated for 40 min in F-12 medium containing fetal calf serum (10%), gentamicin (50 μg/ml), and IPTG (1 mM). Fixation and immunostaining were carried out as above.
RESULTS
Mutation frequencies of E. coli strains increase within J774 macrophages.
Much of the toxicity of the oxygen intermediates is attributed to DNA damage (14). Therefore, we examined the effects of the respiratory burst by monitoring the frequencies of mutagenesis of bacteria within J774 macrophages. J774 macrophage-like cells are extensively used as a model system to study microbe-macrophage interactions. We monitored the appearance of rifampin-resistant (Rifr) bacteria after residing within J774 cells compared to that of untreated bacteria or bacteria recovered from control epithelial HeLa cells. To achieve invasion of HeLa cells by E. coli, the bacteria were transformed with a plasmid expressing the inv gene of Y. pseudotuberculosis, previously shown to enable noninvasive bacteria to invade HeLa cells (15). HeLa cells and J774 macrophage-like cells activated with PMA were infected with E. coli MC4100 cells for 60 min. Thereafter, intracellular bacteria were recovered and grown, and aliquots were plated on rifampin-containing medium. The numbers of rifampin-resistant mutants were normalized to the numbers of viable cells. E. coli MC4100 residing in macrophages exhibited a significant increase in the number of Rifr mutants compared to the untreated control cultures (Table 1). As a control for the experimental design, we assayed mutation frequencies of S. enterica serovar Typhimurium, which is known to survive and multiply in J774 macrophages. S. enterica serovar Typhimurium SL1344 residing in macrophages exhibited almost the same levels of Rifr mutants as the untreated cultures did (Table 1). We also compared the mutation frequencies of isogenic smooth and rough strains of S. enterica serovar Typhimurium LT2 (LT2 and LT2 galE, respectively) after residing in J774 macrophages. The smooth and rough variants of S. enterica serovar Typhimurium were equally resistant to the macrophage-induced DNA damage (Table 1).
TABLE 1.
Rates of spontaneous and intracellularly induced mutagenesis
| Straina | No. of bacteria/109 cells
|
Fold increase relative to control | ||
|---|---|---|---|---|
| Untreated | HeLa | J774 | ||
| Rifr mutants | ||||
| E. coli strainsb | ||||
| MC4100c | 10.2 ± 7.0 | 9.2 ± 9.1 | 124.5 ± 126.2 | 12–14 |
| CC102 | 6.5 ± 6.5 | 48.6 ± 19.0 | 7.5 | |
| CC104 | 8.7 ± 5.6 | 70.7 ± 38.3 | 8 | |
| Salmonella strainsc | ||||
| SL1344 | 5.6 ± 4.3 | 5.3 ± 3.9 | 7.9 ± 5.3 | 1.5 |
| LT2 | 4.4 ± 0.9 | 5.0 ± 1.8 | 1.0 | |
| LT2 galE | 6.2 ± 1.6 | 6.1 ± 2 | 1.0 | |
| Lac+ revertants | ||||
| E. coli strains | ||||
| CC102 | 11.0 ± 6.4 | 40.2 ± 21.2 | 3.6 | |
| CC104 | 8.3 ± 5.7 | 30.0 ± 11.2 | 3.6 | |
SL1344 and LT2 are smooth strains, while LT2 galE and the E. coli strains are rough.
P < 0.0005 was calculated for MC4100 (Rifr), CC102 (Rifr and Lac+), and CC104 (Rifr and Lac+) (Mann-Whitney).
To enable the invasion of HeLa cells, E. coli MC4100 was transformed with plasmid pBF1001 expressing the inv gene of Yersinia pseudotuberculosis (15). The number of J774-induced Rifr mutants obtained with E. coli MC4100 without plasmid pBF1001 was 132.8 ± 100.7 (n = 21 colonies).
The E. coli lacZ strains CC102 and CC104 allow rapid detection of base substitutions by monitoring the number of lacZ+ revertant colonies (7). CC102 allows the detection of GC-to-AT transitions, and CC104 allows the detection of the low-occurrence GC-to-TA transversions, which are typical of oxidative damage. We examined the rates of the appearance of both Rifr mutants and lacZ+ revertants of these strains after residing in macrophages. Both CC102 and CC104 exhibited a significant increase in the rate of lacZ+ revertants due to the intracellular J774 environment (Table 1). As expected, the increase in the frequency of Rifr colonies was higher, similar to that observed with E. coli MC4100 cells (Table 1). These results indicate that J774 macrophages have strong mutagenic activity that results in both transitions and transversions and that E. coli cells are susceptible to the macrophage-intracellular mutagenic environment.
J774-induced DNA damage is typical of oxidative-stress damage.
Oxidative damage to DNA results in a number of typical lesions including the modification oxo8dG. To examine whether the damage occurring inside macrophages is due to reactive oxygen species, we monitored the appearance of oxo8dG in E. coli plasmid DNA directly by high-performance liquid chromatography with electrochemical detection. We found that the number of oxo8dG molecules increased by ≈20-fold after residing in macrophages (128 ± 51 oxo8dG/plasmid) compared to that in the control untreated bacteria (6.6 ± 0.7 oxo8dG/plasmid). This result indicates that the DNA damage induced in macrophages includes oxidative damage due to reactive oxygen species.
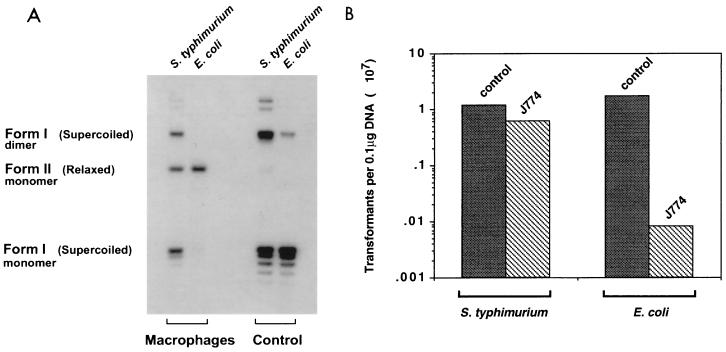
In addition to causing single-point mutations, hydroxyl radicals generated by the reduction of hydrogen peroxide cause single-strand breaks in DNA. To further characterize the damage occurring within macrophages, we monitored the plasmid DNA topology of the intracellular bacteria shortly after they were taken up by macrophages. E. coli MC4100 cells carrying a small reporter plasmid were used to infect J774 macrophages. Shortly after infection, the cells were lysed and the plasmid DNA was extracted from intracellular bacteria and analyzed on agarose gels containing chloroquine. Upon macrophage entry, the reporter plasmid within E. coli undergoes a dramatic change in topology, such that the majority is in a nicked circular form (Fig. 1A). Analysis of plasmid DNA extracted from intracellular S. enterica serovar Typhimurium showed that the majority of this DNA remained intact, in a negative supercoiled form (Fig. 1A).
FIG. 1.
(A) Plasmid DNA topology within macrophages. J774 macrophages were activated and infected with E. coli MC4100 or S. enterica serovar Typhimurium SL1344 bearing pKK177-3. Plasmid DNA was extracted by the alkali lysis procedure before infection (control) and from intracellular bacteria 30 min after infection (Macrophages). Equal amounts of DNA, as quantified by dot blot analysis using γ-32P-end labeled primer 374 corresponding to pKK177-3 (data not shown), were separated on a 1.4% agarose gel containing 10 μg of chloroquine per ml and transferred to a nylon membrane. The membrane was probed with labeled primer 374. (B) Equal amounts of plasmid DNA from the experiment in panel A were used to transform E. coli and S. enterica serovar Typhimurium by electroporation.
Transformation of nicked circular DNA is less efficient than that of supercoiled DNA. To quantify the damage occurring within the macrophages, the plasmid DNA extracted from intracellular bacteria was introduced into fresh cultures of E. coli and S. enterica serovar Typhimurium. The transformation efficiency with DNA extracted from macrophage-treated E. coli was 200-fold lower than the transformation efficiency with DNA isolated from untreated control E. coli. As for S. enterica serovar Typhimurium, the transformation efficiencies for plasmids extracted from both macrophage-treated and control bacteria were approximately equal (Fig. 1B). These results further confirm that E. coli plasmid DNA within J774 is susceptible to strand breaks.
E. coli undergoes a gene expression arrest within J774 cells.
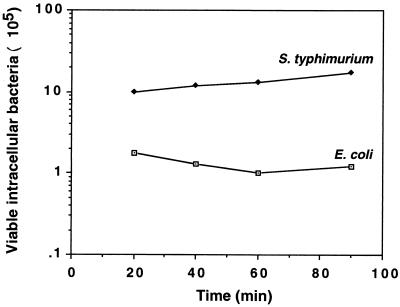
The severe DNA damage observed with E. coli residing in macrophages prompted us to examine the viability of E. coli during the time of infection and its ability to respond to the intracellularly induced stress. To analyze viability, activated J774 cells were infected with E. coli MC4100 or S. enterica serovar Typhimurium SL1344 for 20, 40, 60, and 90 min. Thereafter, macrophages were lysed and aliquots were plated to determine viable intracellular bacteria. The majority of the E. coli cells were found to be viable within the first 90 min of infection (Fig. 2). Staining of intracellular E. coli with propidium iodide, which stains the nucleic acids of dead cells, further confirmed this conclusion (data not shown). However, analysis of intracellular bacteria at 24 h after infection showed that while the number of S. enterica serovar Typhimurium cells increased substantially during infection, E. coli cells could not replicate inside macrophages and the numbers of viable cells were not increased (data not shown).
FIG. 2.
Survival in J774 macrophages. Macrophages were activated, infected with bacterial cultures for 20 min, washed, and lysed, and aliquots were plated to determine the number of viable intracellular bacteria or further incubated for an additional 20, 40, or 70 min in medium containing gentamicin. The results are means of four independent experiments, each carried out with three or four cultures of each strain. Shown are the numbers of viable intracellular bacteria from 20 to 90 min after infection. The number of viable cells at 20 min represents bacterial cells attached to or within macrophages. Since S. enterica serovar Typhimurium actively invades macrophages, the initial number of intracellular S. enterica serovar Typhimurium bacteria is larger than the initial number of intracellular E. coli bacteria.
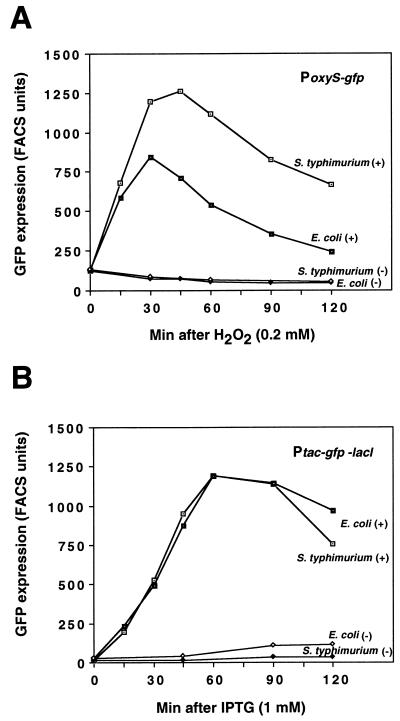
The ability of E. coli and S. enterica serovar Typhimurium to respond to the intracellularly induced stress was examined by monitoring the expression of two transcriptional fusions. In these fusions, the gfp gene encoding GFP was cloned downstream of either the oxyS or the tac promoter. The oxyS promoter is induced by the OxyR transcription factor in response to hydrogen peroxide (1). The tac promoter is under the control of the LacI repressor and can be induced by IPTG. We first tested expression of PoxyS-gfp and Ptac-gfp-lacI in E. coli and S. enterica serovar Typhimurium grown in LB medium and treated with hydrogen peroxide and IPTG, respectively. PoxyS-gfp expression in both E. coli and S. enterica serovar Typhimurium was similar, reaching its highest levels within 30 to 45 min after treatment (Fig. 3A). The pattern of IPTG-dependent GFP expression from Ptac-gfp-lacI was identical between S. enterica serovar Typhimurium and E. coli (Fig. 3B).
FIG. 3.
PoxyS-gfp induction by H2O2 (A) and Ptac-gfp induction by IPTG (B) in LB medium. Exponentially growing S. enterica serovar Typhimurium SL1344 and E. coli MC4100 bearing PoxyS-gfp and Ptac-gfp-lacI were treated with 0.2 mM H2O2 and 1 mM IPTG, respectively. The GFP expression of treated (+) and untreated (−) cultures was analyzed by fluorescence-activated cell sorting. A similar pattern of expression was obtained with the PoxyS E. coli clone (PoxySEc-gfp) (see Materials and Methods), although the levels were reduced twofold (data not shown).
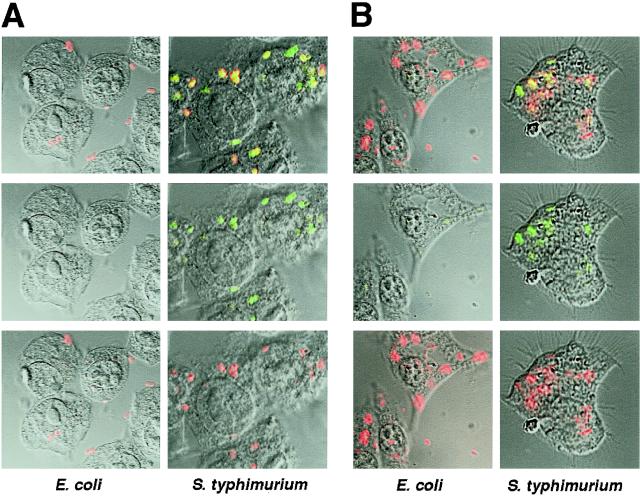
To test for intracellular bacterial GFP expression, J774 macrophage-like cells were infected with E. coli or S. enterica serovar Typhimurium bearing PoxyS-gfp (Fig. 4A) or Ptac-gfp-lacI (Fig. 4B). At 30 min postinfection, the cultures bearing the Ptac-gfp-lacI fusion were further treated with IPTG to activate the promoter Ptac. To localize the bacteria, the infected macrophages were immunostained with anti-E. coli or anti-Salmonella antibodies and analyzed by confocal microscopy. We found that intracellular E. coli did not show expression with either fusion (Fig. 4). In contrast, almost all intracellular S. enterica serovar Typhimurium bacteria colocalized with GFP expression of both fusions (Fig. 4). Longer or shorter periods of infection with E. coli did not result in GFP expression (data not shown). The observation that E. coli was unable to express GFP from both fusions indicates that the macrophage environment leads to an immediate arrest in gene expression of E. coli cells. The enteropathogenic E. coli strain was similarly inactive in macrophages, showing that these effects are not restricted to nonpathogenic E. coli (data not shown).
FIG. 4.
(A) PoxyS-gfp expression within macrophages. J774 cells activated with PMA were infected with S. enterica serovar Typhimurium/PoxyS-gfp or E. coli/PoxyS-gfp. At 30 min after infection, the macrophages were fixed, stained with anti-E. coli or anti-Salmonella antibodies and Cy5-conjugated secondary antibody, and subjected to confocal microscopy. (Bottom) Cy5 conjugate (red); (middle) GFP (green); (top) Cy5 and GFP superimposed (yellow). (B) Ptac-gfp expression within macrophages. J774 cells activated with PMA were infected with S. enterica serovar Typhimurium/Ptac-gfp-lacI or E. coli/Ptac-gfp-lacI. At 30 min after infection, the macrophages were washed and further incubated for 40 min in medium containing gentamicin (50 μg/ml) and IPTG (1 mM). Thereafter, the macrophages were fixed and stained as in panel A and subjected to confocal microscopy. (Bottom) Cy5 conjugate (red); (middle) GFP (green); (top) Cy5 and GFP superimposed (yellow).
DISCUSSION
We have shown that J774 macrophage-like cells can cause severe DNA damage to intracellular bacteria. E. coli residing within J774 cells is susceptible to macrophage-induced mutagenesis and undergoes an immediate gene expression arrest. We have also shown that the mutations generated inside macrophages correspond to typical oxidative lesions including DNA strand breaks and the modification oxo8dG.
Within macrophages, bacteria are exposed to host-induced nutrient limitation, acidification, toxic peptides, and oxidative burst, of which the last is believed to be a major cause of bacterial killing. Knockout mutant mice, incapable of producing the respiratory burst, exhibit impaired survival after bacterial infection (26). Furthermore, the attenuated virulence in mice of an S. enterica serovar Typhimurium recombination-deficient mutant (recBC) (4, 26) indicates that DNA damage is an important consequence of the activities inside macrophages. Weitzman and Stossel showed that human peripheral phagocytes were capable of some mutagenic activity against S. enterica serovar Typhimurium while phagocytes from patients with chronic granulomatous disease were not (32). Using E. coli, we found that J774 macrophage cells have strong mutagenic activity. Since the number of oxo8dG molecules and DNA strand breaks increased inside macrophages, we propose that the macrophage-induced DNA damage is due, at least in part, to oxygen radicals. Given that J774 macrophage-like cells are considered to be less robust than primary macrophages, macrophage-induced DNA damage appears to be even more effective in living organisms.
Upon macrophage entry, E. coli appears to undergo events of a synergistic nature, i.e., immediate transcription-translation arrest and DNA damage. Whether an initial DNA damage results in expression arrest or whether an immediate metabolism arrest renders the cells susceptible to more DNA damage is not clear. The absolute inability of E. coli to respond suggests that within macrophages, E. coli inhibition might be enhanced by the presence of a combination of antimicrobial agents. For example, it has been shown that treatment of E. coli with a combination of nitric oxide (NO) and hydrogen peroxide induces high levels of DNA strand breaks, leading to a dramatic (1,000-fold) increase in hydrogen peroxide-mediated killing (21). That the enteropathogenic E. coli strain was similarly inactive in macrophages suggests that these effects are not restricted to nonpathogenic E. coli.
What makes S. enterica serovar Typhimurium less susceptible to DNA damage within macrophages is not clear. It has been observed that S. enterica serovar Typhimurium mutants with mutations in genes known to affect the oxidative stress response, such as oxyR, ahpC, katG, katE, sodA, and soxS, are not more sensitive to killing by human neutrophils or murine macrophages and do not show attenuated virulence in mice (5, 8, 11, 22, 29, 30). It is possible that protection against oxidative stress results from parallel and redundant activities, as has been observed with a newly discovered sodC gene. S. enterica serovar Typhimurium mutants lacking both of the sodC genes were found to be less lethal for mice than were mutants lacking either gene alone (10). It is also possible that not all S. enterica serovar Typhimurium cells are exposed to the respiratory burst. It was recently shown that the type III protein secretion system encoded by pathogenicity island 2 allows S. enterica serovar Typhimurium to avoid NADPH oxidase-dependent killing by macrophages (31). We observed that the majority of the S. enterica serovar Typhimurium cells residing within macrophages expressed GFP from the hydrogen peroxide-inducible oxyS promoter, suggesting that under the conditions used, most S. enterica serovar Typhimurium cells were exposed to the macrophage respiratory burst. In addition, it is possible that S. enterica serovar Typhimurium possesses distinct defenses that protect against macrophage-induced DNA damage. These defenses could result from enhanced activities of scavenging and/or repair enzymes or could be due to DNA binding proteins protecting the S. enterica serovar Typhimurium chromosome. For example, it has been shown that the nonspecific DNA binding protein Dps reduces DNA strand breaks and point mutations by direct protection of the DNA (19). The crystal structure of Dps revealed that the protein is a ferritin homolog, suggesting that it may protect against DNA damage by sequestering iron (12). Whether S. enterica serovar Typhimurium harbors a novel function or whether protection results from a unique assembly of known functions is an important subject for future studies.
It is also intriguing to speculate that some macrophage mutagenic activity could actually help in bacterial evolution. The finding of a high incidence of mutators among isolates of pathogenic bacteria (16) supports the notion that the mutagenic activity of macrophages could also assist with bacterial evolution, leading to rapid adaptation to escape immune surveillance.
ACKNOWLEDGMENTS
We thank Yael Altuvia for performing the statistical tests. We thank R. Kolter, S. Miller, and K. Sanderson for strains.
This work was supported by grant number 95-00092 from the United States-Israel Binational Science Foundation and by the Human Frontier Science Program and by The Israel Science Foundation founded by The Academy of Sciences and Humanities Centers of Excellence Program (SA).
REFERENCES
- 1.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci USA. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier N A, Lipps C J, So M Y, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 5.Buchmeier N A, Libby S J, Xu Y, Loewen P C, Switala J, Guiney D G, Fang F C. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Investig. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 7.Cupples C G, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Densen P, Mandell G L. Granulocytic phagocytes. In: Mandell G L, Douglas R G, Bennet J E, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone, Inc.; 1995. pp. 78–101. [Google Scholar]
- 10.Fang F C, DeGroote M A, Foster J W, Baumler A J, Ochsner U, Testerman T, Bearson S, Giard J C, Xu Y, Campbell G, Laessig T. Virulent Salmonella typhimurium has two periplasmic Cu,Zn-superoxide dismutases. Proc Natl Acad Sci USA. 1999;96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang F C, Vazquez-Torres A, Xu Y. The transcriptional regulator SoxS is required for resistance of Salmonella typhimurium to paraquat but not for virulence in mice. Infect Immun. 1997;65:5371–5375. doi: 10.1128/iai.65.12.5371-5375.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant R, Filman D, Finkel S, Kolter R, Hogl J. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 13.Hoiseth K S, Stocker B A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 14.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 15.Isberg R R, Leong J M. Multiple β 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell. 1990;60:861–871. doi: 10.1016/0092-8674(90)90099-z. [DOI] [PubMed] [Google Scholar]
- 16.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 17.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maloy R S, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 19.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouy R, Fischer A, Vilmer E, Seger R, Griscelli C. Incidence, severity, and prevention of infections in chronic granulomatous disease. J Pediatr. 1989;114:555–560. doi: 10.1016/s0022-3476(89)80693-6. [DOI] [PubMed] [Google Scholar]
- 21.Pacelli R, Wink D A, Cook J A, Krishna M C, DeGraff W, Friedman N, Tsokos M, Samuni A, Mitchell J B. Nitric oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J Exp Med. 1995;182:1469–1479. doi: 10.1084/jem.182.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papp-Szabo E, Firtel M, Josephy P D. Comparison of the sensitivities of Salmonella typhimurium oxyR and katG mutants to killing by human neutrophils. Infect Immun. 1994;62:2662–2668. doi: 10.1128/iai.62.7.2662-2668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safe A F, Maxwell R T, Howard A J, Garcia R C. Relapsing Salmonella enteritidis infection in a young adult male with chronic granulomatous disease. Postgrad Med J. 1991;67:198–201. doi: 10.1136/pgmj.67.784.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanderson K E, Roth J R. Linkage map of Salmonella typhimurium edition VL. Microbiol Rev. 1983;47:410–453. doi: 10.1128/mr.47.3.410-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandstrom B E, Svoboda P, Granstrom M, Harms-Ringdahl M, Candeias L P. H2O2-driven reduction of the Fe3+-quin2 chelate and the subsequent formation of oxidizing species. Free Radic Biol Med. 1997;23:744–753. doi: 10.1016/s0891-5849(97)00058-0. [DOI] [PubMed] [Google Scholar]
- 26.Shiloh M U, MacMicking J D, Nicholson S, Brause J E, Potter S, Marino M, Fang F, Dinauer M, Nathan C. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 27.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 28.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 29.Taylor P D, Inchley C J, Gallagher M P. The Salmonella typhimurium AhpC polypeptide is not essential for virulence in BALB/c mice but is recognized as an antigen during infection. Infect Immun. 1998;66:3208–3217. doi: 10.1128/iai.66.7.3208-3217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsolis R M, Baumler A J, Heffron F. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect Immun. 1995;63:1739–1744. doi: 10.1128/iai.63.5.1739-1744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez-Torres A, Xu Y, Jones-Carson J, Holden D W, Lucia S M, Dinauer M C, Mastroeni P, Fang F C. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH-oxidase. Science. 2000;287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 32.Weitzman S A, Stossel T P. Mutation caused by human phagocytes. Science. 1981;212:546–547. doi: 10.1126/science.6259738. [DOI] [PubMed] [Google Scholar]