Abstract
An investigation of species of the genus Aspergillus present in arthropod, freshwater, and soil led to the discovery of seven undescribed species in Korea. Based on their morphological characteristics and molecular phylogeny analyses using a combined data set of β-tubulin (BenA) and calmodulin (CaM) sequences, the isolated strains CNUFC IGS2-5, CNUFC YJ1-19, CNUFC WD27, CNUFC U8-70, CNUFC AS2-24, CNUFC S32-1, and CNUFC U7-48, were identified as Aspergillus brunneoviolaceus, A. capensis, A. floccosus, A. inflatus, A. parvulus, A. polyporicola, and A. spelaeus, respectively. In the present study, the detailed morphological descriptions and phylogenetic relationships of these species are provided.
Keywords: Aspergillus, arthropod, freshwater, soil, morphology, phylogeny
1. Introduction
The genus Aspergillus (class: Eurotiomycetes; order: Eurotiales; family: Aspergillaceae) was first identified as asexual fungi for conidiophores resembling an aspergillum by Micheli in 1729 [1]. This genus classified into 6 subgenera and 27 sections [2,3]. Members of this genus are mainly environmental saprobes, acting as decomposers of organic materials and can also be found in vegetation, fruits, foods, indoor environments, water, soil, and air. Some Aspergillus species are of economic importance, producing itaconic acid used in polymer manufacturing and the cholesterol-lowering drug lovastatin [4], whereas others produce mycotoxins, cause food spoilage, promote development of allergies and other health problems, and also causes infections in humans and animals [3].
Aspergillus species identification presently relies on standardized methods based on morphological characteristics, multiloci DNA sequence analyses, and extrolite characterization. Molecular DNA markers are involved in sequencing of the internal transcribed spacer, β-tubulin (BenA), calmodulin (CaM), and the RNA polymerase II second largest subunit (RPB2) sequences. Currently, this genus consists of 446 species [2], only two of which is registered in Korea [5,6]. Furthermore, about 76 species of Aspergillus have been reported from Korea, in comparison to the recent publications of new species that have been discovered from other countries [7–10].
Therefore, the present study aimed to identify and provide a brief description of seven undescribed species belonging to five different sections of Aspergillus in Korea, that is, A. brunneoviolaceus, A. capensis, A. floccosus, A. inflatus, A. parvulus, A. polyporicola, and A. spelaeus, based on their morphological and molecular analyses. This study contributes to the knowledge on biodiversity of Aspergillus species in Korea.
2. Materials and methods
2.1. Sample collection and isolation
The samples collected from various locations, as listed in Table 1, were placed in sterile plastic bags and 50-mL falcon tubes and transferred to the laboratory. Serial dilutions were prepared for the isolations from freshwater and soil samples following the method described by Pangging et al. [11]. The body surface of arthropod was cut and placed onto potato dextrose agar (PDA; DifcoTM Becton, Dickinson and Co., Sparks, MD, USA) supplemented with penicillin (50 mg/L) and streptomycin (50 mg/L) to inhibit the growth of bacteria.
Table 1.
Information of isolates used in this study.
| Species | Strain no. | Source | Location |
|---|---|---|---|
| A. brunneoviolaceus | CNUFC IGS2-5 = QWJQFGC000000441 | Arthropod | Imgok-dong, Gwangsan-gu, Gwangju, Korea (35°13′05.2″ N 126°44′43.7″ E) |
| A. capensis | CNUFC YJ1-19 = NNIBRFG9303 | Freshwater | Jukrim-ri, Sora-myeon, Yeosu-si, Jeonnam Province, Korea (34°45′40.0″ N 127°37′21.8″ E) |
| A. floccosus | CNUFC WD27 = NNIBRFG9304 | Freshwater | Jeongdo-ri, Gugyedeung, Wando, Korea (34°18′46.0″ N 126°45′20.0″ E) |
| A. inflatus | CNUFC U8-70 = QWJQFGC000000299 | Rhizosphere soil | Hyeonpo-ri, Buk-myeon, Ulleung Island, Korea (37°31′13.9″ N 130°48′57.5″ E) |
| A. parvulus | CNUFC AS2-24 = IMYKFGC000000017 | Dry soil | Anmyeon-eup, Taean-gun, Anmyeondo, Korea (36°44′43.5″ N 126°17′54.0″ E) |
| A. polyporicola | CNUFC S32-1 = IMYKFGC000000060 | Rhizosphere soil | Miryang, Gyeongnam Province, Korea (35°29′48.4″ N 128°45′39.9″ E) |
| A. spelaeus | CNUFC U7-48 = QWJQFGC000000300 | Wildgrapes rhizosphere soil | Gitdaebong, Ulleung Island, Korea (37°30′22.7″ N 130°51′25.0″ E) |
Pure isolates were maintained in 20% glycerol at −80 °C and PDA slant tubes at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea as CNUFC IGS2-5, CNUFC YJ1-19, CNUFC WD27, CNUFC U8-70, CNUFC AS2-24, CNUFC S32-1, and CNUFC U7-48, as long-term preservations. Moreover, CNUFC IGS2-5, CNUFC U8-70, CNUFC AS2-24, CNUFC S32-1, and CNUFC U7-48 were deposited at the Collection of National Institute of Biological Resources (NIBR), Incheon, Korea. CNUFC YJ1-19 and CNUFC WD27 were deposited at the Culture Collection of the Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Korea.
2.2. Morphological characteristics
The seven undescribed species were cultured onto Czapek yeast autolysate agar (CYA), Malt extract autolysate agar (MEA) and Yeast extract sucrose agar (YES) [12] and further incubated at 25 °C in the dark for 7 days. An Olympus BX51 microscope with differential interference contrast optics (Olympus, Tokyo, Japan) was used to capture digital image fragments of mycelia that were removed from the cultures and placed on microscope slides with lactic acid (60%).
2.3. DNA extraction, PCR, and sequencing
Fungal isolates were cultured on PDA at 25 °C for 5–7 days. Genomic DNA was extracted using the Solg TM Genomic DNA Preparation Kit (Solgent Co. Ltd., Daejeon, Korea). The primer pairs Bt2a/Bt2b, T10/Bt2b [13], Ben2f/T22 [14] for BenA; Cmd5/Cmd6 [6], CF1/CF4 [15] for CaM were used for amplification. PCR amplification was performed according to the conditions described by Visagie et al. [12]. Thereafter, the PCR products were purified with an Accuprep PCR Purification Kit (Bioneer Corp., Daejeon, South Korea). Sequencing was done using the same primer pairs and then analyzed using ABI PRISM 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
2.4. Phylogenetic analysis
Sequences for the selected strains were aligned with reference sequences obtained from GenBank using Clustal_X version 2.1 [16] and were edited manually with Bioedit version 7.2.6.0 [17]. Maximum likelihood (ML) phylogenies were constructed using MEGA version X [18]. The sequence of Talaromyces flavus CBS 310.38 T was used as an out group. The sequences of the isolates in this study were deposited in the NCBI database under the accession numbers listed in Table 2.
Table 2.
GenBank accession numbers for fungal strains used in this study.
| GenBank Accession no. |
|||
|---|---|---|---|
| Species | Collection no. | BenA | CaM |
| A. acidohumus | DTO 340-H1 (T) | KX423623 | KX423634 |
| A. aculeatinus | CBS 121060 (T) | EU159220 | EU159241 |
| A. aculeatus | NRRL 5094 (T) | HE577806 | AJ964877 |
| A. alabamensis | CBS 125693 (T) | KP987049 | EU147583 |
| A. alboluteus | CBS 145855 (T) | MW478497 | MW478511 |
| A. allahabadii | NRRL 4539 (T) | EF669531 | EF669559 |
| A. ambiguus | NRRL 4737 (T) | EF669534 | EF669564 |
| A. ardalensis | CBS 134372 (T) | HG916683 | HG916725 |
| A. aureoterreus | NRRL 1923 (T) | EF669524 | EF669538 |
| A. barbosae | URM 5930 (T) | LR031377 | LR031392 |
| A. brunneo-uniseriatus | NRRL 4273 (T) | EF652123 | EF652138 |
| A. brunneoviolaceus | CBS 621.78 (T) | EF661105 | EF661147 |
| A. brunneoviolaceus | CNUFC IGS2-5 | OP168874 | OP168867 |
| A. capensis | DTO 179-E6 (T) | KJ775072 | KJ775279 |
| A. capensis | CNUFC YJ1-19 | OP168879 | OP168872 |
| A. carbonarius | NRRL 369 (T) | EF661099 | EF661167 |
| A. carneus | NRRL 527 (T) | EF669529 | EF669569 |
| A. cervinus | NRRL 5025 (T) | EF661251 | EF661261 |
| A. chaetosartoryae | NRRL 5501 (T) | EF652117 | EF652129 |
| A. christenseniae | CBS 122.56 (T) | FJ491639 | FJ491608 |
| A. chrysellus | NRRL 5084 (T) | EF652109 | EF652136 |
| A. citrinoterreus | CBS 138921 (T) | LN680657 | LN680685 |
| A. costaricaensis | CBS 115574 (T) | FJ629277 | FN594545 |
| A. cremeus | NRRL 5081 (T) | EF652120 | EF652125 |
| A. croceus | CCF 4405 (T) | LN873944 | LN873957 |
| A. dimorphicus | NRRL 3650 (T) | EF652111 | EF652135 |
| A. ellipticus | CBS 70779 (T) | AY585530 | EF661170 |
| A. elsenburgensis | CMV 011G4 (T) | MK451215 | MK451513 |
| A. europaeus | CCF 4409 (T) | LN909006 | LN909007 |
| A. flaschentraegeri | NRRL 5042 (T) | EF652113 | EF652130 |
| A. flavipes | NRRL 302 (T) | EU014085 | EF669549 |
| A. floccosus | CBS 116.37 (T) | FJ491714 | KP987066 |
| A. floccosus | CNUFC WD27 | OP168878 | OP168871 |
| A. floridensis | NRRL 62478 (T) | HE984412 | HE984429 |
| A. fumigatiaffinis | CMV 001G1 (T) | MK450913 | MK451390 |
| A. giganteus | NRRL 10 (T) | EF669789 | EF669857 |
| A. gorakhpurensis | NRRL 3649 (T) | EF652114 | EF652126 |
| A. hortai | NRRL 274 (T) | FJ491706 | KP987054 |
| A. hydei | KUMCC 18-0196 (T) | MT161679 | MT178247 |
| A. iizukae | NRRL 3750 (T) | EU014086 | EF669555 |
| A. indologenus | CBS 11480 (T) | AY585539 | AM419750 |
| A. inflatus | CBS 682.70 (T) | FJ531008 | FJ531090 |
| A. inflatus | CNUFC U8-70 | OP168877 | OP168870 |
| A. inusitatus | CBS 147044 (T) | MW478502 | MW478517 |
| A. iranicus | DTO 203-D7 (T) | KP987045 | KP987060 |
| A. itaconicus | NRRL 161 (T) | EF652118 | EF652140 |
| A. japonicus | CBS 114.51 (T) | HE577804 | FN594551 |
| A. lanuginosus | NRRL 4610 (T) | EU014080 | EF669562 |
| A. kanagawaensis | CBS 538.65 (T) | FJ491640 | FJ491597 |
| A. koreanus | EML-GSNP1-1 (T) | KX216530 | KX216528 |
| A. luppii | NRRL 6326 (T) | EU014079 | EF669575 |
| A. melleus | NRRL 5103 (T) | EF661326 | EF661391 |
| A. microcysticus | NRRL 4749 (T) | EF669515 | EF669565 |
| A. micronesiensis | DTO 267D5 (T) | KJ775085 | KP987067 |
| A. movilensis | CCF 4410 (T) | HG916697 | HG916740 |
| A. neoafricanus | NRRL 2399 (T) | EF669516 | EF669543 |
| A. neoflavipes | CBS 260.73 (T) | EU014084 | EF669572 |
| A. neoindicus | CBS 444.75 (T) | EF669532 | EF669574 |
| A. neoniger | CBS 115656 (T) | FJ491691 | FJ491700 |
| A. neoniveus | CBS 261.73 (T) | EU014098 | EF669570 |
| A. niger | NRRL 326 (T) | EF661089 | EF661154 |
| A. niveus | CBS 115.27 (T) | EF669528 | EF669573 |
| A. novoguineensis | CBS 906.96 (T) | FJ491641 | FJ491605 |
| A. nutans | NRRL 4364 (T) | EF661249 | EF661262 |
| A. olivimuriae | NRRL 66783 (T) | MH492010 | MH492011 |
| A. okavangoensis | CBS 147420 (T) | MW480789 | MW480707 |
| A. ostianus | NRRL420 (T) | EF661324 | EF661385 |
| A. oxumiae | CCDCA 11546 (T) | MN521388 | MN531842 |
| A. parvulus | NRRL 4753 (T) | EF661247 | EF661259 |
| A. parvulus | CNUFC AS2-24 | OP168873 | OP168866 |
| A. polyporicola | NRRL 32683 (T) | EU014088 | EF669553 |
| A. polyporicola | NRRL 58570 | LM644274 | LM644252 |
| A. polyporicola | CNUFC S32-1 | OP168875 | OP168868 |
| A. pseudodeflectus | CMV 005H9 | MK451064 | MK451498 |
| A. pseudoterreus | NRRL 4017 (T) | EF669523 | EF669556 |
| A. purpureocrustaceus | CMV 008B3 (T) | MK451138 | MK451515 |
| A. recifensis | URM 6605 (T) | LR031370 | LR031385 |
| A. saccharolyticus | CBS 127449 (T) | HM853553 | HM853554 |
| A. serratalhadensis | URM 7866 (T) | LT993222 | LT993223 |
| A. sigurros | CMV 005I4 (T) | MK451066 | MK451512 |
| A. spelaeus | CCF 4425 (T) | HG916698 | HG916741 |
| A. spelaeus | EMSL 4874 | MW478506 | MW478525 |
| A. spelaeus | CNUFC U7-48 | OP168876 | OP168869 |
| A. stromatoides | CBS 500.65 (T) | FJ531038 | EF652127 |
| A. subnutans | CBS 129386 (T) | KX528454 | KX528455 |
| A. suttoniae | UTHSCSA DI14-215 (T) | LT899536 | LT899589 |
| A. tardus | CBS 433.93 (T) | FJ531001 | FJ531084 |
| A. templicola | DTO 270 C-6 (T) | KJ775092 | KJ775394 |
| A. terreus | CBS 601.65 (T) | EF669519 | EF669544 |
| A. transcarpathiucs | CBS 423.68 (T) | FJ491632 | FJ491610 |
| A. trinidadensis | NRRL 62479 (T) | HE984420 | HE984434 |
| A. tubingensis | NRRL 4875 (T) | EF661086 | EF661151 |
| A. urmiensis | CBS 139558 (T) | KP987041 | KP987056 |
| A. uvarum | ITEM 4834 (T) | AM745751 | AM745755 |
| A. violaceofuscus | CBS 123.27 (T) | FJ491685 | FJ491698 |
| A. wentii | NRRL 375 (T) | EF652106 | EF652131 |
| A. wisconsinensis | CBS 413.64 (T) | FJ491638 | FJ491609 |
Bold letters indicate isolates and accession numbers determined in our study.
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CCDCA: culture collection at Federal University of Lavras, Minas Gerais, Brazil; CCF: Culture Collection of Fungi at the Department of Botany of Charles University in Prague; CMV: working collection housed at the PPRI; CNUFC: Chonnam National University Fungal Collection (Gwangju, South Korea); DTO: Internal collection of Dept. Applied and Industrial Mycology housed at CBS; ITEM: Microbial Culture Collection, Institute of Sciences of Food Production, Bari, Italy; KUMCC: Culture collection of Kunming Institute of Botany, Yunnan, China; NRRL: ARS culture collection, Peoria, IL, USA; URM: Padre Camille Torrend Herbarium, South America; UTHSCSA: Collection of Fungus Testing Laboratory, University of Texas, Health Science Center, San Antonio, USA; T: ex-type strain.
3. Results
3.1. Phylogenetic analysis
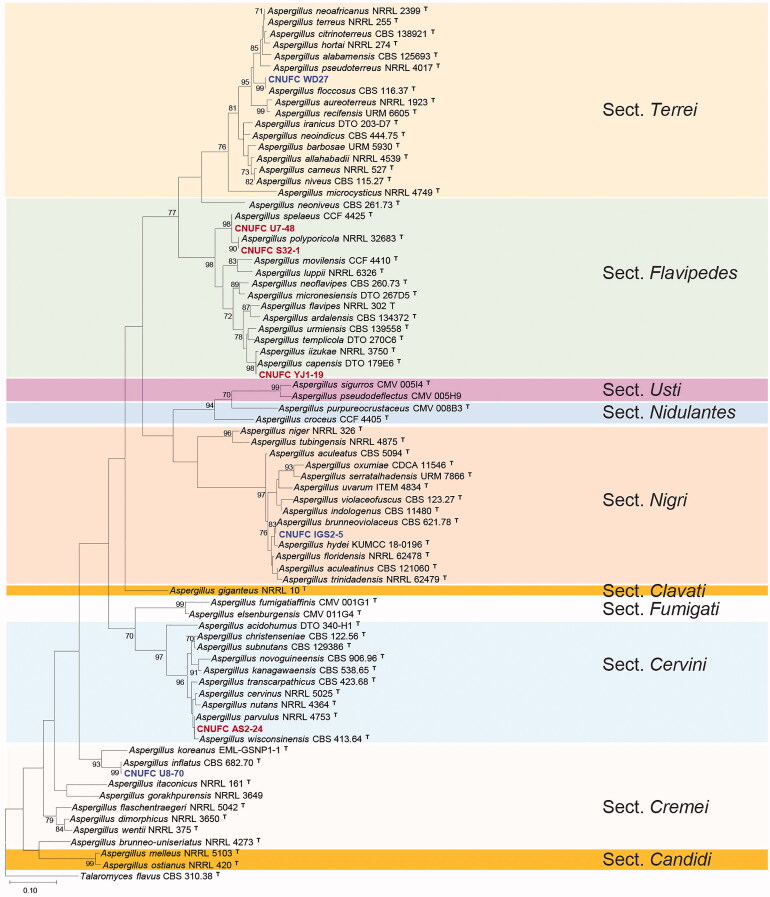
A BLASTn search of the BenA regions of CNUFC IGS2-5, CNUFC YJ1-19, CNUFC WD27, CNUFC-U8-70, CNUFC AS2-24, CNUFC S32-1, and CNUFC U7-48, revealed similarities of 100% (574/574 bp), 97.5% (503/516 bp), 100% (528/528 bp), 99.8% (465/466 bp), 99.4% (511/514 bp), 99.6% (517/519 bp), and 100% (519/519 bp), with A. brunneoviolaceus (MH614578), A. capensis (KJ775072), A. floccosus (FJ491714), A. inflatus (FJ531007), A. parvulus (KX423625), A. polyporicola (EU014088), and A. spelaeus (LT798972), respectively. Similarly, BLASTn using CaM regions of CNUFC IGS2-5, CNUFC YJ1-19, CNUFC WD27, CNUFC U8-70, CNUFC AS2-24, CNUFC S32-1, and CNUFC U7-48, revealed similarities of 99.8% (473/474 bp), 98.4% (499/507 bp), 99.6% (538/540 bp), 99.4% (476/479 bp), 100% (448/448 bp), 99.5% (729/733 bp), and 100% (517/517 bp), with A. brunneoviolaceus (EF661147), A. capensis (KJ775279), A. floccosus (MH292833), A. inflatus (FJ531094), A. kanagawaensis (FJ491592), A. polyporicola (LM644252), and A. spelaeus (HG916745), respectively. Moreover, the ML tree for combined BenA and CaM sequences revealed that the strains, CNUFC IGS2-5, CNUFC YJ1-19, CNUF WD27, CNUFC U8-70, CNUFC AS2-24, CNUFC S32-1, and CNUFC U7-48, were placed in clade with A. brunneoviolaceus, A. capensis, A. floccosus, A. inflatus, A. parvulus, A. polyporicola, and A. spelaeus, in their respective five sections in Aspergillus (Figure 1).
Figure 1.
Phylogenetic tree of Aspergillus brunneoviolaceus CNUFC IGS2-5, A capensis CNUFC YJ1-19, A. floccosus CNUFC WD27, A. inflatus CNUFC U8-70, A. parvulus CNUFC AS2-24, A. polyporicola CNUFC S32-1, and A. spelaeus CNUFC U7-48, and related species based on ML analysis of the combined BenA and CaM sequences. Numbers at the nodes indicate the bootstrap values (≥70%) from 1000 replicates. The bar indicates the number of substitutions per nucleotide. The study isolates are presented in bold and are represented by different colors.
3.2. Taxonomy
3.2.1. Taxonomy of CNUFC IGS2-5
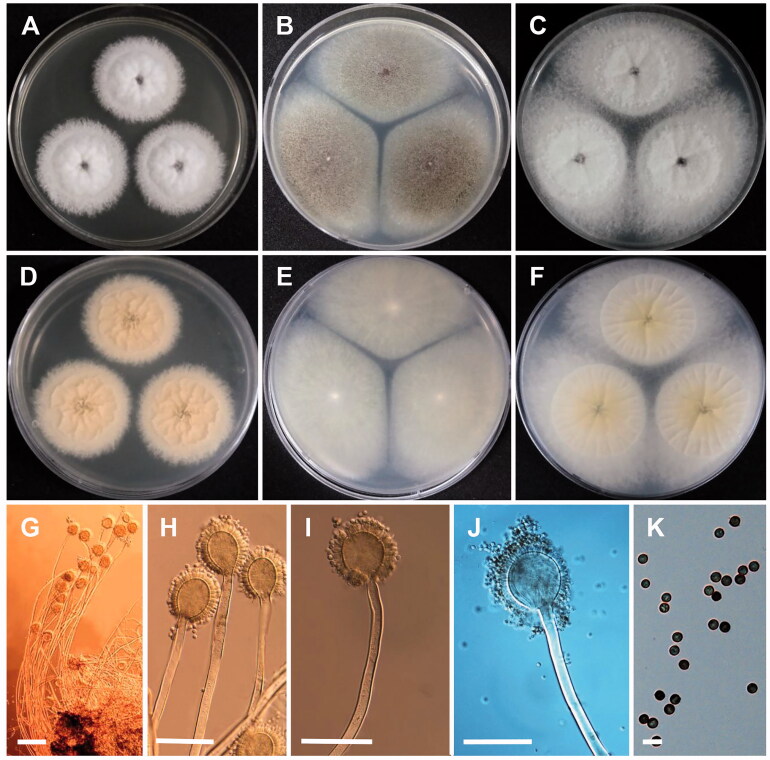
A. brunneoviolaceus Bat. & H. Maia, Anais Soc. Biol. Pernambuco 13: 91 (1955) [MB#292838] (Figure 2)
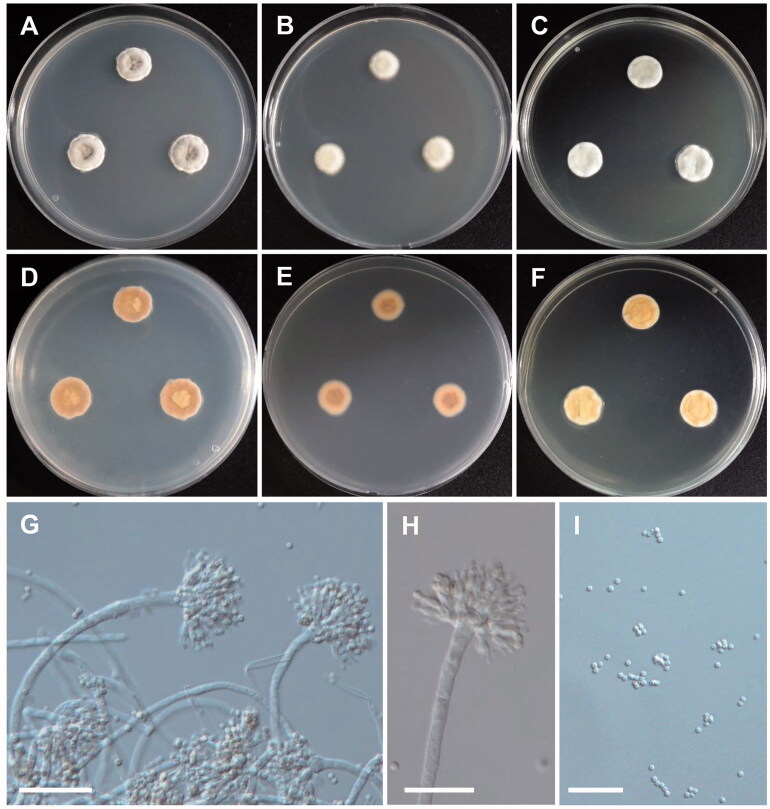
Figure 2.
Morphology of Aspergillus brunneoviolaceus. (A,D) Colonies on CYA. (B,E) Colonies on MEA. (C,F) Colonies on YES. (A–C, Obverse view; D–F, reverse view). (G–J) Conidiophores; (K) Conidia (scale bars: G = 100 μm, H, I = 50 μm, J = 80 μm, and K = 10 μm).
Colony characteristics: On CYA, the colonies initially appeared as white with flat mycelia and then turned brown, followed by reverse pale orange, and eventually reached 70–80 mm in diameter after 7 days at 25 °C. On MEA, colonies were dark brown, sporulation, widespread, and turned reverse colorless to light yellow, and further reached 82–85 mm in diameter after 7 days at 25 °C. On YES, colonies were initially cream with aerial mycelia, and further turned dark brown to black, followed by reverse ivory at margins to pale yellow toward center, and eventually reached 75–80 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores uniceriate, simple, smooth-walled, straight, occasionally sinuous, 232.4–1136.5 µm long. Vesicles spherical, subspherical, 33.5–60.8 × 43.9–63.5 µm. Phialides ampulliform, 6.8–10.4 × 2.8–4.6 µm. Conidia globose, often subglobose, rough, and echinulate on the surface, 3.6–5.8 × 3.8–5.7 µm in diameter.
3.2.2. Taxonomy of CNUFC YJ1-19
A. capensis Visagie, Hirooka & Samson, Studies in Mycology 78: 105 (2014) [MB#809193] (Figure 3)
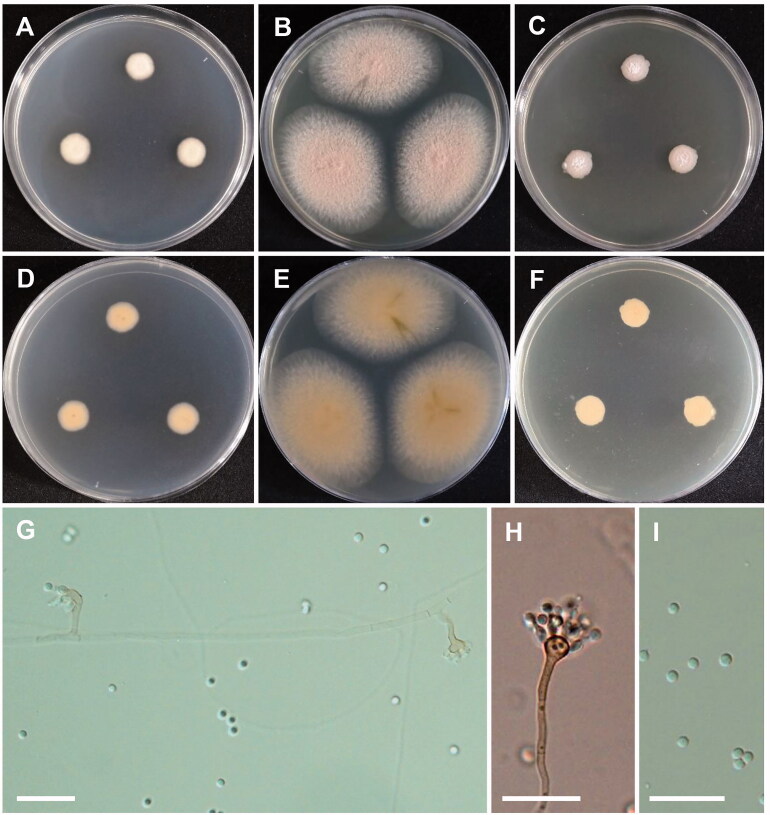
Figure 3.
Morphology of Aspergillus capensis. (A,D) Colonies on CYA. (B,E) Colonies on MEA. (C,F) Colonies on YES. (A–C: obverse view, D–F: reverse view). (G,H) Conidiophores; (I) Conidia (scale bars: G–I = 20 μm).
Colony characteristics: On CYA, the colonies were floccose, with white mycelium, yellowish sporulation at the periphery, brown soluble pigment, and reverse pale brown, and eventually reached 20–24 mm in diameter after 7 days at 25 °C. On MEA, the colonies were floccose, mycelial areas were yellowish white to pale yellow, moderate sporulation, soluble pigment was absent, followed by reverse brown to dark brown coloration, and eventually reached 19–21 mm in diameter after 7 days at 25 °C. On YES, the colonies were floccose, with moderate sporulation, pale yellow mycelia, followed by reverse pale brown, and eventually reached 21–22 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biseriate, 189–990 × 4.2–9.5 µm. Vesicles globose to elongated, 11–29 µm in diameter. Metulae, 5.3–9.4 × 3.8–4.1 µm. Phialides ampulliform, 3.7–5.8 × 2.5–3.6 µm. Conidia globose to subglobose, smooth, 2.3–3.1 × 2.3–3.1 µm in diameter. Sclerotia absent.
3.2.3. Taxonomy of CNUFC WD27
A. floccosus (Y.K. Shih) Samson, S.W. Peterson, Frisvad & Varga, Studies in Mycology 69: 45 (2011) [MB#560393] (Figure 4).
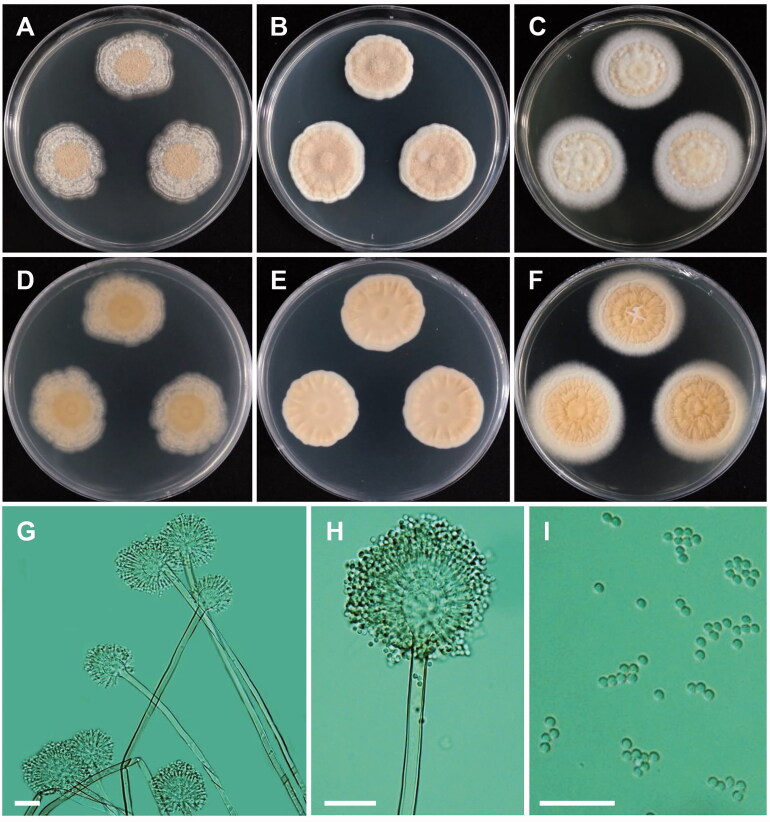
Figure 4.
Morphology of Aspergillus floccosus. (A,D) Colonies on CYA. (B,E) Colonies on MEA. (C,F) Colonies on YES. (A–C: obverse view, D–F: reverse view). (G,H) Conidial heads, Conidiophores; (I) Conidia (scale bars: G–I = 20 μm).
Colony characteristics: On CYA, the colonies were floccose, wrinkled, pale white, with no soluble pigment, moderate sporulation, followed by reverse pale yellow coloration, and eventually reached 25–28 mm in diameter after 7 days at 25 °C. On MEA, the colonies were floccose, regular, lemonade pink, with no soluble pigment, strong sporulation, reverse yellowish orange, and eventually reached 22–27 mm in diameter after 7 days at 25 °C. On YES, the colonies were plane, wrinkled, pale white, with moderate sporulation, no soluble pigment, reverse pale brown, and eventually reached 27–32 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidial heads long, densely columnar, 45–95 µm in diameter. Conidiophores biseriate, 150–375 × 4.5–5.2 µm. Vesicles globose, 12–16 µm in diameter. Metulae closely packed, 5.5–8.8 × 1.8–2.1 µm. Phialides, 4.6–6.5 × 1.8–2.1 µm. Conidia globose, elliptical, 2.0–2.6 µm in diameter.
3.2.4. Taxonomy of CNUFC U8-70
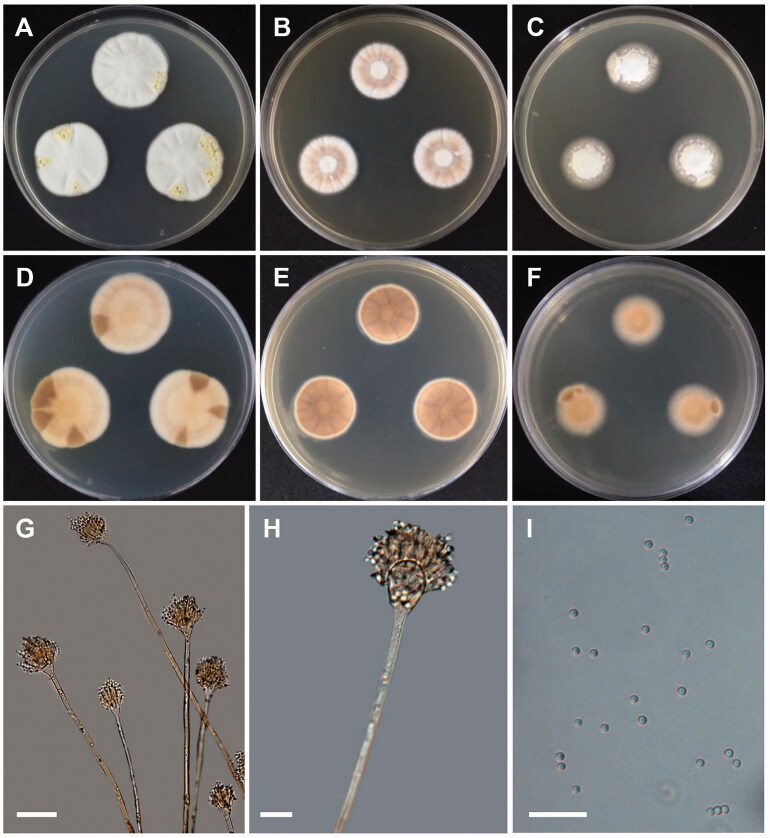
A. inflatus (Stolk & Malla) Samson, Frisvad, Varga, Visagie & Houbraken, Studies in Mycology 78: 155 (2014) [MB#809590] (Figure 5)
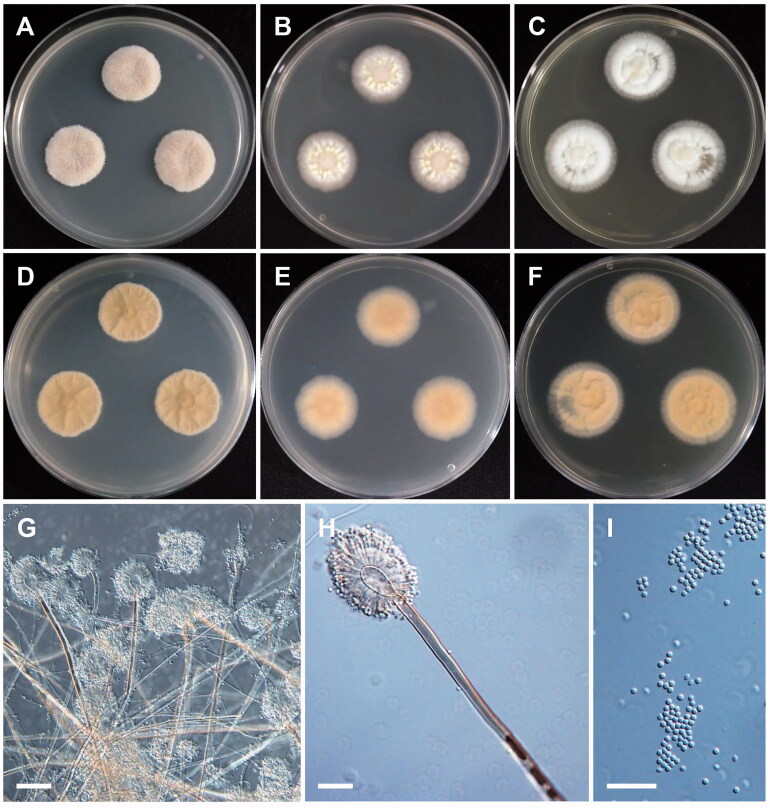
Figure 5.
Morphology of Aspergillus inflatus. (A,D) Colonies on CYA. (B,E) Colonies on MEA. (C,F) Colonies on YES. (A–C: obverse view, D–F: reverse view). (G,H) Conidiophores; (I) Conidia (scale bars: G–I = 20 μm).
Colony characteristics: On CYA, the colonies were furrowed, wrinkled, grayish green, with no soluble pigment, moderate sporulation, reverse yellowish brown coloration, and eventually reached 15–18 mm in diameter after 7 days at 25 °C. On MEA, the colonies were plane, regular, pale yellow, with no soluble pigment, reverse pale brown, and eventually reached 15–17 mm in diameter after 7 days at 25 °C. On YES, the colonies were plane, wrinkled toward center, grayish blue, with moderate sporulation, no soluble pigment, followed by reverse pale yellow coloration, and eventually reached 16–18 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biseriate, smooth walled, 120–480 × 1.6–3.0 µm. Vesicles pyriform, 3.0–6.1 µm in diameter. Metulae, 4.2–9.2 × 1.6–2.0 µm. Phialides ampulliform, 5.2–7.5 × 2–3 µm. Conidia mostly globose, subglobose, 1.5–2.4 µm in diameter.
3.2.5. Taxonomy of CNUFC AS2-24
A. parvulus G. Sm., Transactions of the British Mycological Society 44(1): 45 (1961) [MB#121074] (Figure 6)
Figure 6.
Morphology of Aspergillus parvulus. (A,D) Colonies on CYA. (B,E) Colonies on MEA. (C,F) Colonies on YES. (A–C: obverse view, D–F: reverse view). (G,H) Conidiophores; (I) Conidia (scale bars: G–I = 20 μm).
Colony characteristics: On CYA, the colonies were plane, regular, pale yellow, with no soluble pigment, moderate sporulation, reverse pale yellow coloration, and eventually reached 15–17 mm in diameter after 7 days at 25 °C. On MEA, the colonies were plane, regular, light purple, with no soluble pigment, good sporulation, reverse pale yellow, and eventually reached 22–26 mm in diameter after 7 days at 25 °C. On YES, the colonies were plane, brownish yellow, slightly wrinkled toward center, with moderate sporulation, no soluble pigment, followed by reverse pale yellow coloration, and eventually reached 16–17 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores uniseriate, bent, smooth, 12–72 × 2.5–3.2 µm. Vesicles globose, occasionally subclavate, 6–11 µm in diameter. Phialides ampulliform, 4–6 × 2–3 µm. Conidia globose, 2.6–3.6 µm in diameter.
3.2.6. Taxonomy of CNUFC S32-1
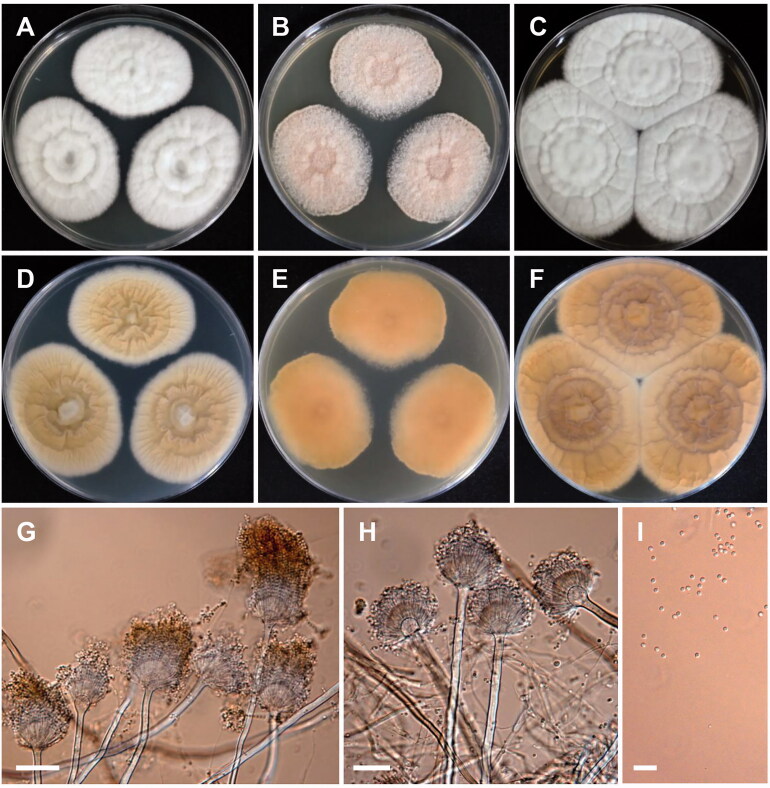
A. polyporicola Hubka, A. Nováková, M. Kolařík, S.W. Peterson, Mycologia 107(1): 194 (2015) [MB#808145] (Figure 7)
Figure 7.
Morphology of Aspergillus polyporicola. (A,D) Colonies on CYA. (B,E) Colonies on MEA. (C,F) Colonies on YES. (A–C: obverse view, D–F: reverse view). (G,H) Conidiophores; (I) Conidia (scale bars: G–I = 20 μm).
Colony characteristics: On CYA, the colonies were floccose, grayish brown, granular, with moderate sporulation, followed by reverse pale brown coloration, and eventually reached 19–21 mm in diameter after 7 days at 25 °C. On MEA, the colonies were granulose, pale yellow to yellowish toward center, with soluble pigment, reverse pale yellow coloration, and eventually reached 19–20 mm in diameter after 7 days at 25 °C. On YES, the colonies were plane, white mycelia, wrinkled toward center, with moderate sporulation, reverse white to pale yellow toward center, and eventually reached 21–22 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biseriate, smooth walled, 270–820 × 3.2–6.0 µm. Vesicles globose to subglobose, pyriform, 7–17 µm in diameter. Metulae, 4.2–8.5 µm. Phialides, 3–5 µm. Conidia globose to subglobose, 2.1–3.1 µm in diameter. No ascospores or ascomata observed.
3.2.7. Taxonomy of CNUFC U7-48
A. spelaeus A. Nováková, Hubka, M. Kolařík, S.W. Peterson, Mycologia 107(1): 194 (2015) [MB#808146] (Figure 8)
Figure 8.
Morphology of Aspergillus spelaeus. (A,D) Colonies on CYA. (B,E) Colonies on MEA. (C,F) Colonies on YES. (A–C: obverse view, D–F: reverse view). (G,H) Conidiophores; (I) Conidia (scale bars: G–I = 20 μm).
Colony characteristics: On CYA, the colonies were floccose, light grayish yellowish brown, with no soluble pigment, followed by reverse pale yellow coloration, and eventually reached 20–22 mm in diameter after 7 days at 25 °C. On MEA, the colonies were plane, delicately granular to granular, with moderate sporulation, abundant small colorless or pale yellow droplets on the colony surface, no soluble pigment, reverse light orange, and eventually reached 19–22 mm in diameter after 7 days at 25 °C. On YES, the colonies were plane, wrinkled toward center, pale white, with no soluble pigment, reverse pale brown coloration, and eventually reached 21–23 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biseriate, 231–890 × 4.1–7.6 µm. Vesicles pyriform, 9.9–26.1 µm in diameter. Metulae mostly covering the entire surface of the vesicle, 5.0–11.2 × 3.1–4.0 µm. Phialides, 3.5–7.2 × 2–3 µm. Conidia smooth, mostly globose, few subglobose, 2.5–3.1 µm in diameter. No ascospores or ascomata observed.
4. Discussion
To date, there have been few reports on undescribed Aspergillus species in Korea despite having a cosmopolitan distribution. Moreover, several new Aspergillus species have been introduced worldwide; therefore, collecting and expanding samples from different habitats is needed for identification of Aspergillus species from the Korean peninsula owing to their economic benefits. The present study provides a comprehensive account of the occurrence and distribution of Aspergillus species in Korea, particularly A. brunneoviolaceus, A. capensis, A. floccosus, A. inflatus, A. parvulus, A. polyporicola, and A. spelaeus. In this study, seven Aspergillus species in five different sections were identified and compared to their most closely related species. Analysis of the combined BenA and CaM datasets revealed that the strains CNUFC IGS2-5, CNUFC YJ1-19, CNUFC WD27, CNUFC U8-70, CNUFC AS2-24, CNUFC S32-1, and CNUFC U7-48 were placed into their respective type species of A. brunneoviolaceus, A. capensis, A. floccosus, A. inflatus, A. parvulus, A. polyporicola, and A. spelaeus.
As shown in Figure 1, CNUFC IGS2-5 aligned with A. brunneoviolaceus NRRL4912 (ex-type strain) in section Nigri. Morphologically, the isolated strains present similar characters with type strain NRRL 4912 of A. brunneoviolaceus described by Batista and da Silva [19]. These include good sporulation with dark brown conidia; uniseriate conidiophores; globular, subglobular, and spherical vesicle, (30–)35–70(–90) μm; and conidia globose to ellipsoidal, smooth, and slightly roughened, 3.5–4.5(–6)×3.5–4.5(–5) μm. Moreover, section Nigri, known as black aspergilli includes species with smooth conidiophores and hyaline or pigmentation below the vesicle; globose, subglobose, and pyriform vesicles; typically radiating conidial heads; or divergent columns in certain species [20]. These aspergilli have been isolated from contaminated materials, indoor air environments, soil samples, and plants [21]. In general, 27 species were accepted in this section [22]. Three additional new species, A. hydei, A. oxumiae, and A. labruscus, were discovered from air under Quercus variabilis, in soil cultivated with Agave sisalana, and on the surface of grape berries [23–25]. A. brunneoviolaceus is a rare member of the group of black aspergilli, which has utmost significance in the industry [26]. To date, A. brunneoviolaceus was isolated from soil (CBS 313.89), thumb nail (PW4048), bronchoalveolar lavage (PW4122), sputum (PW4213), wound (PW4049), Lactuca sativa (CBS 119.49), guano (IHEM 18675), corneal scraping keratitis (IHEM 18675), dropping of Coenobita sp. (IHEM 4062), industrial material (CCF 108), and indoor environment (ITEM 14794, ITEM 14799, and ITEM 14802) [14,27–29]. This is first study to isolate A. brunneoviolaceus from a spider in Korea, thereby revealing its significance as a member of the ecosystem of an arthropod.
Based on the phylogeny, CNUFC YJ1-19 clustered with A. capensis DTO 179-E6 (ex-type strain); CNUFC S32-1 with A. polyporicola NRRL32683 (ex-type strain); and CNUFC U7-48 with A. spelaeus CCF4425 (ex-type strain), in section Flavipedes (Figure 1). The isolate CNUFC YJ1-19 were morphologically similar to A. capensis, as described by Visagie et al. [12], although the length of conidiophores differed. The conidiophores described by Visagie et al. [12] were 235–1400 × 6.5–11 μm in length, whereas the isolate in the present study was 189–990 × 4.2–9.5 µm in length. The morphological characteristics of the isolates A. polyporicola and A. spelaeus in this study were consistent with those previously described by [30]. Section Flavipedes was expanded to include informal A. flavus group species [20,31,32]. Species belonging to section Flavipedes and Terrei are related phenotypically, and moreover, some of the species in the section Terrei were earlier placed in section Flavipedes due to overlapping cultural and morphological characteristics [30,32,33]. The genomic sequences have been useful in providing a robust tool for appropriate identification and delineation of species boundaries [30,34–36]. About 21 species were accepted in the section Flavipedes [37–40]. Members in this section are reported from foods, as endophytes from soils and rhizospheres, from indoor and cave environments, and occasionally as clinical specimens. A. capensis was reported from house dust samples [12], and a healthy plant of oilseed rape (Brassica napus L.) and produced three antifungal metabolites namely, methyl dichloroasterrate, penicillither, and rosellichalasin [41]. These metabolites exhibit antifungal activity toward major plant pathogens such as Botrytis cinerea, Monilinia fructicola, Sclerotinia sclerotiorum, and S. trifoliorum. Rosellichalasin produced by Aspergillus sp. has revealed anticancer activities against human tumor cell lines, including A549, Hela, BEL-7402, and RKO [42]. The taxonomy of A. capensis and A. iizukae needs careful attention. More isolates with DNA sequences to be generated that would be helpful for a better resolution in identification of these two species.
Furthermore, CNUFC U8-70 clustered with A. inflatus in section Cremei (Figure 1). The isolate revealed similar morphological characters as that of A. inflatus CBS682.70 (ex-type strain) [22]. Section Cremei (known as the A. cremeus group) was first described by Raper and Fennell [32] with five species. Recently, about 13 species were included in this section [5,43]. Species belonging to this section are characterized by their yellowish–brown to brown or gray–green colony color, biseriate conidial heads, long conidiophores, and pale gray–green to yellow-brown conidia [22]. Species in this section are frequently found in soil and foods associated with spoilage of cereals and nuts. A. inflatus is reported to produce sterigmatocystin—a precursor to highly potent compounds, namely aflatoxins [44]. A. inflatus isolates were found in root surface of Picea abies, forest soil under Quercus rubra, as well as in scalp and sputum of humans [45].
The isolate CNUFC WD27 was phylogenetically related to the type of A. floccosus clade belonging to section Terrei (Figure 1). Moreover, morphological characters of the isolate were consistent with those of A. floccosus described by Samson et al. [46]. Section Terrei was introduced by Gams et al. [20], for Raper and Fennell (A. terreus group) [32] having buff to brown columnar conidial heads. They have a cosmopolitan distribution and are particularly important in fermentation industries [46]. Two new species were introduced in this section recently, and thus, the accepted number of species increased to 19 in total [4]. A. floccosus was earlier named as Aspergillus terreus var. floccosus, isolated from waste cloth from Wuchang, China, and was used as a clinical specimen in immunocompromised patients [46,47]. In the present study, A. floccosus was isolated from freshwater samples. A. floccosus was found to produce extrolites, aszonalenin, austalides, butyrolactones, hepatotoxic citrinin, decaturin, dihydrocitrinone, isocoumarin, and serantrypinone [46].
In the phylogeny, CNUFC AS2-24 aligned with A. parvulus clade in section Cervini (Figure 1). The morphological characters of the isolate were consistent with those of A. parvulus, as described by Chen et al. [48]. Section Cervini was established by Gams in 1985 for species with radiate or short columnar, fawn colored, uniseriate conidial heads. This section is economically less important, less studied in comparison to other sections, and comprises 10 species [48]. A. parvulus was originally isolated from different soil environments in USA, UK, The Netherlands, and feed ingredients from Argentina [48–50]. Furthermore, in this study, the isolate was obtained from rhizosphere soil. Previous studies reported that A. parvulus exhibits a wide spectrum of antibiotic activities against various bacteria [51], phytotoxic activities [52], produces parvulenone [53], naphthalenone [54], and asparvenone derivatives [55]. Species of section Cervini have not been found to be important human pathogens; however, Hubka et al. [56] reported an isolate (closely related to A. parvulus) as the possible cause of human onychomycosis.
Our study presents undescribed species of Aspergillus from different environmental habitats as well as new sources of isolation from arthropods populations. Further studies should focus on investigating more unique habitats and on sampling across Korea. Our work needs to be coupled with antifungal and antibacterial activity of the discovered species to produce novel metabolites for industrial applications.
Funding Statement
This work was supported by the Project on Survey and Discovery of Indigenous Fungal Species of Korea, funded by the NIBR [202102201], and by the Project on Discovery of Fungi from Freshwater and Collection of Fungarium, funded by the NNIBR of the Ministry of Environment [202101203]. It was also supported in part by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2022R1I1A3068645).
Disclosure statement
No potential conflict of interest is reported by the authors.
References
- 1.Micheli PA. Nova plantarum genera juxta tournefortii methodum disposita. Florence (Italy: ): Typis Bernardi Paperinii; 1729. [Google Scholar]
- 2.Houbraken J, Kocsubé S, Visagie CM, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sklenář F, Jurjević Ž, Peterson SW, et al. Increasing the species diversity in the Aspergillus section Nidulantes: Six novel species mainly from the indoor environment. Mycologia. 2020;112(2):342–370. [DOI] [PubMed] [Google Scholar]
- 4.Correia ACR, Barbosa RN, Frisvad JC, et al. The polyphasic re-identification of a Brazilian Aspergillus section Terrei collection led to the discovery of two new species. Mycol Prog. 2020;19(9):885–903. [Google Scholar]
- 5.Hyde KD, Hongsanan S, Jeewon R, et al. Fungal diversity notes 367-491: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80(1):1–270. [Google Scholar]
- 6.Hong SB, Cho HS, Shin HD, et al. Novel Neosartorya species isolated from soil in Korea. Int J Syst Evol Microbiol. 2006;56(Pt 2):477–486. [DOI] [PubMed] [Google Scholar]
- 7.Kim HJ, Kim JS, Cheon KH, et al. Species list of Aspergillus, Penicillium and Talaromyces in Korea, based on ‘one fungus one name’ system. Kor J Mycol. 2016;44:207–219. [Google Scholar]
- 8.Nguyen TTT, Pangging M, Bangash NK, et al. Five new records of the family Aspergillaceae in Korea, Aspergillus europaeus, A. pragensis, A. tennesseensis, Penicillium fluviserpens, and P. scabrosum. Mycobiology 2020;48(2):81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen TTT, Noh JK, Lee HB.. New species and eight undescribed species belonging to the families Aspergillaceae and Trichocomaceae in Korea. Mycobiology 2021;49(6):534–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JW, Kim SH, You YH, et al. Four unrecorded Aspergillus species from the rhizosphere soil in South Korea. Mycobiology 2021;49(4):346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pangging M, Nguyen TTT, Lee HB.. New records of four species belonging to Eurotiales from soil and freshwater in Korea. Mycobiology 2019;47(2):154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visagie CM, Hirooka Y, Tanney JB, et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014;78:63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass NL, Donaldson GC.. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubka V, Kolarik M.. β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia 2012;29:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson SW, Vega F, Posada F, et al. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia 2005;97(3):659–666. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 18.Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batista AC, da Silva MH.. Alguns aspergillales de contaminação. Anais da Sociedade de Biologia de Pernambuco. 1955;13:91–100. [Google Scholar]
- 20.Gams W, Christensen M, Onions AH, et al. Infrageneric taxa of Aspergillus. In: Samson RA, Pitt JI, editors. Advances in Penicillium and Aspergillus systematics. New York (NY): Plenum Press; 1985. p. 55–62. [Google Scholar]
- 21.Serra R, Cabañes FJ, Perrone G, et al. Aspergillus ibericus: a new species of section Nigri isolated from grapes. Mycologia 2006;98(2):295–306. [DOI] [PubMed] [Google Scholar]
- 22.Samson RA, Visagie CM, Houbraken J, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78:141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doilom M, Guo JW, Phookamsak R, et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front Microbiol. 2020;11:585215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crous PW, Wingfield MJ, Chooi YH, et al. Fungal planet description sheets: 1042–1111. Persoonia 2020;44:301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fungaro MHP, Ferranti LS, Massi FP, et al. Aspergillus labruscus sp. nov., a new species of Aspergillus section Nigri discovered in Brazil. Sci Rep. 2017;7(1):6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ismail MA. Incidence and significance of black aspergilli in agricultural commodities: a review, with a key to all species accepted to-date. Eur J Biol Res. 2017;7:207–222. [Google Scholar]
- 27.Varga J, Frisvad JC, Kocsubé S, et al. New and revisited species in Aspergillus section Nigri. Stud Mycol. 2011;69(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrickx M, Beguin H, Detandt M.. Genetic re-identification and antifungal susceptibility testing of Aspergillus section Nigri strains of the BCCM/IHEM collection. Mycoses. 2012;55(2):148–155. [DOI] [PubMed] [Google Scholar]
- 29.Jurjević Ž, Peterson SW, Stea G, et al. Two novel species of Aspergillus section Nigri from indoor air. IMA Fungus. 2012;3(2):159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubka V, Nováková A, Kolařík M, et al. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia. 2015;107(1):169–208. [DOI] [PubMed] [Google Scholar]
- 31.Thom C, Church MB.. 1926. The aspergilli. Baltimore: Williams & Wilkins. p. 272. [Google Scholar]
- 32.Raper KB, Fennell DI.. The genus Aspergillus. Baltimore (MD): Williams & Wilkins; 1965. [Google Scholar]
- 33.Samson RA. A compilation of the aspergilli described since 1965. Stud Mycol. 1979;18:1–40. [Google Scholar]
- 34.Varga J, Tóth B, Kocsubé S, et al. Evolutionary relationships among Aspergillus terreus isolates and their relatives. Antonie Leeuwenhoek. 2005;88(2):141–150. [DOI] [PubMed] [Google Scholar]
- 35.Peterson SW. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In Samson RA, Pitt JI, editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam: Harwood Academic Publishers; 2000. p. 163–178. [Google Scholar]
- 36.Peterson SW. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 2008;100(2):205–226. [DOI] [PubMed] [Google Scholar]
- 37.Crognale S, Pesciaroli L, Felli M, et al. Aspergillus olivimuriae sp. nov., a halotolerant species isolated from olive brine. Int J Syst Evol Microbiol. 2019;69(9):2899–2906. [DOI] [PubMed] [Google Scholar]
- 38.Sklenář F, Jurjević Z, Houbraken J, et al. Re-examination of species limits in Aspergillus section Flavipedes using advanced species delimitation methods and description of four new species. Stud Mycol. 2021;99:100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visagie CM, Goodwell M, Nkwe DO.. Aspergillus diversity from the gcwihaba cave in Botswana and description of one new species. Fungal Syst Evol. 2021;8:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang XC, Zhuang WY. New species of Aspergillus (Aspergillaceae) from tropical islands of China. J Fungi 2022;8:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin J, Lyu A, Zhang QH, et al. Strain identification and metabolites isolation of Aspergillus capensis CanS-34A from Brassica napus. Mol Biol Rep. 2019;46(3):3451–3460. [DOI] [PubMed] [Google Scholar]
- 42.Xiao L, Liu H, Wu N, et al. Characterization of the high cytochalasin E and rosellichalasin producing-Aspergillus sp. nov. F1 isolated from marine solar saltern in China. World J Microbiol Biotechnol. 2013;29(1):11–17. [DOI] [PubMed] [Google Scholar]
- 43.Hubka V, NovaKova A, Samson RA, et al. Aspergillus europaeus sp. nov., a widely distributed soil-borne species related to A. wentii (section Cremei). Plant Syst Evol. 2016;302(6):641–650. [Google Scholar]
- 44.Rank C, Nielsen KF, Larsen TO, et al. Distribution of sterigmatocystin in filamentous fungi. Fungal Biol. 2011;115(4-5):406–420. [DOI] [PubMed] [Google Scholar]
- 45.Stolk AC, Malla DS.. Penicillium inflatum sp. nov. Persoonia 1971;6:197–200. [Google Scholar]
- 46.Samson RA, Peterson SW, Frisvad JC, et al. New species in Aspergillus section Terrei. Stud Mycol. 2011;69(1):39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoran T, Sartori B, Sappl L, et al. Azole-resistance in Aspergillus terreus and related species: an emerging problem or a rare phenomenon. Front Microbiol. 2018;9:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen AJ, Varga J, Frisvad JC, et al. Polyphasic taxonomy of Aspergillus section Cervini. Stud Mycol. 2016;85:65–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magnoli C, Dalcero AM, Chiacchiera SM, et al. Enumeration and identification of Aspergillus group and Penicillium species in poultry feeds from Argentina. Mycopathologia 1998;142(1):27–32. [DOI] [PubMed] [Google Scholar]
- 50.Smith G. Some new and interesting species of micro-fungi. British Mycol Soc Trans. 1961;44:12–50. [Google Scholar]
- 51.Tsyganenko KS, Zaĭchenko OM.. Antibiotic and phytotoxic properties of some Aspergillus parvulus Smith strains. Mikrobiol Z. 2004;66(5):62–67. [PubMed] [Google Scholar]
- 52.Tsyganenko KS, Zaichenko OM.. Antibiotic properties of some species of genus Aspergillus MICH. Mikrobiolohichnyĭ Zhurnal. 2004;66:56–61. [PubMed] [Google Scholar]
- 53.Chao PD, Schifff P Jr, Slatkin DJ.. Metabolites of aspergilli. 4. New naphthalenones and 6-ethyl-7-methoxyjuglone from Aspergillus parvulus. J Chem Res. 1979;236. [Google Scholar]
- 54.Bartman CD, Campbell IM.. Naphthalenone production in Aspergillus parvulus. Can J Microbiol. 1979;25(2):130–137. [DOI] [PubMed] [Google Scholar]
- 55.Bös M, Canesso R, Inoue-Ohga N, et al. O-methylasparvenone, a nitrogen-free serotonin antagonist. Bioorg Med Chem. 1997;5(12):2165–2171. [DOI] [PubMed] [Google Scholar]
- 56.Hubka V, Kubatova A, Mallatova N, et al. Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Med Mycol. 2012;50(6):601–610. [DOI] [PubMed] [Google Scholar]