Abstract
Pseudomonas aeruginosa OprF forms 0.36-nS channels and, rarely, 2- to 5-nS channels in lipid bilayer membranes. We show that a protein comprising only the N-terminal 162-amino-acid domain of OprF formed the smaller, but not the larger, channels in lipid bilayers. Circular dichroism spectroscopy indicated that this protein folds into a β-sheet-rich structure, and three-dimensional comparative modeling revealed that it shares significant structural similarity with the amino terminus of the orthologous protein Escherichia coli OmpA, which has been shown to form a β-barrel. OprF and OmpA share only 15% identity in this domain, yet these results support the utility of modeling such widely divergent β-barrel domains in three dimensions in order to reveal similarities not readily apparent through primary sequence comparisons. The model is used to further hypothesize why porin activity differs for the N-terminal domains of OprF and OmpA.
OprF is a major outer membrane protein in Pseudomonas aeruginosa that has been studied extensively due to its proposed utility as a vaccine component, its role in antimicrobial drug resistance, and its porin function (3, 6, 7, 13, 20). It has been shown to be required for cell growth in low-osmolarity medium and for the maintenance of cell shape (21). Through epitope-mapping experiments and linker insertion mutagenesis, we originally proposed a 16-β-stranded membrane topology model for OprF (19). However, on the basis of deletion studies and secondary structure predictions, we recently proposed a revised model with the N-terminal half of the protein forming an eight-stranded β-barrel domain that is inserted into the outer membrane. The C-terminal half was proposed to form a domain that is exposed and available to monoclonal antibody binding on the cell surface (9) and binds peptidoglycan in the periplasm (15). These two domains are linked by a proline-rich hinge-and-loop region that contains two disulfide bonds.
A somewhat analogous structure has been proposed for the Escherichia coli outer membrane protein OmpA (5, 12, 18), and these proteins, which also share some functional similarities, are considered orthologs. Consistent with this concept, significant amino acid sequence similarity has been detected between OprF and OmpA, but only in their C-terminal domains (39% identity, 56% similarity). However, secondary structure predictions indicate that the N-terminal domains may also be similar, despite their lack of substantial sequence identity (15% identity, with no regions of similarity identified using BLAST2 with an “expect value” cutoff of 1,000). Recently, Pautsch and Schulz (12) solved a crystal structure for the N-terminal half of OmpA that was mutated at residues 23, 34, and 107 (in order to obtain crystals) and had been reconstituted from inclusion bodies. Although the positioning of surface loop regions could not be defined with certainty, the remainder of the N terminus was shown to form an eight-stranded β-barrel. Pautsch and Schulz (12) also reported evidence (obtained through structural analysis and black lipid bilayer studies) that this domain of OmpA did not form a membrane pore, although recently Arora et al. (1) reported identifying very small channels, 0.05 to 0.08 nS, with this domain (using a protein containing four Trp-to-Phe mutations and purified from outer membrane preparations under denaturing conditions and refolded). Channels approximately 0.25 to 0.4 nS in size have been reported for the full-length OmpA protein (1, 16, 17). Since OprF has been previously shown reproducibly to form small (0.36- to 0.38-nS) channels and, rarely, large (2- to 5-nS) channels, and since in vivo experiments support this porin activity (2, 3, 11, 20), we wondered whether the N-terminal domain of OprF formed channels, and if so, whether the channels were of a size similar to that of native OprF.
We therefore examined the pore-forming ability of a protein comprising only the N-terminal domain of OprF (OprF1–162) that had been purified under nondenaturing conditions from outer membranes. We showed that this protein does indeed form small channels consistent with those previously observed for native OprF, although no large channels were observed. Circular dichroism (CD) spectral analysis and three-dimensional modeling support a β-barrel conformation for this domain, and the modeling reveals similarities between OprF and OmpA in this domain that are not apparent through primary sequence analysis. These structural analyses permit us to hypothesize why the porin activity differs for these domains of OprF and OmpA.
Purification of a protein comprising the N-terminal domain of OprF, OprF1–162.
For this study, native OprF protein was purified as previously described (2) from outer membrane preparations of E. coli DH5α expressing OprF from plasmid pRW5 (15). OprF1–162 was expressed in E. coli DH5α from the previously constructed plasmid pER163 (15). We found we were able to purify the OprF1–162 by using the same procedure as for native OprF. The identity of both proteins was confirmed through Western blot analysis as described previously (10) using the monoclonal antibody MA7-1, which is specific for an epitope within the N terminus of OprF.
Planar lipid bilayer analysis of OprF and OprF1–162.
OprF and OprF1–162 were assessed for pore-forming ability through planar lipid bilayer experiments, which were performed as previously described (3). Briefly, a 1 M KCl solution was placed within two compartments separated by a 0.1-mm3 circular hole that had been covered with a membrane formed from a solution of 1.5% oxidized cholesterol in n-decane. Electrodes were inserted into the KCl solutions in each compartment and a voltage of 50 mV was applied. When either an OprF or OprF1–162 protein sample (in 0.1% Triton X-100) was added to one of the compartments, stepwise increases in conductance were observed, indicating that channels were being formed in the membrane. Approximately 100 single-channel events were recorded for each experiment (Fig. 1).
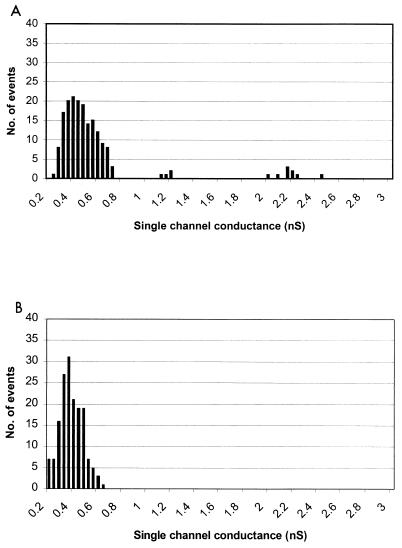
FIG. 1.
Histograms of single-channel conductance measurements showing channel size distributions for OprF (A) and OprF1–162 (B).
For native OprF, small channels in the size range of those reported previously were observed (predominantly 0.4 nS) and, rarely (approximately 5% of events), larger channels (1 to 3 nS) were identified (Fig. 1). These larger channels were observed closely under conditions of low frequency of channel formation to ensure that they were not simply a reflection of multiple smaller channel events occurring simultaneously. For OprF1–162, small channel sizes in the range of 0.36 nS were frequently measured, but no channels of the larger size were observed (Fig. 1). These results suggest that the N-terminal half of OprF is able to form a pore and that the remainder of the protein, or a portion thereof, is required for the formation of the larger channels (perhaps through formation of alternate protein conformations). It should be noted that very small channels with a conductance of 0.04 to 0.08 nS were also frequently seen with both the OprF and OprF1–162 protein preparations (data not shown). However, similar-sized channels were also noted for a negative control sample that comprised a vector-only clone sample (E. coli with pUCP19) (14) after mock purification in the same manner as the OprF and OprF1–162 proteins.
Structural analysis by indirect immunofluorescence of surface epitopes, and CD spectroscopy.
To confirm that the OprF1–162 protein was folding in a conformation similar to that of the equivalent domain in wild-type OprF in our studies and was correctly localized to the cell surface, indirect intact E. coli/pER163 cells expressing OprF1–162 were examined by indirect immunofluorescence labeling using the monoclonal antibody MA7-1, which binds to a surface-exposed epitope, amino acids 55 to 62 in the OprF N terminus (14), as previously described (9). Cells expressing OprF1–162 were highly fluorescent, consistent with surface exposure of this epitope, while cells not expressing any OprF protein sequences showed no fluorescence (data not shown).
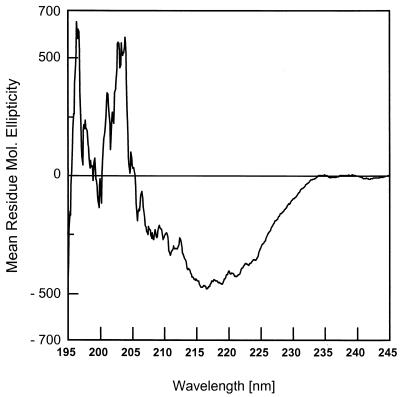
To evaluate the secondary structure of this OprF N-terminal domain, CD spectroscopy was performed on purified OprF1–162 by using a model J-70 spectropolarimeter (Jasco, Tokyo, Japan) connected to a Jasco data processor, using a quartz cell with a 1-mm path length. CD spectra were measured at 25°C, between 190 and 250 nm at a scanning speed of 10 m/min in 10 mM sodium phosphate buffer (pH 7.0) with 0.1% sodium dodecyl sulfate. The resulting spectrum (Fig. 2) was highly similar to that observed for antiparallel β-sheet-rich proteins (4), with a characteristic minimum at 217 nm. This is consistent with the proposal that this domain forms a β-barrel.
FIG. 2.
CD spectral analysis of OprF1–162 in 0.1% sodium dodecyl sulfate.
Three-dimensional modeling.
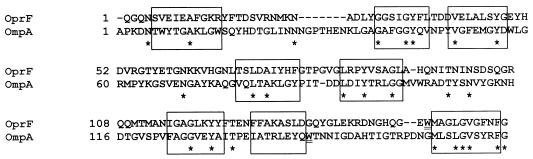
The N-terminal half of OprF, OprF1–162, shares only 15% identity with the corresponding region of OmpA, OmpA1–171, yet secondary structure prediction algorithms, CD spectroscopy results, and other data (15) are consistent with these proteins sharing similar β-sheet secondary structures and thus indicate that OprF1–162 may form a β-barrel. Similarity of OprF and OmpA in the C terminus further supports an orthologous relationship between these proteins. We therefore attempted to model the OprF N terminus using the published OmpA N-terminus crystal structure (12). We visually aligned the N-terminal 160 amino acids of OprF with the corresponding N-terminal 171 amino acids of OmpA used for crystallization (Protein Data Bank Identification no. 1BXW) (12). We used amino acid hydrophobicity, rather than identity, as a guide for constructing the alignment. The alignment was further modified after a first round of modeling revealed that one putative transmembrane β-sheet strand was misaligned because a charged residue was pointing out of the central barrel region into a region of the lipid bilayer (final alignment shown in Fig. 3). Previous studies of OmpA (8) and crystal structures of other outer membrane proteins strongly indicate that charged residues are not permissive in such a location in a β-barrel protein. Using the Insight II (version 97.2) molecular modeling program “Homology” (Molecular Simulations Inc., San Diego, Calif.), the OprF1–162 sequence was threaded to the OmpA1–171 structure, constraining regions that aligned with the β-sheet regions of OmpA and allowing more freedom in the formation of loop regions (which were not precisely positioned in the OmpA model). The entire structure was then energy minimized using the “Discover” program of Insight II (Fig. 4). The model is available from the authors as a Protein Data Bank file, and animations and other images of the model may be viewed as supplementary data at http://www.cmdr.ubc.ca/bobh/oprfmodel.html to aid visualization of its three-dimensional structure.
FIG. 3.
Alignment of the sequences of OprF1–162 and OmpA1–171, according to sequence hydrophobicity and location of charged residues (see the text). Predicted transmembrane regions are boxed, and stars mark identical residues. The two underlined tryptophans are examples of residues conserved in location in three-dimensional space (according to our modeling) but not conserved in location along the sequence.
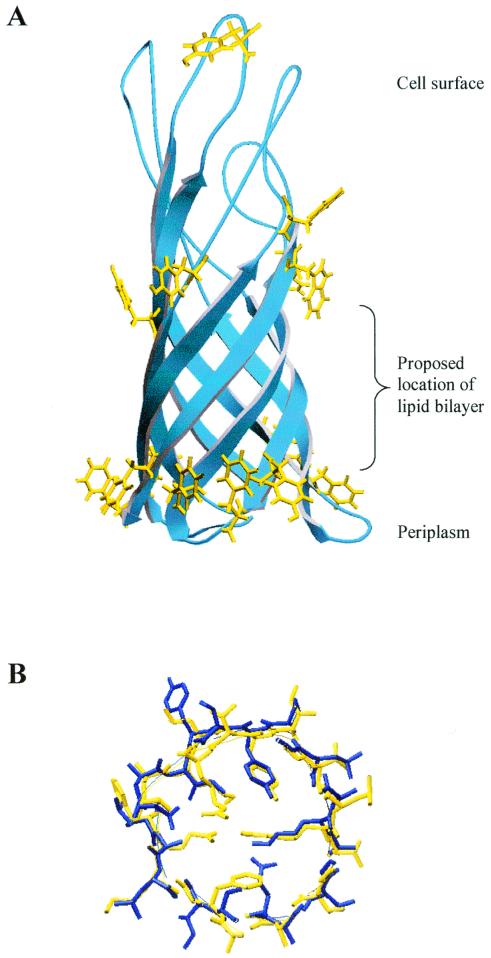
FIG. 4.
Three-dimensional model of OprF1–162, constructed by threading the sequence of OprF1–162 on a crystal structure of OmpA1–171. (A) Overview of the molecule, highlighting all aromatic residues on the outside of the protein (yellow). Note the striking rings of aromatic residues at the proposed water-lipid interfaces. (B) A slice horizontally through the barrel of the proposed model of OprF (blue), overlaid on the structure of OmpA (yellow), illustrating how residues previously proposed to form a barrier to pore formation in OmpA (12) are not conserved in OprF and allow for a larger channel with no salt bridge in that region.
It was apparent from this model that OprF and OmpA share significant structural similarity, particularly in terms of the conservation of the hydrophobicity of residues pointing toward the outside of the barrel, and also rings of aromatic residues at the proposed lipid-water interface of the molecule (Fig. 4). In fact, a number of residues were found to be conserved between the structures in three-dimensional space, though these residues were not in the same location in the primary sequence (for example, the underlined residues in Fig. 3). The degree of structural similarity was striking, considering the marked lack of identity between these two proteins in this domain. This disparity between structural sequence similarity in this region supports the belief that orthologous β-barrel structures diverge quickly in primary sequence from each other over time (relative to other common protein folds) due to a lack of primary sequence constraints while they remain structurally very similar.
There was one notable difference between the structure of OmpA1–171 and the three-dimensional model of OprF1–162 which we hypothesize could explain the fact that no channels, or only very small channels (0.05 to 0.08 nS), have been observed for the OmpA N-terminal domain, whereas we observed channels of predominantly 0.36 nS with OprF1–162. Residues previously implicated in blocking channel formation in the OmpA N-terminal domain (12), or at minimum providing a constriction in the pore, were noticeably not conserved in OprF, and more significantly, the residues that replaced them in OprF permitted the formation of a possible channel (Fig. 4B; see also supplementary data). The previous study reporting this barrier in the pore (12) also presented an alignment of OmpA orthologs, suggesting that this barrier was conserved and that the OmpA β-barrel domain was more conserved than is noted for most porins. However, their analysis was based on phylogenetically very similar organisms. Our analysis of OprF suggests that this proposed constriction is not as conserved as previously thought and that this β-barrel domain is not more conserved in primary sequence than has been observed for other porins.
The evidence presented here and in previous studies (15) strongly suggests that the N-terminal half of OprF can form a β-barrel. A three-dimensional model for the N terminus of OprF is proposed, and we support the benefit and utility of modeling proposed orthologous outer membrane proteins in three-dimensional space, even if they share little sequence identity. There is currently a need for better transmembrane β-strand prediction algorithms for outer membrane proteins. Based on our experience studying outer membrane proteins and on the studies of others, we propose that an amphipathicity plot that pays particular attention to the location of hydrophobic residues and to preferential placement of aromatic residues at the membrane-solvent interface, as well as some of the specific residue constraints reported by Koebnik (8), may be the most effective way to identify transmembrane β-strands. This is particularly important given the significant lack of sequence identity constraints required by a β-barrel structure.
Acknowledgments
We thank Annett Rozek for helpful comments regarding the three-dimensional modeling studies.
R.E.W.H. was a recipient of the Medical Research Council of Canada (MRC) Distinguished Scientist Award and received funding from MRC. This work was funded in part by the Canadian Cystic Fibrosis Foundation.
REFERENCES
- 1.Arora A, Rinehart D, Szabo G, Tamm L K. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J Biol Chem. 2000;275:1594–1600. doi: 10.1074/jbc.275.3.1594. [DOI] [PubMed] [Google Scholar]
- 2.Bellido F, Martin N L, Siehnel R J, Hancock R E W. Reevaluation, using intact cells, of the exclusion limit and role of porin OprF in Pseudomonas aeruginosa outer membrane permeability. J Bacteriol. 1992;174:5196–5203. doi: 10.1128/jb.174.16.5196-5203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz R, Hancock R E W. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim Biophys Acta. 1981;646:298–308. doi: 10.1016/0005-2736(81)90336-9. [DOI] [PubMed] [Google Scholar]
- 4.Brahms S, Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J Mol Biol. 1980;138:149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 5.Chen R, Schmidmayr W, Kramer C, Chen-Schmeisser U, Henning U. Primary structure of major outer membrane protein II (ompA protein) of Escherichia coli K-12. Proc Natl Acad Sci USA. 1980;77:4592–4596. doi: 10.1073/pnas.77.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock R E W, Siehnel R, Martin N. Outer membrane proteins of Pseudomonas. Mol Microbiol. 1990;4:1069–1075. doi: 10.1111/j.1365-2958.1990.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 7.Knapp B, Hundt E, Lenz U, Hungerer K D, Gabelsberger J, Domdey H, Mansouri E, Li Y, von Specht B U. A recombinant hybrid outer membrane protein for vaccination against Pseudomonas aeruginosa. Vaccine. 1999;17:1663–1666. doi: 10.1016/s0264-410x(98)00420-4. [DOI] [PubMed] [Google Scholar]
- 8.Koebnik R. Membrane assembly of the Escherichia coli outer membrane protein OmpA: exploring sequence constraints on transmembrane beta-strands. J Mol Biol. 1999;285:1801–1810. doi: 10.1006/jmbi.1998.2405. [DOI] [PubMed] [Google Scholar]
- 9.Martin N L, Rawling E G, Wong R S, Rosok M, Hancock R E W. Conservation of surface epitopes in Pseudomonas aeruginosa outer membrane porin protein OprF. FEMS Microbiol Lett. 1993;113:261–266. doi: 10.1111/j.1574-6968.1993.tb06524.x. [DOI] [PubMed] [Google Scholar]
- 10.Mutharia L M, Hancock R E W. Surface localization of Pseudomonas aeruginosa outer membrane porin protein F by using monoclonal antibodies. Infect Immun. 1983;42:1027–1033. doi: 10.1128/iai.42.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikaido H, Nikaido K, Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J Biol Chem. 1991;266:770–779. [PubMed] [Google Scholar]
- 12.Pautsch A, Schulz G E. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 13.Piddock L J V, Hall M C, Bellido F, Bains M, Hancock R E W. A pleiotropic, posttherapy, enoxacin-resistant mutant of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1057–1061. doi: 10.1128/aac.36.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawling E G, Martin N L, Hancock R E W. Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect Immun. 1995;63:38–42. doi: 10.1128/iai.63.1.38-42.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawling E G, Brinkman F S L, Hancock R E W. Role of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J Bacteriol. 1998;180:3556–3562. doi: 10.1128/jb.180.14.3556-3562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saint N, De E, Julien S, Orange N, Molle G. Ionophore properties of OmpA of Escherichia coli. Biochim Biophys Acta. 1993;1145:119–123. doi: 10.1016/0005-2736(93)90388-g. [DOI] [PubMed] [Google Scholar]
- 17.Sugawara E, Nikaido H. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J Biol Chem. 1994;269:17981–17987. [PubMed] [Google Scholar]
- 18.Vogel H, Jahnig F. Models for the structure of outer-membrane proteins of Escherichia coli derived from raman spectroscopy and prediction methods. J Mol Biol. 1986;20:191–199. doi: 10.1016/0022-2836(86)90292-5. [DOI] [PubMed] [Google Scholar]
- 19.Wong R S Y, Jost H, Hancock R E W. Linker-insertion mutagenesis of Pseudomonas aeruginosa outer membrane protein OprF. Mol Microbiol. 1993;10:283–292. [PubMed] [Google Scholar]
- 20.Woodruff W A, Hancock R E W. Construction and characterization of Pseudomonas aeruginosa protein F-deficient mutants after in vitro and in vivo insertion mutagenesis of the cloned gene. J Bacteriol. 1988;170:2592–2598. doi: 10.1128/jb.170.6.2592-2598.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodruff W A, Hancock R E W. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J Bacteriol. 1989;171:3304–3309. doi: 10.1128/jb.171.6.3304-3309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]