Abstract
O antigen is part of the lipopolysaccharide present in the outer membrane of gram-negative bacteria. Escherichia coli and Salmonella enterica each have many forms of O antigen, but only three are common to the two species. It has been found that, in general, O-antigen genes are of low GC content. This deviation in GC content from that of typical S. enterica or E. coli genes (51%) is thought to indicate that the O-antigen DNA originated in species other than S. enterica or E. coli and was captured by lateral transfer. The O-antigen structure of Salmonella enterica O35 is identical to that of E. coli O111, commonly found in enteropathogenic E. coli strains. This O antigen, which has been shown to be a virulence factor in E. coli, contains colitose, a 3,6-dideoxyhexose found only rarely in the Enterobacteriaceae. Sequencing of the O35-antigen gene cluster of S. enterica serovar Adelaide revealed the same gene order and flanking genes as in E. coli O111. The divergence between corresponding genes of these two gene clusters at the nucleotide level ranges from 21.8 to 11.7%, within the normal range of divergence between S. enterica and E. coli. We conclude that the ancestor of E. coli and S. enterica had an O antigen identical to the O111 and O35 antigens, respectively, of these species and that the gene cluster encoding it has survived in both species.
Lipopolysaccharide, an important component of the outer membrane of gram-negative bacteria, usually consists of three distinct regions: lipid A, core oligosaccharide, and O-specific polysaccharide (O antigen). O antigens consist of repeats of an O unit of generally two to six sugars. The genes for O-antigen synthesis are normally grouped together on the chromosome in a gene cluster which maps close to gnd in both Escherichia coli and Salmonella enterica. We, among others, have undertaken an extensive study of the genetic basis of O-antigen variation by sequencing and identifying the O-antigen genes, mostly in S. enterica and E. coli (see article by Reeves [34, 35] for review). It has been found that, in general, O-antigen genes are of low G+C content (usually less than 40%). We suggested that this deviation in G+C content from that of typical S. enterica or E. coli genes (51%) indicates that the O-antigen DNA originated in species other than S. enterica or E. coli and was captured by lateral transfer (16). By sequencing and comparison of O-antigen gene clusters, we and others have previously found evidence that DNA recombination events between O-antigen gene clusters within S. enterica (9, 47) and between E. coli and Klebsiella (41) played a role in the formation of new O-antigen forms. We also found evidence for an interspecies transfer of an entire O-antigen gene cluster (J. G. Shepherd, L. Wang, and P. R. Reeves, submitted for publication).
The O antigens of S. enterica and E. coli are extremely diverse, with 54 and 190 known forms, respectively, recognized in their typing schemes (23, 32) (includes Shigella strains in E. coli on the basis of high sequence similarity [8, 10, 13, 28, 33]). In a few instances, there is a serological cross-reaction between E. coli and S. enterica serotypes due to the presence of the same or a similar sugar in their O antigens (11). However, there are only three cases in which the O-antigen structures have been shown to be identical in the two species: E. coli O111 and S. enterica O35 (18), E. coli O55 and S. enterica O50 (18, 22), and E. coli O157 and S. enterica O30 (31). It is noteworthy that in E. coli, all three O antigens are associated with enteropathogenic E. coli (EPEC) and sometimes enterohemorrhagic E. coli (EHEC) strains. Nonetheless, there is no obvious relationship between the three O-antigen structures other than that O111 and O55 both contain colitose, and there is no close relationship between the gene clusters for E. coli O111 and O157. (The sequence of O55 is not known.)
The low proportion of forms present in both species suggests that there has been extensive turnover of O antigens, presumably by lateral gene transfer since divergence of the two species, because the majority of O antigens, found in only one of the species, must have been gained or lost by one of them since divergence. This may well be due to selection by the host immune system, because the surface-exposed O antigen is highly immunogenic and the antibody has been shown to be protective, providing strong selection for O antigens not previously seen by the usual host of any given clone. This may account both for the maintenance of many different O-antigen forms and, due to changes in environmental circumstances, for the turnover of forms present in a species.
It is believed that E. coli and S. enterica diverged from a common ancestor about 140 million years ago (29, 30). The very small proportion of O antigens common to both suggests that most of the polymorphism arose after the divergence. We present here the genetic basis for the identity of O antigens shared between S. enterica O35 and E. coli O111. There are three possible explanations for the presence of a common O antigen, and one could distinguish between them by comparison of the sequences of the two O-antigen gene clusters. The first possibility is that the two gene clusters are derived from a common ancestral gene cluster present in the ancestor of E. coli and S. enterica: one would expect the two gene clusters to share overall organization and have a high level of identity, with sequence divergence in the range of that for E. coli and S. enterica housekeeping genes. The second possibility is that the two gene clusters were assembled separately after the divergence of E. coli and S. enterica or acquired from different sources: the two gene clusters need not have the same gene cluster structure and could be highly divergent in sequence. The third possibility is that the two gene clusters were separately transferred from a common source into E. coli and S. enterica after the species diverged or were recently transferred from one to the other: the two gene clusters would have a common organization of genes and a higher level of DNA identity. We have already cloned and sequenced the E. coli O111 gene cluster (5, 6, 44). In this study, we sequenced the O-antigen gene cluster and flanking genes from an O35 S. enterica serovar Adelaide strain and compared this sequence with that of E. coli O111.
Sequencing the O-antigen gene cluster of S. enterica O35 (serovar Adelaide).
Oligonucleotides which bind to the 5′ end of the gnd gene (5′-CACTGCCATACCGACGACGCCGATCTGTTGCTTGG) and the middle of the JUMPstart sequence (5′-ATTGGTAGCTGTAAGCCAAGGGCGGTAGCGT) were used for PCR amplification of the O-antigen gene cluster from S. enterica serovar Adelaide strain M274. Long PCR was carried out by using the Expand Long Template PCR System from Boehringer. The PCR cycles were as follows: denaturation at 94°C for 10 s, annealing at 65°C for 30 s, and extension at 68°C for 15 min. The 39-bp JUMPstart sequence is present upstream of many polysaccharide gene clusters (15), and the gnd gene is present downstream of typical O-antigen gene clusters of E. coli and S. enterica.
A PCR fragment of about 13 kb was obtained and subjected to DNase I digestion, and fragments were cloned into pGEM-T to make a bank by the previously described method (46). To limit the effect of PCR errors, 10 individual PCR products were pooled before making the bank. Forty clones were sequenced from one end. DNA templates for sequencing were prepared by using the 96-well-format plasmid DNA miniprep kit from Advanced Genetic Technologies Corp and the procedure developed in The Institute for Genomic Research (43). Sequencing was performed with an Applied Biosystem 377 automated DNA sequencer. Sequences from these 40 clones were assembled into seven contigs by using the Australian National Genomic Information Service, which incorporates several sets of programs (24–26). These seven contigs were readily aligned with the E. coli O111 sequence (GenBank accession no. AF078736), and the gap lengths between adjacent contigs were estimated. Gaps and regions of inadequate coverage were then sequenced from PCR products amplified from chromosomal DNA with specific primers.
The galF gene is located upstream of the O-antigen gene cluster in S. enterica (37): we also sequenced the region from about 650 bp downstream of the start of galF through the intergenic region to the O-antigen gene cluster. Oligonucleotides which bind to the O111 galF gene (5′-CGAAAAACCGGATCAGCCGCAGACGCT) and the first gene in the S. enterica serovar Adelaide O-antigen gene cluster (5′-TCTGCAAACAATCTTCCC) were used for PCR amplification, and the PCR walking procedure was carried out to sequence this region.
Combining the two regions, a sequence of 13,650 bases was obtained, which covers the DNA from galF to the start of gnd. The galF gene extends from position 1 to position 273; DNA from position 13,647 to position 13,650 encodes the first four bases of gnd. Thus, DNA from position 274 to position 13,646 comprises the O35 O-antigen gene cluster and adjacent intergenic regions.
Comparison of the two O-antigen gene clusters.
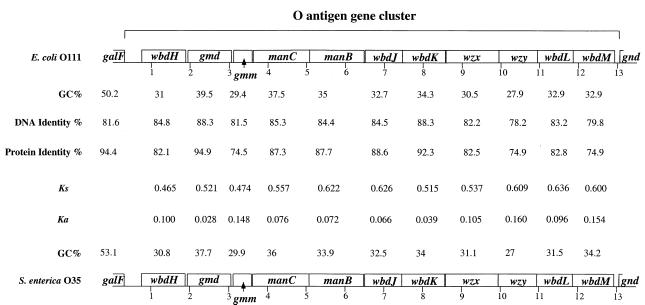
The O antigens of E. coli O111 and S. enterica O35 contain colitose, d-glucose, d-galactose, and n-acetyl-d-glucosamine (18). There are 11 genes in the E. coli O111 gene cluster (Fig. 1) (44), and in the S. enterica O35 gene cluster, we found the same 11 genes in the same order (Fig. 1). There are five genes (manB, manC, gmd, wbdK, and wbdJ) of the putative GDP-colitose pathway. ManB, ManC, and Gmd have functions established biochemically which lead to 4-keto-6-deoxy GDP-mannose, an intermediate in other pathways, while WbdK and WbdJ are proposed on the basis of homologies to complete the synthesis of GDP-colitose (44). There are three presumptive sugar transferase genes (wbdH, wbdL, and wbdM) for synthesis of the O unit: the O-unit flippase gene (wzx), the O-antigen polymerase gene (wzy), and gmm (originally named wbdI) (44).
FIG. 1.
Comparison of the E. coli O111 and S. enterica O35 gene clusters.
The wzx and wzy genes of the E. coli O111 gene cluster were originally identified by comparison of the hydrophobic profiles of their deduced amino acid sequences with those of other Wzx and Wzy proteins (44). However, there is some ambiguity in using only the hydrophobic profile for these genes, because proteins of unrelated function can have similar profiles. The level of sequence similarity is often very low for both Wzx and Wzy, and we have confirmed their identification by Motif (14) and PSI-BLAST (1) searches. Each of the four potential Wzx or Wzy proteins (of both E. coli O111 and S. enterica O35) was grouped with known or putative Wzx or Wzy proteins, and motifs were generated. These motifs were then used to search databases with PSI-BLAST. The Wzy proteins of E. coli O111 and S. enterica O35 grouped with putative Wzy proteins of E. coli O157 (GenBank entry AF061251), S. enterica B (GenBank entry M60066), and E. coli O16 (GenBank entry U09876), as well as distantly related putative Wzy proteins, but no other proteins were retrieved (E-value, ≤4 × 10−35) after several iterations. The Wzx proteins of E. coli O111 and S. enterica O35 B grouped with putative Wzx proteins of E. coli O157 (GenBank entry AF061251), S. enterica B (GenBank entry X56793), E. coli O16 (GenBank entry U09876), and E. coli O113 (GenBank entry AF172324), and also distantly related Wzx proteins, but no other proteins were retrieved (E-value, ≤4 × 10−20) after several iterations. The S. enterica B wzy and wzx genes have been identified biochemically, and the motif searches give very strong support to the original identifications.
With the exception of gmm, these genes are those expected for synthesis of the repeat unit plus the expected flippase and polymerase genes. The gmm gene encodes GDP-mannosyl hydrolase (12), which would remove GDP-mannose from the GDP-colitose pathway. Notably, in addition to manB, manC, and gmd, gmm is found in the gene cluster for E. coli O157 (46) and the colanic acid gene clusters in E. coli and S. enterica (39; G. Stevenson, R. Lon, and P. R. Reeves, submitted for publication), which all contain fucose. The GDP-fucose pathway has been fully characterized. It starts with fructose-6-phosphate, which is converted by the enzyme encoded by manA (a housekeeping gene) into mannose-6-phosphate. Enzymes encoded by manB and manC then convert mannose-6-phosphate into GDP-mannose. The next enzyme in the GDP-fucose pathway is Gmd (GDP-d-mannose 4, 6-dehydratase), which produces 4-keto-6-deoxymannose, and the final steps to make GDP-l-fucose are catalyzed by Fcl. The GDP-colitose synthesis and GDP-fucose pathways diverge after gmd, and so share the first three steps (44). This distribution suggests that gmm is involved in some form of regulation in E. coli and S. enterica GDP-sugar pathways that continue beyond GDP-mannose. However, gmm is not present in the GDP-fucose pathway of the Yersinia enterocolitica O8 pathway (48). All enzymes in the GDP-fucose pathway have previously been defined biochemically (2, 39).
Comparison of the proteins encoded by the two gene clusters (Fig. 1) showed that the proteins of the six GDP-colitose-related genes had identity levels of between 87.3 and 94.9%; the three potential transferases, WbdH, WbdL, and WbdM, had identity levels of 74.9 to 82.8%; and the O-antigen flippase and polymerase had identity levels of 82.5 and 74.9%, respectively.
The intergenic regions between galF and the first O-antigen gene (wbdH) are 540 and 519 bp in length, respectively, in S. enterica O35 and E. coli O111. These two sequences share about 64% identity. There are stronger similarities between the two sequences in the 39-bp JUMPstart sequence (located about 200 bp 5′ to wbdH) and a region of 20 bp just upstream of wbdH. The JUMPstart sequence is involved in the expression of O-antigen gene clusters (45), and there are only 4 base substitutions in this region. Two segments of 20 bp, located 5 and 4 bp upstream of the start of wbdH in S. enterica O35 and E. coli O111, respectively, are identical between these two strains, except for a single base present in O35, but absent in O111.
The intergenic regions between the last O-antigen genes (wbdM) and gnd are 180 and 177 bp in length in S. enterica O35 and E. coli O111, respectively, and share about 74% identity. The last 49-bp fragments of these two regions share 91% identity. This 49-bp segment is part of the regulatory region for gnd and was previously reported to be conserved among E. coli strains and between E. coli and S. enterica strain LT2 (3, 4).
Evolutionary origins of these two gene clusters.
Like many O-antigen gene clusters, the O111 cluster is of low GC content and is thought to have been introduced into E. coli by lateral gene transfer (5, 44). The S. enterica O35 gene cluster encoding the same O antigen also has a low GC content, and the same argument applies. The two gene clusters also have the same organization and are clearly closely related.
E. coli and S. enterica diverged about 140 million years ago (29, 30), and typical homologous proteins are, on average, 93% identical in the two species, ranging from 76.3 to 100% (38). The identity between proteins encoded by the E. coli O111 and S. enterica O35 antigen genes ranges from 74.5 to 94.9%, so our sequence data are consistent with the two gene clusters being in the common ancestor of E. coli and S. enterica.
The two O-antigen gene clusters are of low GC content, the averages for E. coli O111 being 44.55, 32.73, and 22.98% for codons 1 (P1), 2 (P2), and 3 (P3) and 44.65, 32.17, and 21.85% for O35, with overall GC contents being 0.334 and 0.329, respectively. It is most likely that the ancestral gene clusters were assembled in a low-GC-content organism and later underwent lateral gene transfer to the ancestor of E. coli and S. enterica. We attempted to use the expected changes in GC content after lateral transfer to provide an independent estimate of time since transfer to E. coli or S. enterica. Bacterial species display wide variation in overall GC content, but the genes within any particular species are generally similar in base composition. For example, Sueoka (40) found that in E. coli, more than 82% of genes have a similar GC content (20), and Muto and Osawa (27) presented plots for the GC content of each codon base (P1, P2, and P3) against the genomic GC content, using average data for each species. The three plots are quite different, because each codon position is under different constraints. DNA transferred by lateral transfer would begin with the base composition of the donor genome at the time of the transfer, but would then be subject to the same mutational process affecting all genes in the recipient genome and over time would ameliorate to reflect the DNA composition of the new genome. The bases at the three codon positions would adjust at different rates due to different constraints on base change, and during this adjustment process, known as amelioration, they would not conform to the usual relationship for GC content at the three codon positions.
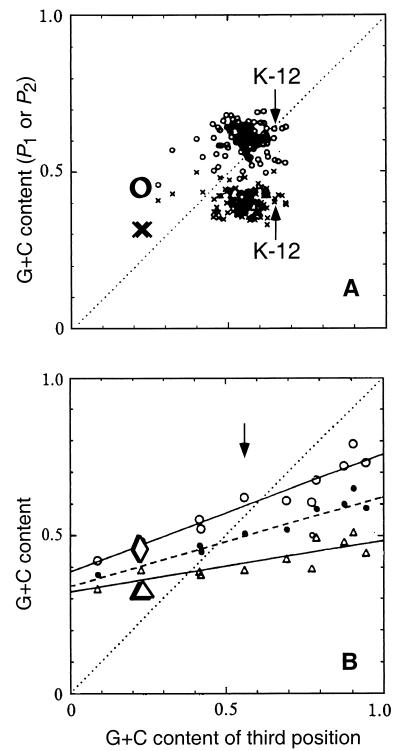
This principle can be used to estimate time since lateral transfer. Sueoka (40) (using data from Muto and Osawa) looked at genes from species with different GC contents and plotted regression of P1 and P2 against P3. The value for P3 was taken to represent the equilibrium value for that species, because at most codons, the third base can be substituted for by the transition alternative without a change of amino acid, and hence in general without a significant effect on fitness. The regression lines reflect the long-term equilibrium between mutational drive for change in GC content and the constraints of the effect on amino acid composition. We superimposed the data for the O111 and O35 gene clusters on the Sueoka (40) regression plots (Fig. 2). The value of P3 is higher than predicted by the Sueoka regression line for the observed value for P1 or P2. This is as expected for genes which have had time for random genetic drift after transfer from a low-GC-content species to a species with GC content of about 0.5, as found in E. coli, S. enterica, and, presumably, their ancestor.
FIG. 2.
(A) Plot of G+C content of the first (small circles) and second (small crosses) codon positions against that of the third codon position for all individual E. coli K-12 genes (taken from Sueoka [40]) with the data for the E. coli O111 and S. enterica O35 gene clusters superimposed. The data represent the average for the 11 O-antigen genes sequenced for both E. coli and S. enterica. (B) Average G+C content of the first (P1; open circles), second (P2; open triangles), and combined (P1 and P2; solid circles) codon positions plotted against the third (P3) codon position for a range of bacterial species (40). The arrow indicates the E. coli data. The values for codon positions 1 and 2 (larger open diamond and triangle, respectively) for the E. coli O111 and S. enterica O35 gene clusters are superimposed on the Sueoka graph (42). The data represent the average for the 11 O-antigen genes sequenced for both E. coli and S. enterica.
We also tried to estimate the time of transfer of the gene clusters to E. coli or S. enterica by the method described by Lawrence and Ochman (19) for “back-amelioration,” by using the program kindly supplied by Lawrence. The GC contents of the three codon positions can each be back-ameliorated from a known sequence until the sequence best conforms to the relationship proposed by Muto and Osawa (27), providing estimates both of the time of introduction of the genes and the GC content of the donor genome (19). Unfortunately, the program did not give meaningful results with our data, although we could repeat the analyses reported by Lawrence and Ochman (19). We found that back-amelioration of either the E. coli O111 or S. enterica O35 data does not bring the values of P1, P2, and P3 to a better fit to the Muto and Osawa plots. After the first cycle, there was a worse fit to the Muto and Osawa plots and there were progressively worse fits with subsequent cycles. There are two possible explanations. There is considerable variation in the values of P1, P2, and P3 within a species, and the program will not work well for genes which deviate much from the average from which the three Muto and Osawa plots were obtained. Alternatively, if the parameters used for the amelioration program were not correct for this case, then again back-amelioration would not work. It is perhaps notable that in the original application of this amelioration algorithm, there was a similar failure to obtain meaningful results for 11 of 33 groups of genes in E. coli thought on the basis of GC content to have been transferred to E. coli (19).
The synonymous substitution rate (Ks) and nonsynonymous substitution rate (Ka) were calculated by the method of Li et al. (21). The mean Ks value for these 11 genes is 0.56, which is about half the average value reported by Sharp, although well within the range observed. The average value of Ka was 0.095, which is higher than that observed by Sharp, but again is within the range observed. The relatively high value for Ka/Ks suggests that there may have been adaptive changes after divergence of E. coli and S. enterica, perhaps related to ongoing adaptation after lateral transfer from a distantly related species, rather than adaptation related to differences in the niches of E. coli and S. enterica.
We conclude that the most probably relationship between the S. enterica O35 and E. coli O111 gene clusters is that they are derived from a gene cluster in the common ancestor. Our conclusion is based on the identity of gene order in the two clusters and the level of divergence between them. It seems clear that they do have a common ancestor and are not independently assembled gene clusters. We do not support the alternative explanation that the gene cluster transferred from E. coli to S. enterica, or vice versa, although we cannot exclude the possibility that this occurred soon after species divergence. It also seems highly unlikely that the gene cluster transferred to the two species from other species which by chance have levels of divergence similar to those of E. coli and S. enterica, but again we cannot formally exclude that alternative. We were unable to confirm the date of lateral transfer by back-amelioration, but this simply says that current methods cannot always give answers to this question. Although the conclusion is to some extent tentative, it gives the first indication of the time frame for survival of an O-antigen gene cluster in a species, because at least one of the three found in both E. coli and S. enterica has all of the hallmarks of a survivor from the common ancestor.
Distribution of the E. coli O111 or S. enterica O35 O antigen.
It is interesting that the E. coli O111 or S. enterica O35 O antigen, which we now believe to have been in both E. coli and S. enterica since divergence, is currently known for its association with EPEC and EHEC pathogenic forms of E. coli, which are generally thought to have arisen recently in evolutionary terms, with key genes involved in pathogenicity present on plasmids. It seems probable that the O111 antigen was in E. coli before the arrival of these plasmids and hence was not in strains occupying a niche involved in the EPEC or EHEC mode of pathogenesis.
The situation for O35 of S. enterica is also interesting. There are 46 different O antigens present in 2,422 serovars of S. enterica (32, 36). Seven subspecies in S. enterica have been defined by biotyping, and, with few exceptions, phylogenetic trees constructed for individual genes match the subspecies tree (7, 42). The distribution of each O antigen among the subspecies varies, with many found predominantly in one or two subspecies (36). Fifty-four of the 2,422 serovars carry the O35 O antigen, with 22, 7, 5, and 20 serovars in subspecies I, II, IIIa, and IIIb, respectively. Taking into account that there are 1,430, 478, 95, and 319 serovars, respectively, in the four subspecies, it is clear that O35 has a major presence in subspecies IIIa and IIIb, where it is found in 5 and 6% of serotypes, respectively. This may indicate that the O35 antigen has been in S. enterica subspecies IIIa and IIIb serovars for a long time and only recently was transferred into subspecies I and II serovars.
Subspecies IIIa and IIIb serovars are generally associated with cold-blooded vertebrates, and subspecies I and II are associated with warm-blooded animals. It appears that antigen 35 has relatively recently transferred to subspecies I and II, where it is a significant antigen, with serovar Adelaide the 45th most frequently isolated S. enterica subspecies I serovar around the worldwide in a survey of isolates from 1934 to 1975 (17).
General conclusion.
Comparison of the gene clusters for the O antigen known as O35 in the S. enterica scheme and O111 in the E. coli scheme indicates that this O antigen was present in the common ancestor of E. coli and S. enterica and has been retained in both species, while most O antigens are present in one or the other species only. It remains for further work to establish if the other two O antigens present in both species were also in the common ancestor. The current distribution of this antigen in S. enterica suggests that it is involved in relatively recent adaptations, so that its long-term survival does not imply continuity of any given niche over the long period of time that this O antigen has been present in this lineage.
Acknowledgments
We thank Kanella Andrianopoulos for technical assistance.
This study was supported by the Australian Research Council.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3398–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrianopoulos K, Wang L, Reeves P R. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998;180:998–1001. doi: 10.1128/jb.180.4.998-1001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker H V, II, Wolf R E., Jr Essential site for growth rate dependent regulation within the Escherichia coli gnd structural gene. Proc Natl Acad Sci USA. 1984;81:7669–7673. doi: 10.1073/pnas.81.24.7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barcak G J, Wolf R E., Jr Growth-rate-dependent expression and cloning of gnd alleles from natural isolates of Escherichia coli. J Bacteriol. 1988;170:365–371. doi: 10.1128/jb.170.1.365-371.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastin D A, Reeves P R. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 6.Bastin D A, Romana L K, Reeves P R. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol Microbiol. 1991;5:2223–2231. doi: 10.1111/j.1365-2958.1991.tb02152.x. [DOI] [PubMed] [Google Scholar]
- 7.Boyd E F, Nelson K, Wang F-S, Whittam T S, Selander R K. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenner D J, Fanning G R, Miklos G V, Steigerwalt A G. Polynucleotide sequence relatedness among Shigella species. Int J Syst Bacteriol. 1973;23:1–7. [Google Scholar]
- 9.Curd H, Liu D, Reeves P R. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J Bacteriol. 1998;180:1002–1007. doi: 10.1128/jb.180.4.1002-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewing W H. Serological relationships between Shigella and coliform cultures. J Bacteriol. 1953;66:333–340. doi: 10.1128/jb.66.3.333-340.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing W H. Edwards and Ewing's identification of the Enterobacteriaceae. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. [Google Scholar]
- 12.Frick D N, Townsend B D, Bessman M J. A novel GDP-mannose mannosyl hydrolase shares homology with the MutT family of enzymes. J Biol Chem. 1995;270:24086–24091. doi: 10.1074/jbc.270.41.24086. [DOI] [PubMed] [Google Scholar]
- 13.Goullet P. Esterase electrophoretic pattern between Shigella species and Escherichia coli. J Gen Microbiol. 1980;117:493–500. doi: 10.1099/00221287-117-2-493. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff S, Henikoff J G, Alford W J, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs M, Reeves P R. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 16.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 17.Kelterborn E. On the frequency of occurrence of Salmonella species. An analysis of 1.5 million strains of Salmonella isolated in 109 countries during the period 1934–1975. Zentbl Bakteriol Hyg A. 1979;243:289–307. [PubMed] [Google Scholar]
- 18.Kenne L, Lindberg B, Soderholm E, Bundle D R, Griffith D W. Structural studies of the O-antigens from Salmonella greenside and Salmonella adelaide. Carbohydr Res. 1983;111:289–296. doi: 10.1016/0008-6215(83)88313-x. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence J G, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 20.Lawrence J G, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W-H. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 22.Lindberg B, Lindh F, Longren J, Lindberg A A, Svenson S B. Structural studies of the O-specific side-chain of the lipopolysaccharide from Escherichia coli O55. Carbohydr Res. 1981;97:105–112. doi: 10.1016/s0008-6215(00)80528-5. [DOI] [PubMed] [Google Scholar]
- 23.Lior H. Classification of Escherichia coli. In: Gyles C L, et al., editors. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 31–72. [Google Scholar]
- 24.Littlejohn T G. Bioinformatics: the essential ingredient. Today's Life Sci. 1996;8:28–33. [Google Scholar]
- 25.Littlejohn T G, Bucholtz C A, Campbell R M M, Gata B A, Huynh C, Kim S H. Computing for biotechnology—WebANGIS. Australas Biotechnol. 1996;6:211–217. [Google Scholar]
- 26.L'vov V L, Shashkov A S, Dmitriev B A, Kochetrov N K. Structural studies of the O-specific side chain of the lipopolysaccharide from Escherichia coli O:7. Carbohydr Res. 1984;126:249–259. doi: 10.1016/0008-6215(84)85382-3. [DOI] [PubMed] [Google Scholar]
- 27.Muto A, Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci USA. 1987;84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman H, Whittam T S, Caugant D A, Selander R K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 29.Ochman H, Wilson A C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 30.Ochman H, Wilson A C. Evolutionary history of enteric bacteria. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1649–1654. [Google Scholar]
- 31.Perry M B, MacLean L, Griffith D W. Structure of the O-chain polysaccharide of the phenol-phase soluble lipopolysaccharide of Escherichia coli O:157:H7. Biochem Cell Biol. 1986;64:21–28. doi: 10.1139/o86-004. [DOI] [PubMed] [Google Scholar]
- 32.Popoff M Y, Minor L L. Antigenic formulas of the Salmonella serovars, 7th revision. WHO Collaborating Centre for Reference and Research on Salmonella. Paris, France: Institut Pasteur; 1997. [Google Scholar]
- 33.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves P R. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 35.Reeves P R. Biosynthesis and assembly of lipopolysaccharide. In: Neuberger A, van Deenen L L M, editors. Bacterial cell wall, new comprehensive biochemistry. Vol. 27. Amsterdam, The Netherlands: Elsevier Science Publishers; 1994. pp. 281–314. [Google Scholar]
- 36.Reeves P R. Role of O-antigen variation in the immune response. Trends Microbiol. 1995;3:381–386. doi: 10.1016/s0966-842x(00)88983-0. [DOI] [PubMed] [Google Scholar]
- 37.Sanderson K E, Hessel A, Rudd K E. Genetic map of Salmonella typhimurium, edition VIII. Microbiol Rev. 1995;59:241–303. doi: 10.1128/mr.59.2.241-303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp P M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci USA. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb* gene cluster of Klebsiella O3 into E. coli via recombination. J Bacteriol. 1998;180:2775–2778. doi: 10.1128/jb.180.10.2775-2778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thampapillai G, Lan R, Reeves P R. Molecular evolution in the gnd locus of Salmonella enterica. Mol Biol Evol. 1994;11:813–828. doi: 10.1093/oxfordjournals.molbev.a040165. [DOI] [PubMed] [Google Scholar]
- 43.Utterback T R, McDonald L A, Fuldner R A. A reliable, efficient protocol for 96-well plasmid DNA miniprep with rapid DNA quantification for high-throughput automated DNA sequencing. Genome Sci Technol. 1995;1:1–8. [Google Scholar]
- 44.Wang L, Curd H, Qu W, Reeves P R. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J Clin Microbiol. 1998;36:3182–3187. doi: 10.1128/jcm.36.11.3182-3187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Jensen S, Hallman R, Reeves P R. Expression of the O antigen gene cluster is regulated by RfaH through the JUMPstart sequence. FEMS Microbiol Lett. 1998;165:201–206. doi: 10.1111/j.1574-6968.1998.tb13147.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Reeves P R. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang S-H, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Radziejewska-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O8. Mol Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]