Abstract
Atherosclerosis is an important pathological basis of coronary heart disease, and the antisense non-coding RNA in the INK4 locus (ANRIL) is located in the genetically susceptible segment with the strongest correlation with it - the short arm 2 region 1 of chromosome 9 (Chr9p21). ANRIL can produce linear, circular and other transcripts through different transcriptional splicing methods, which can regulate the proliferation and apoptosis of related cells and closely related to the development of atherosclerotic plaques. Linear ANRIL can regulate proliferation of vascular smooth muscle cells (VSMCs) in plaques by chromatin modification, as well as affecting on proliferation and the apoptosis of macrophages at the transcriptional level; circular ANRIL can affect on proliferation and apoptosis of VSMCs by chromatin modification as well as interfering with rRNA maturation. In this review we describe the evolutionary characteristics of ANRIL, the formation and structure of transcripts, and the mechanism by which each transcript regulates the proliferation and apoptosis of vascular cells and then participates in atherosclerosis.
Keywords: Long non-coding RNA, ANRIL, Atherosclerosis, Apoptosis, Cell proliferation
1. Introduction
Coronary heart disease is an important cause of death in the world, and its important pathological basis is atherosclerosis. In the early stage of atherosclerosis, long-term mechanical change in blood flow, high-fat and high-glucose environment, inflammatory infection damage, etc. The pathological factors of injury and pro-apoptotic factors increase, leading to the increase of endothelial cell injury and apoptosis, and the destruction of vascular endothelial integrity [1]. Subsequently, macrophages are activated to recognize and phagocytose oxidized lipoproteins accumulated under the endothelium through their surface receptors to form foam cells, which constitute an important core of necrotic lipids [2]. Activated macrophages can also produce a large number of interstitial collagen fibers to participate in the construction of plaque fibrous caps, and maintain plaque stability by removing apoptotic cells. Insufficient macrophage proliferation and increased apoptosis can lead to insufficient plaque fibrous cap strength and easy rupture, and apoptotic cells that are not cleared in time can further activate thrombin, induce intraplaque thrombosis, and then lead to acute coronary events [3]. Activated endothelial cells and macrophages can promote the transition of adjacent vascular smooth muscle cells from a quiescent and tiling telescopic differentiation state to a dedifferentiated state by paracrine various growth factors, and migrate to the vascular intima under the action of various chemokines. Smooth muscle cells can secrete a large number of extracellular matrix components such as collagen and polysaccharide molecules [4], which further accelerates the accumulation of plaques.
Non-coding RNA (ncRNA) refers to RNAs that are not translated into proteins, mainly including microRNA (miRNAs), long non-coding RNA (lncRNA), and circular RNA (circRNA) [5,6]. They are transcribed from the genome but not translated into proteins, and perform their respective biological functions at the RNA level [7]. NcRNAs bind to many molecular targets to form regulatory networks, which in turn initiate specific cellular biological responses, with the function of regulating gene expression, influencing intracellular signaling, participating in epigenetic modifications and other life activities, and thus playing a role in the occurrence and development of tumors and other diseases [8,9].
LncRNAs are a class of RNAs with transcripts longer than 200bp that do not encode proteins [[10], [11], [12], [13]]. LncRNAs were considered to be “transcriptional noise” with no biological function in the early days, and are by-products of RNA polymerase II transcription, which have no biological function [[14], [15], [16], [17]]. LncRNAs are characterized by their large number, variety and mode of action. At present, there is no uniform classification standard for lncRNAs [[18], [19], [20]]. According to the localization of lncRNAs in cells, they can be divided into cytoplasmic lncRNAs and cytosolic lncRNAs, some of which are located in both the nucleus and the cytoplasm [21,22]. LncRNAs may play different regulatory functions according to their different cellular localization (Table 1) [23,24]. In the cytoplasm, lncRNAs can act as competing endogenous RNAs (ceRNAs) to compete with miRNAs for binding and contribute to the release of target mRNAs.
Table 1.
Role of lncRNAs in atherosclerosis.
| LncRNA | Pathophysiological Effects | Indirect targets & Signaling pathway | Ref. |
|---|---|---|---|
| ANRIL | Slows cell cycle gene expression | [[25], [26], [27], [28], [29]] | |
| MALAT1 | Migratory behavior, increases AKT pathway behavior in endothelial cells | CXCR2 and AKT, AKT pathway | [[30], [31], [32], [33], [34], [35]] |
| LOC100129973 | Suppress apoptosis of endothelial cells | API5 and BCL2L12 | [36] |
| MEG3 | Suppresses migration, proliferation and tube formation of endothelial cells | RhoB and PTEN | [37,38] |
| LEENE | Regulates eNOS expression and EC function | [39] | |
| LISPR1 | Angiogenesis, vascular stability and permeability | S1P signaling pathway | [40] |
| SMILR | VSMC proliferation | HAS2 | [41] |
| LincRNA-p21 | Reduces cell proliferation and increases apoptosis | p53 feed forward loop | [42] |
| SENCR | Promotes migration, proliferation and tube formation of endothelial cells | CCL5, CEACAM1, and CX3CL1 (migratory and angiogenic genes) | [43,44] |
| MYOSLID | Activates VSMC contractile phenotype | TGFβ/SMAD, MYOCD/SRF pathways | [45] |
| H19 | Reduces autophagy, apoptosis and reactive oxygen species in endothelial cells | MAPK and NF-kβ pathways | [[46], [47], [48]] |
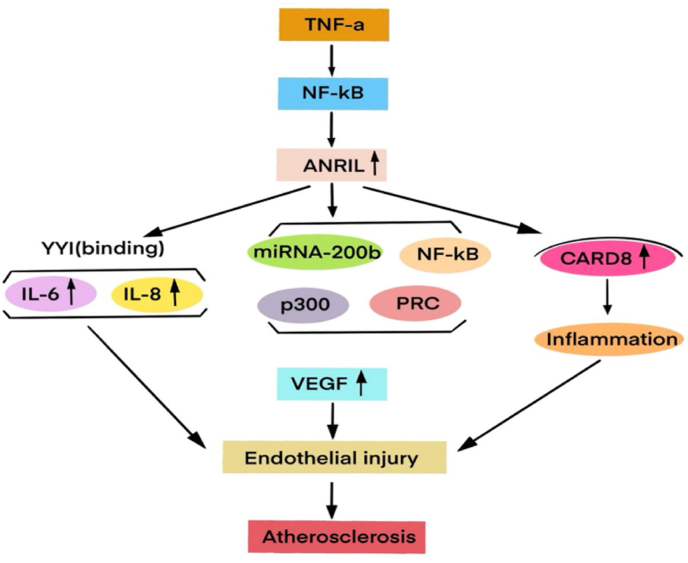
The antisense non-coding RNA (antisense non-coding RNA in the INK4 locus, ANRIL) in the INK4 locus originated in placental mammals and acquired additional exons during evolution, and then part of the exons was gradually lost, and finally It is fully formed in apes [49]. ANRIL is located in the most susceptibility gene segment of human cardiovascular disease - the short arm 2 region 1 of chromosome 9 (Chr9p21), which can be distributed in the cytoplasm and nucleus [50]. Studies have shown that ANRIL is closely related to the occurrence of atherosclerosis [51,52]. Through the genome-wide association analysis study and subsequent studies of 15 596 patients with coronary heart disease and 34 992 control samples in Europe and Asia, it was found that Chr9p21 has the most cardiovascular disease-related single nucleotide polymorphism mutations [53]. It is the most genetically susceptible segment associated with coronary heart disease. Hu et all. conducted a meta-analysis on the correlation between ANRIL polymorphisms and coronary heart disease risk in different regions and ethnic groups in Asia, Europe, North America, etc., and found that a variety of mononucleotides such as rs1333040, rs1333049 and rs2383207 on ANRIL were more abundant [54]. There is a correlation between the state of the disease and the risk of coronary heart disease. By selectively preserving the transcriptional splicing process of different exons, ANRIL can combine the editing of precursor mRNA to generate a variety of transcripts, including linear ANRIL transcripts and circular ANRIL transcripts [55]. Coronary heart disease-related single nucleotide polymorphism risk mutation at Chr9p21 locus can regulate the alternative splicing of ANRIL and affect the expression level of different ANRIL transcripts [50]. Linear RNA is a class of non-coding linear transcripts longer than 200 nucleotides, which can be involved in endothelial cell dysfunction, inflammatory response, lipid and lipid in atherosclerosis by mediating cell signaling, chromatin modification, transcription and translation regulation, etc. [[56], [57], [58]]. Different from the traditional transcriptional splicing method of linear RNA, circular RNA can resist exonuclease digestion and degradation, and its structure is more stable [59]. Circular RNA can participate in a variety of cell proliferation, apoptosis and inflammatory signaling pathways by regulating the transcription and expression of target genes, acting as protein bridging molecules and other mechanisms, thereby affecting the occurrence and development of atherosclerosis [[60], [61], [62]]. Different ANRIL transcripts play an important role in promoting and protecting the occurrence and evolution of atherosclerosis (Fig. 1) [[50], [51], [52]].
Fig. 1.
Role of lncRNA ANRIL in atherosclerosis.
This article reviews the research progress of ANRIL transcripts with different structures that affect the occurrence and development of atherosclerosis by regulating the proliferation and apoptosis of vascular cells.
2. ANRIL affects cell proliferation and apoptosis by interfering with ribosomal RNA maturation process
Last few years the function of circular ANRIL has also been gradually explored, and with the progression of atherosclerosis, the expression level of circular ANRIL in plaque lesion tissue gradually decreases [63,64]. Proteomic studies have found that overexpression of cyclic ANRIL can significantly increase the expression levels of 32 proteins, among which the nucleolar protein (pescadillo), which plays an important role in the processing of ribosomal RNA precursors and ribosome assembly, is closely related to the expression of cyclic ANRIL [65]. Cyclic ANRIL weakens the binding of ribosomal RNA precursors to exonuclease through competitive binding to nucleolar proteins, which affects the maturation of ribosomes, resulting in decreased smooth muscle cell proliferation and increased apoptosis [66]. Immunofluorescence staining showed that overexpression of cyclic ANRIL in smooth muscle cells can reduce a large number of nucleoli, nuclear damage, and increase apoptosis in smooth muscle cells, confirming that cyclic ANRIL can affect the process of ribosomal RNA maturation and processing by interfering with it [67]. Proliferation and apoptosis of smooth muscle cells can slow down the progression of atherosclerosis by removing excessively proliferating smooth muscle cells from plaques [68].
3. ANRIL affects cell proliferation and apoptosis through other mechanisms
Cyclic ANRIL can also be used as a molecular scaffold for chromatin modification complexes, affecting target gene expression by regulating the covalent modification of histones, thereby affecting cell proliferation and apoptosis; or as a dynamic scaffold, binding, storing or transport transcription factors to specific subcellular locations such as mitochondria, affecting cell proliferation and metabolism [69,70]. There are many miRNA complementary binding sites on some circular RNAs, which can capture miRNA and produce a “sponge effect”, reducing its negative gene regulation effect on target mRNA [71]. Whether cyclic ANRIL also has such an effective miRNA binding site remains to be further bioinformatic analysis. In addition, circular RNAs can also bind to effector proteins and affect their related signaling pathways [72]. Whether cyclic ANRIL can affect cell proliferation and apoptosis through the above mechanisms and participate in the occurrence and development of atherosclerosis is also worthy of our exploration and verification.
3.1. Anril affects cell proliferation through chromatin modification
Meseure et al. found by RNA co-immunoprecipitation method that linear ANRIL binds efficiently to polycomb family proteins such as CBX7 and SUZ12 [73]. Polycomb family proteins can regulate gene expression by initiating and maintaining epigenetic modifications of chromatin [74,75]. Knockout of linear ANRIL can disrupt the binding of polycomb family protein SUZ12 to Chr9p21 site, resulting in increased expression of cyclin-dependent kinase 2 inhibitor B (CDKN2B) gene at Chr9p21 site, and increased vascular smooth muscle expression. decreased cell proliferation [76]. Knockout of linear ANRIL can also reduce the methylation level of an important protein component of chromatin, that is, histone H3 methylation level, and the cyclin-dependent kinase 2 inhibitor A (CDKN2A) gene expression level is increased, which leads to a decrease in the level of vascular cell proliferation [67]. In addition, linear ANRIL can also mediate the binding of SUZ12 to the p15INK4b site, affecting the proliferation and metabolic activity of vascular smooth muscle cells [66]. All of these studies have shown that linear ANRIL can affect the proliferation of vascular smooth muscle cells through chromatin modification, thereby accelerating plaque accumulation in the late stage of atherosclerosis.
3.2. Anril affects cell proliferation and apoptosis by interfering with transcription
Linear ANRIL can affect the transcriptional expression levels of multiple genes and increase the risk of atherosclerosis by interfering with the transcription of target genes. Overexpression of linear ANRIL in macrophage cell lines can induce AEBP2, EZH2, Jumonji/Jmj Jarid2 C-domain family protein, MEL18 DNA-binding protein, YY1 transcriptional repressor proteins, Gli-Kruppel family proteins with transcription regulation functions, such as COREST/REST, and these proteins are enriched in Alu repeats with the most active transcription of genes in the chromosomal segment [77,78]. Alu repeats are important regulators of trans-action, which can activate gene transcription by binding to target gene promoters such as cyclins, and affect the level of target gene transcription and expression. Cytologically, it is manifested as increased macrophage proliferation, enhanced metabolic activity, and decreased apoptosis [79]. Holdt et al. overexpressed linear ANRIL after 25%, 33%, and 100% mutational disruption of the Alu transcription regulator sequence and found that overexpressed ANRIL enhanced macrophage proliferation and inhibited apoptosis with Alu degradation [75]. Also, linear ANRIL affects transcriptional expression of target genes, such as cyclins, by regulating the level of transcription, thereby affecting the proliferation and apoptosis of macrophages. Studies have found that the expression of linear ANRIL in peripheral blood mononuclear cells of 2880 subjects is closely related to cell proliferation and apoptosis and the transcriptional expression level of target genes, and It is confirmed that linear ANRIL affects the proliferation and apoptosis of macrophages by interfering with transcription, thereby affecting the stability of atheromatous plaques [80].
4. Conclusions
More and more evidences show that ANRIL plays an important role in atherosclerotic cardiovascular disease, but what mechanism affects cell proliferation and apoptosis and participates in the occurrence and development of atherosclerosis still needs further research. With the continuous emergence of new technologies, studies have discovered the existence of more ANRIL transcripts, and there are more means to detect the transcriptional expression levels of target genes and protein molecules in different pathways, which provides an important guarantee for more effective mechanism exploration. However, due to the presence of many common exon fragments among different ANRIL transcripts, it is difficult to distinguish it, and the expression level of ANRIL in peripheral blood is low, and the degradation rate of linear ANRIL in vitro is high. These factors can become a serious obstacle to the process of studying the effect of ANRIL on the mechanism of the onset and development of atherosclerosis. Therefore, it is necessary to create a more stable system and a more accurate measurement method to further study the mechanism of action of ANRIL.
Author contributions
Ilgiz Gareev, Valentin Kudriashov and Albert Sufianov conceptualized and designed the study. All authors participated in the acquisition, analysis and interpretation of the data. Sema Begliarzade, Tatiana Ilyasova and Yanchao Liang drafted the manuscript. Ozal Beylerli contributed to critical revisions of the manuscript. All authors agreed on the journal to which the article would be submitted, gave final approval for the version to be published, and agreed to be accountable for all aspects of the work.
Funding
None.
Declaration of competing interest
The authors declare that no conflicts of interest exist.
Contributor Information
Ilgiz Gareev, Email: ilgiz_gareev@mail.ru.
Valentin Kudriashov, Email: vkudryashov.uro@gmail.com.
Albert Sufianov, Email: sufianov@gmail.com.
Sema Begliarzade, Email: semanagiyeva@yandex.ru.
Tatiana Ilyasova, Email: Iltanya67@yandex.ru.
Yanchao Liang, Email: liangyanchao@hrbmu.edu.cn.
Ozal Beylerli, Email: obeylerli@mail.ru.
References
- 1.Kim S.M., Huh J.W., Kim E.Y., et al. Endothelial dysfunction induces atherosclerosis: increased aggrecan expression promotes apoptosis in vascular smooth muscle cells[J] Bmb Rep. 2019;52(2):145–150. doi: 10.5483/Bmbrep.2019. 52. 2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaftenaar F., Frodermann V., Kuiper J., et al. Atherosclerosis: the interplay between lipids and immune cells [J] Curr. Opin. Lipidol. 2016;27(3):209–215. doi: 10.1097/Mol.0000000000000302. [DOI] [PubMed] [Google Scholar]
- 3.Linton M.F., Babaev V.R., Huang J., et al. Macrophage apoptosis and efferocytosis in the pathogenesis of atherosclerosis [J] Circ. J. 2016;80(11):2259–2268. doi: 10.1253/Circj.Cj-16-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allahverdian S., Chaabane C., Boukais K., et al. Smooth muscle cell fate and plasticity in atherosclerosis [J] Cardiovasc. Res. 2018;114(4):540–550. doi: 10.1093/Cvr/Cvy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer T.R., Munro T., Mattick J.S. The potential of long noncoding RNA therapies. Trends Pharmacol. Sci. 2022;43:269–280. doi: 10.1016/j.tips.2022.01.008. [DOI] [PubMed] [Google Scholar]
- 6.He C., Wang K., Gao Y., Wang C., Li L., Liao Y., Hu K., Liang M. Roles of noncoding RNA in reproduction. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.777510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Tian P., Huang T., Huang J. Noncoding-RNA-Mediated regulation in response to macronutrient stress in plants. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 2021;20:629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L., Wang R., Shen D., Cheng S., Wang H., Lu Z., Zheng Q., Wang L., Xia L., Li G. Role of noncoding RNA in drug resistance of prostate cancer. Cell Death Dis. 2021;12:590. doi: 10.1038/s41419-021-03854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gareev I., Beylerli O., Liang Y., Xiang H., Liu C., Xu X., Yuan C., Ahmad A., Yang G. The role of MicroRNAs in therapeutic resistance of malignant primary brain tumors. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.740303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W., Li H., Yu Q., Xiao W., Wang D.O. LncRNA-mediated DNA methylation: an emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 2022;41:100. doi: 10.1186/s13046-022-02319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J., Sun Z., Gareev I., Yan T., Chen X., Ahmad A., Zhang D., Zhao B., Beylerli O., Yang G., Zhao S. Exosomal miR-2276-5p in plasma is a potential diagnostic and prognostic biomarker in glioma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.671202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L., Li L.P., Yi H.C. DeepWalk based method to predict lncRNA-miRNA associations via lncRNA-miRNA-disease-protein-drug graph. BMC Bioinf. 2022;22:621. doi: 10.1186/s12859-022-04579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Zhang F. The role of microRNA in the pathogenesis of glial brain tumors. Noncoding RNA Res. 2022;7(2):71–76. doi: 10.1016/j.ncrna.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nojima T., Proudfoot N.J. Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol. 2022 doi: 10.1038/s41580-021-00447-6. [DOI] [PubMed] [Google Scholar]
- 16.Wu J., Al-Zahrani A., Beylerli O., Sufianov R., Talybov R., Meshcheryakova S., Sufianova G., Gareev I., Sufianov A. Circulating miRNAs as diagnostic and prognostic biomarkers in high-grade gliomas. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.898537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandes M., Marques H., Teixeira A.L., Medeiros R. miRNA- and lncRNA-based therapeutics for non-Hodgkin's Lymphoma: moving towards an RNA-guided precision medicine. Cancers. 2021;13 doi: 10.3390/cancers13246324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gareev I., Gileva Y., Dzidzaria A., Beylerli O., Pavlov V., Agaverdiev M., Mazorov B., Biganyakov I., Vardikyan A., Jin M., Ahmad A. Long non-coding RNAs in oncourology. Noncoding RNA Res. 2021 Aug 26;6(3):139–145. doi: 10.1016/j.ncrna.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang J., Fang X., Chen J., Zhang H., Tang Z. Long non-coding RNA (lncRNA) in oral squamous cell carcinoma: biological function and clinical application. Cancers. 2021;13 doi: 10.3390/cancers13235944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Guang Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022 Feb 25;7(2):66–70. doi: 10.1016/j.ncrna.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui X.Y., Zhan J.K., Liu Y.S. Roles and functions of antisense lncRNA in vascular aging. Ageing Res. Rev. 2021;72 doi: 10.1016/j.arr.2021.101480. [DOI] [PubMed] [Google Scholar]
- 22.Beylerli O., Gareev I., Pavlov V., Chen X., Zhao S. The role of long noncoding RNAs in the biology of pituitary adenomas. World Neurosurg. 2020 May;137:252–256. doi: 10.1016/j.wneu.2019.10.137. [DOI] [PubMed] [Google Scholar]
- 23.Kong H., Sun M.L., Zhang X.A., Wang X.Q. Crosstalk among circRNA/lncRNA, miRNA, and mRNA in osteoarthritis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.774370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sufianov A., Begliarzade S., Ilyasova T., Liang Y., Beylerli O. MicroRNAs as prognostic markers and therapeutic targets in gliomas. Noncoding RNA Res. 2022 Jul 6;7(3):171–177. doi: 10.1016/j.ncrna.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas A.A., Feng B., Chakrabarti S. ANRIL regulates production of extracellular matrix proteins and vasoactive factors in diabetic complications. Am. J. Physiol. Endocrinol. Metabol. 2018;314(3):E191. doi: 10.1152/ajpendo.00268.2017. –E200. [DOI] [PubMed] [Google Scholar]
- 26.Rahimi E., Ahmadi A., Boroumand M.A., Mohammad Soltani B., Behmanesh M. Association of ANRIL expression with coronary artery disease in type 2 diabetic patients. Cell J. 2018;20(1):41–45. doi: 10.22074/cellj.2018.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bochenek G., Hasler R., El Mokhtari N.E., Konig I.R., Loos B.G., Jepsen S., Rosenstiel P., Schreiber S., Schaefer A.S. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 2013;22(22):4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- 28.Holdt L.M., Hoffmann S., Sass K., Langenberger D., Scholz M., Krohn K., Finstermeier K., Stahringer A., Wilfert W., Beutner F., Gielen S., Schuler G., Gabel G., Bergert H., Bechmann I., Stadler P.F., Thiery J., Teupser D. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9(7) doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X., Han X., Wittfeldt A., Sun J., Liu C., Wang X., Gan L.M., Cao H., Liang Z. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-kappaB pathway. RNA Biol. 2016;13(1):98–108. doi: 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Z., He Y., Li D., Fang X., Shang T., Zhang H., Zheng X. Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR-21. Am. J. Transl Res. 2017;9(7):3326–3335. [PMC free article] [PubMed] [Google Scholar]
- 31.Michalik K.M., You X., Manavski Y., Doddaballapur A., Zornig M., Braun T., John D., Ponomareva Y., Chen W., Uchida S., Boon R.A., Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 32.Tang Y., Jin X., Xiang Y., Chen Y., Shen C.X., Zhang Y.C., Li Y.G. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett. 2015;589(20 Pt B):3189–3196. doi: 10.1016/j.febslet.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 33.Luo F., Sun B., Li H., Xu Y., Liu Y., Liu X., Lu L., Li J., Wang Q., Wei S., Shi L., Lu X., Liu Q., Zhang A. A MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis. Oncotarget. 2016;7(5):5769–5787. doi: 10.18632/oncotarget.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Man H.S., Yan M.S., Lee J.J., Marsden P.A. Epigenetic determinants of cardiovascular gene expression: vascular endothelium. Epigenomics. 2016;8(7):959–979. doi: 10.2217/epi-2016-0012. [DOI] [PubMed] [Google Scholar]
- 35.Deng Q.J., Xie L.Q., Li H. Overexpressed MALAT1 promotes invasion and metastasis of gastric cancer cells via increasing EGFL7 expression. Life Sci. 2016;157:38–44. doi: 10.1016/j.lfs.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 36.Lu W., Huang S.Y., Su L., Zhao B.X., Miao J.Y. Long noncoding RNA LOC100129973 suppresses apoptosis by targeting miR-4707–5p and miR-4767 in vascular endothelial cells. Sci. Rep. 2016;6 doi: 10.1038/srep21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Wang T.T., Li Y., Shi M.M., Li H.M., Yuan H.X., Mo Z.W., Chen J., Zhang B., Chen Y.X., Wang J.F., Dai W.P., Xu Y.Q., Wang Z.P., Zhang X., Ou Z.J., Ou J.S. High density lipoprotein from coronary artery disease patients caused abnormal expression of long non-coding RNAs in vascular endothelial cells. Biochem. Biophys. Res. Commun. 2017;487(3):552–559. doi: 10.1016/j.bbrc.2017.04.082. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C.Y., Yu M.S., Li X., Zhang Z., Han C.R., Yan B. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumour Biol. 2017;39(6) doi: 10.1177/1010428317701311. 1010428317701311. [DOI] [PubMed] [Google Scholar]
- 39.Miao Y., Ajami N.E., Huang T.S., Lin F.M., Lou C.H., Wang Y.T., Li S., Kang J., Munkacsi H., Maurya M.R., Gupta S., Chien S., Subramaniam S., Chen Z. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat. Commun. 2018;9(1):292. doi: 10.1038/s41467-017-02113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Josipovic I., Pfluger B., Fork C., Vasconez A.E., Oo J.A., Hitzel J., Seredinski S., Gamen E., Heringdorf D.M.Z., Chen W., Looso M., Pullamsetti S.S., Brandes R.P., Leisegang M.S. Long noncoding RNA LISPR1 is required for S1P signaling and endothelial cell function. J. Mol. Cell. Cardiol. 2018;116:57–68. doi: 10.1016/j.yjmcc.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Ballantyne M.D., Pinel K., Dakin R., Vesey A.T., Diver L., Mackenzie R., Garcia R., Welsh P., Sattar N., Hamilton G., Joshi N., Dweck M.R., Miano J.M., McBride M.W., Newby D.E., McDonald R.A., Baker A.H. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation. 2016;133(21):2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G., Cai J., Han Y., Chen J., Huang Z.P., Chen C., Cai Y., Huang H., Yang Y., Liu Y., Xu Z., He D., Zhang X., Hu X., Pinello L., Zhong D., He F., Yuan G.C., Wang D.Z., Zeng C. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130(17):1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell R.D., Long X., Lin M., Bergmann J.H., Nanda V., Cowan S.L., Zhou Q., Han Y., Spector D.L., Zheng D., Miano J.M. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler. Thromb. Vasc. Biol. 2014;34(6):1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulberdaa M., Scott E., Ballantyne M., Garcia R., Descamps B., Angelini G.D., Brittan M., Hunter A., McBride M., McClure J., Miano J.M., Emanueli C., Mills N.L., Mountford J.C., Baker A.H. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol. Ther. 2016;24(5):978–990. doi: 10.1038/mt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J., Zhang W., Lin M., Wu W., Jiang P., Tou E., Xue M., Richards A., Jourd’heuil D., Asif A., Zheng D., Singer H.A., Miano J.M., Long X. MYOSLID is a novel serum response factor-dependent long noncoding RNA that amplifies the vascular smooth muscle differentiation program. Arterioscler. Thromb. Vasc. Biol. 2016;36(10):2088–2099. doi: 10.1161/ATVBAHA.116.307879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kallen A.N., Zhou X.B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.S., Zhang H., Min W., Bennett A.M., Gregory R.I., Ding Y., Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z., Gao W., Long Q.Q., Zhang J., Li Y.F., Liu D.C., Yan J.J., Yang Z.J., Wang L.S. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci. Rep. 2017;7(1):7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan J.X. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017;21(2):322–328. [PubMed] [Google Scholar]
- 49.Pasmant E., Sabbagh A., Vidaud M., et al. Anril, A long, noncoding Rna, is an unexpected major Hotspot in Gwas [J] Faseb. J. 2011;25(2):444–448. doi: 10.1096/Fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 50.Davignon J., Ganz P. Role of endothelial dysfunction in atherosclerosis. Circulation. 2004;109:Iii27–32. doi: 10.1161/01.Cir.0000131515.03336.F8. [DOI] [PubMed] [Google Scholar]
- 51.Huang T., Zhao Hy, Zhang Xb, Gao Xl, Peng Wp, Zhou Y., Zhao Wh, Yang Hf. Lncrna anril regulates cell proliferation and migration via sponging Mir-339-5p and regulating Frs2 expression in atherosclerosis. Eur. Rev. Med. Pharmacol. Sci. 2020 Feb;24(4):1956–1969. doi: 10.26355/Eurrev_202002_20373. [DOI] [PubMed] [Google Scholar]
- 52.Holdt Lm, Teupser D. Long noncoding Rna anril: Lnc-Ing genetic variation at the chromosome 9p21 locus to molecular mechanisms of atherosclerosis. Front Cardiovasc Med. 2018 Nov 6;5:145. doi: 10.3389/Fcvm.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ibc 50k Cad Consortium Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011 Sep;7(9) doi: 10.1371/Journal.Pgen.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu L., Su G., Wang X. The roles of anril polymorphisms in coronary artery disease: a meta-analysis. Biosci. Rep. 2019 Dec 20;39(12) doi: 10.1042/Bsr20181559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fica Sm, Tuttle N., Novak T., Ns Li, Lu J., Koodathingal P., Dai Q., Staley Jp, Piccirilli Ja. Rna catalyses nuclear pre-Mrna splicing. Nature. 2013 Nov 14;503(7475):229–234. doi: 10.1038/Nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gareev I., Gileva Y., Dzidzaria A., Beylerli O., Pavlov V., Agaverdiev M., Mazorov B., Biganyakov I., Vardikyan A., Jin M., Ahmad A. Long non-coding RNAs in oncourology. Noncoding RNA Res. 2021 Aug 26;6(3):139–145. doi: 10.1016/j.ncrna.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Guang Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022 Feb 25;7(2):66–70. doi: 10.1016/j.ncrna.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beylerli O., Gareev I., Pavlov V., Chen X., Zhao S. The role of long noncoding RNAs in the biology of pituitary adenomas. World Neurosurg. 2020 May;137:252–256. doi: 10.1016/j.wneu.2019.10.137. [DOI] [PubMed] [Google Scholar]
- 59.Clarke Mc, Figg N., Maguire Jj, et al. Apoptosis of vascular smooth muscle cells induces features of plaque vulnerability in atherosclerosis. Nat. Med. 2006;12:1075–1080. doi: 10.1038/Nm1459. [DOI] [PubMed] [Google Scholar]
- 60.Beilerli A., Gareev I., Beylerli O., Yang G., Pavlov V., Aliev G., Ahmad A. Circular RNAs as biomarkers and therapeutic targets in cancer. Semin. Cancer Biol. 2022 Aug;83:242–252. doi: 10.1016/j.semcancer.2020.12.026. [DOI] [PubMed] [Google Scholar]
- 61.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., Zhu P., Chang Z., Wu Q., Zhao Y., Jia Y., Xu P., Liu H., Shan G. Exon-intron circular Rnas regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015 Mar;22(3):256–264. doi: 10.1038/Nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 62.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular Rnas. Mol. Cell. 2018;71(3):428–442. doi: 10.1016/J.Molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 63.Xu T., Wu J., Han P., et al. Circular Rna expression profiles and features in human tissues:A study using Rna-Seq data. BMC Genom. 2017;18 doi: 10.1186/S12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan W.L., Lim B.T., Anene-Nzelu C.G., et al. A Landscape of circular Rna expression in the human heart. Cardiovasc. Res. 2017;113(3):298–309. doi: 10.1093/Cvr/Cvw250. [DOI] [PubMed] [Google Scholar]
- 65.Holdt L.M., Stahringer A., Sass K., et al. Circular non-coding Rna anril modulates ribosomal Rna maturation and atherosclerosis in humans. Nat. Commun. 2016;7 doi: 10.1038/Ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren C., Smith S.G., Yap K., et al. Structure-guided discovery of selective antagonists for the chromodomain of polycomb repressive protein Cbx7. ACS Med. Chem. Lett. 2016;7(6):601–605. doi: 10.1021/Acsmedchemlett.6b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lillycrop K., Murray R., Cheong C., et al. Anril promoter Dna methylation:A perinatal marker for later adiposity. EBioMedicine. 2017;19:60–72. doi: 10.1016/J.Ebiom.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dubland J.A., Francis G.A. So much cholesterol:the unrecognized importance of smooth muscle cells in atherosclerotic foam cell formation. Curr. Opin. Lipidol. 2016;27(2):155–161. doi: 10.1097/Mol.0000000000000279. [DOI] [PubMed] [Google Scholar]
- 69.Holdt L.M., Kohlmaier A., Teupser D. Molecular roles and function of circular Rnas in eukaryotic cells. Cell. Mol. Life Sci. 2018;75(6):1071–1098. doi: 10.1007/S00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altesha M.A., Ni T., Khan A., et al. Circular Rna in cardiovascular disease. J. Cell. Physiol. 2019;234(5):5588–5600. doi: 10.1002/Jcp.27384. [DOI] [PubMed] [Google Scholar]
- 71.Kristensen L.S., Andersen M.S., Stagsted L., et al. The biogenesis, biology and characterization of circular Rnas. Nat. Rev. Genet. 2019;20(11):675–691. doi: 10.1038/S41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 72.Du W.W., Zhang C., Yang W., et al. Identifying and characterizing circrna-protein interaction. Theranostics. 2017;7(17):4183–4191. doi: 10.7150/Thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meseure D., Vacher S., Alsibai K.D., et al. Expression of anril-polycomb complexes-Cdkn2a/B/arf genes in breast tumors:identification of A two-gene (Ezh2/Cbx7) signature with independent prognostic value. Mol. Cancer Res. 2016;14(7):623–633. doi: 10.1158/1541-7786.Mcr-15-0418. [DOI] [PubMed] [Google Scholar]
- 74.Chittock E.C., Latwiel S., Miller T.C., et al. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017;45(1):193–205. doi: 10.1042/Bst20160173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schuettengruber B., Bourbon H.M., Di Croce L., et al. Genome regulation by polycomb and trithorax:70 Years and counting. Cell. 2017;171(1):34–57. doi: 10.1016/J.Cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 76.Kong Y., Hsieh C.H., Alonso L.C. Anril:A Lncrna at the Cdkn2a/B locus with roles in cancer and metabolic disease. Front. Endocrinol. 2018;9:405. doi: 10.3389/Fendo.2018.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simion V., Haemmig S., Feinberg M.W. Lncrnas in vascular biology and disease. Vasc. Pharmacol. 2019;114:145–156. doi: 10.1016/J.Vph.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chi J.S., Li J.Z., Jia J.J., et al. Long non-coding Rna anril in gene regulation and its duality in atherosclerosis. J. Huazhong Univ Sci. Technol. Med. Sci. 2017;37(6):816–822. doi: 10.1007/S11596-017-1812-Y. [DOI] [PubMed] [Google Scholar]
- 79.Hueso M., Cruzado J.M., Torras J., et al. Aluminating the path of atherosclerosis progression:chaos theory suggests A role for Alu repeats in the development of atherosclerotic vascular disease. Int. J. Mol. Sci. 2018;19(6) doi: 10.3390/Ijms19061734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holdt L.M., Hoffmann S., Sass K., et al. Alu elements in anril non-coding Rna at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks [J/ol] PLoS Genet. 2013;9(7) doi: 10.1371/Journal.Pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]