Abstract
Recently, M. Dmitrova et al. (Mol. Gen. Genet. 257:205–212, 1998) described a LexA-based genetic system to monitor protein-protein interactions in an Escherichia coli background. However, the plasmids used in this system, pMS604 and pDP804, were not readily amenable for general use. In this report, we describe modifications of both plasmids that allow fragments of DNA to be fused to either vector in any reading frame. Homodimerization and heterodimerization of full-length proteins involved in polysialic acid synthesis in E. coli K1, as well as heterodimerization between a full-length protein and a protein fragment, demonstrate the usefulness of the modified plasmids for investigating bacterial protein-protein interactions in vivo.
The K1 capsule of Escherichia coli is a linear homopolymer of α-(2,8)-linked N-acetylneuraminic acid (NeuNAc; sialic acid) (15, 17). The capsule is identical to the polysialic acid (polySia) found on human embryonic nerve cell adhesion molecules and is poorly immunogenic due to the molecular mimicry of host structures (17). Surface-displayed sialic acid also confers resistance to the alternative complement pathway, allowing E. coli to establish extraintestinal infections such as neonatal septicemia and meningitis (15, 17).
The genes for the biosynthesis and transport of the K1 capsule are encoded on the 17-kb kps gene cluster, which is functionally divided into three regions (4, 21, 23). The genes in regions 1 and 3, designated kps, are conserved among E. coli strains that synthesize serologically distinct capsules and are involved with transport of the polymer to the bacterial cell surface (4, 21, 23). In contrast, the genes in region 2 are unique to a given capsular type. In E. coli K1, region 2 encodes the neuDBACES genes, which direct the synthesis, activation, and polymerization of NeuNAc (4).
Biosynthesis of polySia in E. coli K1 involves the NeuB-catalyzed condensation of N-acetylmannosamine (ManNAc) and phosphoenolpyruvate to form NeuNAc (20). ManNAc is provided by the neuC gene product, a UDP-GlcNAc 2-epimerase (D. A. Daines, W. F. Vann, D. O. Chaffin, C. E. Rubens, and R. P. Silver, submitted for publication). NeuD is also required for NeuNAc synthesis, although its enzymatic role remains an enigma (5a).
Activation of NeuNAc is performed by NeuA, which adds a nucleotide monophosphate to the sugar to form CMP-NeuNAc (19). NeuS is the sialyltransferase, polymerizing activated CMP-NeuNAc to polySia (16).
The current view of polymer synthesis and assembly assumes that the components of the kps cluster form a hetero-oligomeric protein complex (4, 13). Identification and characterization of the protein-protein interactions involved in this complex are crucial to our understanding of the mechanism of polymer synthesis and transport. Recently, a LexA-based genetic system for studying protein-protein interactions in an E. coli background was reported (6). The LexA repressor is an important component in the regulation of SOS response genes (14). The protein consists of two domains, a DNA-binding domain (DBD) and a dimerization domain. Although a truncated LexA consisting of only the DBD can recognize its operator sequence, it is functional as a transcriptional repressor only in dimeric form. Other domains can be fused in frame to the DBD and will restore the repressor's function if these domains interact. The LexA-based system described by Dmitrova et al. (6) consists of two compatible plasmids carrying either a wild-type (pMS604) or a mutant (pDP804) lexA DBD gene fragment. The mutant LexA DBD contains three amino acid changes that allow it to recognize a mutant operator. The plasmids were used in two E. coli reporter strains, SU101 and SU202. SU101 has a wild-type LexA operator sequence upstream of a lacZ reporter gene expressed by the sulA promoter engineered in the chromosome, while SU202 has the same reporter system but is controlled by a hybrid LexA operator sequence (6). Since sulA is one of the most repressed genes of the SOS response, LexA binds the sulA operator more tightly than the other natural SOS operators (14). A wild-type LexA fusion homodimer recognizes the wild-type operator in SU101 and represses reporter gene transcription, but only a heterodimer of a wild-type and a mutant LexA DBD fusion can recognize the hybrid operator in SU202. This allowed both homo- and hetero-association of protein fusions to be monitored. The system was tested for heterodimerization using the leucine zipper domains of the eukaryotic proteins Jun (pDP804) and Fos (pMS604). Although the focus of the study was protein heterodimerization, Dmitrova et al. (6) also analyzed homodimerization by fusing a Fos zipper region mutated so that it could interact with itself to the wild-type LexA DBD.
Plasmids pMS604 and pDP804 carry the Fos and Jun leucine zipper domains, respectively, and were not readily amenable for general use. In this report we describe modifications of both plasmids that allow fragments of DNA to be fused to either vector in any reading frame. We demonstrate the usefulness of the modified plasmids for investigating bacterial protein-protein interactions in vivo by demonstrating homodimerization of full-length NeuD, NeuA, NeuC, and NeuB as well as heterodimerization between NeuD and NeuB.
Modification of pDP804 and pMS604.
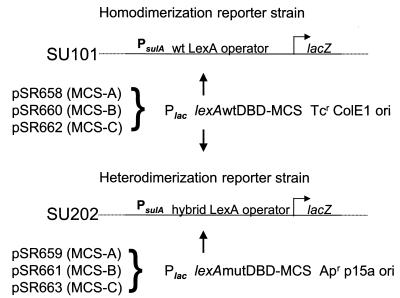
Plasmid pDP804 carries the p15A origin of replication and an ampicillin resistance gene. Plasmid pMS604 has a ColE1 origin of replication and a tetracycline resistance gene (6). In each plasmid, a lac promoter controls expression of the gene encoding the wild-type or mutant LexA DBD fused to the leucine zipper region of the Fos or Jun protein, respectively, and is therefore inducible by isopropyl-β-d-thiogalactopyranoside (IPTG) (6). To remove the Fos leucine zipper fusion, pMS604 was digested with the restriction endonucleases PinAI (AgeI) and XhoI and gel purified by standard procedures (2). Plasmid pDP804 was subjected to identical procedures, except that the restriction endonucleases were BssHII and BamHI. PCR-generated fragments of the multiple cloning sites (MCS) from the vectors pTrcHisA, pTrcHisB, and pTrcHisC (Invitrogen, Carlsbad, Calif.) with compatible restriction sites were ligated to the purified vector DNA. The six plasmids created allow fragments of DNA to be fused to either the wild-type or the mutant LexA DBD in any of three reading frames (Fig. 1). The pMS604 (wild type)-based plasmids are pSR658, pSR660, and pSR662, carrying the MCS of pTrcHisA, -B, and -C, respectively. The pDP804 (mutant)-based plasmids are pSR659, pSR661, and pSR663, which carry MCS-A, MCS-B, and MCS-C, respectively. Details of constructions and sequences of the vectors can be obtained from R. P. Silver. Using the modified plasmids, an entire gene or region of interest can be fused to the wild-type LexA DBD, and homodimerization can be monitored by lacZ reporter gene transcriptional repression in SU101 via β-galactosidase activity assays. Alternatively, genes encoding two different domains or proteins can be fused to the wild-type and mutant LexA DBD in the proper frame, and heterodimerization occurring in SU202 can be quantitated in the same fashion (Fig. 1). In this study we used pSR658, carrying the wild-type LexA DBD, and pSR659, expressing the mutant LexA DBD.
FIG. 1.
Modified LexA-based system for investigating protein-protein interactions. The LexA-based system was based on that of Dmitrova et al. (6). A homodimerizing fusion expressed from one of the wild-type (wt) LexA DBD::MCS plasmids (pSR658, pSR660, or pSR662) will bind to the wild-type LexA operator and repress expression of lacZ in the chromosome of the reporter strain SU101. A heterodimerizing fusion, one subunit expressed from one of the wild-type LexA DBD::MCS plasmids and the other from one of the mutated (mut) LexA DBD::MCS plasmids (pSR659, pSR661, or pSR663), will bind to the hybrid LexA operator and repress expression of lacZ in the chromosome of the reporter strain SU202. The choice of plasmid MCS (A, B, or C) is made according to the reading frame of the desired fusion. Wild-type and mutated DBD correspond to LexA1–87WT and LexA1–87408, respectively (6).
Full-length protein fusions can homodimerize.
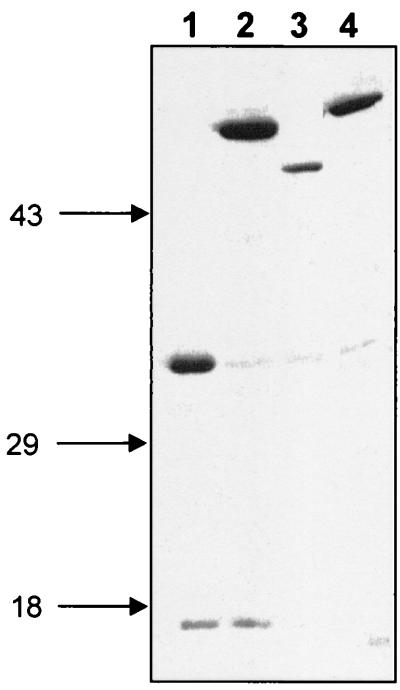
The four genes involved in the synthesis and activation of sialic acid in E. coli K1, neuA, neuB, neuC, and neuD, were fused in frame to the wild-type LexA DBD in pSR658. To determine that the plasmids were expressing protein when induced, we performed a Western blot analysis using induced SU101 cells carrying each plasmid grown to stationary phase in 1 mM IPTG. Twenty-microliter aliquots of cells were pelleted, resuspended in 15 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer with β-mercaptoethanol, and incubated at 95°C for 5 min. The samples were resolved by SDS-PAGE on a 12% polyacrylamide gel. After separation, the proteins were transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, Calif.) and probed with anti-LexA antibody (Invitrogen), using standard procedures (2). The blot was detected using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Piscataway, N.J.). The LexA fusions added approximately 9.6 kDa to the protein of interest. All full-length neu gene fusions were expressed, and their mobilities on an SDS-polyacrylamide gel exhibited the expected pattern (Fig. 2).
FIG. 2.
Immunoblot of LexA-Neu fusions. Full-length E. coli K1 region 2 genes were ligated into pSR658. An immunoblot of crude lysates of induced stationary-phase cultures probed with anti-LexA antiserum (Invitrogen) is shown. The calculated molecular masses in kilodaltons for the wild-type and LexA fusions, respectively, are in parentheses. Lane 1, LexA::NeuD (22.2, 31.8); lane 2, LexA::NeuC (43.1, 52.7); lane 3, LexA::NeuB (38.1, 47.7); lane 4, LexA::NeuA (45.8, 55.4). Molecular mass markers in kilodaltons appear at the left.
To determine whether the LexA fusions were functional, we tested their ability to complement a K-12–K1 hybrid strain with a deletion or interruption in the gene of interest. Each retained the ability to restore capsule synthesis when placed in trans in the corresponding acapsular mutant strain, indicating that the 87-amino-acid LexA DBD fused to the amino terminus of each protein did not interfere with its activity in vivo (data not shown). A capsule-positive phenotype was determined by sensitivity to capsule-specific phage (K1F) and by haloes of precipitated antigen-antibody complexes on agar plates supplemented with horse antiserum raised against the α-(2,8)-polysialic acid capsule (H.46) (22).
Measuring repression of β-galactosidase activity in induced cells of strain SU101 carrying each fusion allowed quantitation of homodimerization of full-length proteins fused to the wild-type LexA DBD in pSR658. For these experiments, only those fusion proteins that repressed expression of β-galactosidase at a level that resulted in a pale colony color of the reporter strain on MacConkey agar plates were considered to be interacting efficiently. Quantitatively, the amount of repression required to display a discernibly different colony color was on average ≥25% of the control level. Cells were grown to stationary phase with the appropriate antibiotics and 1 mM IPTG. Then 100 μl of each culture was used to inoculate 2 ml of fresh Luria-Bertani medium with IPTG and antibiotics. The cultures were grown to log phase and harvested; β-Galactosidase activity was determined as described by Miller (10). The units were calculated from the results of at least three independent cultures assayed in triplicate. Percent repression relative to the control cultures was calculated as [1.0 − (Miller units of sample/Miller units of control)] × 100. The CMP-NeuNAc synthase protein, NeuA, fused to wild-type LexA DBD repressed β-galactosidase activity by an average of 78%, compared to SU101 with pSR658 alone (Table 1). This is consistent with the observation that NeuA is enzymatically active as a dimer (W. F. Vann and D. Stoughton, unpublished data). The NeuC fusion repressed at 68%, while NeuB, NeuNAc synthase, repressed expression of β-galactosidase by 39%. The fusion with NeuD was the most efficient of the capsule synthesis proteins, exhibiting 87% repression. NeuD belongs to a family of acetyl- or acyltransferases that includes LpxA, LacA, and CysE (1, 7, 18). Like other members of this family, NeuD contains a signature hexapeptide repeat motif referred to as an isoleucine patch (7, 18). The crystal structures of three hexapeptide proteins have been determined; they each form trimers and share a unique structural feature known as a left-handed parallel α helix (3, 14). NeuD also forms trimers of identical subunits in vivo (5). These observations are consistent with the high level of NeuD homodimerization observed with the LexA system.
TABLE 1.
Homodimerization of full-length proteins and protein fragments fused to the wild-type LexA DNA-binding domain (DBD) in pSR658
| LexA fusion | Lengtha | % Repressionb (mean ± SD) |
|---|---|---|
| NeuA | 419 | 78 ± 2 |
| NeuA1 | 250 (1–250) | 6 ± 2 |
| NeuB | 346 | 39 ± 6 |
| NeuC | 391 | 65 ± 5 |
| NeuD | 207 | 87 ± 1 |
| NeuD1 | 108 (1–108) | 21 ± 9 |
| NeuD2 | 118 (90–207) | 13 ± 5 |
| NeuD3 | 64 (144–207) | 16 ± 1 |
| Chloramphenicol acetyltransferase | 220 | 94 ± 2 |
Length of amino acid insert encoded by neu insert in pSR658. The amino acid residues of protein fragments are in parentheses.
Relative to pSR658 alone in strain SU101. Average β-galactosidase activity in SU101(pSR658) is 2,482 ± 254 Miller units.
The ability of multimeric fusion proteins to repress β-galactosidase expression in this system was confirmed by fusing a gene encoding the type I chloramphenicol acetyltransferase from Tn9 to the LexA DBD in pSR658. Chloramphenicol acetyltransferase is a well-characterized homotrimer and belongs to the same family of acetyl- or acyltransferases as NeuD. This construct conferred chloramphenicol resistance to DH5α cells, and its expression was detected via immunoblot using anti-LexA antiserum (data not shown). The fusion protein repressed β-galactosidase expression in SU101 by 94% (Table 1). The reporter strain SU101 contains a Tn9 insertion in its chromosomal copy of lacZ and constitutively expresses unfused type I chloramphenicol acetyltransferase subunits. The high level of repression that we observed indicates that the modified LexA-based system is useful in determining interactions between fusion proteins even in the presence of wild-type subunits expressed by the host cell.
Fragments of neu genes do not homodimerize efficiently.
In contrast to the results with full-length protein fusions, fragments of the neu genes did not yield fusion proteins that homodimerized as efficiently. For these experiments, three fragments of neuD (neuD1 to neuD3) and a fragment (neuA1) encoding the amino-terminal 250 amino acids of neuA (Table 1) were amplified by PCR and fused to pSR658 in frame with the LexA DBD. These plasmids were then used to transform SU101. Since the fragment fusions could not be identified by complementation, the transformants were selected via antibiotic resistance and the inserts were confirmed by colony PCR with primers that flanked the region of interest, by restriction endonuclease digests of purified plasmid preparations, and by DNA sequencing. We were unable to detect the fragment fusions on a Western blot probed with anti-LexA antiserum. The inability to detect small fusions on an immunoblot was also reported by Enz et al. in their study of FecA and FecR fragments replacing the Jun and Fos zipper regions in the parent vectors pDP804 and pMS604 (8). However, the ability of the NeuD2 fragment to heterodimerize with NeuB (see below) indicates that the NeuD2 fragment fusion was expressed. Moreover, the NeuA1, NeuD1, and NeuD3 fusions were positive on a dot blot probed with anti-LexA antiserum (data not shown). While this does not confirm the size of the small fragment fusions, it does establish that they are indeed expressed.
Unlike the full-length NeuA fusion, the NeuA1 fusion did not homodimerize, as reflected by its inability to efficiently repress β-galactosidase expression (Table 1). Interestingly, a truncated protein encoding the first 250 amino acids of NeuA displayed approximately 10% of the enzymatic activity of the full-length enzyme in vitro and formed only transient dimers (Vann and Stoughton, unpublished). The NeuD fragment fusions each repressed β-galactosidase expression at a much lower level than full-length NeuD (Table 1). Fragmentation of NeuD likely resulted in the inability of the fusions to fold properly, and as a result they homodimerized poorly.
NeuB and NeuD interact in vivo.
The data in Table 1 indicate that the LexA-based system can be used to study homodimerization of full-length proteins. To investigate the suitability of the modified LexA system to study the protein-protein interactions among different components of the kps complex, we examined the interaction between the NeuD and NeuB proteins. Both proteins are involved in sialic acid synthesis and are likely to form a complex in vivo (13). For these experiments, full-length neuB and neuD genes, as well as fragments of neuD, were fused in frame to either the wild-type (pSR658) or mutant (pSR659) lexA DBD and introduced into the reporter strain SU202. As shown in Table 2, we observed significant interaction between NeuD and NeuB, quantitated as 71% repression of β-galactosidase activity, in cells harboring the NeuB fusion expressed from the high-copy-number vector, pSR658, and a NeuD fusion in the lower-copy-number vector, pSR659.
TABLE 2.
Heterodimerization of NeuB and NeuD
| Fusiona
|
% Repressionb (mean ± SD) | |
|---|---|---|
| pSR658 | pSR659 | |
| neuB | neuD | 71 ± 4 |
| neuB | neuD1 | 29 ± 4 |
| neuB | neuD2 | 66 ± 4 |
| neuB | neuD3 | 33 ± 4 |
| neuD | neuB | 20 ± 4 |
| neuD1 | neuB | 35 ± 5 |
| neuD2 | neuB | 49 ± 6 |
| neuD3 | neuB | 18 ± 1 |
Gene fused to either the wild-type lexA DBD (pSR658) or the mutant lexA DBD (pSR659).
Percent repression of β-galactosidase activity relative to strain SU202 with pSR658 and pSR659 alone. Average activity of the vector control was 4,318 ± 130 Miller units.
The interaction between NeuB and NeuD was further investigated by assaying β-galactosidase expression in SU202 harboring the full-length NeuB fusion in pSR658 and pSR659 carrying fragments of NeuD. Only one NeuD fragment, NeuD2, displayed interaction approaching that of full-length NeuD with the full-length NeuB fusion. Fragment NeuD2, when expressed from pSR659, repressed β-galactosidase expression 66%.
These data suggest that NeuD and NeuB interact in vivo and that amino acid residues 90 to 207 of the NeuD protein are important for this interaction. However, the formation of heterodimers by two proteins is not necessarily straightforward when one of the proteins, like NeuD, is able to form homodimers, and we did observe conflicting results with the reciprocal experiment. When the neuD gene, fused to the lexA DBD in pSR658 was introduced into SU202 harboring neuB fused to the mutant lexA DBD in pSR659, little repression of β-galactosidase activity was seen (Table 2). We postulate that when NeuD is expressed from pSR658, the high-copy-number vector, it is more likely to form homodimers and less likely to interact with NeuB and repress the hybrid promoter in SU202. This view is supported by the observation that the NeuD2 fragment of NeuD, which, in contrast to the full-length protein, is unable to efficiently homodimerize (Table 1), effectively interacted with NeuB even when expressed from the higher-copy-number vector (Table 2). This was reflected in 49% repression of β-galactosidase activity.
Conclusions.
In this communication we describe the use of a modified LexA-based genetic system to investigate, in vivo, the interactions between proteins that comprise a putative hetero-oligomeric complex responsible for K1 capsule synthesis (4, 13). Both homo- and heterodimerization of protein fusions were detected. This system allows determination of the heterodimerization of two fusion proteins even when one protein homodimerizes strongly, as is the case with NeuD. The system can also reveal the interaction of fusion proteins in the presence of unfused subunits expressed by the host cell, as shown by the chloramphenicol acetyltransferase experiment. Only those fusion proteins that repressed expression of β-galactosidase at a level that resulted in a pale colony color of the reporter strain on MacConkey agar plates (≥25% of the controls) were considered to be interacting efficiently. That is not to say, however, that repression below 25% may not be biologically significant. However, the limitations of any LexA-based system include difficulty in deducing weak or transient interactions, steric hindrance by the fusion moiety, and sequestration of the fused moiety into an intracellular complex (9). Moreover, the inability to detect heterodimerization does not preclude association of a given protein with a higher-order multimer of another protein. It is unlikely that such interactions would be detected in this system. The different copy numbers of the vectors, although useful in determining the heterodimerization of a strongly homodimerizing protein by allowing reciprocal fusions, add a level of complexity to the system, and it is recommended that any putative heterodimerizing fusions also be assayed when expressed from the opposite vector pair. It is also possible that overexpression of a wild-type LexA fusion that strongly homodimerizes, may, to some degree, overcome the low affinity of the wild-type LexA DBD for the hybrid operator in SU202. With these limitations in mind, we consider this modified system a fast, economical, and reproducible tool to initially screen for possible interactions and to be suitable for use in conjunction with biochemical methods for demonstrating protein-protein interactions, such as gel filtration and protein cross-linking.
Acknowledgments
We are extremely grateful to M. Granger-Schnarr for her kind gift of plasmids pDP804 and pMS604 and the reporter strains SU101 and SU202. We also thank Virginia Clark, Marty Pavelka, and Lou Passador for critically reviewing the manuscript.
This work was supported by NIH grants AI39615 to R.P.S. D.A.D. was supported by Molecular Pathogenesis of Bacteria and Viruses training grant AI07362 from the Public Health Service.
REFERENCES
- 1.Annunziato P W, Wright L F, Vann W F, Silver R P. Nucleotide sequence and genetic analysis of the neuD and neuB genes in region 2 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1995;177:312–319. doi: 10.1128/jb.177.2.312-319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 3.Beaman T W, Sugantino M, Roderick S L. Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa. Biochemistry. 1998;37:6689–6696. doi: 10.1021/bi980106v. [DOI] [PubMed] [Google Scholar]
- 4.Bliss J M, Silver R P. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol Microbiol. 1996;21:221–231. doi: 10.1046/j.1365-2958.1996.6461357.x. [DOI] [PubMed] [Google Scholar]
- 5.Daines D A. Ph.D. thesis. Rochester, N.Y: University of Rochester; 1999. [Google Scholar]
- 5a.Daines D A, Wright L F, Chaffin D O, Rubens C E, Silver R P. NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol Lett. 2000;189:281–284. doi: 10.1111/j.1574-6968.2000.tb09244.x. [DOI] [PubMed] [Google Scholar]
- 6.Dmitrova M, Younes-Cauet G, Oertel-Buchheit P, Porte D, Schnarr M, Granger-Schnarr M. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol Gen Genet. 1998;257:205–212. doi: 10.1007/s004380050640. [DOI] [PubMed] [Google Scholar]
- 7.Downie J A. The nodL gene from Rhizobium leguminosarum is homologous to the acetyltransferases encoded by lacA and cysE. Mol Microbiol. 1989;3:1649–1651. doi: 10.1111/j.1365-2958.1989.tb00150.x. [DOI] [PubMed] [Google Scholar]
- 8.Enz S, Mahren S, Stroeher U H, Braun V. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J Bacteriol. 2000;182:637–646. doi: 10.1128/jb.182.3.637-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golemis E A, Brent R. Fused protein domains inhibit DNA binding by LexA. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 11.Moxon E R, Kroll J S. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- 12.Raetz C R H, Roderick S L. A left-handed parallel β helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 13.Rigg G P, Barrett B, Roberts I S. The localization of KpsC, S and T, and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: evidence for a membrane-bound complex. Microbiology. 1998;144:2905–2914. doi: 10.1099/00221287-144-10-2905. [DOI] [PubMed] [Google Scholar]
- 14.Schnarr M, Granger-Schnarr M. LexA, the self-cleaving transcriptional repressor of the SOS system. Nucleic Acids and Mol Biol. 1993;7:170–189. [Google Scholar]
- 15.Silver R P, Vimr E R. Polysialic acid capsule of Escherichia coli K1. In: Iglewski B H, Clark V L, editors. Molecular basis of microbial pathogenesis. San Diego, Calif: Academic Press, Inc.; 1990. pp. 39–60. [Google Scholar]
- 16.Steenbergen S M, Wrona T J, Vimr E R. Functional analysis of the sialyltransferase complexes in Escherichia coli K1 and K92. J Bacteriol. 1992;174:1099–1108. doi: 10.1128/jb.174.4.1099-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troy F A. Polysialylation: from bacteria to brains. Glycobiology. 1992;2:5–23. doi: 10.1093/glycob/2.1.5. [DOI] [PubMed] [Google Scholar]
- 18.Vaara M. Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme. FEMS Microbiol Lett. 1992;97:249–254. doi: 10.1016/0378-1097(92)90344-n. [DOI] [PubMed] [Google Scholar]
- 19.Vann W F, Silver R P, Abeijon C, Chang K, Aaronson W, Sutton A, Finn C W, Linder W, Kotsatos M. Purification, properties, and genetic location of Escherichia coli cytidine 5′-monophosphate N-acetylneuraminic acid synthetase. J Biol Chem. 1987;262:17556–17562. [PubMed] [Google Scholar]
- 20.Vann W F, Tavarez J J, Crowley J, Vimr E, Silver R P. Purification and characterization of the Escherichia coli K1 neuB gene product N-acetylneuraminic acid synthetase. Glycobiology. 1997;7:697–701. doi: 10.1093/glycob/7.5.697. [DOI] [PubMed] [Google Scholar]
- 21.Vimr E, Steenbergen S, Cieslewicz M. Biosynthesis of the polysialic acid capsule in Escherichia coli K1. J Ind Microbiol. 1995;15:352–360. doi: 10.1007/BF01569991. [DOI] [PubMed] [Google Scholar]
- 22.Vimr E R, Aaronson W, Silver R P. Genetic analysis of chromosomal mutations in the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1989;172:1106–1117. doi: 10.1128/jb.171.2.1106-1117.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitfield C, Roberts I S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]