Abstract
Bacillus subtilis has been shown to express a cytosolic oxalate decarboxylase (EC 4.1.1.2). The enzyme was induced in acidic growth media, particularly at pH 5.0, but not by oxalate. The enzyme was purified, and N-terminal sequencing identified the protein to be encoded by yvrK. The role of the first oxalate decarboxylase to be identified in a prokaryote is discussed.
Oxalate decarboxylase (EC 4.1.1.2) converts oxalate to formate and CO2 in an O2-dependent reaction (4). It acts exclusively on oxalate, and no other subtype of decarboxylase is capable of catalyzing this reaction. Reports of oxalate decarboxylases have, with the exception of the guinea pig liver enzyme (15, 19), been restricted to fungi. The best characterized enzymes are from the white-rot wood-decaying Collybia velutipes (reclassified as Flammulina velutipes) (12) and from Aspergillus niger (7). Oxalate decarboxylases have been used in the clinical assay of oxalate in blood and urine and could be used to lower oxalate levels in foods and the environment (3). Only the unrelated thiaminepyrophosphate-requiring oxalyl-coenzyme A decarboxylases have been detected in bacteria, such as Oxalobacter formigenes (11).
The oxalate-degrading enzymes, oxalate decarboxylase and oxalate oxidase, belong to the cupin superfamily, which is defined by their conserved motifs and a proposed common β-barrel fold (1, 2). The decarboxylases are members of the bicupin subclass of this superfamily since they contain a duplication of these motifs and are therefore thought to contain two β-barrel domains per polypeptide. Since the Bacillus subtilis gene yxaG was identified as coding for a hypothetical bicupin (2), we and others (3) identified yvrK and yoaN as coding for hypothetical bicupins that share even greater sequence identity with oxalate decarboxylases, particularly within their motifs (Fig. 1). The only other genes coding for similar bicupins are found in Synechocystis sp. and Streptococcus mutans (3). The function of none of these hypothetical genes has been reported previously. The aim of this work was to establish whether B. subtilis is capable of oxalate decarboxylation, under which conditions activity is induced, and to identify which, if any, of the above genes code for the enzyme.
FIG. 1.
Amino acid sequence comparison between the N- and C-terminal domain cupin motifs of C. velutipes oxalate decarboxylase (OxDe), those of the B. subtilis bicupins YvrK, YoaN, and YxaG, barley (Hordeum vulgare) oxalate oxidase (OxOx), and moss (Barbula unguiculata) germin-like manganese-superoxide dismutase (GLSD). The number of intervening amino acid residues between the two parts of each cupin motif is indicated. Conserved residues are highlighted with asterisks. The conserved residues in bold are predicted to ligate a Mn2+ ion in the active site of oxalate oxidase.
pH-dependent induction of B. subtilis oxalate decarboxylase.
B. subtilis 168, whose genome has been determined (8), was grown in Luria broth containing 10 μM MnCl2 in shake flasks at 30°C with agitation at 200 rpm. Cell extracts exhibited appreciable oxalate decarboxylase activity only when the broth was acidified before inoculation. Enzyme activity was determined using the stopped assay of Magro et al. (10), in which the production of formate was linked to the reduction of NAD with formate dehydrogenase. The highest activities, on the basis of both biomass and volume, were obtained when the broth was acidified to pH 5.0 with HCl. Of four fungal enzymes tested, all (6, 10, 17) but one (14) are similarly induced in an acid-dependent manner. In all cases tested (5, 10, 14, 17), the fungal enzymes were induced by oxalate. By contrast, the addition of up to 2.5% potassium oxalate to the medium did not increase the level of bacterial enzyme activity, suggesting a different role for the prokaryotic enzyme.
Similar acid-dependent induction was observed when B. subtilis was grown in New Brunswick 1-liter BioFlo III fermenters maintained at 30% air saturation (Table 1). Despite a relatively long lag phase and a low biomass yield, the highest activity was obtained with cultures maintained at pH 5.0. Enzyme activity was detected throughout growth and reached a maximum during late log growth. Cultures maintained at pH 6.0 also yielded substantial activity that reached a maximum during the stationary phase. When the pH was not maintained by the addition of HCl, the media became more alkaline. This increase in pH resulted in cultures that yielded only moderate to low activity even when started at pH 5.0 or 6.0. The lowest activity was observed with cultures that started at pH 7.0, whether the pH was controlled or not. The maximum growth rates during log phase growth were surprisingly similar in all conditions. The time to stationary phase was largely determined by the duration of the lag phase.
TABLE 1.
Acid induction of B. subtilis oxalate decarboxylase
| Initial pH (maintained with HCl) | Final pH | Maximum activity (U/liter) | Time to stationary phase (h) | Maximum absorbance (600 nm) |
|---|---|---|---|---|
| 5.0 (+) | 5.0 | 23 | 13a | 3.2 |
| 6.0 (+) | 6.0 | 20 | 8 | 3.7 |
| 7.0 (+) | 7.0 | 2 | 7 | 4.6 |
| 5.0 (−) | 8.0 | 9 | 8 | 3.8 |
| 6.0 (−) | 8.0 | 4 | 7 | 4.3 |
| 7.0 (−) | 8.3 | 2 | 6 | 4.6 |
Including a 6-h lag.
Purification of the enzyme.
In order to maximize the volumetric yield of enzyme while also minimizing the total biomass, the organism was grown in a New Brunswick 20-liter fermenter with the pH maintained at 5.0. The cells were harvested during late log phase and frozen in liquid nitrogen. Thawed cells were broken in the presence of 50 mM Tris-HCl (pH 7.0) with three passes through an APV Gaulin homogenizer. After the addition of DNase I, the cell debris was removed by centrifugation. The crude extract was applied to a DEAE-Sepharose FastFlow (Amersham Pharmacia Biotech) column. After unwanted protein was eluted from the column with Tris buffer containing 200 mM NaCl, the decarboxylase was eluted with buffer containing 500 mM NaCl. The enzyme was dialyzed with Tris buffer, applied to a fast-performance liquid chromatography (FPLC) Mono Q HR10/10 column, eluted with a salt gradient in Tris buffer at 250 mM NaCl, and dialyzed with 50 mM citric acid-NaOH (pH 4.0). After clarification by centrifugation, the enzyme was applied to an FPLC MonoS HR 5/5 column and eluted with a salt gradient in citrate buffer at 500 mM NaCl. The enzyme was applied to a Sephadex 200 column and eluted with citrate buffer containing 100 mM NaCl.
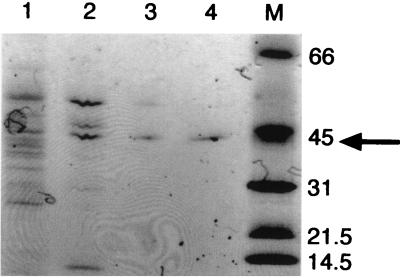
Overall, the enzyme was purified 1,000-fold to yield 100 μg with a specific activity of at least 26 U mg−1. The reported specific activities of the fungal enzymes varied markedly, with 67 to 350 and 28.3 to 80 U mg−1 for the C. velutipes (12, 18) and A. niger (6, 7) enzymes, respectively. The bacterial enzyme was homogeneous according to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2) with Coomassie staining using PhastSystem (Amersham Pharmacia Biotech). The decarboxylase can be frozen in liquid nitrogen and stored at −20°C without loss of activity, provided that the pH is buffered at or near 7.0.
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel (8 to 25% polyacrylamide) showing the purification of B. subtilis oxalate decarboxylase (indicated by the arrow). Lanes: 1, post-DEAE–Sepharose fraction; 2, post-MonoQ fraction; 3, post-MonoS fraction; 4, post-Sephadex 200 fraction; M, molecular mass markers (in kilodaltons).
Properties of the enzyme.
Since the protein was pure, it was possible to determine its N-terminal sequence using an Applied Biosystems 494 Protein Sequencer without additional electrophoretic separation. This sequence, MKKQNDIPQPIRGDK, unequivocally showed the enzyme to be encoded by the gene yvrK, one of the two most likely candidates. The subunit molecular mass was 44 kDa according to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was therefore very close to that of 43,407 Da predicted for YvrK. The C. velutipes and C. versicolor fungal enzymes have somewhat larger subunit sizes of 64 kDa (55 kDa after deglycosylation) (12) and 59 kDa (5), respectively. The enzyme appears to exist as a pentamer in solution since gel filtration yielded an oligomer molecular mass of 220 kDa. However, it is possible that it is actually a hexamer since the related barley oxalate oxidase is known to elute later than expected from gel filtration columns (16). If this is the case, each bacterial decarboxylase oligomer would be composed of 2 × 6 cupin domains rather than 1 × 6, as is the case with the oxidase (20). By contrast, the 560-kDa C. velutipes fungal enzyme appears to be composed of about nine subunits and therefore about 18 cupin domains (12). The pI of the bacterial enzyme was determined using PhastSystem to be 6.1, a little higher than the predicted 5.1. The pH activity profile of the bacterial enzyme with citrate buffers yielded a bell-shaped curve with 70% activity at pH 3.0, a maximum at pH 5.0, and zero activity at pH 7.5. The fungal enzymes have acidic pH optima within the range of 2.0 to 5.2 (7, 13). The UV-visible spectrum of the bacterial decarboxylase, like that of oxalate oxidase (16), showed the essentially typical absorbance of a protein with no additional chromophores.
The activities of all fungal enzymes tested to date (7, 9, 17) have been shown to be O2 dependent. In order to test the O2 dependence of the B. subtilis enzyme, all solutions for the assay were subjected to six vacuum-N2 gas flush cycles and transferred to a Belle Technology glove box (Portesham, Dorset, United Kingdom), maintained at <1 ppm of O2, 1.5 h before commencing assays. The assays conducted in the anaerobic glove box yielded 76% of the enzyme activity of the control assays that were performed in parallel but with exposure to atmospheric O2. For comparison, the C. velutipes (Sigma) and Aspergillus spp. (Boehringer) enzymes retained 71 and 47% of their activities under these conditions, respectively, with the Aspergillus enzyme being the most sensitive to a lack of O2, as expected (7, 17).
With the absence of conserved cysteines in the decarboxylases that would be capable of disulfide bond formation, the universal and curious dependence of activity on O2 suggests the role of a transition metal in catalysis. The related cupins, barley oxalate oxidase (16) and a new type of germin-like superoxide dismutase from a moss (21), have both been shown to contain a Mn ion. The residues predicted to ligate Mn2+ in the oxidase are conserved in the germin-like manganese-superoxide dismutase and both the N- and C-terminal domains of C. velutipes oxalate decarboxylase and those of all three of the B. subtilis bicupins shown in Fig. 1, including the newly identified decarboxylase, YvrK. The presence of a metal ion, the catalytic mechanism of YvrK, and the enzymatic activities of the related bicupins YoaN and YxaG are currently being investigated.
Implications of a cytosolic, acid-induced bacterial oxalate decarboxylase.
Fungi are believed to utilize oxalate in lignin degradation, nutrient availability, pathogenesis, and competition (4). All of these roles involve the secretion of oxalic acid and the acidification of the organism's environment. Therefore, the cytosolic and secreted fungal enzymes are thought to reduce excess oxalic acid levels. Their oxalate-dependent induction is certainly consistent with this. This paper describes the first report of a bacterial oxalate decarboxylase that reveals a previously unknown role of oxalate in B. subtilis. The lack of induction of the bacterial enzyme by exogenous oxalate suggests a role that is different to that in fungi. It is possible that the B. subtilis enzyme is involved in decarboxylative phosphorylation similar to that described for the gram-negative bacterium O. formigenes, in which the antiporting of oxalate and formate are coupled to oxalate decarboxylation to generate a proton-motive gradient (11). Importantly, however, O. formigenes utilizes the unrelated oxalyl-coenzyme A decarboxylase.
There is an alternative role for this novel cytosolic bacterial oxalate decarboxylase which is induced by acid but not by oxalate. We have evidence for the presence of an oxalate-producing glyoxylate dehydrogenase in B. subtilis, which will be described elsewhere. With the absence of isocitrate lyase in B. subtilis, glyoxylate is most likely to be produced by either the transamination or oxidation of glycine. In addition, the genome of this organism is predicted to contain five genes that code for formate dehydrogenases (8). If one considers the net conversion of glyoxylate to 2CO2 with the above enzymes, there is the potential to produce 2ATP from the reducing equivalents with an overall consumption of 1 proton. Therefore, this short pathway could contribute to the raising of cytoplasmic pH when the organism encounters low values of pH in soil and rotting vegetation. The physiological role of YvrK and its regulation will be the subject of future study.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council through a quota studentship for A.T. and the John Innes Centre Competitive Strategic Grant.
We thank Simon Foster, University of Sheffield, for the gift of B. subtilis, Mike Chan for technical assistance, Carol Gormal and Mike Naldrett for the N-terminal sequencing, and Laura Bowater and Gary Sawers for helpful discussions.
REFERENCES
- 1.Dunwell J M. Cupins: a new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol Genet Eng Rev. 1998;15:1–32. doi: 10.1080/02648725.1998.10647950. [DOI] [PubMed] [Google Scholar]
- 2.Dunwell J M, Gane P J. Microbial relatives of seed storage proteins: conservation of motifs in a functionally diverse superfamily of enzymes. J Mol Evol. 1998;46:147–154. doi: 10.1007/pl00006289. [DOI] [PubMed] [Google Scholar]
- 3.Dunwell J M, Khuri S, Gane P J. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev. 2000;64:153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutton M V, Evans C S. Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can J Microbiol. 1996;42:881–895. [Google Scholar]
- 5.Dutton M V, Kathiara M, Gallagher I M, Evans C S. Purification and characterization of oxalate decarboxylase from Coriolus versicolor. FEMS Microbiol Lett. 1994;116:321–325. [Google Scholar]
- 6.Emiliani E, Bekes P. Enzymatic oxalate decarboxylation in Aspergillus niger. Arch Biochem Biophys. 1964;105:488–493. doi: 10.1016/0003-9861(64)90040-2. [DOI] [PubMed] [Google Scholar]
- 7.Emiliani E, Riera B. Enzymatic oxalate decarboxylation in Aspergillus niger: II. Hydrogen peroxide formation and other characteristics of the oxalate decarboxylase. Biochim Biophys Acta. 1968;167:414–421. doi: 10.1016/0005-2744(68)90221-0. [DOI] [PubMed] [Google Scholar]
- 8.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, KlaerrBlanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, Oreilly M, Ogawa K, Ogiwara A, Oudega B, Park S H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 9.Lillehoj E B, Smith F G. An oxalic acid decarboxylase of Myrothecium verrucaria. Arch Biochem Biophys. 1965;109:216–220. [Google Scholar]
- 10.Magro P, Marciano P, Di Lenna P. Enzymatic oxalate decarboxylation in isolates of Sclerotinia sclerotiorum. FEMS Microbiol Lett. 1988;49:49–52. [Google Scholar]
- 11.Maloney P C. Bacterial transporters. Curr Opin Cell Biol. 1994;6:571–582. doi: 10.1016/0955-0674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 12.Mehta A, Datta A. Oxalate decarboxylase from Collybia velutipes: purification, characterization, and cDNA cloning. J Biol Chem. 1991;266:23548–23553. [PubMed] [Google Scholar]
- 13.Micales J A. Oxalate decarboxylase in the brown-rot wood decay fungus Postia placenta. Mater Org. 1995;29:177–186. [Google Scholar]
- 14.Micales J A. Localization and induction of oxalate decarboxylase in the brown-rot wood decay fungus Postia placenta. Int Biodeterior Biodegrad. 1997;39:125–132. [Google Scholar]
- 15.Murthy M S R, Talwar H S, Nath R, Thind S K. Oxalate decarboxylase from guinea pig liver. IRCS Med Sci. 1981;9:683–684. [Google Scholar]
- 16.Requena L, Bornemann S. Barley (Hordeum vulgare) oxalate oxidase is a manganese-containing enzyme. Biochem J. 1999;343:185–190. [PMC free article] [PubMed] [Google Scholar]
- 17.Shimazono H. Oxalic acid decarboxylase, a new enzyme from the mycelium of wood destroying fungi. J Biochem. 1955;42:321–340. [Google Scholar]
- 18.Shimazono H, Hayaishi O. Enzymatic decarboxylation of oxalic acid. J Biol Chem. 1957;227:151–159. [PubMed] [Google Scholar]
- 19.Talwar H S, Murthy M S R, Nath R, Thind S K. Oxalate decarboxylase from guinea pig liver. Indian J Biochem Biophys. 1981;18:105–105. [Google Scholar]
- 20.Woo E J, Dunwell J M, Goodenough P W, Pickersgill R W. Barley oxalate oxidase is a hexameric protein related to seed storage proteins: evidence from X-ray crystallography. FEBS Lett. 1998;437:87–90. doi: 10.1016/s0014-5793(98)01203-4. [DOI] [PubMed] [Google Scholar]
- 21.Yamahara T, Shiono T, Suzuki T, Tanaka K, Takio S, Sato K, Yamazaki S, Satoh T. Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, Barbula unguiculata. J Biol Chem. 1999;274:33274–33278. doi: 10.1074/jbc.274.47.33274. [DOI] [PubMed] [Google Scholar]