Abstract
Background
There is growing evidence for the inherited basis of susceptibility to childhood acute lymphoblastic leukemia (ALL). Genome-wide association studies have identified non-coding ALL risk variants at the ARID5B gene locus, but their exact functional effects and the molecular mechanism linking ARID5B to B-cell ALL leukemogenesis remain largely unknown.
Methods
We performed targeted sequencing of ARID5B in germline DNA of 5008 children with ALL. Variants were evaluated for association with ALL susceptibility using 3644 patients from the UK10K cohort as non-ALL controls, under an additive model. Cis-regulatory elements in ARID5B were systematically identified using dCas9-KRAB–mediated enhancer interference system enhancer screen in ALL cells. Disruption of transcription factor binding by ARID5B variant was predicted informatically and then confirmed using chromatin immunoprecipitation and coimmunoprecipitation. ARID5B variant association with hematological traits was examined using UK Biobank dataset. All statistical tests were 2-sided.
Results
We identified 54 common variants in ARID5B statistically significantly associated with leukemia risk, all of which were noncoding. Six cis-regulatory elements at the ARID5B locus were discovered using CRISPR-based high-throughput enhancer screening. Strikingly, the top ALL risk variant (rs7090445, P = 5.57 × 10–45) is located precisely within the strongest enhancer element, which is also distally tethered to the ARID5B promoter. The variant allele disrupts the MEF2C binding motif sequence, resulting in reduced MEF2C affinity and decreased local chromosome accessibility. MEF2C influences ARID5B expression in ALL, likely via a transcription factor complex with RUNX1. Using the UK Biobank dataset (n = 349 861), we showed that rs7090445 was also associated with lymphocyte percentage and count in the general population (P = 8.6 × 10–22 and 2.1 × 10–18, respectively).
Conclusions
Our results indicate that ALL risk variants in ARID5B function by modulating cis-regulatory elements at this locus.
Acute lymphoblastic leukemia (ALL) is the most common malignancy in children, with the risk highest between 2 and 5 years of age (1,2). Although the vast majority of patients can be cured with combination chemotherapy, the etiology of this cancer remains incompletely understood. Recent work by us and others used the Genome-Wide Association Study (GWAS) approach to identify genetic variants related to ALL susceptibility (3-15). In total, at least 20 genomic loci have been reported, with common polymorphisms conferring 1.17- to 3.7-fold increased risk of developing ALL. Cumulatively, these variants account for 21% of the estimated inheritability of ALL in children (16). ARID5B is among the first ALL risk genes identified by GWAS and consistently exhibited one of the strongest association signals across the genome in diverse racial and ethnic populations (5,8,12). The effects of ARID5B variants on ALL risk vary significantly across molecular subtypes and are most pronounced in those with hyperdiploid karyotypes (3,4,8). However, despite the overwhelming evidence from these epidemiology studies, the biological mechanisms linking ARID5B to normal and malignant hematopoiesis remain largely unknown.
ARID5B belongs to the AT-rich interaction domain (ARID) protein family characterized by a shared DNA-binding ARID domain (17-20). The nuclear localization and specific binding affinity of ARID5B to the A/T-rich consensus sequence (AATA[C/T]) (17,21) point to a potential function as a transcription factor. One of the most prominent phenotypes of Arid5−/− mice is leanness, directly implicating it in adipogenesis (22). ARID5B interacts with PHF2 to form a histone lysine demethylase complex in hepatocytes, which can activate the expression of target genes PEPCK and G6PC and thus regulate glucose metabolism (23). In natural killer (NK) cells, downregulation of ARID5B represses UQCRB expression and decreases mitochondrial membrane potential and mitochondrial oxidative metabolism, along with BCL2 downregulation (24). In the hematopoietic compartment, Arid5b−/− mice exhibit a range of transient defects in lymphocyte development, including a reduction of cellularity in bone marrow, thymus, and spleen and a statistically significant decrease in early T- and B-cell progenitors in bone marrow in 3-week-old mice; most of these abnormalities disappeared at 6 weeks old (25). Interestingly, genetic variants in ARID5B have been linked to susceptibility to autoimmune diseases (eg, rheumatoid arthritis and systemic lupus erythematosus), adding another layer of complexity to its potential functions (26-28).
Recently, an imputation-based fine mapping analysis identified putative functional variants at the ARID5B locus (29). In particular, rs7090445 was described as the candidate causal variant driving the association signal with ALL susceptibility by influencing RUNX3-mediated transcription regulation in-cis. However, the full spectrum of genetic variation at this locus in children with ALL is unknown, and their potential overlap with regulatory DNA elements remains unclear. In this study, we sequenced a multiethnic cohort of 5008 children with ALL to comprehensively describe germline polymorphisms in the ARID5B gene and systematically screened sequences spanning ARID5B gene locus to map cis-regulatory elements (CREs) and functional genetic variants. Moreover, we identified the transcription factor complex directly interacting with leukemia risk alleles and thus regulating ARID5B expression in ALL. Our results provided novel insights into molecular mechanisms by which noncoding variants contribute to genetic susceptibility to ALL.
Methods
Patients
A total of 5008 patients with childhood ALL were enrolled in frontline clinical trials conducted by the Children’s Oncology Group (COG) [AALL0331 (30), AALL0232 (31), and COG9904/9905/9906 (32)] and St. Jude Children’s Research Hospital (St. Jude) [Total Therapy XIIIA (33), XIIIB (34), and XV (35) studies; Supplementary Table 1, available online]. Germline DNA was extracted from bone marrow or peripheral blood during remission. This study was approved by the institutional review boards at St. Jude and COG-affiliated institutions, and informed consent was obtained from parents, guardians, or patients.
ARID5B Sequencing
Illumina dual-indexed libraries were created from germline DNA and pooled in sets of 96 before hybridization with customized Roche NimbleGen SeqCap EZ probes (Roche) to capture a 200-kb region encompassing the exon and open chromatin regions in ARID5B (195 kb) and 3 kb up- and 1 kb downstream of the gene. Quantitative polymerase chain reaction (PCR) was used to define the appropriate capture product titer necessary to efficiently populate an Illumina HiSeq 2000 flow cell for paired-end 2 × 100 bp sequencing. Coverage of greater than 20 × depth was achieved across more than 80% of the targeted regions for nearly all samples. Sequence reads in the BigWig format were mapped and aligned using Integrative Genomics Viewer (36), and variants were called using the GATK pipeline (version 3.1) (37) and annotated using the ANNOVAR program (38) with the annotation databases, including RefSeq (39). All the ARID5B nonsilent variants were manually reviewed in the Integrative Genomics Viewer.
dCas9-KRAB–Mediated Enhancer Interference System (CRISPRi) Screen of Regulatory Elements in ARID5B
A total of 10 495 single-guide RNA (sgRNA) oligos targeting potential regulatory elements in ARID5B were designed for array-based oligonucleotide synthesis, including 10 sgRNAs targeting ARID5B promoter and untranslated region (UTR) and 10 sgRNAs not targeting any location in human genome as negative control (Supplementary Table 2, available online). The oligo pool was amplified by PCR and cloned into LentiGuide-Puro backbone (Addgene #52963). The ARID5BP2A-mCherry reporter cell line was overexpressed with lentiviral dCas9-KRAB followed by infection with virus of pooled sgRNA library at low multiplicity of infection (0.3), followed by blasticidin and puromycin selection. Genomic DNA from mCherryHigh (top 10%) and mCherryLow (bottom 10%) populations was used to amplify sgRNA sequences by PCR, followed by sequencing on MiSeq for single-end 150-bp read length (Illumina). The sequencing primers were: forward: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG; reverse: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTTTAGTTTGTATGTCTGTTGCTATTATGTCTACTATTCTTTC. The sgRNA library was described in Supplementary Table 2 (available online).
The FASTQ data were debarcoded and mapped to the original reference sgRNA library. The differentially enriched sgRNAs were identified by comparing normalized counts between the top 10% and the bottom 10% of mCherry-expressing bulk populations using DESeq2 (40). Four independent screenings were performed.
Details of other experimental methods and analytical procedures are provided in the Supplementary Methods (available online).
Results
ARID5B Variant Discovery and Association With ALL Susceptibility
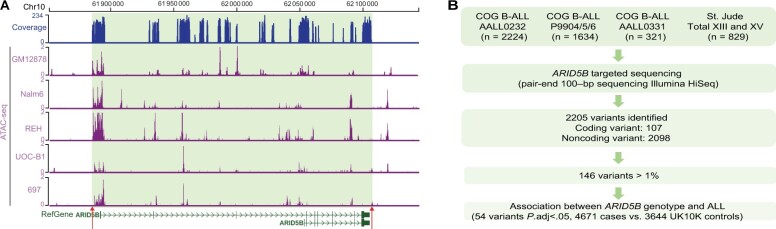
To fully examine genetic variation in ARID5B, we performed targeted sequencing of ARID5B exons, flanking sequences of 5′ and 3′UTR, as well as putative CREs at this locus (inferred from assay of transposase accessible chromatin sequencing (ATAC-seq) data from lymphoblastoid cell line GM12878, and ALL cell lines [Nalm6, REH, UOCB-1, and 697 cells]) (Figure 1, A). In 5008 children with newly diagnosed B-ALL, we identified 2205 variants, including 107 coding and 2098 noncoding single nucleotide polymorphisms (SNPs). Among these, 2 coding and 144 noncoding variants with a minor allele frequency greater than 1% were subsequently tested for association with ALL. Using an additive logistic regression model with genetic ancestry as covariates, we tested the effects of these variants on ALL risk by comparing genotype frequency in the ALL cohort vs that in 3644 patients in the UK10K cohort as non-ALL controls (41). We identified 54 leukemia risk variants that met the statistical significance threshold of adjusted P value less than .05 using the Benjamini-Hochberg procedure (Figure 1, B; Supplementary Table 3, available online). We also performed the genetic association analysis in individuals of European descent with similar results (Supplementary Table 3, available online).
Figure 1.
ARID5B targeted sequencing in 5008 children acute lymphoblastic leukemia (ALL) patients. A) Read density and coverage of the ARID5B target region. Regions 3 kb upstream of the 5' untranslated region (UTR) and 1 kb downstream of the 3'UTR (indicated by arrows and shaded backgroud), all coding regions, and putative open chromatin regions (based on assay of transposase accessible chromatin sequencing [ATAC-seq] data from GM12878, Nalm6, REH, UOC-B1, and 697 cells) were included in targeted sequencing. B) CONSORT flow diagram for discovering ARID5B risk variant. All of the common and rare variants of ARID5B were discovered in 5008 children with ALL by targeted sequencing. A total of 146 variants with sufficient frequency were subjected to association analysis after adjusting ancestry, and 54 variants were statistically significantly associated with ALL susceptibility (adjusted P < .05, estimated by the Benjamini-Hochberg procedure).
These ALL risk variants are distributed across promoter or CREs predicted from histone marks and/or ATAC-seq signals (Supplementary Figure 1, A, available online). We also explored the National Human Genome Research Institute-European Bioinformatics Institute GWAS catalog and identified 4 ARID5B SNPs (rs7090445, rs4245595, rs10821936, rs7089424) reported in previous ALL GWAS studies (P < 5 × 10–8). All of them are located in intron 3 of ARID5B gene (Supplementary Figure 1, A, available online). Interestingly, the cluster of ARID5B variants with the most statistically significant association with ALL, namely, rs4948492, rs7090445, rs4245595, rs10821936, rs10821937, and rs7896246, was mapped to a lymphocyte-specific open chromatin region observed only in common lymphoid progenitors, CD4 T cell, CD8 T cell, NK cell, and B cell (hg38 chr10: 61 960 967–61 962 170) (Supplementary Figure 1, A, available online).
CRISPRi Screen to Identify Regulatory Elements and Functional Variants in ARID5B
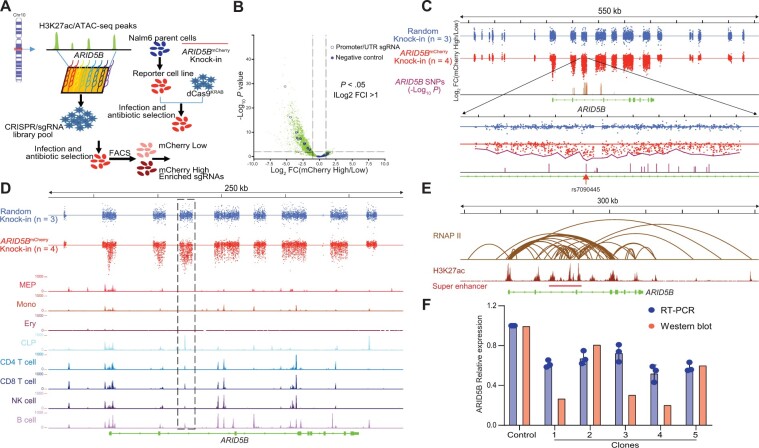
To experimentally evaluate putative enhancers in a scalable fashion, we engineered a reporter cell line in which predicted regulatory elements at the ARID5B locus were perturbed systematically to test their effects on ARID5B transcription. Using CRISPR/Cas9 genome editing, we first inserted mCherry at the 3′ end of the ARID5B coding frame in human ALL cell line Nalm6 (Supplementary Figure 2, available online) such that mCherry expression directly reflected ARID5B transcription and could be used to measure the effects of CREs at this locus. Then, we used the CRISPRi in which transcription repressor KRAB is targeted to each enhancer by sgRNA, and the effect of enhancer disruption on ARID5B-mCherry expression is measured by flow cytometry. To this end, we designed 10 497 sgRNAs tiling across 21 putative CREs predicted based on H3K27ac marks and ATAC-seq data in hematopoietic cells (Figure 2, A), with sgRNAs targeting ARID5B promoter and nonspecific regions as the positive and negative controls, respectively. This sgRNA library was lentivirally transduced into the Nalm6 reporter cells at a low titer to ensure each cell expressed no more than 1 sgRNA. After transduction, cells with the top 10% and bottom 10% with respect to ARID5B-mCherry expression were harvested by flow cytometry and subjected to massively parallel sequencing to identify sgRNAs present in each population (Figure 2, A). A total of 1561 sgRNA exhibited statistically different prevalence between these 2 groups, 98% of which were enriched in cells with low ARID5B expression; that is, disruption of these CREs resulted in decreased transcription (Figure 2, B). As a control, Nalm6 cells in which mCherry was randomly inserted into the genome were also transduced with the same sgRNA library, and, not surprisingly, we did not observe a statistically significant correlation between any sgRNA and mCherry intensity (Figure 2, C).
Figure 2.
CRISPR/dCas9-KRAB library screening for the interrogation of CREs of ARID5B. A) An overall schema of the design strategy. A single-guide RNA (sgRNA) library was designed using GM12878 H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) data and normal human hematopoietic cell assay of transposase accessible chromatin sequencing (ATAC-seq) data for reference. A single-cell clone reporter cell line stably expressing dCas9-KRAB was established then infected with CRISPR/sgRNA library pool virus at low multiplicity of infection (0.2) to ensure that each cell expressed no more than 1 sgRNA. Flow sorting was used to differentiate the mCherry-low population (the bottom 10%) and the mCherry-high population (the top 10%). Genomic DNA from these 2 populations was then sequenced to calculate the enrichment of each sgRNA. B) A volcano plot was generated to show the enrichment of sgRNAs in the mCherry-high or the mCherry-low populations. In total, 10 497 sgRNAs were designed, including 10 sgRNAs (open blue circles) that target the ARID5 B’s promoter and untranslated region, and 10 negative sgRNAs (solid blue circles) that do not target any location in the human genome. P values of .05 and |log2 FC| = 1 were used as cutoffs. C) The CRISPR/dCas9-KRAB library screening results. A Nalm6 single-cell clone with random knock-in mCherry was used as a negative control (top). For ARID5B mCherry knock-in cells (middle), sgRNAs were overrepresented mainly in predicted enhancers, including the ARID5B promoter, and were less represented in other regions. In contrast, for the negative control, sgRNAs were distributed almost equally across the whole ARID5B gene (data were derived in triplicate). Six cis-regulatory elements (CREs) were nominated out of 21 segments targeted by the sgRNA library from upstream to downstream of the ARID5B gene by 2-sided t test (P < .001) via comparing each segment of ARID5BmCherry knock-in with that of the random knock-in. The highlighted region shown encompasses SNPs having the strongest associations with ALL. In particular, SNP rs7090445 is juxtaposed with the most highly enriched sgRNA. D) Alignment of CRISPRi screening results with human hematopoietic cells ATAC-seq data. The most statistically significant CRE overlapped with lymphoid lineages (common lymphoid progenitors, CD4 T cell, CD8 T cell, natural killer cell, and B cell) specific ATAC-seq peaks (highlighted in the dashed box). E) A public GM12878 chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) dataset was used to interrogate the CRISPR/dCas9-KRAB library data. There were strong interactions between the ARID5B promoter and CREs nominated based on CRISPRi screening, as shown by the DNA loop generated from RNA polymerase II ChI A-PET data. The most statistically significant CRE is located in a super enhancer. F) Part of the highlighted region (chr10:61 960 755–61 962 481, 1.7 kb) in panel C was deleted using CRISPR/Cas9, resulting in a statistically significant decrease in ARID5B expression at both the protein and mRNA levels throughout all 5 single-cell clones.
Because each regulatory region encompasses multiple sgRNAs, we estimated a summary P value to indicate each CRE’s impact on ARID5B expression. Of the 21 predicted CREs, 6 were statistically significant at the P < .0001 level (Supplementary Table 4, available online). All 6 CREs exhibited strong chromatin interactions with the ARID5B promoter mediated by RNA polymerase II, as suggested by chromatin interaction analysis by paired-end tag sequencing data from the lymphoblastoid cell line GM12878 (Figure 2, E). In particular, the intron 3 CRE (hg38 chr10: 61 957 118– 61 967 424) overlapped with lymphocyte-specific ATAC-seq peaks (Figure 2, D) and a super-enhancer defined by ROSE algorithm based on H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) signal (hg38 chr10: 61 959 470–62 003 808) (42) (Figure 2, E), showing the strongest effects on ARID5B expression among all enhancers tested. This CRE also harbored ARID5B variants, with the strongest association with ALL development (Figure 2, C; Supplementary Figure 1, A, available online), with the sgRNA with the most profound disruption of ARID5B transcription only 16 bp away from the top risk variant, rs7090445. Additionally, among the ARID5B variants with the most statistically significant association with ALL (rs4948492, rs7090445, rs4245595, rs10821936, rs10821937, and rs7896246), rs7090445 was the only one that lies within a lymphoid-specific open chromatin region identified by ATAC-seq (Supplementary Figure 1, B, available online).
Because CRISPRi screen does not necessarily provide single-nucleotide resolution of CRE activity, we performed further validation experiments focusing on rs7090445. Genomic deletion of this intron 3 enhancer led to a 1.64-fold and 2.27-fold decrease in ARID5B expression on transcript and protein levels, respectively (Figure 2, F). Substitution of the reference allele (T) with the ALL risk allele (C) at rs7090445 reduced its enhancer activity by 1.43-fold in a luciferase reporter gene assay (Supplementary Figure 3, A, available online). Together, these results suggest that rs7090445 is likely the functional variant underlying the ALL susceptibility association signal at this locus.
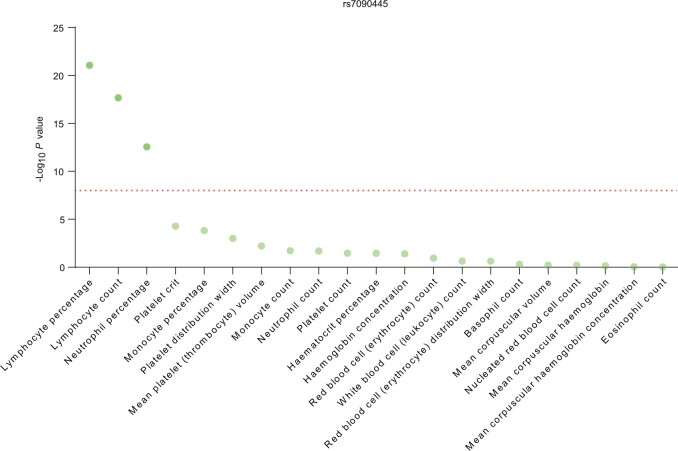
To explore the effects of rs7090445 on hematopoiesis generally, we examined 21 blood cell phenotypes in 349 861 individuals in the UK Biobank cohort (43). As shown in Figure 3, rs7090445 was most statistically significantly associated with lymphocyte percentage, lymphocytes count, and neutrophile percentage (P = 8.6 × 10–22, 2.1 × 10–18, and 2.7 × 10–13, respectively), and ALL risk allele C was always linked to an expansion of lymphocytes.
Figure 3.
Association of rs7090445 with normal blood traits. Association of genotype at rs7090445 with 21 blood cell phenotypes was tested in 349 861 individuals included in the UK Biobank cohort. The dashed line corresponds to P = 5 × 10–8 (P value estimated by Wald’s test). rs7090445 was statistically significantly associated with lymphocyte percentage, lymphocyte count, and neutrophile percentage.
Transcription Factor (TF) Regulating of the rs7090445 Enhancer Element and ARID5B Transcription
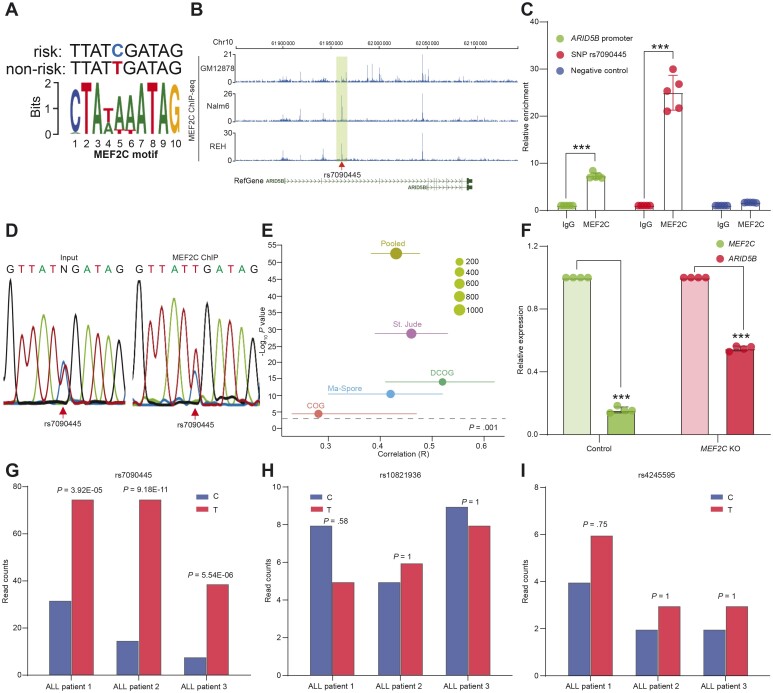
We hypothesized that rs7090445 causes differential TF binding, which in turn results in deregulation of ARID5B expression. Through analyzing ENCODE ChIP-seq data, we identified that ChIP-seq peaks of 10 transcription factors were overlapping with rs7090445 and also reported in HaploReg (v4.1) (44) (Supplementary Table 5, available online). Among these 10 transcriptional factors, we predicted that MEF2C (myocyte-specific enhancer factor 2C) shows the highest motif matching score, and its binding is statistically significantly altered by rs7090445, with the motif match score changing from 7.3 for reference allele (T) to 2.26 for the risk allele (C) (Figure 4, A). Second, we performed MEF2C ChIP-seq in lymphoblastoid cell line GM12878 as well as 2 ALL cell lines, Nalm6 and REH, and identified multiple binding sites at this locus (Figure 4, B), including the intron 3 enhancer described above. MEF2C binding to both this enhancer and the ARID5B promoter was confirmed using ChIP-quantitative PCR in Nalm6 cells (Figure 4, C). In fact, in DNA fragments pulled down by the anti-MEF2C antibody, we observed a statistically significant overrepresentation of the reference allele (T) compared with ALL risk allele (C), suggesting an allele-biased binding (Figure 4, D). Moreover, exploring the global expression profile of leukemia blasts in 4 pediatric ALL cohorts [DCOG (n = 190) (45), St. Jude TOTAL XIIIA/XIIIB/XV (n = 533) (46), COG P9906 (n = 207) (47), and Ma-Spore ALL 2010 (n = 231) (48)], we noted a moderately strong correlation between MEF2C and ARID5B transcripts (Figure 4, E; Supplementary Figure 3, C-F, available online). In Nalm6 cells, knocking out MEF2C using CRISPR editing resulted in a statistically significant downregulation of ARID5B, again pointing to a potentially direct regulation from MEF2C to ARID5B (Figure 4, F). ALL cells devoid of both MEF2C and rs7090445 enhancer showed the most severe ARID5B downregulation (Supplementary Figure 3, B, available online), although we cannot rule out the contribution of other CREs at this locus or the role of other TFs. Finally, we explored allelic chromatin accessibility in 3 ALL patients with heterozygous genotypes at rs7090445 and other ALL risk SNPs at this locus. As shown in Figure 4, G, there was a statistically significant overrepresentation of the reference allele (T) from the ATAC-seq signal compared with the risk allele (C), suggesting a reduced transcriptional activity associated with the risk allele (C). By contrast, at other SNPs in linkage disequilibrium with rs7090445 (rs10821936 and rs4245595), reference and variant alleles contributed equally to ATAC-seq signal, as shown in Figure 4, H and I, respectively. Taken together, these results suggested that rs7090445 genotype influenced MEF2C binding and thus MEF2C-mediated regulation of ARID5B transcription.
Figure 4.
MEF2C as a transcriptional regulator of ARID5B. A) Motif analysis of the region around the single nucleotide polymorphism (SNP) rs7090445 identified MEF2C as a potential regulator of ARID5B. B) MEF2C chromatin immunoprecipitation sequencing (ChIP-seq) data from GM12878, Nalm6, and REH cells confirmed the binding of MEF2C to the SNP region (shaded box) as well as to other regions of the ARID5B locus. C) In Nalm6 cells, MEF2C ChIP-quantitative PCR further confirmed the binding of MEF2C to the SNP region (middle 2 bars) as well as to the promoter of ARID5B (left 2 bars). Primers targeting upstream regions devoid of ATAC-seq or histone mark in acute lymphoblastic leukemia (ALL) cell lines were included as negative control (right 2 bars).***P < .001. Error bar = mean + SD. P value was estimated by 2-sided t test. D) Sanger sequencing was performed on MEF2C antibody-ChIPed DNA, demonstrating statistically significant overrepresentation of the reference allele (T) compared with the ALL risk allele (C). E) Positive correlation between the expression of MEF2C and ARID5B in ALL blasts, as determined by gene expression array from cohorts and presented as a meta-analysis based on a random effect model using the inverse-variance method. The correlation coeffecient was estimated using Pearson correlation. F) CRISPR/Cas9 was used to knock down MEF2C, and a concomitant decrease of ARID5B expression was confirmed by real-time PCR (RT-PCR). ***P < .001. Error bar = mean + SD. P value was estimated by 2-sided t test. G) ATAC-seq data for ALL PDX samples that were heterozygous for rs7090445 were interrogated. There were far fewer read counts for the risk allele (left bars) compared with the reference allele (right bars). Two SNPs in linkage disequilibrium (LD) with rs7090445 (rs10821936 and rs4245595) were also analyzed in the same way, but these SNPs showed no statistically significant difference in the read counts between the risk (left bars) and reference allele (right bars), as shown in panels H and I, respectively.
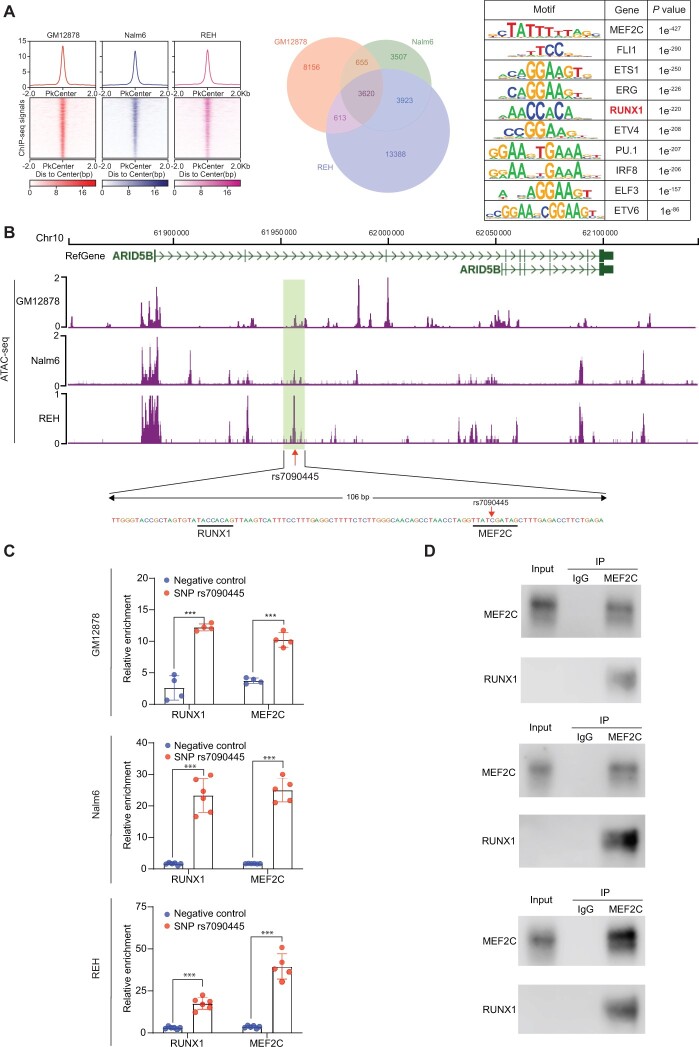
TFs often form a complex with cofactors to exert regulatory function to control gene transcription. To explore this, we performed MEF2C ChIP-seq in Nalm6, REH, and GM12878 cells to map MEF2C binding sites across the genome. In total, we identified 3620 regions consistently occupied by MEF2C in all 3 cell lines. As expected, the MEF2C/MEF2D motif was most statistically significantly enriched within these regions (TTATCGATAG, P = 1 × 10–427). Interestingly, sequences within MEF2C binding sites also exhibited marked overrepresentation of motifs for ETS-domain TFs (P = 1 × 10–290) and RUNX1 (TACCACAG, P = 1 × 10–220), suggesting possible cooccupancy of these TFs (Figure 5, A). Focusing on the CRE encompassing rs7090445, we indeed identified a putative RUNX1 binding site 50 bp from the MEF2C binding site (Figure 5, B). Performing ChIP-qPCR in GM12878, Nalm6, and REH cells using primers targeting this enhancer element, we confirmed direct binding of RUNX1, along with MEF2C (Figure 5, C). However, the proximal colocalization of these 2 proteins at rs7090445 does not necessarily result in TF interaction. To validate the presence of MEF2C and RUNX1 in the same protein complex, we conducted co-immunoprecipitation with MEF2C antibody using total proteins extracted from GM12878, Nalm6, and REH cells. As shown in Figure 5, D, RUNX1 was statistically significantly enriched in the MEF2C protein complex compared with either IgG control consistently across 3 cell line models. Collectively, these results pointed to RUNX1 as a potential cofactor of MEF2C to regulate transcription of ARID5B.
Figure 5.
RUNX1 as a cofactor of MEF2C in regulating ARID5B transcription. A) MEF2C chromatin immunoprecipitation sequencing (ChIP-seq) was performed in GM12878, Nalm6, and REH cell lines to identify MEF2C binding sites across the genome. In the heatmap, each row represents a genomic region with MEF2C binding centering around the summit of the ChIP-seq peak, with the color indicating signal intensity. The aggregation plot (top) shows the average of MEF2C binding signal for all peaks identified in that cell line. The Venn diagram indicates overlap of MEF2C binding sites among 3 cell lines. The right panel describes motif enrichment analysis in MEF2C ChIP-seq peaks common in 3 cell lines, with enrichment P value for the top 10 motif (P value was estimated by Fisher Exact test). B) Examining assay of transposase accessible chromatin sequencing (ATAC-seq) signal around rs7090445 in GM12878, Nalm6, and REH cells (top), we identified a putative RUNX1 binding site signified by the TACCACAG motif, which is 50 bp from that of MEF2C (TTATCGATAG), as shown in the bottom panel. C) RUNX1 binding to the cis-regulatory element (CRE) encompassing rs70904455 was confirmed using ChIP-quantitative PCR. Across 3 cell lines (GM12878, Nalm6, and REH), DNA sequence specific to this cis-regulatory element (CRE) was enriched in the pull-down lysates using RUNX1 or MEF2C antibodies compared with sequences outside of this CRE (negative controls). All values shown were normalized to IgG control. For each experiment (one cell line, RUNX1 or MEF2C binding), we compared the binding between 2 groups, that is, negative control (region outside of CRE) vs the rs7090445 region, using a 2-sample 2-sided t test. Error bar = mean + SD. The significance threshold was set as .0083 considering 6 experiments and multiple testing. ***P < .001. D) Direct interaction between MEF2C and RUNX1 was confirmed by co-immunoprecipitation experiments. Antibody specific for MEF2C was used to pull down its interacting proteins, and the presence of RUNX1 in this complex was determined using immunoblotting, with IgG as control. This was done in GM12878, Nalm6, and REH cell lines (top, middle, and bottom panels).
Discussion
The hypothesis of a genetic basis for leukemia susceptibility in children originally arose from familial studies (49-51). However, the impact of common genetic variants on ALL risk was not definitively established until GWAS was applied to systematically identify risk loci (3,4). Among these, the genomic region encompassing the ARID5B gene on 10q21.2 is one of the strongest GWAS hits (5,6,8,9,16). Despite the overwhelming evidence from these molecular epidemiology studies, the mechanisms by which genetic polymorphism transcriptionally influences ARID5B expression remain poorly understood. Therefore, our study addressed this knowledge gap by comprehensively mapping CREs and thus annotating ALL risk variant function in ARID5B. Our results pointed to rs7090445 as the likely causal variant responsible for the association signal at this locus, and this polymorphism leads to ablated transcriptional activation of ARID5B by MEF2C.
Previous fine-mapping studies of the ARID5B locus highlighted rs7090445 as the potential causal variant for ALL susceptibility (29), focusing on the hyperdiploid subtype. It was shown that the CRE encompassing this variant is tethered together with the ARID5B promoter, but the molecular machinery responsible for maintaining the 3-dimensional chromatin interaction at this locus was unclear. The same study also pointed to RUNX3 as the transcription factor with differential activity, with the risk vs reference allele at this variant using reporter gene assay (29). However, our data pointed to MEF2C as the TF whose binding affinity is most greatly affected by rs7090445. We also showed that knocking down MEF2C directly led to ARID5B downregulation, and the expression of these 2 genes is highly correlated in ALL blasts; there is initial evidence that MEF2C may recruit RUNX1 in regulating ARID5B transcription in B cells. That said, we could not definitively rule out the involvement of RUNX3 in ARID5B regulation given the similarity in binding motifs of different RUNX paralogs. Future work is needed to systematically identify the members of this complex and their relative contribution to the enhancer activity at this locus. Recently, Kachuri et al. (52) reported that ARID5B variant rs4245597 (which is in very strong linkage disequilibrium with rs7090445) was also associated with ratios of lymphocytes to other blood cell types. However, only a very small proportion of the effect on leukemia susceptibility was mediated by blood cell traits, supporting the potential pleiotropic effects of the ARID5B locus.
MEF2C belongs to the myocyte enhancer factor 2 family of TFs involved in a wide spectrum of physiological processes, particularly developmental differentiation (53,54). MEF2C expression varies statistically significantly across hematopoietic compartments, with the highest level in common lymphoid progenitor cells, and remains high during B-cell maturation and is absent in T cells (55). Mef2c deficiency led to profound defects in the production of B cells and also defective BCR signaling and immune response to antigen in these cells (56). Some studies suggest that MEF2C and MEF2D jointly coordinate the transcriptional network critical for early B-cell development (57). MEF2C can also interact with a variety of cofactors to form TF complexes, for example, p300 and E2A (58,59), although interaction with RUNX1 has not been reported previously.
This study is distinct from previous work in a number of aspects: 1) we comprehensively and experimentally characterized regulatory elements at the ARID5B locus using ALL cell models, whereas this was explored using only ENCODE and other publicly available datasets from other tissue types in the past; and 2) we directly resequenced this genomic region to discover variants, whereas the previous studies relied on linkage disequilibrium-based imputation to infer untyped variants and test their association. In fact, of 54 ALL risk variants that met the adjusted P value less than .05 cutoff in this study, only 4 were found in the National Human Genome Research Institute-European Bioinformatics Institute GWAS catalogue for ALL susceptibility. Because of direct sequencing, we also identified many rare variants that were not examined previously using the common variant-rich SNP arrays. For example, we examined rare variants in the coding region of ARID5B in ALL cases vs UK10K controls but did not observe any statistically significant difference in variant burden in the SKAT analysis (Supplementary Figure 1, C, available online). However, our sample size was still insufficient to reliably evaluate these rare variants, and therefore we could not definitively ascertain their impact on ALL susceptibility. Additionally, future studies should explore massively parallel reporter assays to directly determine the degree to which each variant affects the activity of the enhancer it resides in, as described for other genes.
In conclusion, we described a comprehensive annotation of ALL risk variants at the ARID5B locus and mechanistically explored the role of ARID5B in hematopoiesis. Our results shed light on the biological basis for the increased leukemia risk conferred by noncoding variants in ARID5B, further implicating this gene in normal and malignant B-cell development.
Funding
This work was supported by the National Institutes of Health (GM115279, CA234490, CA176063, and CA114766) and by the American Lebanese Syrian Associated Charities (ALSAC).
Notes
Role of the funder: The funder had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: The authors declare no competing financial interests.
Author contributions: J.J.Y and C.L.: Conceptualization, Methodology, Writing-Review & Editing, Supervision, Funding acquisition. X.Z., C.G.: Investigation, Visualization. Writing-Original Draft. M.Q., W.Y., Y.Z., and W.Y.: Formal analysis. Y.Z., C.L., B.X., C.T., P. W., M.V.R, M.L., C.H.P., S.H., E.R., M.D., and D.S.: Resources, Funding acquisition.
Acknowledgements: We thank the patients and parents who participated in the clinical trials included in this study. We thank Dr Ruopeng Feng for data interpretation. We thank Dr Keith A. Laycock, for scientific editing of the article.
Disclaimers: The content is solely the responsibility of the authors and does not necessarily represent the official views of the U.S. National Institutes of Health.
Supplementary Material
Contributor Information
Xujie Zhao, Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Maoxiang Qian, Institute of Pediatrics and Department of Hematology and Oncology, Children’s Hospital of Fudan University, National Children’s Medical Center, and the Shanghai Key Laboratory of Medical Epigenetics, Institutes of Biomedical Sciences, Fudan University, Shanghai, China.
Charnise Goodings, Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Yang Zhang, Department of Tumor Cell Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Wenjian Yang, Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Ping Wang, Department of Genome Technologies, The Jackson Laboratory for Genomic Medicine, Farmington, CT, USA.
Beisi Xu, Center for Applied Bioinformatics, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Cheng Tian, Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Ching-Hon Pui, Department of Oncology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Stephen P Hunger, Department of Pediatrics and The Center for Childhood Cancer Research, The Children’s Hospital of Philadelphia and The Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Elizabeth A Raetz, Department of Pediatrics and Perlmutter Cancer Center, New York University Langone Medical Center, New York, NY, USA.
Meenakshi Devidas, Department of Global Pediatric Medicine, St Jude Children’s Research Hospital, Memphis, TN, USA.
Mary V Relling, Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Mignon L Loh, Department of Pediatrics, Benioff Children’s Hospital and the Helen Diller Family Comprehensive Cancer Center, University of California San Francisco, San Francisco, CA, USA.
Daniel Savic, Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Chunliang Li, Department of Tumor Cell Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Jun J Yang, Department of Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA; Department of Oncology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Data Availability
ARID5B sequencing data and other molecular profiling results are deposited in NCBI GEO (GSE195831).
References
- 1. Pui CH, Robison LL, Look AT.. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030-1043. [DOI] [PubMed] [Google Scholar]
- 2. Hunger SP, Mullighan CG.. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541-1552. [DOI] [PubMed] [Google Scholar]
- 3. Trevino LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang W, Trevino LR, Yang JJ, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia. 2010;24(4):894-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang JJ, Xu H, Yang WJ, et al. Genome-wide association study identifies a novel susceptibility locus at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. Blood. 2012;120(21):877. [Google Scholar]
- 7. Perez-Andreu V, Roberts KG, Harvey RC, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet. 2013;45(12):1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu H, Cheng C, Devidas M, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. JCO. 2012;30(7):751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu H, Yang WJ, Perez-Andreu V, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. 2013;105(10):733-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vijayakrishnan J, Kumar R, Henrion MY, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia. 2017;31(3):573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prasad RB, Hosking FJ, Vijayakrishnan J, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115(9):1765-1767. [DOI] [PubMed] [Google Scholar]
- 12. Healy J, Richer C, Bourgey M, et al. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica. 2010;95(9):1608-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans TJ, Milne E, Anderson D, et al. Confirmation of childhood acute lymphoblastic leukemia variants, ARID5B and IKZF1, and interaction with parental environmental exposures. PLoS One. 2014;9(10):e110255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Migliorini G, Fiege B, Hosking FJ, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122(19):3298-3307. [DOI] [PubMed] [Google Scholar]
- 15. Walsh KM, de Smith AJ, Chokkalingam AP, et al. Novel childhood ALL susceptibility locus BMI1-PIP4K2A is specifically associated with the hyperdiploid subtype. Blood. 2013;121(23):4808-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vijayakrishnan J, Qian M, Studd JB, et al. Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat Commun. 2019;10(1):5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whitson RH, Huang T, Itakura K.. The novel Mrf-2 DNA-binding domain recognizes a five-base core sequence through major and minor-groove contacts. Biochem Biophys Res Commun. 1999;258(2):326-331. [DOI] [PubMed] [Google Scholar]
- 18. Yuan YC, Whitson RH, Itakura K, et al. Resonance assignments of the Mrf-2 DNA-binding domain. J Biomol NMR. 1998;11(4):459-460. [DOI] [PubMed] [Google Scholar]
- 19. Yuan YC, Whitson RH, Liu Q, et al. A novel DNA-binding motif shares structural homology to DNA replication and repair nucleases and polymerases. Nat Struct Biol. 1998;5(11):959-964. [DOI] [PubMed] [Google Scholar]
- 20. Zhu L, Hu J, Lin D, et al. Dynamics of the Mrf-2 DNA-binding domain free and in complex with DNA. Biochemistry. 2001;40(31):9142-9150. [DOI] [PubMed] [Google Scholar]
- 21. Watanabe M, Layne MD, Hsieh CM, et al. Regulation of smooth muscle cell differentiation by AT-rich interaction domain transcription factors Mrf2alpha and Mrf2beta. Circ Res. 2002;91(5):382-389. [DOI] [PubMed] [Google Scholar]
- 22. Whitson RH, Tsark W, Huang TH, et al. Neonatal mortality and leanness in mice lacking the ARID transcription factor Mrf-2. Biochem Biophys Res Commun. 2003;312(4):997-1004. [DOI] [PubMed] [Google Scholar]
- 23. Baba A, Ohtake F, Okuno Y, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 2011;13(6):668-675. [DOI] [PubMed] [Google Scholar]
- 24. Cichocki F, Wu CY, Zhang B, et al. ARID5B regulates metabolic programming in human adaptive NK cells. J Exp Med. 2018;215(9):2379-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lahoud MH, Ristevski S, Venter DJ, et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 2001;11(8):1327-1334. [DOI] [PubMed] [Google Scholar]
- 26. Okada Y, Terao C, Ikari K, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012;44(5):511-516. [DOI] [PubMed] [Google Scholar]
- 27. Tomer Y, Hasham A, Davies TF, et al. Fine mapping of loci linked to autoimmune thyroid disease identifies novel susceptibility genes. J Clin Endocrinol Metab. 2013;98(1):E144-E152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang W, Tang H, Zhang Y, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am J Hum Genet. 2013;92(1):41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Studd JB, Vijayakrishnan J, Yang M, et al. Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2. Nat Commun. 2017;8:14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maloney KW, Devidas M, Wang C, et al. Outcome in children with standard-risk b-cell acute lymphoblastic leukemia: results of children's oncology group trial AALL0331. J Clin Oncol. 2020;38(6):602-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larsen EC, Devidas M, Chen S, et al. Dexamethasone and high-dose methotrexate improve outcome for children and young adults with high-risk B-acute lymphoblastic leukemia: a report from children’s oncology group study AALL0232. J Clin Oncol. 2016;34(20):2380-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borowitz MJ, Devidas M, Hunger SP, et al. ; Children’s Oncology Group. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children's Oncology Group study. Blood. 2008;111(12):5477-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pui CH, Mahmoud HH, Rivera GK, et al. Early intensification of intrathecal chemotherapy virtually eliminates central nervous system relapse in children with acute lymphoblastic leukemia. Blood. 1998;92(2):411-415. [PubMed] [Google Scholar]
- 34. Pui CH, Sandlund JT, Pei D, et al. ; Total Therapy Study XIIIB at St Jude Children’s Research Hospital. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children’s Research Hospital. Blood. 2004;104(9):2690-2696. [DOI] [PubMed] [Google Scholar]
- 35. Pui CH, Campana D, Pei D, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360(26):2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2018. doi: 10.1101/201178:201178. [DOI] [Google Scholar]
- 38. Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733-D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walter K, Min JL, Huang J, et al. UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liam Abbott SB, Churchhouse C, Ganna A, et al. ; The Hail team. UK BIOBANK GWAS. http://www.nealelab.is/uk-biobank/. Accessed 2018.
- 44. Ward LD, Kellis M.. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930-D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10(2):125-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paugh SW, Bonten EJ, Savic D, et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat Genet. 2015;47(6):607-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harvey RC, Mullighan CG, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116(23):4874-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qian MX, Zhang H, Kham SKY, et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017;27(2):185-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garber JE, Goldstein AM, Kantor AF, et al. Follow-up study of twenty-four families with Li-Fraumeni syndrome. Cancer Res. 1991;51(22):6094-6097. [PubMed] [Google Scholar]
- 50. Hemminki K, Jiang Y.. Risks among siblings and twins for childhood acute lymphoid leukaemia: results from the Swedish Family-Cancer Database. Leukemia. 2002;16(2):297-298. [DOI] [PubMed] [Google Scholar]
- 51. Greaves MF, Maia AT, Wiemels JL, et al. Leukemia in twins: lessons in natural history. Blood. 2003;102(7):2321-2333. [DOI] [PubMed] [Google Scholar]
- 52. Kachuri L, Jeon S, DeWan AT, et al. Genetic determinants of blood-cell traits influence susceptibility to childhood acute lymphoblastic leukemia. Am J Hum Genet. 2021;108(10):1823-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhu B, Gulick T.. Phosphorylation and alternative pre-mRNA splicing converge to regulate myocyte enhancer factor 2C activity. Mol Cell Biol. 2004;24(18):8264-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cante-Barrett K, Pieters R, Meijerink JP.. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014;33(4):403-410. [DOI] [PubMed] [Google Scholar]
- 55. Stehling-Sun S, Dade J, Nutt SL, et al. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat Immunol. 2009;10(3):289-296. [DOI] [PubMed] [Google Scholar]
- 56. Wilker PR, Kohyama M, Sandau MM, et al. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat Immunol. 2008;9(6):603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Herglotz J, Unrau L, Hauschildt F, et al. Essential control of early B-cell development by Mef2 transcription factors. Blood. 2016;127(5):572-581. [DOI] [PubMed] [Google Scholar]
- 58. Youn HD, Sun L, Prywes R, et al. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286(5440):790-793. [DOI] [PubMed] [Google Scholar]
- 59. Youn HD, Liu JO.. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity. 2000;13(1):85-94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ARID5B sequencing data and other molecular profiling results are deposited in NCBI GEO (GSE195831).