Abstract
The current vaccine development strategies for the COVID-19 pandemic utilize whole inactive or attenuated viruses, virus-like particles, recombinant proteins, and antigen-coding DNA and mRNA with various delivery strategies. While highly effective, these vaccine development strategies are time-consuming and often do not provide reliable protection for immunocompromised individuals, young children, and pregnant women. Here, we propose a novel modular vaccine platform to address these shortcomings using chemically synthesized peptides identified based on the validated bioinformatic data about the target. The vaccine is based on the rational design of an immunogen containing two defined B-cell epitopes from the spike glycoprotein of SARS-CoV-2 and the universal T-helper epitope PADRE. The epitopes were conjugated to short DNA probes and combined with a complementary scaffold strand, resulting in sequence-specific self-assembly. The immunogens were then formulated by conjugation to gold nanoparticles by three methods or by co-crystallization with epsilon inulin. BALB/C mice were immunized with each formulation, and the IgG immune responses and virus neutralizing titers were compared. The results demonstrate that this assembly is immunogenic and generates neutralizing antibodies against wildtype SARS-CoV-2 and the Delta variant.
Keywords: Variants of concerns (VOC), SARS-CoV-2, Peptide, Vaccine, PADRE, Epitope
Graphical abstract
DNA hybridization was used to assemble linear B cell epitopes from SARS-CoV2 spike with the artificial T helper epitope PADRE. After formulation the assembly was immunogenic and produced antibodies with affinity to the whole glycoprotein spike and neutralizing activity against the wild type virus as well as the Delta variant. The selected epitopes are located in highly conserved regions of the virus and have no mutations in currently identified variants, including Omicron.

Background
The ongoing SARS-CoV-2 pandemic has demonstrated the need for vaccine technologies to ensure a rapid and efficient response to novel pathogens. Current vaccines for SARS-CoV-2 are effective,1 but their inflammatory nature limits the use in individuals with immune disorders.2., 3., 4., 5., 6., 7. Moreover, herd immunity towards novel pathogens has proven unreliable and difficult to achieve, leaving immunocompromised individuals at risk.8., 9. Immunocompromised individuals are at greater risk of infection and may remain infectious for longer than the immunocompetent, increasing the rate of community spread.10., 11., 12. This highlights the need for vaccine platforms that can be individualized and adapted for immunocompromised individuals and individuals with autoimmune disorders.13
Conventional vaccines are composed of whole pathogens or their components. As a result, they can cause unwanted side effects, such as vaccine-enhanced infection, interfering neutralization antibodies, Hoskins effect, toxicity, and pathogen de-attenuation.14., 15., 16., 17., 18. These issues put the immunocompromised vaccine recipients at even greater risk and further hinder vaccine development. Additionally, some current SARS-CoV-2 vaccine recipients experience cardiovascular complications such as coagulopathies and myocarditis.19., 20.
Finally, vaccine development, production, and distribution are time-consuming and expensive, limiting their utility in fighting novel pathogens21., 22.. The currently approved vaccines for SARS-CoV-2 are built upon prior developed SARS-CoV and Middle East Respiratory Syndrome (MERS) vaccines. These seemingly quickly developed vaccines depended on prior knowledge about the immunopathological effects of targeting the nucleocapsid protein,23 as well as the 2p mutation used to stabilize the spike glycoprotein in the most immunogenic form.24
With these limitations in mind, we aim to develop a novel approach in vaccinology, a proof-of-concept modular epitope vaccine against SARS-CoV-2.
Our strategy is based on short peptide epitopes that generate immune responses precisely aimed at the critical components of the SARS-CoV-2, limiting the risk of adverse effects.25., 26., 27. The immunogen is assembled on a long single-stranded DNA scaffold to which short DNA probes are annealed in a sequence-specific manner. Each DNA probe carries a specific epitope covalently attached to the end of the probe. We used two peptides corresponding to the well-identified linear epitopes from the S-protein of SARS-CoV-2.28 The Pan HLA DR-binding Epitope (PADRE)29 was introduced via attachment to one of the probes to endow the vaccine with antigenicity. Each probe-epitope conjugate and the scaffold DNA are prepared individually and can be modified or replaced, making the immunogen truly modular.
Several formulations of the immunogen were prepared, and mice were immunized to evaluate the spike- and peptide-specific immune responses. We found that our novel vaccine design generated immune responses in mice without any adverse events, and the serum from these mice could neutralize the wildtype and the Delta variant of SARS-CoV-2.
Results
Preparation of the vaccine
The immunogen is prepared from three synthetic peptides, three short DNA strands (probes), and one long DNA strand, which is complementary to the three probes and acts as a sequence-specific scaffold for assembly.
Each DNA strand is synthesized chemically. A copper-free click chemistry dibenzocyclooctyne (DBCO) handle is introduced at the 5′ of each probe strand for attachment to the epitopes, and a thiol is introduced to the 5′ of the scaffold for downstream use during formulation.
B-cell epitopes (peptides P2 and P3) are selected from the reported linear neutralizing epitopes of the SARS-CoV-2 spike28. The T-cell epitope is the artificial universal T helper epitope PADRE (peptide P1), ensuring robust immunostimulatory signaling29. Here, the peptides are synthesized with an N-terminal azidolysine for conjugation to the appropriate DBCO-labeled probes via click chemistry. The resulting probe-epitope conjugates and the scaffold DNA are the individual modules of our immunogen. Upon equimolar mixing, they spontaneously assemble into the immunogen by sequence-specific hybridization of the probes with the corresponding regions of the scaffold. The resulting immunogen complex is shown in Fig. 1a.
Fig. 1.

Schematic representation of immunogen and its validation. Three epitopes as peptides (P1–P3) are assembled on a DNA scaffold containing thiol for coupling with gold. P1 is the universal T helper cell epitope PADRE, P2 is an S-protein-derived peptide (residues 1148–1159), important for cell membrane fusion, and P3 is a peptide located after RBD (residues 553–564), important for receptor recognition (A). Immunogen assembly was validated using spectrophotometric melting by 260 nm absorbance change. The melting curve was taken at a temperature range from 10 to 85 °C (B). Immunogen‑gold formulations generated by different methods were characterized by agarose gel electrophoresis. From top to bottom: free gold nanoparticles, freezing-based conjugation of immunogen (FR), acid-based conjugation (PA), salt-based conjugation (SC) (C).
The assembly was validated by UV spectrophotometric melting experiments. The melting profile for the construct is shown in (Fig. 1b). The sharp increase in absorbance near 55 °C corresponds to the experimental melting temperature. This confirms the presence of a DNA duplex, and the absence of a hyperchromic effect at temperatures below 40 °C confirms stability at physiological temperatures.
Selecting the optimal method for formulation
Conjugation to gold nanoparticles (GNP) was utilized to improve the uptake and biological stability of the construct.30 We have used three methods to conjugate the immunogen to GNP: a) freezing-based conjugation (FR), b) pH-assisted conjugation (PA), and c) salt concentration-based conjugation (SC), as described in the methods section.
The GNP-immunogen formulations are negatively-charged particles with the net charge of particles defined by the density of coverage of the nanoparticle with immunogens. Therefore, we used gel electrophoresis to characterize the formulations (Fig. 1c). The PA formulation exhibited the highest mobility towards the positive electrode, followed by the FR formulation, which had slightly lower mobility. The SC formulation produced a diffuse band with low electrophoretic mobility. These results suggested that the immunogen conjugation with FR and PA methods produced particles with a higher immunogen density per GNP than the SC method. A fourth gold-free formulation (IN) based on cocrystallization with microcrystalline inulin was chosen since inulin is known to activate complement and improve antigen uptake.31 This formulation served as a control for the GNP formulations.
We utilized AFM topographic imaging and dynamic light scattering (DLS) to provide additional characterization of the vaccine formulations. In the AFM experiments, the samples of formulations, along with the unconjugated gold nanoparticles used as a control, were diluted to the concentration 0.5 nM, deposited on functionalized mica32 (APS-mica) for 2 min, rinsed with deionized water, dried, and imaged with AFM operating in tapping mode. Images of gold nanoparticles are shown in Fig. S1A. Spherical particles are clearly seen in the images. The sizes of each particle (diameter) were measured using the cross-section tool of the AFM software. The histogram of the diameter distribution is shown in Fig. S1B. The histogram is fit by a Gaussian with the maximum 35.4 nm ± 0.1 nm (SEM). The results of DLS data for the same sample are shown in Fig. S2A, and they produce a mean value of 24 nm. The data in Fig. S2B show the Zeta potential values obtained for the same samples.
Similar characterization was done for the vaccine formulations; the results for each of these studies are shown in Figs. S3–S5. The measurements for all samples are combined as histograms in Fig. 2 . The results demonstrate that the conjugation method using PA resulted in the largest diameter of 71 nm, followed by SC at 50 nm, FR at 33 nm and Au (gold nano particles) at 24 nm. The PA conjugation method also resulted in the greatest negative values zeta potential. AFM data compared with the DLS ones show lower values for particle sizes, which is consistent with the drying of the sample for AFM measurements. The Zeta potentials for PA and FR formulations illustrate higher values than the control, which is consistent with the gel-electrophoresis data illustrating the negative charge increase by coating with the negatively charged DNA backbone of the vaccine.
Fig. 2.
Comparison between conjugation methods characterized on DLS, AFM and Zeta potential. Zeta potential data are shown in grey bars, whereas DLS and AFM data depicted in blue and blue bars, respectively. Error bars are shown in all histograms.
We also tested the cytotoxicity of the samples. The results are assembled in Fig. S6. The cytotoxicity of the four formulations was measured in Vero-E6 cells with or without adjuvant using a standard colorimetric MTT assay that measures cell proliferation and viability twice. The percent viability of the cells is plotted. All the conjugated immunogens have minimal or <10 % cytotoxicity in Vero-E6. The CpG adjuvant-only treated cells have zero toxicity in Vero-E6 cells.
These four immunogen formulations were further tested for their ability to induce an antigen-specific immune response in mice.
Comparing the formulations based on the peptide-specific IgG response strengths
Four groups of BALB/C mice were immunized with the FR, PA, SC, and IN immunogen formulations (25 mg/injection) with a class B CpG adjuvant (100 μl/injection). A control group of mice received the adjuvant alone. All five groups of mice were immunized with three doses of the respective immunogen as prime dose (0 days), 1st boost (14 days,) and 2nd boost (28 days), as shown in schema (Fig. 3a). Sera were collected on days 0, 14, 28, 61, and tested by ELISA for IgG against peptides P1 and P2, as well as the whole protein S or its receptor-binding domain (RBD) fragment. Plates were coated separately with P1 or P2 (0.1 μg/well) to determine the peptide-specific IgG response. Sera from each time point were applied in 5-fold serial dilutions from 1:25 to 1:78125. Sera from all the four vaccinated groups (FR, PA, SC, and IN) indicated the presence of detectable IgG immune response against P1 and P2 at 1:25 on day 61 (Fig. 3b & c). However, mice immunized with immunogens formulated by FR and PA methods elicited higher IgG immune response with a maximal immune response on day 61. The immunogen formulated by the SC method elicited a lower IgG immune response against both P1 and P2 (Fig. 3b & c). On the other hand, mice immunized with immunogen formulated with IN exhibited a higher IgG immune response against P1 than against P2 (Fig. 3b & c). The observed difference in IgG immune response to immunogens produced by FR and PA methods versus SC method correlated with the density of immunogen on the gold nanoparticles produced by each method (Fig. 3c). These results indicate that GNP-conjugated immunogens with high-density coverage of the nanoparticles elicit a stronger response, while the inulin formulation does not efficiently co-deliver the P1 and P2, leading to poor T helper activation of P2 specific B cells.
Fig. 3.
IgG immune response against P1 and P2 in mice immunized with GNP-conjugated vaccine candidates. BALB/c mice were immunized with GNP-conjugated vaccine candidates at different time intervals (Day 0, 14, 28, and 61), and blood and spleen were collected as shown in schema (A). High binding 96 well plates were separately coated with P1 and P2 (100 ng/well) overnight at 4 °C. Sera prepared from different time points of immunization were diluted at 1:25 and incubated for 1 h at room temperature. IgG immune response against P1 (B) and P2 (C) was determined using HRP-conjugated goat anti-mouse IgG in the presence of TMB as substrate. *P < 0.05 is considered significant.
Binding of the whole spike in ELISA
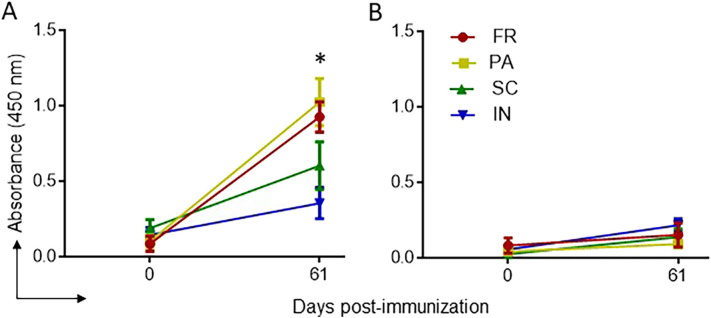
To confirm that the peptide-specific antibodies can bind the same regions in the native spike, we used ELISA with plates coated with stabilized whole spike glycoprotein. The receptor-binding domain (RBD) protein was used as a negative control since it includes neither of our peptides in its sequence. The sera were applied in 5-fold serial dilutions, starting with 1:25 and up to 1:78125. The immune responses were similar to the previous results against P2. FR and PA immunogens elicited a higher immune response against spike after the 2nd booster at 1:25 dilutions (Fig. 4a). SC formulation elicited a detectable but far weaker IgG immune response. In contrast, the IN formulation had comparable signals against negative control (RBD) and whole spike (Fig. 4b). The position of the P2 and P3 on the spike are depicted in Supplementary Fig. S7.
Fig. 4.
IgG immune response against SARS-CoV-2 whole spike and RBD in mice immunized with GNP-conjugated vaccine candidates. High binding 96 well plates were separately coated with whole spike or RBD (1 μg/ml) overnight at 4 °C. Sera prepared from different time immunization points were diluted at 1:25 and incubated for 1 h at room temperature. IgG immune response against the spike (A) and the RBD (B) was determined, as explained above. *P < 0.05 is considered significant.
Virus neutralization with the generated sera
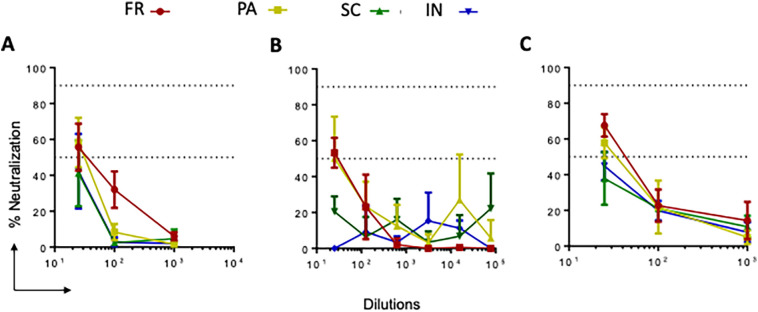
The neutralization activity of pooled sera from each group was tested against three SARS-CoV-2 strains: SARS-CoV-2 isolate USA-WA1/2020 (Fig. 5a), SARS-CoV-2 isolate USA-WI1/2020 (Fig. 5b), and SARS-CoV-2 isolate hCoV-19/USA/PHC658/2021 (Delta Variant) (Fig. 5c). We found that at 1:25 dilution, total sera from PA and FR groups could neutralize both wild-type and delta variants of SARS-CoV-2 (>50 % neutralization), and higher dilutions of sera showed <50 % neutralization titers (Fig. 5). These data suggest that GNP-conjugated vaccine candidates elicit antigen-specific immune responses with neutralization potential.
Fig. 5.
SARS-CoV-2 live virus neutralization ability of sera from mice immunized with GNP-conjugated vaccine candidates. Sera (Day 61) from all groups of immunized mice were evaluated for live virus neutralization ability against SARS-CoV-2 WT, WI, and delta variants. Sera (5× diluted, starting from 1:25) were separately incubated with WT, WI, and delta viruses at 104 PFU for 1 h. Next, the virus-sera mix was added to the cells and incubated for 24 h. Then cells were fixed and permeabilized and stained with the combination of anti-SARS-CoV-2 spike protein antibodies (rabbit mAb) and Alexa Fluor 488 goat anti-rabbit as primary and secondary antibodies, respectively. The nuclei were stained using Hoechst 33342 and plates were read using Operetta Imager. The percent neutralization was calculated based on the differential intensity of the fluorescence.
Flow cytometry analysis of CD20+ B cells and CD4+/CD8+ T cells memory phenotypes in GNP-conjugated vaccine groups
Using a flow cytometric analysis, we performed a phenotypic assessment of CD4+ and CD8+ T cells and B cells in the spleen and peripheral blood in control and GNP-conjugated vaccine immunized mice at necropsy. Here, CD4+ and CD8+ T cells phenotypic assessment was performed based on CD44 and CD62L (l-selectin) surface expression as memory phenotype and activation markers. The splenocytes and blood cells were stained with fluorochrome-labeled anti-CD4, anti-CD8, anti-CD20, anti-CD44, and anti-CD62L antibodies, and representative gating was used to obtain CD4+ and CD8+ T cells to analyze the expression of CD44 and CD62L (Supplementary Fig. S8). Although gated CD4+ and CD8+ T cells express comparable levels of CD44 and CD62L, no difference was observed in the percentage of naïve (CD44−CD62L+), TCM (CD44+CD62L+), and TE/EM (CD44+CDL−) in splenocytes and blood cells across immunized groups compared to control (Supplementary Fig. S9). Further, splenocytes and blood cells from GNP-conjugated vaccine immunized mice did not exhibit the difference in the percentage of CD20+ B cells compared to control (Supplementary Fig. S10).
Discussion
This study demonstrated that peptides assembled on the DNA scaffold resulted in an efficient immunogen, producing a robust and specific immune response. Using nucleic acids as an assembly scaffold permits stoichiometrically and spatially controlled assembly of separate epitopes into a single immunogen. The rigid DNA duplex enables the segregation of individual epitopes and prevents the formation of unwanted conformational epitopes. The formulation of the immunogen is a crucial component for vaccine development. The DNA complementarity permitted the incorporation of the polyA-thiol tail, which is necessary for high-efficiency conjugation with the gold nanoparticles. The modular vaccine was assembled by chemical conjugation to gold nanoparticles (GNPs) using three conjugation strategies or co-crystallization with inulin microcrystals. The use of GNPs offers a simple formulation procedure with low toxicity and improved immunogen stability and uptake. The electrophoresis data suggests that the density of DNA coverage of the gold nanoparticle is the primary factor affecting the immunological efficiency of the GNP-immunogen construct. The thiol‑gold coupling method defines the final product's density, consistent with the reported differences.33 The acidic pH-based conjugation (PA) seemingly produced the highest coverage, closely followed by freezing-based (FR) conjugation. At the same time, the salt concentration-based method (SC) resulted in smearing and low mobility, suggesting low coverage and/or aggregation.
We tested the immunogenicity of the resulting vaccines and found differences between each formulation. The inulin-based formulation had high anti-P1 antibody levels and a significantly weaker response to P2, suggesting dissociation of the immunogen molecule and inefficient immunological synapse formation. P1 acts as B-cell and T-helper epitope simultaneously and thus can form immunological synapses regardless of formulation and assembly. Salt concentration-based SC formulation had a poor immune response to both peptides, suggesting poor uptake or processing by antigen-presenting cells due to aggregation or loss of antigen during dialysis. Finally, acid- (PA) and freezing-based (FR) formulations had almost identical antibody levels against both P1 and P2, indicating the stability of the formulation.
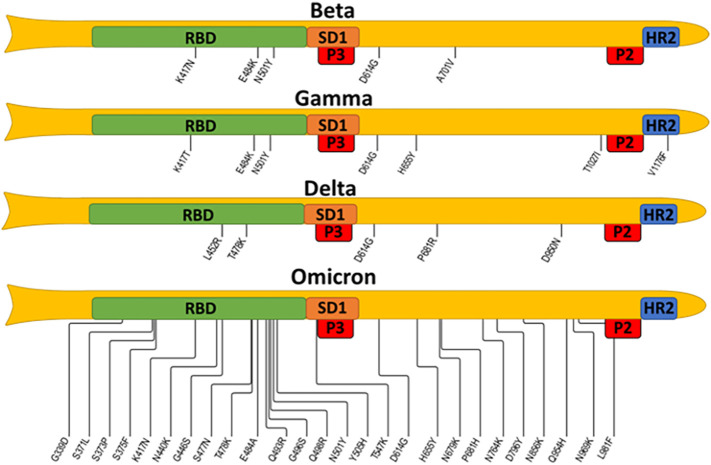
The immune responses against the full-length spike or RBD (negative control) confirmed the findings of peptide ELISA. Strong immune responses to spike with minimal immune responses to RBD were found in sera generated by acid- (PA) and freezing-based (FR) formulations and showed neutralization titers against wildtype SARS-CoV-2 and delta variant.34 The selected epitopes do not carry any of these mutations, as shown in Fig. 6 , in which all mutations were mapped in the currently monitored VOCs. This finding suggests that our vaccine should be equally efficient for all circulating variants, including the omicron. Furthermore, the selected epitopes are in highly conserved or invariant regions,35., 36. opening an avenue for a universal pan beta coronavirus vaccine. Our data contrasts with the non-neutralizing and spike non-reactive antibodies generated when the peptides we use are converted into an immunogen by the more traditional keyhole limpet hemocyanin (KLH) technology.37 This inefficacy of KLH-conjugated vaccines was reported by the same group that first identified the B-cell epitopes we used in this current vaccine design. We believe the differences in the immune responses are related to the carrier-mediated folding of the epitopes into non-native conformations. On the other hand, the highly hydrophilic and rigid DNA-based carrier does not permit such interactions between the peptides and the carrier itself, thus resulting in a more native-like behavior of the peptides. The intrinsic stability of DNA, along with a strong negative charge, lends high stability to the vaccines. The gold-based formulations can be freeze-thawed 3 or more times with no aggregation and stored in a refrigerator for at least 6 months.
Fig. 6.
SARS-CoV-2 VOCs map of mutations spanning from RBD to heptad repeat 2 (HR2).
The four spike protein variants: beta, gamma, delta, and omicron BA1, are represented schematically; note that the N-terminal domain and mutations localized within are omitted. Selected epitopes are expected to disrupt the activity of the subdomain 1 (SD1) and heptad repeat 2 (HR2), thus blocking viral attachment and cellular entry. The information about mutations is taken from the covdb.stanford.edu database.
The developed peptide-array approach has several important features. The accessibility and affordability of solid-phase peptide synthesis of short peptide epitopes simplify the manufacturing of our vaccine.38 We use programmed-complementarity DNA-based assembly of separate epitopes into a modular vaccine, which further improves adaptability and simplifies chemical synthesis and formulation. Most epitope vaccines utilize chemical conjugation of the epitopes to a carrier protein, such as ovalbumin or KLH, or combine the epitopes into a single fusion protein like beads on a string. Since the epitopes incorporated in such immunogens have no explicit borders, they can go through different patterns of proteolytic processing and produce a variety of new, uncharacterized, and unwanted epitopes, known as neoepitopes.39., 40. The folding of these immunogens produces additional conformational neoepitopes, complicates storage, and results in a large batch-to-batch variability should the artificial protein have several stable structures.41 Using a rigid DNA duplex for immunogen assembly keeps the epitopes in our vaccine structurally and spatially separated, minimizing the risk of neoepitope formation. Finally, DNA has low antigenicity and does not lead to carrier-induced epitope suppression.42., 43. An epitope vaccine assembled on our platform can generate a precise immune response with high efficacy and no detectable shifts in lymphocyte populations, as was demonstrated by flow cytometry Figs. S8–S10. As epitope vaccines in general,44, 45., 46., 47. this modular vaccine may provide protective antibodies with little or no therapeutically inefficient or harmful antibodies. This is critical if targeting some parts of the pathogen can lead to immune enhancement of the disease or adverse reactions. Further investigation of this vaccine platform is necessary to introduce additional B cell and T helper epitopes, increase immunogenicity, and determine the therapeutic potential in a virus challenge model systems. Moreover, cytotoxic T-cell epitopes need to be introduced and their protective potential characterized.
In conclusion, to our knowledge, this is the first epitope vaccine made through DNA hybridization-based assembly of separate epitopes into a single molecule. Using nucleic acids is a very convenient strategy since they are non-toxic and easy to synthesize and characterize. Most importantly, they are highly programmable, allowing for reproducible and predictable assembly and long-term storage in ambient conditions. This assembly can then co-deliver the components to the antigen-presenting cells and facilitate the formation of immunological synapses. Our newly developed platform also greatly simplifies the fusion of immunomodulatory ligands, such as TLR agonists, allowing for direct delivery of these ligands into the immune cells. The use of fused TLR agonists is known to significantly improve vaccine efficacy,48 allowing for T-cell-independent B-cell maturation.49 This is especially important for the elderly and immunocompromised patients and can minimize side effects,50 improving the vaccine coverage in adverse event-prone populations, like individuals with autoimmune diseases.
Methods
Vaccine preparation
Following N-terminal azidolysine-labeled peptides at 95 % purity were purchased from Genscript (Piscataway, NJ, USA).
-
1.
P1 is the pan HLA DR-binding epitope PADRE and is designed to stimulate T helper cells in genetically diverse populations. The peptide sequence is (LysN3)-GSAKFVAAWTLKAAA.
-
2.
P2 is derived from residues 1148–1159 of the full spike. This region is located right before the heptad repeat 2, which is important for membrane fusion. The peptide sequence is (LysN3)-FKEELDKYFKNH.
-
3.
P3 is derived from residues 553–564 of the full spike. This region is located on the S1, right after the RBD. The peptide sequence is (LysN3)-TESNKKFLPFQQ.
The DNA oligonucleotides (ssDNA) were synthesized using MerMade12 DNA synthesizer with the help of the standard phosphoramidite chemistry with details provided in the supplement. The following set of oligonucleotides was synthesized:
Strand 1 DBCO-TATACAGCCTACTCACTATA for coupling with P1;
Strand 2 DBCO-TATACTGAGCTAGTCGTATA for coupling with P2;
Strand 3 DBCO-TATACCTTCATCCTTATATA for coupling with P3;
Scaffold strand: (SH)-A9-TATAGTGAGTAGGCTGTATA-TATACGACTAGCTCAGTATA-TATATAAGGATGAAGGTATA.
Citrate-coated gold nanoparticles, 15 nm in diameter and at optical density (OD) 50 were purchased from Luna Nanotech (Markham, ON, Canada) and used for formulation.
All materials were purchased pyrogen-free and sterile and handled aseptically.
Peptide-DNA coupling
Each peptide and DNA strand were dissolved in 6 M guanidinium hydrochloride in the presence of sodium phosphate buffer (50 mM, pH 7.0) at the final concentrations of 200 μM ssDNA and 400 μM peptides. The reaction was carried out for 24 h at 4 °C, followed by dialysis to remove excess peptides. Reaction completion was verified by the disappearance of 308 nm absorbance shoulder on UV–Vis, corresponding to the unreacted DBCO. Coupling and dialysis were performed aseptically.
Assembly of the antigen
Peptide-coupled DNA strands (1–3) were combined in 0.5 × PBS pH 7.4 at a concentration of 150 μM and 140 μM of the complement. The mixture was heated to 95 °C and gradually cooled to 4 °C (2 h). The annealed product was diluted to 1.2 OD at 260 nm in isotonic PBS. These steps were performed aseptically. The assembly was verified by the melting experiments using UV–Vis spectrophotometer Varian Cary 50 Bio (Varian Palo Alto, Ca, USA) with a temperature range of 10–85 °C, data interval 0.1 °C, temperature ramp rate 2 °C/min, signal averaging time 0.1. The melting temperature was compared to the theoretically melting temperature predicted by OligoAnalyzer™ from Integrated DNA Technologies (IDTDNA.com). Vaccine formulation.
Vaccine candidates were formulated by the following four methods:
In the FR method, the annealing product was diluted in water 4-fold (<20 mM Na+) and added to the GNP stock solution (136 nM) at a molar ratio of 150:1. The mixture was placed in a −20 °C freezer for 1 h. Then, the product was allowed to thaw, and 10 × PBS was added to adjust the tonicity of the final product to isotonic. The solution was diluted to 50 nM (as GNP) in PBS, aliquoted at 100 μl/dose, and frozen for use as an immunogen.
In the PA method, the annealing product was combined with GNP stock solution at a molar ratio of 50:1 and incubated for 5 min. Then, pH was adjusted to 3.0 with 1 M citric acid, incubated for 3 min, and neutralized with NaOH to pH 6.5. Heat annealing was performed as described previously. The conjugate was dialyzed against isotonic PBS, diluted, and aliquoted as in the FR method.
In the SC method, the initial procedure was identical to the FR method. Instead of freezing, saturated NaCl (6.15 M) was gradually added to a 1 M final concentration (48 h). The conjugate was dialyzed against isotonic PBS using a 50 kDa molecular weight cut-off membrane, diluted, and aliquoted as in the FR method.
For IN co-crystallization method, inulin microcrystals (epsilon form) were prepared as previously described.31 The microcrystals were washed five times via centrifugation and resuspension in 45 °C water. After the final centrifugation, the wet pellet was resuspended in the annealed product at final proportions of 300 mg of wet pellet and 5 nmol of annealed product per 1 ml isotonic PBS. The mixture was then heated up to 55 °C and allowed to cool down slowly. The resulting microcrystals were aliquoted at 100 μl/dose and stored at 4 °C until further use.
All formulation-related procedures were performed aseptically.
Agarose gel electrophoresis
Agarose (2 %) (Sigma-Aldrich, Inc. St. Louis, MO) gel was prepared in sodium borate buffer (10 mM, pH 8.0) and used for electrophoresis of unconjugated and conjugated GNP prepared by the FR, PA, and SC methods. The nanoparticles were loaded directly in buffer without loading dye at 10 μl/well, and electrophoresis was carried out at 15 V/cm (300 V).
AFM imaging
Samples for AFM scanning were diluted for each conjugation method to 0.5 nM before depositing onto 1-(3-aminopropyl)-silatrane (APS) functionalized mica. The samples were incubated for 2 min to bind to the APS mica, followed by gentle washing with DI water and drying with a slow argon gas flow. The samples were then dried in a vacuum chamber overnight.
Once dried, the samples were scanned on a Multimode AFM/Nanoscope IIId system with TESPA probes (Bruker Nano Inc., Camarillo, CA). The images captured were 3 × 3 μm in size with 1536 pixels/line.
The images were analyzed using Femtoscan software (Advance Technologies Center, Moscow, Russia). A cross-sectional line was drawn over the center of each particle and a diameter in nm was obtained. Once all the particles on each image were analyzed, histograms were generated using Origin software.
Dynamic light scattering
The Dynamic Light Scattering (DLS) and zeta potential scans were completed on a Malvern NanoZS meter. The DLS samples were diluted 100× (0.5 nM) before filling the cuvette for DLS measurements. The computer software from Malvern Instruments, Ltd., automatically generated the graphs. The DLS experiments were completed in triplicate, and the runs were overlayed on the same graph. The Zeta potential measurement samples were diluted 50× (1 nM), and the Malvern Instruments, Ltd., software fitted the different zeta potential on an aggregate graph, allowing for easy comparison between the ten different runs. If a run was an extreme outlier from the other runs, it was excluded from the results.
Cytotoxicity assay
Vero-E6 cells were seeded at 10,000 cells/well in a 96-well plate containing 100 μl complete media specific for each cell type. For adherence, cells were incubated overnight at 37 °C in a humidified 5 % CO2 incubator. After overnight incubation, the media was replaced with fresh media, and Vero-E6 cells were treated with the formulations at 50 nM with or without adjuvant. Untreated cells were considered a negative control. After the treatment, cells were incubated at 37 °C in humidified 5 % CO2 incubator. 48 h post-treatment, 20 μl of MTT substrate (5 mg/ml) was added to each well and incubated for 4 additional hours at 37 °C in the dark. Then the culture media was carefully removed, the blue formazan crystals were dissolved in 200 μl of DMSO, and the purple color was read at 590 nm with a reference filter of 620 nm.
Cells and viral titer determination
Vero E6 (ATCC® CRL-1586™) and Vero-STAT1 knockout cells (ATCC® CCL-81-VHG™) were cultured in DMEM containing 10 % fetal bovine serum, 2 mM l-glutamine, penicillin (100 units/ml), streptomycin (100 units/ml), and 10 mM HEPES. Calu-3 cell (ATCC 184HTB-55) were cultured in Eagle's 188 Minimum Essential Medium (EMEM) (ATCC 30-2003) containing 10 % FBS. SARS-CoV-2 isolates USA-WI1/2020 (BEI; cat# NR-52384), USA-WA1/2020 (BEI; cat# NR-NR-52281) were passaged in Vero-STAT1 knockout cells, whereas hCoV-19/USA/PHC658/2021 (Delta Variant) (BEI; cat# NR-55672) was passaged in Calu-3 cells. The viral titer was determined using the plaque assay as described previously in.51 In brief, Vero E6 cells (2.5 × 105) were seeded in 6-well plates and incubated for 24 h. After 24 h, cells were washed with sterile 1 × PBS, and the virus stock was ten-fold serially diluted in serum-free OptiMEM media and then added to the cells in duplicate. The plates were incubated at 37 °C for 1 h with slight shaking every 15 min. Then, 2 ml of 0.5 % agarose in minimal essential media (MEM) containing 5 % FBS and antibiotics, penicillin (100 units/ml), and streptomycin (100 units/ml), were added to each well and incubated at 37 °C for 72 h. The cells were fixed with 4 % paraformaldehyde overnight, followed by removing the overlay and staining with 0.2 % crystal violet to visualize plaque-forming units (PFU). All assays were performed in a BSL-3 laboratory setting.
Animal experiments
BALB/c mice (males, 6 to 8 weeks old) were purchased from Jackson Laboratories, housed in micro isolator cages, and maintained at 12-h light-dark cycle at 22.2 °C, and 30–40 % humidity. Mice were given feed and sterile water daily, and they were acclimated to the environment for approximately 1–2 weeks to determine that they were healthy and suitable for experiments. Four groups of mice (n = 5 each group) were immunized subcutaneously with GNP-conjugated vaccines (100 μl) by different methods, FR, PA, SC, and IN. The immunogens were diluted in LPS-free water and mixed with 25 μg ODN 1826 Class B CpG oligonucleotide (a murine TLR9 ligand) as an adjuvant. Another group of mice immunized with only 25 μg ODN 1826 Class B CpG oligonucleotide serves as the control. Baseline blood samples were collected from the submaxillary vein on day 0 (before the immunization). Blood samples were collected every 14 days with boosts of the same doses were given. All mice were immunized with 2 boosts and finally necropsied at day 61. Blood samples and spleens were collected at necropsy for flow cytometry. All the animal procedures, including housing, are approved by the University of Nebraska Medical Center (UNMC) Institutional Animal Care and Use Committee (IACUC) protocol # 21-020-04-EP. and are conducted according to the National Institutes of Health guidelines. Furthermore, this study is reported according to ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines).
Enzyme-linked immunosorbent assay (ELISA)
Ninety-six-well high binding plates were separately coated with peptide P1, P2 (100 ng/well), whole spike, and spike-RBD glycoproteins (0.1 μg/well) 1 × phosphate buffered saline (PBS) and incubated at 4 °C overnight. The next day, the plates were washed three times and blocked with 1 % BSA in 1 × PBS containing 0.1 % Tween 20 (PBST) for 1 h at 37 °C. Sera samples were diluted five-fold serially starting at 1:25 in 1 % BSA containing PBST, then added to the plates and incubated at 37 °C for 1 h. The plates were washed three times with PBST, then HRP-conjugated goat anti-mice IgG Human ads-HRP (SouthernBiotech1:4,000 dilution) (cat#103005) was added and incubated for 1 h at room temperature. Plates were washed five times with PBST before adding 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution. The reaction was stopped after 5 min by adding 0.16 M sulfuric acid. The OD at 450 nm was measured with a Bio-Rad microplate reader. A graph was plotted as Absorbance at 450 nm vs. dilutions.
Immunofluorescence-based neutralization assay
Neutralization assays were performed against USA_WI/2020, USA-WA1/2020, and hCoV-19/USA/PHC658/2021 strains. Sera samples were serially diluted starting from 1: 25 (5-fold) in serum-free Dulbecco's modified Eagle's medium (DMEM) in triplicate wells and incubated with 20,000 focus-forming units of SARS-CoV-2 virus at 37 °C for 1 h. The serum-virus mixture was added to Vero E6 cell (C1008, ATCC, no. CRL-1586) monolayers seeded in 96 healthy blackout plates and incubated at 37 °C for 1 h. The inoculum was removed and replaced with complete DMEM and incubated at 37 °C for 24 h. After 24 h, DMEM was removed, and cells were washed twice with PBS and fixed with 4 % paraformaldehyde in PBS for 30 min at room temperature. Following fixation, plates were washed thrice with 1 × PBS and permeabilized with 50 μl/well of 0.1 % Triton X-100 (Fisher BP151-100) in PBS for 10 min. After permeabilization, cells were washed thrice with 1 × PBS and blocked with 3 % BSA for 30 min. Then, cells were stained with 50 μl/well of primary antibody, anti-SARS-CoV-2 spike (rabbit mAb, Sino Biologicals MA14AP0204) at 1:1000 diluted in 3 % BSA-PBS and incubated overnight at 4 °C on a shaker. The next day, cells were washed three times with 1 × PBS and stained with a secondary antibody (Alexa Fluor 488 Goat anti-rabbit) at 1:2000 dilution in 3 % BSA-PBS. Finally, cells were incubated at room temperature in a shaker for 1 h, washed three times with 1 × PBS, and stained the nuclei using Hoechst 33342 (Invitrogen H3570) and Cell Mask (Invitrogen C10046) at 1:20,000 diluted in 1 × PBS. The plate was shaken for 15 min after proper sealing with aluminum foil/sealer and taken for reading using Operetta Imager. Percentage neutralization was calculated based on the difference in fluorescent intensity.
Analysis of B cells and CD4+/CD8+ memory phenotypes by flow cytometry
Blood samples and spleen tissues were collected from immunized mice at necropsy. Single-cell suspension from spleen tissues was prepared, and RBCs were lysed using RBC lysis buffer. Cells (5 × 106) were washed and suspended in staining buffer, and Fc block was performed with purified anti-mouse CD16/32. Dead cells were discriminated by adding Zombie Aqua fixable viability dye. Fluorochrome-labeled mouse-specific antibodies were diluted with staining buffer into cocktails and added to cells at 100 μl per sample. Cells were incubated for 30 min at room temperature and washed twice with staining buffer. Then, cells were fixed using 2 % PFA and acquired using the Becton Dickson Fortessa X450 flow cytometer. Fluorescence minus one (FMO) control was performed in parallel, and FMO determined subsequent gating. All data were analyzed using Flowjo version 10.6 (Trees Star Inc., Ashland, Oregon, USA) software.
Statistical analysis
Two-way ANOVA with Tukey's multiple comparison test was performed to assess statistical significance (P values < 0.05 is considered significant) using (GraphPad PRISM 6.07, San Diego, CA) software.
Credit authorship contribution statement
Conceptualization and study design: YLL and SNB.
Methodology: KZ, KP, RR, OAA, AA, AB, MJ and SF.
Data analysis: KZ, KP, RR and OAA.
Visualization: KZ, KP, RR, YLL and SNB.
Supervision: YLL and SNB.
Funding acquisition: YLL and SNB.
Intellectual input: YLL, SNB and KZ.
Project administration: YLL and SNB.
Writing (original draft): KZ, KP, RR, YLL and SNB.
Writing (review and editing): All authors.
Declaration of competing interest
Authors declare no competing interests.
Acknowledgments
The work was supported by grants to YLL from NSF (MCB 1515346) and NIH (5U54GM115458-05) to YLL and SNB. The authors thank T. Stormberg and other members of the Lyubchenko lab for useful insights into the data interpretation. This work was partially supported by the National Institute of Allergy and Infectious Diseases grants R01 AI113883, and DA052845. SNB acknowledges independent research and development (IRAD) funding from the National Strategic Research Institute (NSRI) and Nebraska Research Initiative (NRI) grants at the University of Nebraska. We also acknowledge the University of Nebraska Medical Center (UNMC BSL-3) core facility for allowing us to perform all in vitro experiments involving SARS-CoV-2 and the COBRE Nebraska Center for Nanomedicine supported by the National Institute of General Medical Science (NIGMS) grant 5P30 GM127200. The UNMC BSL-3 core facility is administered through the Office of the Vice-Chancellor for Research and is supported by the Nebraska Research Initiative (NRI). The following reagents were deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: (i) USA-WI1/2020 (BEI; cat# NR-52384) ii) USA-WA1/2020 (BEI; cat#NR-52281) iii) hCoV-19/USA/PHC658/2021 (Delta Variant) (BEI; cat# NR-55672, iv) Spike Glycoprotein (Stabilized) from SARS-CoV-2, Wuhan-Hu-1 (BEI; cat# 53524) and v) Spike Glycoprotein RBD from SARS-CoV-2, Wuhan-Hu-1 (BEI; cat# 53800).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nano.2022.102604.
Appendix A. Supplementary data
Supplementary data contains figures S1-S10.
References
- 1.Self W.H., Tenforde M.W., Rhoads J.P., et al. Comparative effectiveness of Moderna, Pfizer-BioNTech, and Janssen (Johnson & Johnson) vaccines in preventing COVID-19 hospitalizations among adults without immunocompromising conditions — United States, March–August 2021. MMWR Morb Mortal Wkly Rep. 2021;70(38):1337–1343. doi: 10.15585/mmwr.mm7038e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vuille-Lessard É., Montani M., Bosch J., Semmo N. Autoimmune hepatitis triggered by SARS-CoV-2 vaccination. J Autoimmun. 2021;123 doi: 10.1016/j.jaut.2021.102710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waqar S.H.B., Khan A.A., Memon S. Thrombotic thrombocytopenic purpura: a new menace after COVID bnt162b2 vaccine. Int J Hematol. 2021;114(5):626–629. doi: 10.1007/s12185-021-03190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velikova T., Georgiev T. SARS-CoV-2 vaccines and autoimmune diseases amidst the COVID-19 crisis. Rheumatol Int. 2021;41(3):509–518. doi: 10.1007/s00296-021-04792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masset C., Kervella D., Kandel-Aznar C., Fantou A., Blancho G., Hamidou M. Relapse of IgG4-related nephritis following mRNA COVID-19 vaccine. Kidney Int. 2021;100(2):465–466. doi: 10.1016/j.kint.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karabulut K., Andronikashvili A., Kapici A.H. Recurrence of thrombotic thrombocytopenic purpura after mRNA-1273 COVID-19 vaccine administered shortly after COVID-19. Konstantopoulos K, ed. Case Rep Hematol. 2021;2021:1–4. doi: 10.1155/2021/4130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz G.A., Parsons G.T., Gering S.K., Meier A.R., Hutchinson I.V., Robicsek A. Myocarditis and pericarditis after vaccination for COVID-19. JAMA. 2021;326(12):1210. doi: 10.1001/jama.2021.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomson H. Herd immunity to covid-19 may not be attainable in the UK A high vaccination rate will save many lives, but it probably won't be enough to stop covid-19 becoming a seasonal disease, finds Helen Thomson. New Sci. 2021;251(3348):17. doi: 10.1016/S0262-4079(21)01448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aschwanden C. Five reasons why COVID herd immunity is probably impossible. Nature. 2021;591(7851):520–522. doi: 10.1038/d41586-021-00728-2. [DOI] [PubMed] [Google Scholar]
- 10.Avanzato V.A., Matson M.J., Seifert S.N., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183(7):1901–1912. doi: 10.1016/j.cell.2020.10.049. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbasi J. Researchers tie severe immunosuppression to chronic COVID-19 and virus variants. JAMA. 2021;325(20):2033. doi: 10.1001/jama.2021.7212. [DOI] [PubMed] [Google Scholar]
- 12.Kaila V., Sirkeoja S., Blomqvist S., et al. SARS-CoV-2 late shedding may be infectious between immunocompromised hosts. Infect Dis. 2021;53(11):880–882. doi: 10.1080/23744235.2021.1939891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez Y., Novelli L., Rojas M., et al. Autoinflammatory and autoimmune conditions at the crossroad of COVID-19. J Autoimmun. 2020;114 doi: 10.1016/j.jaut.2020.102506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vennema H., de Groot R.J., Harbour D.A., et al. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J Virol. 1990;64(3):1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicasio M., Sautto G., Clementi N., et al. Neutralization interfering antibodies: a “novel” example of humoral immune dysfunction facilitating viral escape? Viruses. 2012;4(9):1731–1752. doi: 10.3390/v4091731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weingartl H., Czub M., Czub S., et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox G.J., Orlova M., Schurr E. Tuberculosis in newborns: the lessons of the “Lübeck Disaster” (1929–1933) PLoS Pathog. 2016;12(1) doi: 10.1371/journal.ppat.1005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeVries A.S., Harper J., Murray A., et al. Vaccine-derived poliomyelitis 12 years after infection in minnesota. N Engl J Med. 2011;364(24):2316–2323. doi: 10.1056/NEJMoa1008677. [DOI] [PubMed] [Google Scholar]
- 19.Ostrowski S.R., Søgaard O.S., Tolstrup M., et al. Inflammation and platelet activation after COVID-19 vaccines - possible mechanisms behind vaccine-induced immune thrombocytopenia and thrombosis. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.779453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matta A., Kunadharaju R., Osman M., et al. Clinical presentation and outcomes of myocarditis post mRNA vaccination: a meta-analysis and systematic review. Cureus. 2021;13(11) doi: 10.7759/cureus.19240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heaton P.M. Challenges of developing novel vaccines with particular global health importance. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.517290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Excler J.L., Saville M., Berkley S., Kim J.H. Vaccine development for emerging infectious diseases. Nat Med. 2021;27(4):591–600. doi: 10.1038/s41591-021-01301-0. [DOI] [PubMed] [Google Scholar]
- 23.Yasui F., Kai C., Kitabatake M., et al. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J Immunol Baltim Md 1950. 2008;181(9):6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 24.Kirchdoerfer R.N., Wang N., Pallesen J., et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep. 2018;8(1):15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wisnewski A.V., Redlich C.A., Liu J., et al. Immunogenic amino acid motifs and linear epitopes of COVID-19 mRNA vaccines. Ito E, ed. PLoS ONE. 2021;16(9) doi: 10.1371/journal.pone.0252849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palatnik-de-Sousa C.B., Rosa D.S., Soares I.da S. Editorial: epitope discovery and synthetic vaccine design. Front Immunol. 2018;9:826. doi: 10.3389/fimmu.2018.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chauhan V., Rungta T., Goyal K., Singh M.P. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci Rep. 2019;9(1):2517. doi: 10.1038/s41598-019-39299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Yun Lai D., Nan Zhang H., et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell Mol Immunol. 2020;17(10):1095–1097. doi: 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosa D. The pan HLA DR-binding epitope improves adjuvant-assisted immunization with a recombinant protein containing a malaria vaccine candidate. Immunol Lett. 2004;92(3):259–268. doi: 10.1016/j.imlet.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Mateu Ferrando R., Lay L., Polito L. Gold nanoparticle-based platforms for vaccine development. Drug Discov Today Technol. 2021 doi: 10.1016/j.ddtec.2021.02.001. Published online February 26. [DOI] [PubMed] [Google Scholar]
- 31.Cooper P.D., Barclay T.G., Ginic-Markovic M., Petrovsky N. The polysaccharide inulin is characterized by an extensive series of periodic isoforms with varying biological actions. Glycobiology. 2013;23(10):1164–1174. doi: 10.1093/glycob/cwt053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shlyakhtenko L.S., Gall A.A., Lyubchenko Y.L. In: Cell Imaging Techniques. Taatjes D.J., Roth J., editors. Vol. 931. Humana Press; 2012. Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy; pp. 295–312. Methods in Molecular Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B., Liu J. Freezing Directed Construction of Bio/Nano Interfaces: Reagentless Conjugation, Denser Spherical Nucleic Acids, and Better Nanoflares. J Am Chem Soc. 2017;139(28):9471–9474. doi: 10.1021/jacs.7b04885. [DOI] [PubMed] [Google Scholar]
- 34.Kannan S.R., Spratt A.N., Cohen A.R., et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun. 2021;124 doi: 10.1016/j.jaut.2021.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia S., Zhu Y., Liu M., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17(7):765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pokhrel S., Kraemer B.R., Burkholz S., Mochly-Rosen D. Natural variants in SARS-CoV-2 Spike protein pinpoint structural and functional hotspots with implications for prophylaxis and therapeutic strategies. Sci Rep. 2021;11(1):13120. doi: 10.1038/s41598-021-92641-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Liang Ma M., Lei Q., et al. Linear epitope landscape of the SARS-CoV-2 Spike protein constructed from 1,051 COVID-19 patients. Cell Rep. 2021;34(13):108915. doi: 10.1016/j.celrep.2021.108915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skwarczynski M., Toth I. Peptide-based synthetic vaccines. Chem Sci. 2016;7(2):842–854. doi: 10.1039/C5SC03892H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avci F., Berti F., Dull P., et al. Glycoconjugates: what it would take to master these well-known yet little-understood immunogens for vaccine development. Papasian CJ, ed. mSphere. 2019;4(5) doi: 10.1128/mSphere.00520-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero P., Donda A., Hubbell J.A. An optimized antigen–protein fusion. Nat Biomed Eng. 2020;4(6):583–584. doi: 10.1038/s41551-020-0572-3. [DOI] [PubMed] [Google Scholar]
- 41.Koch C., Jensen S.S., Øster A., Houen G. A comparison of the immunogenicity of the native and denatured forms of a protein. APMIS. 1996;104(1–6):115–125. doi: 10.1111/j.1699-0463.1996.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 42.Liu X., Xu Y., Yu T., et al. A DNA Nanostructure Platform for Directed Assembly of Synthetic Vaccines. Nano Lett. 2012;12(8):4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jegerlehner A., Wiesel M., Dietmeier K., et al. Carrier induced epitopic suppression of antibody responses induced by virus-like particles is a dynamic phenomenon caused by carrier-specific antibodies. Vaccine. 2010;28(33):5503–5512. doi: 10.1016/j.vaccine.2010.02.103. [DOI] [PubMed] [Google Scholar]
- 44.De Groot A.S., Moise L., McMurry J.A., Martin W. In: Clinical Applications of Immunomics. Falus A., editor. Springer; US: 2009. Epitope-based immunome-derived vaccines: a strategy for improved design and safety; pp. 39–69. [DOI] [Google Scholar]
- 45.Oscherwitz J. The promise and challenge of epitope-focused vaccines. Hum Vaccines Immunother. 2016;12(8):2113–2116. doi: 10.1080/21645515.2016.1160977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell A.W., McCluskey J., Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6(5):404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- 47.Rajčáni J., Szathmary S. Peptide vaccines: new trends for avoiding the autoimmune response. Open Infect Dis J. 2018;10(1):47–62. doi: 10.2174/1874279301810010047. [DOI] [Google Scholar]
- 48.Huleatt J.W., Jacobs A.R., Tang J., et al. Vaccination with recombinant fusion proteins incorporating Toll-like receptor ligands induces rapid cellular and humoral immunity. Vaccine. 2007;25(4):763–775. doi: 10.1016/j.vaccine.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Pihlgren M., Silva A.B., Madani R., et al. TLR4- and TRIF-dependent stimulation of B lymphocytes by peptide liposomes enables T cell–independent isotype switch in mice. Blood. 2013;121(1):85–94. doi: 10.1182/blood-2012-02-413831. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z.B., Xu J. Better adjuvants for better vaccines: progress in adjuvant delivery systems, modifications, and adjuvant-antigen codelivery. Vaccines. 2020;8(1):128. doi: 10.3390/vaccines8010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendoza E.J., Manguiat K., Wood H., Drebot M. Two detailed plaque assay protocols for the quantification of infectious SARS-CoV-2. Curr Protoc Microbiol. 2020;57(1) doi: 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data contains figures S1-S10.