Abstract
The role of ssgA in cell division and development of streptomycetes was analyzed. An ssgA null mutant of Streptomyces coelicolor produced aerial hyphae but failed to sporulate, and ssgA can therefore be regarded as a novel whi gene. In addition to the morphological changes, antibiotic production was also disturbed, with strongly reduced actinorhodin production. These defects could be complemented by plasmid-borne ssgA. In the wild-type strain, transcription of ssgA was induced by nutritional shift-down and was shown to be linked to that of the upstream-located gene ssgR, which belongs to the family of iclR-type transcriptional regulator genes. Analysis of mycelium harvested from liquid-grown cultures by transmission electron microscopy showed that septum formation had strongly increased in ssgA-overexpressing strains in comparison to wild-type S. coelicolor and that spore-like compartments were produced at high frequency. Furthermore, the hyphae were significantly wider and contained irregular and often extremely thick septa. These data underline the important role for ssgA in Streptomyces cell division.
Streptomycetes are gram-positive, filamentous soil bacteria that have become a major focus for the study of microbial development. Streptomyces growth on solid media is started by the development of a complex vegetative mycelium of branching hyphae. Environmental signals such as nutrient depletion cause the development of almost aseptate aerial hyphae that partially parasitize the substrate mycelium. Elongation of the cell wall takes places at the tips of the hyphae, and occasional septation leads to multinucleoid compartments separated by cross walls. Exponential growth is achieved by branching of the vegetative hyphae, resulting in an intricate mycelial network. Eventually, the aerial hyphae become subdivided into uninucleoid cells that develop into chains of hydrophobic spores (10). One of the striking features of streptomycetes and other actinomycetes is their ability to produce a wide variety of secondary metabolites, including many antibiotics, which are produced at about the same time as the onset of morphological differentiation in surface-grown cultures (19, 31).
The process leading to sporulation on solid media has been well documented, helped by the availability of a wide variety of developmental mutants (reviewed in references 8 and 26). In principle, these mutants can be divided into two classes: the bald (bld) mutants, which fail to produce the fuzzy aerial mycelium, and the white (whi) mutants, which produce aerial hyphae but cannot form the grey-pigmented spores. The whi genes are further subdivided into early and late whi genes, depending on the developmental state of the aerial hyphae. The early whi genes, including whiA, whiB, whiG, and whiH, are involved in the regulatory cascade involving the early stages of sporulation and fail to produce spore compartments even after prolonged incubation (15, 36). The late whi genes, including whiD and sigF, are involved in the final stages of sporulation and spore maturation (10, 33).
Some Streptomyces species, including S. albus (12), S. griseus (27), S. roseosporus (21), and S. venezuelae (17), have the capacity to produce spores in liquid cultures. This process is often elicited by nutritional shift-down from a rich medium to a defined minimal medium (14, 27), indicating a positive control by the stringent response and suggesting a possible correlation between sporulation and secondary metabolism. Interestingly, S. roseosporus was also shown to sporulate when grown in rich media.
Little is known about the processes underlying submerged sporulation. One of the best-characterized proteins involved is factor C, which was identified as a 34-kDa protein that restores submerged sporulation to an S. griseus mutant. Although antibodies against factor C cross-react with proteins in a wide variety of prokaryotic and eukaryotic organisms, no homologue has yet been identified in any of the databases (5, 6). More recently, a mutant of S. griseus (designated SY1) that produced submerged spores in rich as well as in minimal liquid media was identified. Introduction of a DNA fragment harboring the ssgA gene into SY1 suppressed submerged sporulation (23, 24). ssgA encodes an approximately 15-kDa protein of unknown function. Recently, data from the S. coelicolor genome sequencing project (www.sanger.ac.uk/projects/S_coelicolor) revealed an open reading frame (ORF) highly homologous to ssgA, but analysis of genomes from many other eubacteria, including other gram-positive bacteria such as Bacillus subtilis and the related actinomycetes Mycobacterium leprae or M. tuberculosis, did not reveal a similar ORF. This indicates that ssgA might be limited to the genus Streptomyces. Introduction of a multicopy plasmid harboring ssgA into S. griseus resulted in fragmentation of the mycelium and suppressed submerged sporulation, while it inhibited development on agar plates. Western blot analysis with polyclonal antibodies raised against SsgA revealed that timing of ssgA expression in S. griseus correlates to the onset of sporulation in liquid cultures (25).
These data suggested a possible involvement of SsgA in cell division and sporulation, although no direct evidence has been presented. Here we show that S. griseus strain SY1 is not mutated in the ssgA gene and describe a defined knockout mutant of the S. coelicolor homologue, which has a Whi phenotype. We have also analyzed the cytological effect of overexpression of ssgA and show by electron microscopy (EM) that SsgA in fact enhances cell division by stimulating septum formation in liquid-grown cultures of S. coelicolor.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
Escherichia coli K-12 strains JM109 (30) and ET12567 (28) were used for propagating plasmids. The strains were grown and transformed by standard procedures (37); transformants were selected in L broth containing 1% (wt/vol) glucose and ampicillin at a final concentration of 200 μg ml−1. L broth with 1% (wt/vol) glucose and 30 μg of chloramphenicol ml−1 was used to grow ET12567.
S. griseus (ATCC 23345) was obtained from the American Type Culture Collection, and S. griseus mutant strain SY1 was described previously (23). S. coelicolor A3(2) M145 (prototrophic, SCP1− SCP2−), obtained from the John Innes Centre strain collection, was used for transformation and propagation of Streptomyces plasmids. Protoplast preparation and transformation were performed as described by Hopwood et al. (20). SFM (16) was used to make spore suspensions. R2YE (20) was used for regenerating protoplasts and, after addition of the appropriate antibiotic, for selecting recombinants. For liquid culturing of Streptomyces YEME (20), tryptone soy broth (Difco) containing 10% (wt/vol) sucrose (TSBS) or standard minimal medium (MM [20]) with 1% (wt/vol) mannitol as the carbon source was used. For nutritional shift-down, S. coelicolor M145 was grown in TSBS to an optical density at 550 nm (OD550) of 0.7, washed, and transferred to MM.
Plasmids pUC18 (42), pIJ2925 (22), and pSET152 (4) were used for cloning experiments. While pSET152 is a conjugative shuttle plasmid, in the experiments described in this study the plasmid and its derivatives were introduced by standard protoplast transformation. The E. coli-Streptomyces shuttle vector pWHM3 (39) was used as a high-copy-number vector (approximately 100 copies per chromosome) in S. coelicolor. Plasmid pWHM3-E is a derivative of pWHM3 harboring the 300-bp EcoRI-BamHI fragment containing the ermE promoter (PermE) (3) in pWHM3. Standard procedures were used to isolate plasmid DNA from E. coli (37), and to isolate plasmid and total DNAs from actinomycetes (20).
PCR conditions.
PCRs were performed in a minicycler (MJ Research, Watertown, Mass.) using Pfu polymerase (Stratagene, La Jolla, Calif.) and the buffer provided by the supplier, in the presence of 5% (vol/vol) dimethyl sulfoxide and 200 μM deoxynucleoside triphosphate (dNTP). No additional Mg2+ was added to the reaction mixture. The following PCR program was used for 30 cycles: 45 s of melting at 94°C, 1 min of annealing at 54°C, and 90 s of extension at 72°C. The reaction was completed by an additional 10-min incubation at 72°C. Oligonucleotides used for PCR are shown in Table 1.
TABLE 1.
Oligonucleotides used in PCR
| Primer | Sequencea | Positionb (nt) |
|---|---|---|
| ssg1 | 5′ GGCGAATTCGAACAGCTACGTGGCGAAGTCGCCA 3′ EcoRI | −194 to −170 |
| ssg2 | 5′ GTGGGATCCGTGCTCGCGGCGCTGGTCGTCTC 3′ BamHI | +539 to +517 |
| ssg3 | 5′ GGGAATTCCATATGCGCGAGTCGGTTCAAGCA 3′ EcoRI NdeI | −30 to −10 |
| ssgN3 | 5′ GGGAATTCCATATGATGACGTTCCTCGTCTCCG 3′ EcoRI NdeI | +1 to +22 |
| ssg4 | 5′ GTGGATCCCCGGTCAGCCGGCGTTCTG 3′ BamHI | +412 to +394 |
| Q1 | 5′ CTGAATTCTAGCATCGAGGGCAGGACATCA 3′ EcoRI | −1450 to −1427 |
| Q6 | 5′ CTGAAGCTTAACGACCGGCCCAGGTGGCGAC 3′ HindIII | +520 to +541 |
| Q10 | 5′ CTGAATTCGGCTCAGCGGTTGCAGAACGAG 3′ EcoRI | −196 to −175 |
| Q11 | 5′ ATAGGGATCCCGGGTCTCGTAGCGCAGC 3′ BamHI | +75 to +48 |
| Q14 | 5′ CTGGATCCTGGTGCCGCTGGGGCAGGAGG 3′ BamHI | +310 to +330 |
| Q15 | 5′ CTGAAGCTTGAGACGGGTTACGGCCACGATGC 3′ HindIII | +1870 to +1850 |
Underlined nucleotides indicate nonhomologous sequences added to create restriction sites (in italics) at the ends of the PCR fragments.
Location with respect to the first ucleotide (+1) of the ATG translational start codon of S. griseus ssgA of S. coelicolor ssgA for oligonucleotides designated “ssg” or “Q”, respectively.
Construction of the ssgA deletion mutant.
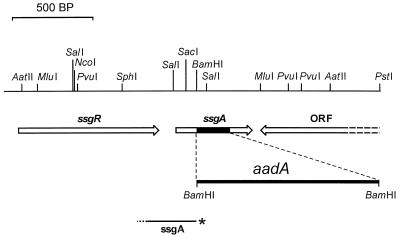
Two DNA fragments of approximately 1.5 kb were amplified from the S. coelicolor M145 chromosome by PCR with oligonucleotides Q1 plus Q11 and Q14 plus Q15 (Table 1). Digestion of these PCR fragments with the appropriate enzymes resulted in an EcoRI-BamHI fragment and a BamHI-HindIII fragment, encompassing nucleotides (nt) −1450 to +75 and +310 to +1870, respectively, relative to the translational start (+1) of ssgA. These fragments were ligated together into EcoRI-HindIII-digested pUC18. Subsequently, the aadA gene, conferring resistance to spectinomycin and streptomycin (34), was inserted into the BamHI site in the plasmid-borne and truncated ssgA, and the aacC4 gene, conferring apramycin resistance (7), was inserted into the HindIII site, resulting in the disruption construct pΔssgA. As a result, the construct has the +75–+310 region of ssgA replaced by aadA. Apramycin resistance, the selectable marker for the plasmid, is present after integration of the plasmid in the chromosome but should be lost after a second mutational crossover event. Therefore, after transformation of the plasmid to S. coelicolor M145 and a double-crossover event between pΔssgA and the chromosome, the desired mutant is expected to be resistant to spectinomycin and streptomycin and sensitive to apramycin.
Constructs for the expression of ssgA.
A 750-bp DNA fragment containing the S. griseus ssgA gene (accession no. D50051) was amplified from the S. griseus chromosome by PCR, using primers ssg1 and ssg2 (Table 1). The PCR fragment was cloned as an EcoRI-BamHI fragment in pIJ2925, giving pGWS1. The insert of pGWS1 was cloned behind PermE in pWHM3-E, and the PermE-ssgA cassette was transferred to pSET152, resulting in pGWS4 (Table 2). From earlier work we know that the ribosome binding site of S. ramocissimus tuf1 is efficiently recognized by ribosomes and hence typically results in high expression (40). We therefore replaced the upstream region of S. griseus ssgA in some of the constructs by that of S. ramocissimus tuf1, to allow higher expression of the gene. To achieve this, a 560-bp fragment was amplified by PCR using oligonucleotides ssgN3 and ssg2 and cloned as an EcoRI-HindIII fragment into pIJ2925, giving pGWS5 (Table 2). The ssgA insert of pGWS5 was then cloned as an NdeI-BglII fragment into EcoRI-BamHI-digested pUSRT3-3 containing the tuf1 ribosome binding site (40) after filling in the 5′ protruding ends of the NdeI and EcoRI sites, using the Klenow fragment of DNA polymerase I and dNTPs according to standard procedures (37). The resulting clone was designated pGWS1-SD. From this clone, the derivative pGWS4-SD was made similarly as described for pGWS1 and pGWS4 (Table 2).
TABLE 2.
Plasmids and strains used in this study
| Plasmid or strain | Description | Reference |
|---|---|---|
| Plasmids | ||
| pUSRT3-3 | Derivative of pUC18, harboring S. ramocissimus tuf3 behind the S. ramocissimus tuf1 ribosome binding site | 40 |
| pWHM3-E | pWHM3 with the constitutive PermE | This study |
| pGWS1 | pIJ2925 containing the 750-bp S. griseus ssgA PCR (ssg1-ssg2) product | This study |
| pGWS1-SD | pGWS1 with the upsream region of ssgA replaced by nt −1 to −70 of S. ramocissimus tuf1 | This study |
| pGWS4 | pSET152 containing ssgA behind PermE | This study |
| pGWS4-SD | pGWS4 with the upstream region of ssgA replaced by nt −1 to −70 of S. ramocissimus tuf1 | This study |
| pGWS5 | pIJ2925 with 560-bp S. griseus ssgA PCR (ssgN3-ssg2) product | This study |
| pGWS6 | pIJ2925 with 280-bp PCR (Q10-Q6) product of S. coelicolor ssgA | This study |
| pGWS7 | S. coelicolor ssgA behind PermEin pSET152 | This study |
| pGWS7-SD | pGWS7 with the upstream region of ssgA replaced by nt −1 to −70 of S. ramocissimus tuf1 | This study |
| pΔssgA | Construct for disruption of S. coelicolor ssgA | This study |
| Streptomyces strains | ||
| M145 | Wild-type S. coelicolor A3(2) | 20 |
| GSA2 | M145 harboring pGWS4-SD | This study |
| GSA3 | ssgA knockout mutant of M145 | This study |
| GSA4 | GSA3 with pGWS7 integrated into the chromosome | This study |
| B2682 | Wild-type S. griseus NRRL B2682 | |
| SY1 | Mutant of S. griseus B2682 that sporulates in rich liquid media | 23 |
For homologous expression of ssgA in S. coelicolor and for complementation of the ssgA null mutant, we amplified the ssgA gene from the S. coelicolor M145 chromosome by PCR with oligonucleotides Q10 and Q6, designed to encompass nt −196 to −175 and +520 to +541, respectively, relative to the start of the gene. The PCR fragment was cloned in pIJ2925 or behind PermE in pWHM3-E, giving pGWS6 or pGWS7, respectively.
Western analysis of SsgA.
Protein extracts were prepared by ultrasonication of the mycelium on ice, at 30 W for 300 s in standard buffer (10 mM Tris-HCl [pH 7.6], 60 mM NH4Cl, 10 mM magnesium acetate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Samples were then centrifuged at 30,000 × g for 30 min. The resulting S30 protein extract (supernatant) was submitted to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. In all lanes, approximately 5 μg of protein was loaded. Gels were either stained with Coomassie brilliant blue or blotted onto Hybond-C Super nylon membranes (Amersham) and immunostained with antibodies raised against SsgA (25).
Nuclease S1 protection assays.
RNA was purified as described by Hopwood et al. (20), except that DNase I treatment was used in addition to salt precipitation to eliminate DNA from the nucleic acid preparations. The concentration and the integrity of the RNA were checked by spectrophotometry and by gel electrophoresis. For each nuclease S1 protection assay, about 0.02 pmol (approximately 104 Cerenkov cpm) of labeled probe was hybridized to 30 μg of RNA in NaTCA buffer (32) at 45°C overnight after denaturation at 70°C for 15 min. All subsequent steps were carried out as described previously (38), using an excess of probe. Experiments were carried out twice on independently isolated RNA. The 330-bp ssgA probe (Fig. 1) for mapping ssgA transcripts was generated by PCR amplification using the universal primer (17-mer) and 32P-end-labeled Q11 and with pGWS6 as the template. The probe contains an approximately 50-nt nonhomologous extension at the 3′ end, to allow discrimination between DNA-RNA hybrids and reannealed probe.
FIG. 1.
Restriction map of S. coelicolor ssgA and flanking regions and representation of the ssgA disruption mutant. The dark area in the ssgA gene is replaced by aadA (conferring resistance to spectinomycin and streptomycin; presented as a thick line and not exactly to scale) in the ssgA knockout mutant GSA3. The probe (ssgA) used in nuclease S1 mapping experiments is shown; the asterisk represents the 32P-labeled 5′ end, and the dotted part represents an approximately 50-nt nonhomologous extension at the 3′ end, all drawn to scale. ORF, gene encoding a putative membrane protein (accession no. Q9X9U1); ssgR, gene encoding an IclR-like regulatory protein (accession no. Q9X9U3).
Phase-contrast microscopy.
Cultures were examined by light microscopy using a Zeiss standard 25 phase-contrast microscope. For photography, we used a Zeiss MC80 camera.
EM.
Samples of M145 and GSA2 for EM were prepared as follows. Mycelium was washed in phosphate-buffered saline, centrifuged at 2,300 rpm for 1 min, and resuspended in a fixative containing 1.5% (wt/vol) glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4; 360 mosmol) at room temperature for 20 h. Mycelium was pelleted, rinsed twice in phosphate-buffered saline, and postfixed in 1% (wt/vol) osmium tetroxide in Millonig phosphate buffer (pH 7.3; 330 mosmol) at room temperature for 20 h. After rinsing, the samples were resuspended in 2% Bacto Agar at 60°C, centrifuged at 11,000 rpm for 2 min, cut in 1-mm3 blocks, and dehydrated in a graded series of ethanol. After incubation in a graded series of epoxy resin LX-112 (Ladd Research Industries, Burlington, Vt.) in propylene oxide, the blocks were placed in capsules filled with epoxy resin and polymerized at 60°C for 72 h. Ultrathin sections (70 nm) were cut on an ultramicrotome (Reichert OM U3), collected on copper grids, stained with uranyl acetate and lead hydroxide, and examined in a Philips EM410 transmission electron microscope.
Computer analysis.
The BLAST search engines BLASTN, BLASTP, and BLASTX (1) were used to perform database searches, and the Wisconsin Package (13) was used for DNA and protein sequence alignments. Figure 2 was produced using the Boxshade program (www.ch.embnet.org/software/box_form.html).
FIG. 2.
Alignment of amino acid sequences of SsgA and SsgA-like proteins of S. griseus and S. coelicolor. SsgAsco and SsgAsgr refer to the SsgA homologues from S. coelicolor and S. griseus, respectively. 5F2a, 5H1, E19A, and L2 refer to the cosmids on which the four SsgA-like proteins of S. coelicolor were located. The protein accession numbers are CAB40672, CAB42965, CAB51005, and CAB70943, respectively. Amino acid identities/similarities to S. coelicolor SsgA are as follows: SsgAsgr, 78/82%; L2, 45/52%; E19a, 42/50%; 5H1, 33/39%; 5F2a, 32/38%.
Nucleotide sequence accession numbers.
The sequences shown in Fig. 1 have been assigned GenBank accession no. Q9X9U1 and Q9X9U3.
RESULTS
Genomic organization around S. coelicolor ssgA.
The sequence of S. griseus ssgA was published previously (accession no. D50051). Extensive recent searches of both the translated nucleotide and protein databases using the BLAST search engines BLASTX and BLASTP identified one ssgA homologue, which is located on cosmid Q11 (accession no. AL096823) of the S. coelicolor cosmid library (35), with the predicted gene product showing 78% amino acid identity to S. griseus SsgA. The organization around S. coelicolor ssgA is shown in Fig. 1. Upstream of ssgA lies a gene encoding an IclR regulator-like protein (241 amino acids), with significant end-to-end homology to several regulatory proteins, for example, 28% amino acid identity (38% similarity) to S. coelicolor GylR (accession no. P15360) and Acinetobacter calcoaceticus PobR (accession no. Q43992). Experiments described below provide evidence that transcription of ssgA is linked to this upstream ORF, and we therefore tentatively call the gene ssgR. Downstream of ssgA lies a gene encoding a putative membrane protein (513 amino acids), positioned in the opposite direction.
The S. coelicolor genome sequencing project (www.sanger.ac.uk/projects/S_coelicolor) revealed several more ORFs with relevant homology to ssgA (see the legend to Fig. 2 for accession numbers), with their predicted gene products showing between 30 and 45% amino acid identity to S. griseus SsgA. The functions of these ORFs, which, like ssgA itself, have no relevant homology to any other gene in the databases, are unknown.
An alignment of the SsgA homologues of S. griseus and S. coelicolor and the four SsgA-like proteins identified on the S. coelicolor genome is shown in Fig. 2.
S. griseus SY1 is not an ssgA mutant.
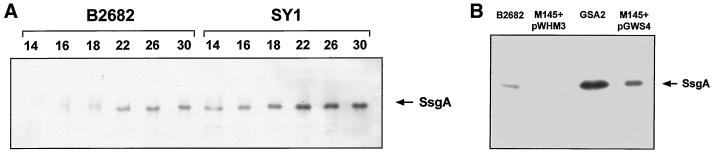
A mutant of S. griseus NRRL B2682, designated SY1, is able to produce submerged spores not only in minimal media but also in rich media (23). This phenotype could be suppressed by the introduction of a DNA fragment harboring ssgA. While introduction of plasmid-borne ssgA did not restore the wild-type phenotype to this mutant, we sought to ascertain that the aberrant phenotype of SY1 was not due to a mutation in the chromosomal ssgA. Therefore, we cloned the ssgA gene from SY1 by PCR with oligonucleotides ssg1 and ssg2 (Table 1) and determined its sequence. The DNA sequence was shown to be identical to that of the wild-type gene. We also analyzed the expression of ssgA in both wild-type S. griseus NRRL B2682 and its mutant SY1 by Western analysis with antibodies raised against SsgA (25). In solid-grown cultures (MM or R2YE) or in liquid minimal medium cultures, conditions that allow either strain to sporulate, SsgA protein levels increased with the culture age, reaching a peak at a time correlated to sporulation (25). In contrast to the wild-type strain, S. griseus SY1 also sporulates in rich liquid media. Under these conditions, SsgA was produced abundantly in SY1 but at significantly lower levels in B2682, although the two strains are similar in growth phase dependence of ssgA (Fig. 3A). Apparently, a certainly threshold level of SsgA is required to stimulate submerged sporulation. These data show that the mutation in SY1 does not lie in the ssgA gene, although expression of the gene is increased in rich liquid cultures, allowing S. griseus to sporulate under these conditions.
FIG. 3.
Western analysis of SsgA expression. (A) Expression of ssgA in S. griseus B2682 and S. griseus SY1 during normal growth in rich medium (TSBS). Samples were taken 14, 16, 18, 20, 26, and 30 h after inoculation of the cultures. Under these conditions, spores emerged in SY1 cultures after 18 h; B2682 failed to produce submerged spores in TSBS. (B) Production of SsgA in S. griseus B2682 and S. coelicolor M145 harboring pWHM3 (control plasmid), GSA2 (high-level expression), and pGWS4 (low-level expression) during growth in TSBS (mycelium was harvested 30 h after inoculation).
These data prompted the creation of an ssgA null mutant. Since S. coelicolor is genetically by far the best-characterized Streptomyces strain, with the genome sequencing project almost completed, we decided to analyze the role of ssgA in this strain.
Construction of an ssgA null mutant of S. coelicolor.
To establish the role of ssgA in the development of S. coelicolor, we created an ssgA null mutant of this species. For this purpose, nt +75 to +310 of ssgA (relative to the start of the gene) were replaced by the aadA gene, conferring resistance to spectinomycin and streptomycin. The construct is shown in Fig. 1. S. coelicolor M145 was transformed with the disruption construct pΔssgA (see Materials and Methods), and colonies resistant to spectinomycin and streptomycin were selected. Three of these did not show resistance to apramycin (the marker for the plasmid), indicative of the absence of the plasmid due to a second crossover event. Since aadA was apparently still present, these colonies were considered to be possible ssgA disruption mutants. The putative ssgA mutants were screened by three Southern hybridizations, one with a probe recognizing the aadA gene, one with a probe recognizing ssgA, and one with a probe confirming the absence of aacC4 (hybridization data not shown). All three had the +75–+310 part of ssgA replaced by the aadA gene and lacked the aacC4 gene, showing that they were indeed true ssgA disruption mutants. Since the ORF located downstream of ssgA, which encodes a putative membrane protein, is oriented in the opposite direction, effects of the ssgA disruption on its expression are unlikely. One of the ssgA knockout mutants was selected and was designated GSA3.
ssgA is essential for sporulation of S. coelicolor M145.
The S. coelicolor ssgA mutant GSA3 was plated on several solid media, including R2YE, SFM, and MM with mannitol as the carbon source. On R2YE, SFM, and MM, aerial mycelium was produced normally, but the hyphae failed to produce spores within 2 weeks (Fig. 4A). After prolonged incubation (more than 2 weeks), colonies remained white on R2YE plates; some spores were produced on SFM and MM with mannitol as the carbon source, although titers were low in comparison to the parental strain M145. Interestingly, antibiotic production was disturbed in GSA3, with almost complete absence of the blue-pigmented actinorhodin.
FIG. 4.
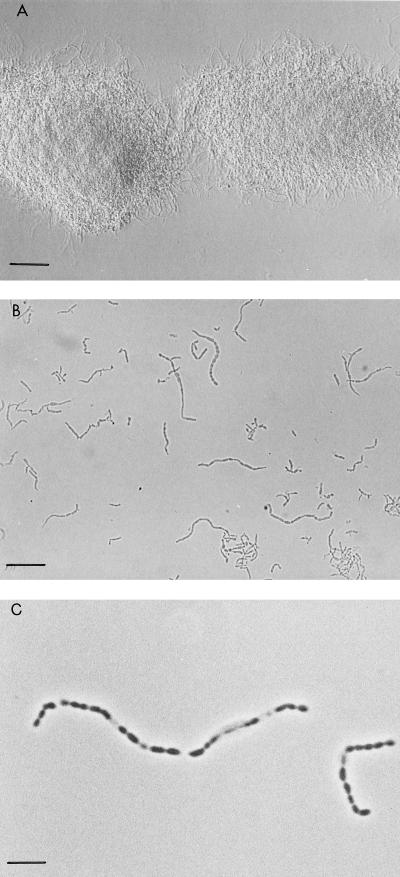
(A) Phenotype of the ssgA null mutant of S. coelicolor M145 on SFM medium. GSA3 is the ssgA mutant of M145; GSA4 is the ssgA mutant complemented with pGWS7. The white appearance of GSA3 is due to the failure of the mutant to produce the grey-pigmented spores. GSA4 consistently produces more spores than M145. (B and C) Impression prints of the surfaces of solid-grown cultures of S. coelicolor M145 (B) and its ssgA null mutant GSA3 (C). While M145 sporulates abundantly, aerial hyphae of GSA3 show typical coiling, but no spores (bars = 10 μm).
To verify that the phenotypes described above were solely due to the deletion of ssgA, we complemented mutant strain GSA3 by transformation with plasmid pGWS7, an integrative vector harboring ssgA behind PermE. This plasmid allows constitutive expression of ssgA in S. coelicolor. One of several integrants with the same phenotype was selected and designated GSA4. As shown in Fig. 4A, sporulation is fully restored to the complemented strain, showing that the Whi phenotype of GSA3 was indeed due to the deletion of the ssgA knockout. In GSA4, production of actinorhodin was also restored to levels similar to those in the wild-type strain, indicating that disturbance of the regulation of antibiotic biosynthesis could also be ascribed to the ssgA deletion. Interestingly, GSA4 consistently produced more spores than M145, as judged by the denser layer of spores (determined by microscopy) and the increased brown-grey pigmentation of the colonies. This apparent increase in sporulation is most likely due to the constitutive and enhanced expression of ssgA in the complemented mutant (Fig. 3B). Finally, GSA4 shows increased fragmentation in submerged culture, which again can be ascribed to the increased expression of ssgA in this strain.
The sporulation mutants are categorized in various classes, depending on the stage in which they are blocked in their development; while some are blocked in an earlier phase, some have differentiated so far that coiling of their tips is normal, and chains of immature spores are produced (9, 14). To analyze the developmental state of the aerial hyphae, we made impressions of the top layer of 7-day-old surface-grown cultures of S. coelicolor M145 and GSA3 by gently pressing a moist coverslip onto the surface and analyzed these by phase-contrast microscopy. While the wild-type strain produced large amounts of spores after 7 days (Fig. 4B), the ssgA mutant GSA3 showed typical coiling of the tips of the aerial hyphae but failed to sporulate (Fig. 4C). This indicates that the arrest occurs in a later stage of aerial hyphae development, as early whi mutants fail to coil. We plan to compare the phenotype of the ssgA mutant to that of other whi mutants by scanning EM, to determine what mutant class it falls into.
ssgA and ssgR are transcriptionally linked.
To analyze the transcription of ssgA in S. coelicolor and establish whether ssgA is transcribed from a promoter directly upstream of the gene, we performed nuclease S1 mapping experiments on RNA isolated from liquid cultures of S. coelicolor M145, using the 330-bp ssgA probe resulting from PCR with 32P-end-labeled oligonucleotide Q11 and the universal primer on pSCF6 DNA. The generated probe harbors 280 nt homologous to the ssgA locus and 50 nt of nonhomologous extension at the 3′ end to discriminate between DNA-RNA hybrids and reannealing of the probe. The location of the probe is shown in Fig. 1.
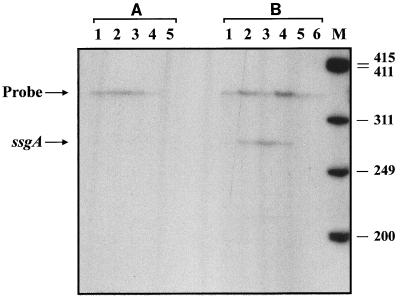
We failed to identify transcripts when RNA isolated from cultures grown as a so-called normal growth curve in liquid MM was used (Fig. 5A). Since the Whi phenotype of GSA3 strongly suggests that ssgA is involved in sporulation (Fig. 4), and the protein appears shortly after nutrient depletion in liquid cultures of S. griseus (25), we subjected S. coelicolor to nutritional shift-down conditions. The burst of ppGpp production following shift-down is generally regarded as an important signal for the onset of morphological differentiation (2) and elicits submerged sporulation in S. griseus (27). Cultures of S. coelicolor M145 were allowed to grow in TSBS to an OD550 of 0.7, washed in MM, and transferred to MM with mannitol as the carbon source. Cultures were incubated at 30°C, and RNA was isolated after 0, 15, 30, 60, 120, and 240 min. The RNA was analyzed by nuclease S1 mapping, again using the ssgA probe.
FIG. 5.
Transcriptional analysis of ssgA. (A) Analysis of samples taken from normal growth curve in MM. Lanes 1 to 5 refer to samples taken at an OD550 of 0.3 (early log), 0.6 (mid-log), 0.9 (late log), 1.2 (transition phase), and 1.5 (stationary phase), respectively. (B) Analysis of samples taken after nutritional shift-down. Lanes 1 to 6 refer to samples taken 0, 15, 30, 60, 120, and 240 min after shift-down, respectively. Lane M, 32P-end-labeled HaeIII-digested ϕX174 size markers; numbers on the right refer to sizes of the marker bands (in nucleotides). Positions of the ssgA transcript and reannealed probe are shown on the left.
While at the time just before shift-down no transcript could be identified, within 15 min afterward a band of approximately 280 nt appeared, corresponding to full-length protection of the probe (Fig. 5B). This strongly suggests that transcription of ssgA is initiated from a promoter either inside or upstream of ssgR. A linkage of transcription of ssgA and ssgR indicates that the regulatory gene ssgR may also play a role in the ssgA-mediated regulation of cell division. The ssgRA transcript levels reached a peak at around 30 min after shift-down; 2 h after shift-down, no transcripts could be detected.
High-level overexpression of ssgA in S. coelicolor.
The Whi phenotype of the ssgA null mutant (Fig. 4) and the enhanced fragmentation of Streptomyces strains harboring multiple copies of ssgA (24) both suggest possible involvement of SsgA in cell division, prompting analysis of the cytological effects of high-level overexpression of ssgA in S. coelicolor.
To optimize expression of ssgA, we fused the coding region to the ribosome binding site of S. ramocissimus tuf1 and placed it under the control of PermE. Cloning into the integrative vector pSET152 resulted in pGWS4-SD. Since S. coelicolor ssgA was not discovered until recently, we performed most of our expression studies with the S. griseus homologue, which has been available for several years. We recently also created plasmid pGWS7-SD, which contains S. coelicolor ssgA but is otherwise identical to pGWS4-SD, and confirmed that results obtained with either homologue were indistinguishable. The expression construct pGWS4-SD was introduced into S. coelicolor M145, and integrants were checked for the presence of ssgA by PCR. All transformants were correct integrants and showed the same phenotype. One was selected and designated GSA2.
SsgA protein levels in GSA2 were compared to those in the wild-type strain by Western analysis, using polyclonal antibodies raised against SsgA (see Materials and Methods). Both strains were grown at 30°C in liquid TSBS medium, and S30 extracts were prepared from mycelium grown until early exponential, mid-exponential, and late exponential phases (OD550 0.4, 0.8, and 1.2, respectively). We failed to detect a band corresponding to SsgA in extracts from S. coelicolor M145, while a strong band with an apparent molecular mass of approximately 16 kDa was observed for all growth phases of liquid-grown cultures of GSA2 integrants, indicative of significant overproduction of SsgA in this strain (Fig. 3B).
SsgA inhibits branching and induces sporulation in liquid cultures of S. coelicolor.
The morphologies of the S. coelicolor ssgA null mutant and its parental strain M145 were compared by phase-contrast microscopy to those of S. coelicolor transformants overexpressing ssgA. Wild-type (i.e., harboring a single copy of ssgA) S. coelicolor produced large mycelial lumps in rich and minimal liquid cultures. The ssgA knockout mutant GSA3 formed even denser mycelial clumps, indicative of the formation of highly branched mycelial networks in the absence of ssgA. In contrast, hyphae of S. coelicolor containing a plasmid expressing S. coelicolor ssgA from P-ermE (pGWS4, giving increased expression of ssgA [Fig. 3B]) showed strongly reduced branching in complex and minimal medium cultures. This resulted in clearly less dense mycelial lumps, in which the (often long) individual hyphae can easily be discerned. Furthermore, small fragments appeared approximately 16 h after inoculation, and fragmentation increased over time. Not only did liquid-grown cultures of GSA2, expressing ssgA at a high level (Fig. 3B), show fragmented growth and strongly reduced branching, but the hyphae displayed a strangely swollen appearance, comparable to the tips of S. griseus hyphae at the time of sporulation in submerged culture (Fig. 6B). Surprisingly, chains of spore-like bodies were often observed in GSA2 cultures, most likely as the result of the SsgA-induced increase in the frequency of septation (see below and Fig. 7C). While these compartments normally did not show physical separation, we sometimes observed hyphae that had developed into chains of immature spores. These were highly irregular, and many of the spore-like compartments appeared to be empty (Fig. 6C). The abnormal length of these submerged spore chains, comprising sometimes more than 20 compartments, suggests that separation of the spores had not been completed.
FIG. 6.
Phase-contrast micrographs of liquid-grown cultures of S. coelicolor M145 (wild type) and GSA2 (overexpressing ssgA). Cultures were grown for 24 h in TSBS. (A) S. coelicolor M145 forming a typical lump-like mycelial network (bar = 50 μm). (B) S. coelicolor GSA2. The mycelium is strongly fragmented, with hyphae that are significantly wider than in the parental S. coelicolor M145. Clearly visible are the spore-like bodies that are produced by this strain (bar = 25 μm). (C) Closeup of immature spore chains of S. coelicolor GSA2. Note that the spores have irregular shapes, and several appear to be empty (bar = 5 μm).
FIG. 7.
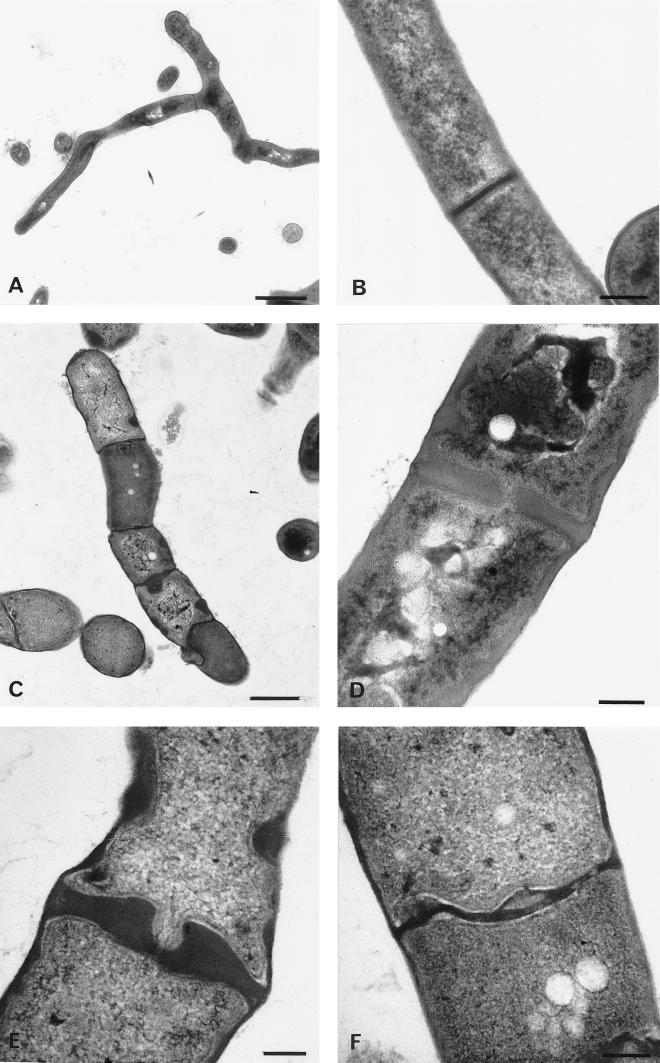
Transmission electron micrographs of S. coelicolor M145 (harboring control plasmid pSET152) and of S. coelicolor GSA2. Cultures were grown for 40 h in liquid TSBS medium. (A) Image showing wild-type vegetative hyphae with cross walls (bar = 1.0 μm). (B) High magnification of wild-type cross wall (bar = 0.2 μm). (C) Image of GSA2 showing the effect of SsgA overexpression on the frequency of septum formation and hyphal morphology (bar = 1.0 μm). (D to F) High magnification of abnormally shaped septa in GSA2 hyphae (bar = 0.2 μm).
Analysis of S. coelicolor GSA2 by TEM reveals increased septation and ectopic deposits of cell wall material.
To assess the effect of ssgA overexpression on mycelial morphology in more detail, we analyzed the hyphae of liquid-grown cultures of GSA2 by transmission EM (TEM), with M145 harboring pSET152 as the control. The wild-type strain showed normal morphology, with few thin cross walls (Fig. 7A and B). Conversely, as was also observed by phase-contrast microscopy (Fig. 6B and C), GSA2 displayed an extremely high frequency of septation, forming small compartments separated by cross-wall-like structures sometimes more than 10 times thicker than wild-type cross walls (Fig. 7C to F). Furthermore, the compartments are oddly misshapen, resulting in hyphae that resemble a string of spores rather than the typical filaments with occasional cross walls. While in S. coelicolor M145 hyphae cross walls appear with a frequency of approximately one per 8 μm (as determined by TEM), hyphae of the ssgA-overexpressing strain GSA2 produce small compartments that are misshapen, with many having a size comparable to that of spores (approximately 1 μm). Lumps of cell wall material are often deposited at opposite sides of the hyphal walls, perhaps indicative of unfinished septum formation. Another striking feature is the clearly increased thickness of the hyphae, which are 1.5 to 3 times wider than wild-type hyphae, as was confirmed by light microscopy.
DISCUSSION
The data presented here show that the putative cell division gene ssgA is a novel whi gene and is essential for the proper sporulation of S. coelicolor A3(2). On solid media, the white, fluffy aerial mycelium is formed abundantly by the ssgA null mutant, as in the wild-type strain. However, spores were produced only on particular media after prolonged incubation, and at a low level. Such a conditional phenotype is typical of several bld mutants, several of which sporulate on minimal media containing mannitol as the carbon source, but is to our knowledge the first example of a conditional whi mutant. The coiling of the aerial hyphae shows that the ssgA mutant does not fall in the category of the early whi mutants. However, a more thorough analysis of the mutant, and particularly comparison to well-characterized mutants (9, 33), is required to further determine its developmental state.
Another interesting phenotype of the ssgA mutant is that actinorhodin production is strongly reduced, while undecylprodigiosin production is at least as strong as in the wild-type strain. Since actinorhodin is produced later in development than undecylprodigiosin (2), this phenotype may be the result of an arrest in development at a time where actinorhodin production has not yet been initiated.
Transcription of ssgA could not be detected during normal growth in liquid MM or TSBS but was induced by nutritional shift-down, which is known to elicit the so-called stringent response and antibiotic production in streptomycetes, and in particular submerged sporulation in S. griseus. Interestingly, ssgA was not transcribed from a promoter directly upstream of the gene under the chosen conditions but linked to that of ssgR, a member of the family of iclR-type transcriptional regulator genes. We therefore speculate that ssgR itself, like ssgA, may also be involved in the regulation of cell division. We are currently working on the creation of a knockout mutant of ssgR to provide more insight into its role in the Streptomyces life cycle.
The gene dosage of ssgA plays a decisive role in determining the hyphal morphology in submerged cultures of S. coelicolor. While the ssgA null mutant GSA3 produces extremely dense disk-like mycelia, increased expression of ssgA results in restricted branching of the hyphae, with the branches often reduced to tiny bulges, similar to the so-called sporulation branches previously described for S. griseus (18). Interestingly, S. coelicolor GSA2, which is optimized for the stable overexpression of ssgA, produces very small mycelial fragments and even chains of immature spores. Stimulation of sporulation was also observed when the developmental sigma factor WhiG was overproduced in S. coelicolor, although the level and frequency of spore formation were lower in that case (11). This suggests that the machinery required for submerged sporulation is present in S. coelicolor. However, the occurrence of many empty compartments and the impaired spore separation indicate that other conditions need to be met to produce mature submerged spores. The observation that SsgA triggers submerged sporulation seems in conflict with earlier experiments in S. griseus, which showed that an increased gene dosage of ssgA inhibits submerged sporulation in S. griseus SY1, a strain which normally sporulates in rich and minimal liquid media. However, since the genotype of the latter mutation could not be ascertained, it is difficult to speculate on a fitting explanation.
Cross walls are often believed to provide extra stability to the hyphae, like the steps of a ladder. However, the opposite may be true. For example, the ftsZ null mutant produces no cross walls at all, but long hyphae are still formed in liquid culture, without any obvious sign of excessive breakage of the hyphae (29). Furthermore, strains that produce cross walls at higher frequency lyse rapidly in the presence of lysozyme and show increased sensitivity to high sucrose and/or glycine concentrations (e.g., several strains that sporulate in liquid culture but also S. coelicolor GSA2 [G. P. van Wezel, unpublished results]). Thus, the increased frequency of septation might explain the enhanced fragmentation of strains overexpressing ssgA.
Analysis of GSA2 by TEM shows how dramatic the effect of SsgA overproduction is on the frequency and morphology of the septa. While in S. coelicolor M145 hyphae cross walls appear at low frequency (one per 8 μm), hyphae of GSA2 produce compartments with a size comparable to that of spores (approximately 1 μm). There is a distinct difference between vegetative cross walls and sporulation septa; cross walls form semipermeable boundaries between different compartments, while sporulation septa lead to actual cell division (41). Considering the hyperfragmenting phenotype of strains overexpressing ssgA and the inhibition of sporulation on solid media in the ssgA null mutant, it is likely that SsgA is involved in the formation of sporulation septa. Furthermore, overproduction of SsgA (in GSA2) stimulates the formation of lumps of cell wall material at opposite sides of the hyphal walls, which might reflect the initiation of spore septation. Other observations linking ssgA to sporulation are the significant widening of the hyphae in this strain, resulting in a diameter similar to that of spores, and the emergence of immature spore chains in liquid-grown cultures of S. coelicolor GSA2.
The molecular mode of action of SsgA is still unclear. The absence of an apparent DNA binding motif in the protein suggests that SsgA does not function as a transcriptional regulator. An intriguing possibility is that SsgA might function by directly interacting with one or more proteins belonging to the cell division machinery. The availability of purified SsgA protein will allow the search for putative interaction partners for SsgA; these experiments are in progress.
In conclusion, the gene dosage of ssgA has a dramatic effect on Streptomyces hyphal morphology and particularly on sporulation. While an ssgA knockout mutant fails to sporulate on solid media, overexpression of the gene results in strongly increased septum formation and production of spore-like bodies by clearly widened hyphae in submerged culture, indicative of pleiotropic changes in the mycelial buildup. The regulation of ssgA expression as a tool for modifying the hyphal morphology may be of great value for applications in biotechnological fermentations.
ACKNOWLEDGMENTS
We are grateful to E. Taal and C. Rousseau for excellent technical assistance, to G. Hoogvliet for providing S. coelicolor shift-down RNA samples, to K. Findlay for advise on electron microscopic techniques, and to E. Vijgenboom and K. F. Chater for discussions.
This work was supported by a grant from the Netherlands Research Council for Chemical Sciences (CW), with financial aid from the Netherlands Technology Foundation (STW).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibb M J. The regulation of antibiotic production in Streptomyces coelicolorA3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M J, White J, Ward J M, Janssen G R. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraeais translated in the absence of a conventional ribosome-binding site. Mol Microbiol. 1994;14:533–545. doi: 10.1111/j.1365-2958.1994.tb02187.x. [DOI] [PubMed] [Google Scholar]
- 4.Bierman M, Logan R, Obrien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomycesspp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 5.Biro S, Bekesi I, Vitalis S, Szabo G. A substance effecting differentiation in Streptomyces griseus. Eur J Microbiol. 1980;103:359–363. doi: 10.1111/j.1432-1033.1980.tb04322.x. [DOI] [PubMed] [Google Scholar]
- 6.Birko Z, Sumegi A, Vinnai A, van Wezel G P, Szeszak F, Vitalis S, Szabo P T, Kele Z, Janaki T, Biro S. Characterization of the gene for factor C, an extracellular signal protein involved in morphological differentiation of Streptomyces griseus. Microbiology. 1999;145:2245–2253. doi: 10.1099/00221287-145-9-2245. [DOI] [PubMed] [Google Scholar]
- 7.Blondelet-Rouault M H, Weiser J, Lebrihi A, Branny P, Pernodet J L. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene. 1997;190:315–317. doi: 10.1016/s0378-1119(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 8.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 9.Chater K F. Taking a genetic scalpel to the Streptomycescolony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 10.Chater K F, Losick R. The mycelial life-style of Streptomyces coelicolor A3(2) and its relatives. In: Shapiro J H, Dworkin M, editors. Bacteria as multicellular organisms. New York, N.Y: Oxford University Press; 1997. pp. 149–182. [Google Scholar]
- 11.Chater K F, Bruton C J, Plaskitt K A, Buttner M J, Mendez C, Hellman J D. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell. 1989;59:133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- 12.Daza A, Martin J F, Dominguez A, Gil J A. Sporulation of several species of Streptomycesafter nutritional downshift. J Gen Microbiol. 1989;135:2483–2491. doi: 10.1099/00221287-135-9-2483. [DOI] [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ensign J C. Physiological regulation of sporulation of Streptomyces griseus. In: Okami Y, Beppu T, Ogawara H, editors. Biology of actinomycetes 1988. Tokyo, Japan: Scientific Societies Press; 1988. pp. 308–315. [Google Scholar]
- 15.Flardh K, Findlay K C, Chater K F. Association of early sporulation genes with developmental decision points in Streptomyces coelicolorA3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 16.Floriano B, Bibb M J. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolorA3(2) Mol Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- 17.Glazebrook M A, Doull J L, Stuttard C, Vining L C. Sporulation of Streptomyces venezuelaein submerged cultures. J Gen Microbiol. 1990;136:581–588. doi: 10.1099/00221287-136-3-581. [DOI] [PubMed] [Google Scholar]
- 18.Hao J, Kendrick K E. Visualization of penicillin-binding proteins during sporulation of Streptomyces griseus. J Bacteriol. 1998;180:2125–2132. doi: 10.1128/jb.180.8.2125-2132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopwood D A. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology. 1999;145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- 20.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 21.Huber F M, Piper R L, Mertz F P. Sporulation of Streptomyces roseosporusin submerged culture. J Ind Microbiol. 1987;2:235–241. [Google Scholar]
- 22.Janssen G R, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia colicolonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 23.Kawamoto S, Ensign J C. Isolation of mutants of Streptomyces griseusthat sporulate in nutrient rich media: cloning of DNA fragments that suppress the mutations. Actinomycetologica. 1995;9:124–135. [Google Scholar]
- 24.Kawamoto S, Ensign J C. Cloning and characterization of a gene involved in regulation of sporulation and cell division in Streptomyces griseus. Actinomycetologica. 1995;9:136–151. [Google Scholar]
- 25.Kawamoto S, Watanabe H, Hesketh A, Ensign J C, Ochi K. Expression of the ssgA gene product, associated with sporulation and cell division in Streptomyces griseus. Microbiology. 1997;143:1077–1086. doi: 10.1099/00221287-143-4-1077. [DOI] [PubMed] [Google Scholar]
- 26.Kelemen G H, Buttner M J. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 27.Kendrick K, Ensign J C. Sporulation of Streptomyces griseusin submerged culture. J Bacteriol. 1983;155:357–366. doi: 10.1128/jb.155.1.357-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Analysis of Streptomyces avermitilisgenes required for avermectin biosynthesis utilising a novel integration vector. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 29.McCormick J R, Su E P, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 30.Messing J, Crea R, Seeburg P H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyadoh S. Research on antibiotic screening in Japan over the last decade: a producing microorganisms approach. Actinomycetologica. 1993;7:100–106. [Google Scholar]
- 32.Murray M G. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal Biochem. 1986;158:165–170. doi: 10.1016/0003-2697(86)90605-6. [DOI] [PubMed] [Google Scholar]
- 33.Potuckova L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. A new RNA polymerase sigma factor, sigma F, is required for the late stages of morphological differentiation in Streptomycesspp. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 34.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 35.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolorA3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 36.Ryding N J, Bibb M J, Molle V, Findlay K C, Chater K F, Buttner M J. New sporulation loci in Streptomyces coelicolorA3(2) J Bacteriol. 1999;181:5419–5425. doi: 10.1128/jb.181.17.5419-5425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Strauch E, Takano E, Baylis H A, Bibb M J. The stringent response in Streptomyces coelicolorA3(2) Mol Microbiol. 1991;5:289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 39.Vara J, Lewandowska-Skarbek M, Wang Y-G, Donadio S, Hutchinson C R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vijgenboom E, Woudt L P, Heinstra P W H, Rietveld K, van Haarlem J, van Wezel G P, Shochat S, Bosch L. Three tuf-like genes in the kirromycin producer Streptomyces ramocissimus. Microbiology. 1994;140:983–998. doi: 10.1099/00221287-140-4-983. [DOI] [PubMed] [Google Scholar]
- 41.Wildermuth H, Hopwood D A. Septation during sporulation in Streptomyces coelicolor. J Gen Microbiol. 1970;60:51–59. doi: 10.1099/00221287-60-1-51. [DOI] [PubMed] [Google Scholar]
- 42.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]