Abstract
Sertraline is a commonly used SSRI antidepressant drug, metabolized by CYP2C19 and CYP2B6, that exhibits a substantial interindividual variation in clinical response, of which only a part can be attributed to known genetic variants. In the current study we have examined the role of a newly discovered ultrarapid CYP2C:TG haplotype and CYP2B6 variants in order to identify the possible missing heritability for such variation in sertraline response in a large patient population (n = 840). Compared to the reference group (CYP2C19*1/*1, n = 160), sertraline exposure was increased by 128% in CYP2C19 PMs (n = 29, p < 0.001) but decreased by about 20% in CYP2C19 ultrarapid metabolizers (Ums) (homozygous carriers of CYP2C19*17 and/or CYP2C:TG haplotype) with the diplotypes CYP2C19*17/*17, CYP2C:TG/TG, or CYP2C19*17/CYP2C:TG (n = 135, p < 0.003, p = 0.022, p < 0.003, respectively). Interestingly, in patients carrying the increased function CYP2B6*4 allele, and also carrying the CYP2C19*17 and CYP2C:TG alleles (n = 10), sertraline exposure was 35.4% lower compared to the reference group, whereas in subjects being poor metabolizers (PM) in both the CYP2C19 and CYP2B6 gene, the sertraline concentrations were raised by 189%. In summary, the CYP2C19 variants including the CYP2C:TG haplotype had a significant impact on sertraline metabolism, as well as the CYP2B6*4, *6, and *9 alleles. Knowing the CYP2B6 and CYP2C19 genotype, including the CYP2C:TG haplotype status, can prospectively be useful to clinicians in making more appropriate sertraline dosing decisions
Study highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Sertraline is a commonly used antidepressant subjected to metabolism by multiple enzymes with CYP2C19 playing a key role along with CYP2B6 and CYP3A4. Previous pharmacogenetic studies have reported significant effects of CYP2C19 and CYP2B6 genotypes on sertraline concentration, but with inconsistent findings probably reflecting that the concurrent effects of the genotypes have not been investigated in a large patient population. Furthermore, the studies have not accounted for the recently discovered CYP2C:TG haplotype associated with increased CYP2C19‐mediated metabolism.
WHAT QUESTION DID THIS STUDY ADDRESS?
Our study investigated the impact of the novel CYP2C:TG haplotype on sertraline serum concentration in a large population genotyped for CYP2C19 (variants *2, *3, *4, and *17) and CYP2B6 (variants *4, *6, and *9).
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study demonstrates that the novel CYP2C haplotype (CYP2C:TG) is associated with increased rate of CYP2C19 metabolism of sertraline, as previously shown for escitalopram. In addition, patients carrying both the CYP2B6*4 variant and CYP2C19 genotypes encoding ultrarapid CYP2C19 metabolism are at increased risk of underexposure and therapeutic failure when treated with standard recommended doses of sertraline, whereas patients with only inactive or decreased function alleles of both CYP2C19 and CYP2B6 have a lower capacity for sertraline metabolism than CYP2C19 poor metabolizers carrying functional CYP2B6 alleles.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Preemptive genotyping of sertraline patients would include CYP2C:TG, CYP2C19, and CYP2B6 variants to increase the dose precision of the drug in patients suffering from depression and/or anxiety.
INTRODUCTION
Sertraline is a selective serotonin reuptake inhibitor (SSRI), used in the treatment of major depressive, obsessive–compulsive, panic, post‐traumatic stress, and social anxiety disorders. 1 There is an extensive interindividual variability in the pharmacokinetics of sertraline. The individual variability in sertraline pharmacokinetics could relate to differences in activity of the drug‐metabolizing enzymes involved in sertraline metabolism, where the polymorphic enzymes CYP2C19 and CYP2B6 are of predominant importance; however, sertraline is also a substrate of CYP2C9, CYP2D6, and CYP3A4. 2 , 3 Significant effects of CYP2C19 variants on sertraline metabolism have already been observed by several studies, whereas there are no studies of CYP2B6 and sertraline in a large patient population. 2 , 3 , 4 , 5 , 6 By contrast, CYP2D6 and CYP2C9 variants are not predictors of sertraline metabolism, as shown by Braten et al. 5 Regarding CYP3A4, a study with grapefruit juice, 7 which is a potent inhibitor, reported a 1.5‐fold increase in sertraline exposure during juice consumption, suggesting a relevant impact of CYP3A4 on sertraline metabolism alongside CYP2C19 and CYP2B6. 7
CYP2C19 and CYP2B6 are highly polymorphic which influences the metabolism of clinically relevant drugs. The major CYP2C19 variants are the non‐functional alleles CYP2C19*2, *3, *4, and CYP2C19*17, which causes increased enzyme expression. 8 , 9 , 10 , 11 , 12 Among important CYP2B6 allelic variants are CYP2B6*6 (rs3745274, c.516G > T and rs2279343, c.785A > G) and CYP2B6*9 (rs3745274, c.516G > T) associated with decreased enzyme activity and increased exposure of different CYP2B6 substrates, 13 , 14 , 15 while the CYP2B6*4 variant (rs2279343, c.785A > G) has been associated with increased enzyme activity primarily based on in vitro studies. 16 Currently, homozygous carriers of CYP2B6*6 and/or *9 are classified by the as PMs, while homozygous carriers of the CYP2B6*4 allele are assigned as UMs by PharmGKB and the CPIC guidelines. 16 , 17 , 18
A previous study from our research group on 1200 Scandinavian patients demonstrated a significant increase in sertraline concentration in CYP2C19 intermediate metabolizers (IMs) and poor metabolizers (PMs). 5 In that study, however, only a marginally decreased serum concentration of sertraline was observed in CYP2C19 ultrarapid metabolizers (Ums) (patients homozygous for CYP2C19*17) in comparison to normal metabolizers (NMs). Interestingly, a large proportion of the patients defined as CYP2C19 NMs was underexposed to sertraline and required doses above the recommended 50 mg/day, potentially indicating the presence of unknown genetic variants causing increased sertraline metabolism among NMs.
Recently, we discovered a novel common CYP2C‐haplotype defined by the co‐occurrence of rs2860840T and rs11188059G (CYP2C:TG), which was significantly associated with reduced exposure of the CYP2C19 substrate escitalopram. 19 In patients, the presence of the CYP2C:TG haplotype was associated with even lower escitalopram levels as compared to subjects carrying the CYP2C19*17 allele. In the present study we aimed to examine the genetic basis for prediction of sertraline metabolism by investigating both the novel CYP2C:TG haplotype and CYP2B6 alleles in a large patient population.
METHODS
The study encompassed patients who had been CYP‐genotyped between January 1, 2017 and December 31, 2020 and for which the therapeutic drug monitoring (TDM) database at the Center for Psychopharmacology, Diakonhjemmet Hospital serum contained data on concentration of sertraline and metabolites. The patient population included from the TDM database represents individuals treated in primary practice and secondary health care units from all over Norway. The specific diagnoses of the patients are unknown.
The included patients had at least one sertraline concentration measurement at steady state (trough level), i.e., ≥4 days/dose intervals, with blood sample collection 10–30 h after the last dose intake. Exclusion criteria were: (i) comedication with the potent CYP inducers phenobarbital, phenytoin, and carbamazepine; (ii) comedication with the CYP2C19 inhibitors omeprazole, esomeprazole, lansoprazole, pantoprazole, fluoxetine, and fluvoxamine; (iii) measured serum concentration of sertraline and/or the metabolite N‐desmethylsertraline below the lower limit of quantification; and (iv) patient aged below 18 or above 90 years.
Information about sampling time and prescribed daily dose, time of last dose intake, as well as comedications, were retrieved from the requisition forms. The information about comedicated drugs was limited to the list written by the physician on the TDM requisition form, which is usually complete for psychotropic and other centrally acting central nervous system (CNS) drugs, while information on somatic drugs may be incomplete.
The study was approved by the Norwegian Regional Committee for Medical and Health Research Ethics (#2018/1848) and the Investigational Review board at Diakonhjemmet Hospital. All participants selected from the TDM database at the Center for Psychopharmacology were informed about the project and their right to withdraw from the study and the use of their biobanked blood sample for research purposes.
Serum concentration analysis of sertraline
Serum concentration analysis of sertraline was carried out at the Center for Psychopharmacology. In addition, determination of N‐desmethylsertraline concentration is part of the routine TDM assay. However, as this metabolite is mainly formed by CYP3A4, 7 and to a limited extent by CYP2C19, 5 N‐desmethylsertraline concentrations were not focused on in the present study.
During the inclusion period two different liquid chromatography–mass spectrometry (LC–MS) methods were used for routine TDM analysis. The methods were similar in terms of sample preparation (protein precipitation), chromatographic conditions, and type of internal standard. The most recent method was based on ultrahigh‐performance LC (UHPLC) with a high‐resolution accurate mass (HRAM) spectrometry system.
Briefly, serum samples were purified by protein precipitation mixing 200 μl serum with 400 μl acetonitrile–methanol (90/10 vol/vol), which included the internal standard (13C6‐sertraline and 13C6‐desmethylsertraline), followed by centrifugation for 10 min (4000 rpm at 4°C). Subsequently, 4 μl of purified sample were then injected into a Vanquish Binary UHPLC system coupled to a Q Exactive Orbitrap HRAM MS with electrospray ionization operated in positive ionization mode (Thermo Scientific, Waltham, MA, USA). Chromatographic separation was performed on a XBridge BEH C18 column (2.5 μm, 2.1 × 75 mm; Waters). The mobile phase gradient comprised a mixture of acetonitrile and ammonium acetate buffer (pH 4.8). Limits of detection for sertraline and N‐desmethylsertraline were 4 and 8 nmol/L, respectively.
TaqMan real‐time PCR analysis
Genotyping of CYP2C19 had previously been performed with TaqMan genotyping assays (Thermo Fisher Scientific) using real‐time polymerase chain reaction (PCR) implemented for routine pharmacogenetic analysis at the Center for Psychopharmacology. In addition to CYP2C19 genotypes, CYP2C9 and CYP2D6 genotypes were also available in the routine laboratory database for the majority of the patients, as these genes are usually ordered together by the physicians.
The panel for CYP2C19 genotyping included the non‐functional alleles CYP2C19*2 (rs4244285), CYP2C19*3 (rs4986893), and CYP2C19*4 (rs28399504), and the increased function allele CYP2C19*17 (rs12248560). Routine genotyping of CYP2C9 included the decreased function alleles CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910), while routine CYP2D6 genotyping included the no function alleles CYP2D6*3 (rs35742686), CYP2D6*4 (rs3892097), CYP2D6*5 (gene deletion), and CYP2D6*6 (rs5030655), the decreased function alleles CYP2D6*9 (rs5030656), CYP2D6*10 (rs1065852), and CYP2D6*41 (rs28371725), as well as copy number analysis to identify multiplication of the CYP2D6 gene giving rise to ultrarapid metabolism.
Genotyping of the novel CYP2C‐haplotype (rs2860840T and rs11188059G) was performed using predesigned TaqMan‐based real‐time PCR assays (Thermo Fisher Scientific).
Genotyping of CYP2B6 included two CYP2B6 variants rs3745274, c.516G>T and rs2279343, c.785A>G determining three different haplotypes: CYP2B6*4 (c.516G+c.785G), CYP2B6*6 (c.516T+c.785G), and CYP2B6*9 (c.516T+c.785A). Absence of any of these two variants was assigned as CYP2B6*1 (c.516G+c.785A).
Statistics
Impact of the tested CYP2C19 and CYP2B6 genetic variants on sertraline serum concentration was investigated using a multivariate‐linear mixed model. First, the different genotypes of CYP2C19 and CYP2B6 were investigated for their quantitative impact on sertraline serum concentration. Second, patients were divided into genotype‐predicted phenotype subgroups and the impact of the respective phenotype subgroups on sertraline serum concentration were investigated. To account for the variable number of serum concentration measurements per patient, the quantitative effects of the genotypes and genotype predicted phenotypes were estimated using multivariate linear mixed‐model analysis, as this allows for inclusion of multiple measurements per patient. In the two mixed‐model analyses, dose‐adjusted serum concentrations of sertraline were used as the dependent variable, with the patient as the mixed effect (random) variable and genotype, sex, and time between last dose administration and blood sampling as fixed effect variables. When accounting for covariates, the mixed‐model analysis estimated the effects of having any other genotype or phenotype than *1/*1 or NM, respectively. Covariates that differed significantly between the genotype subgroups were added to the mixed‐model analysis if the significance reached p < 0.05. Samples with a harmonized serum concentration of sertraline outside the 95% percentile were not included in the analysis.
To restore normality of dose‐adjusted serum concentrations of sertraline, these measurements were ln‐transformed prior to linear mixed‐model analysis. The natural logarithm of the normalized sertraline concentration levels (c) was used because of its linear dependence on the time between the drug intake and blood sampling (t) and the elimination rate constant (K e ) according to the equation ∆ln(c) = K e .
In the mixed‐model analysis of CYP2B6 genotype‐predicted phenotype subgroups, homozygote and heterozygote carriers of CYP2B6*4 were categorized as ultrarapid metabolizers (UMs), non‐carriers (CYP2B6*1/*1) were set as normal metabolizer (NM), heterozygote carriers of CYP2B6*6 or *9, in combination with *1 or *4, were set as intermediate metabolizers (IM), and homozygote carriers of CYP2B6*6 or *9 as poor metabolizers (PMs). For CYP2C19, heterozygote carriers of non‐functional alleles were defined as intermediate metabolizers (IMs), while homozygote carriers of non‐functional alleles were defined as PMs. Since the novel CYP2C:TG‐haplotype was associated with increased metabolism of the CYP2C19 substrate escitalopram to a similar extent as CYP2C19*17, 19 carriers of CYP2C19*17 and CYP2C:TG, in any combination, were defined as UMs. Patients carrying CYP2C19*17 or CYP2C:TG in combination with CYP2C19*1 were defined as rapid metabolisers (RMs) and CYP2C19*1/*1 as NMs. Genotypes and their assigned phenotype subgroup are provided in Table 1.
TABLE 1.
Patient demographics for each diplotype
| Phenotype | Genotype | Patients, n (%) | Samples, n | Women, n (%) | Age, years, mean (SD) | Drug dosage, mg/day, mean (SD) | Time between last drug intake and blood sampling (h), mean (SD) |
|---|---|---|---|---|---|---|---|
| UM | CYP2C19*17/CYP2C19*17 | 44 (5.2) | 89 | 31 (70.5) | 41.3 (17.2) | 115.5 (60.2) | 20.3 (5.75) |
| CYP2C:TG/CYP2C:TG | 26 (3.1) | 34 | 14 (53.9) | 45.2 (20.0) | 97.1 (35.8) | 21.1 (4.54) | |
| CYP2C:TG/CYP2C19*17 | 65 (7.7) | 108 | 43 (66.2) | 51.1 (19.7) | 120.7 (71.0) | 21.0 (5.64) | |
| RM | CYP2C19*1/CYP2C19*17 | 150 (17.9%) | 286 | 103 (68.7) | 45.8 (18.1) | 145.0 (126.0) | 19.9 (5.31) |
| CYP2C19*1/CYP2C:TG | 142 (16.9) | 222 | 95 (66.9) | 44.2 (17.9) | 111.6 (53.3) | 20.4 (5.21) | |
| NM | CYP2C19*1/CYP2C19*1 (Ref.) | 160 (19.1) | 288 | 99 (61.9) | 44.2 (18.6) | 106.3 (51.4) | 20.2 (5.68) |
| IM | CYP2C19*17/CYP2C19 Null | 55 (6.6) | 93 | 33 (60.0) | 46.4 (18.4) | 113.6 (57.5) | 20.0 (5.74) |
| CYP2C:TG/CYP2C19 Null | 61 (7.3) | 120 | 41 (67.2) | 43.9 (18.7) | 98.0 (48.4) | 18.7 (5.69) | |
| CYP2C19*1/CYP2C19 Null | 108 (12.9) | 167 | 69 (63.9) | 44.9 (18.5) | 100.8 (53.0) | 21.4 (5.10) | |
| PM | CYP2C19 Null/CYP2C19 Null | 29 (3.5) | 75 | 21 (72.4) | 34.0 (14.3) | 110.5 (47.3) | 18.9 (5.46) |
| Total | 840 | 1482 | 549 (65.4) | 44.6 (18.4) | 115.3 (75.3) | 20.2 (5.48) | |
| UM | CYP2B6*1/*4 | 33 (4.2) | 62 | 21 (63.6) | 42.1 (16.0) | 117.3 (61.2) | 20.7 (6.04) |
| CYP2B6*4/*4 | 4 (0.5) | 8 | 3 (75.0) | 63.8 (21.8) | 150.0 (46.3) | 18.3 (5.80) | |
| NM | CYP2B6*1/*1 (Ref.) | 454 (57.3) | 848 | 297 (65.4) | 43.8 (17.9) | 121.7 (87.2) | 20.1 (5.41) |
| IM | CYP2B6*1/*6 | 251 (31.7) | 383 | 162 (64.5) | 42.7 (18.3) | 106.1 (50.9) | 20.5 (5.54) |
| CYP2B6*1/*9 | 3 (0.4) | 7 | 3 (100) | 45.3 (11.2) | 132.1 (23.8) | 24.2 (1.36) | |
| CYP2B6*4/*6 | 7 (0.9) | 13 | 5 (71.4) | 54.5 (22.8) | 107.7 (78.7) | 19.4 (5.90) | |
| PM | CYP2B6*6/*6 | 39 (4.9) | 46 | 24 (61.5) | 45.7 (19.6) | 87.0 (52.2) a | 20.3 (5.47) |
| CYP2B6*9/*9 | 1 (0.1) | 2 | 1 (100) | 63.5 (0.7) | 100.0 (0) | 23.4 (0.14) | |
| Total | 792 | 1369 | 516 (65.2) | 43.7 (18.1) | 116.0 (76.5) | 20.2 (5.48) |
Note: Null = no function allele (i.e., the presence of CYP2C19*2, CYP2C19*3, or CYP2C19*4). Ref. = reference genotype.
Abbreviations: IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
Indicates significant difference (p < 0.05) compared with patients with the reference genotype (CYP2C19*1/*1 or CYP2B6*1/*1), patients not carrying any of the investigated variants.
Mixed‐model estimates are presented as fold‐changes in sertraline concentration (nM per 100 mg/day), which appear when back‐transforming the beta values for each of the fixed effects (genotypes). After estimating the effects of the various genotypes, these were applied to predict the mean values of the dose‐adjusted serum concentration for each genotype, using the mixed‐model algorithm adjusted by mean values of the covariates (sampling time set to 20 h and age <65 years).
Statistical analysis was performed using R, version 4.0.3 and RStudio. 20 , 21 A p‐value of 0.05 was considered to be statistically significant.
RESULTS
In total, 840 patients, with an overall number of 1482 valid TDM measurements, met the inclusion criteria. Patient characteristics and genotype frequencies of CYP2C19, CYP2C:TG, and CYP2B6 are presented in Table 1. All investigated variants (single nucleotide polymorphisms, SNPs) were in Hardy Weinberg equilibrium and the frequencies were in concordance with frequencies observed in Europeans in general.22
Effect of CYP2C19 genotypes on the sertraline concentration
The impact of the CYP2C:TG haplotype and CYP2C19 variants on sertraline serum concentration was investigated using multivariate linear mixed‐model analysis, adjusting for CYP2B6 genotype, age, sex, and time between last dose and blood sampling. CYP2D6 and CYP2C9 genotypes had no significant impact on the dose‐adjusted serum concentrations of sertraline (p > 0.1) and were therefore excluded from the statistical analysis.
Results of the mixed‐model analysis estimating the quantitative effect of the CYP2C:TG haplotype and CYP2C19 variants on sertraline concentration are presented in Table 2 and Figure 1. Compared with the reference group (CYP2C19*1/*1), a lower sertraline serum concentration was observed in CYP2C19*17/*17 (21.6% decrease, n = 44, p = 0.003), CYP2C:TG/CYP2C:TG (21.2% decrease, n = 26, p = 0.022), CYP2C19*17/CYP2C:TG (20.0% decrease, n = 65, p = 0.003), and CYP2C19*1/*17 (17.0% decrease, n = 150, p < 0.001) patients, while no significant impact of CYP2C19*1/CYP2C:TG genotype was detected in this patient population (n = 142, p > 0.1)
TABLE 2.
Impact of CYP2C19 and CYP2B6 genotype on sertraline concentration estimated in the fixed effect mixed‐model analysis
| Phenotype | Genotype | Patients, n (%) | Samples, n | Sertraline | ||
|---|---|---|---|---|---|---|
| Predicted mean serum concentration, nM/100 mg day | Fold difference | p‐Value | ||||
| CYP2C19 | ||||||
| NM | CYP2C19*1/*1 (Ref.) | 160 (19.1) | 288 | 71.6 (62.5–82.13) | ‐ | ‐ |
| UM | CYP2C19*17/CYP2C19*17 | 44 (5.2) | 89 | 56.2 (47.8–66.0) | 0.78 (0.67–0.92) | 0.003 |
| CYP2C:TG/CYP2C:TG | 26 (3.1) | 34 | 56.5 (46.1–69.2) | 0.79 (0.64–0.97) | 0.022 | |
| CYP2C:TG/CYP2C19*17 | 65 (7.7) | 108 | 57.3 (49.4–66.4) | 0.80 (0.69–0.93) | 0.003 | |
| RM | CYP2C19*1/CYP2C19*17 | 150 (17.9) | 286 | 59.5 (53.3–66.4) | 0.83 (0.74–0.93) | <0.001 |
| CYP2C19*1/ CYP2C:TG | 142 (16.9) | 222 | 72.2 (64.5–80.8) | 1.01 (0.90–1.13) | 0.893 | |
| IM | CYP2C19*17/CYP2C19 Null | 55 (6.6) | 93 | 74.1 (63.8–86.1) | 1.03 (0.89–1.20) | 0.661 |
| CYP2C:TG/CYP2C19 Null | 61 (7.3) | 120 | 86.6 (75.0–100.0) | 1.21 (1.05–1.40) | 0.010 | |
| CYP2C19*1/CYP2C19 Null | 108 (12.9) | 167 | 97.8 (86.8–110.3) | 1.37 (1.21–1.54) | <0.001 | |
| PM | CYP2C19Null/CYP2C19 Null | 29 (3.5) | 75 | 163.3 (134.6–198.1) | 2.28 (1.88–2.77) | <0.001 |
| CYP2B6 | ||||||
| NM | CYP2B6*1/*1 (Ref.) | 454 (57.3) | 848 | 71.6 (62.5–82.1) | ‐ | ‐ |
| UM | CYP2B6 (*1/*4, *4/*4) | 37 (4.7) | 70 | 59.2 (50.2–69.7) | 0.83 (0.70–0.97) | 0.022 |
| IM | CYP2B6 (*1/*6, *1/*9, *4/*6) | 261 (33.0) | 403 | 82.3 (76.5–88.5) | 1.15 (1.07–1.24) | <0.001 |
| PM | CYP2B6 (*6/*6, *9/*9) | 40 (5.1) | 48 | 89.4 (75.9–105.3) | 1.25 (1.06–1.47) | 0.008 |
Note: A 1.9‐fold difference (p = 0.019) and a 1.18‐fold difference (p < 0.001) in sertraline concentration were observed for women and patients aged >65 years, respectively.
Null = no function allele (i.e. the presence of CYP2C19*2, CYP2C19*3, or CYP2C19*4).
Ref. = reference genotype.
Genotype is used as fixed effects in the mixed model. The sertraline concentration of patients not carrying any of the analyzed variants of CYP2C19 or CYP2B6 were set as the reference group. Fold difference represents the estimated relative impact of the occurrence of the genotype on sertraline concentration, as compared with the reference group. Serum concentrations are predicted from the mixed‐model analysis, with age <65 years and sampling time 20 h as model determined covariate values.
Abbreviations: CI, confidence interval; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
FIGURE 1.
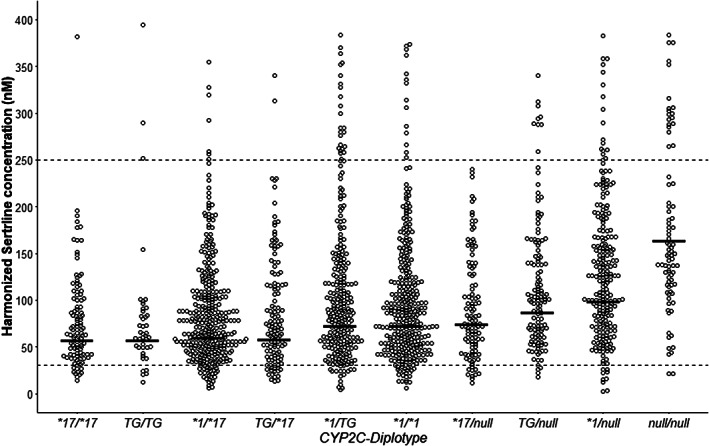
Sertraline serum concentration measurements in patient subgroups carrying different CYP2C19 genotypes in combination with CYP2C:TG, where the crossbar indicates the predicted mean serum concentration for each diplotype. Sertraline serum concentrations were normalized to the dosage of 100 mg/day. The predicted mean serum concentration is based on mixed‐model analyses of the different CYP2C19 genotypes and is presented as crossbar for each CYP2C19 diplotype. Patients with a CYP2C19*1/*1 genotype were used as reference in the mixed‐model analysis. Null = no function allele (i.e., presence of CYP2C19*2, CYP2C19*3, or CYP2C19*4). The dotted line indicates the therapeutic reference range (30–250 nM)
Compared to the reference group (CYP2C19*1/*1), patients homozygous for the non‐functional CYP2C19 alleles (*2, *3, or *4) had a 2.3‐fold (n = 29, p < 0.001) increase in serum concentrations of sertraline (Table 2). Compared to the reference group, patients heterozygous for the non‐functional alleles of CYP2C19 in combination with either CYP2C:TG or CYP2C19*17 had 1.21‐fold (n = 61, p = 0.01) and 1.37‐fold (n = 55, p < 0.001) increased sertraline concentration, respectively (Table 2)
Effect of CYP2B6 genotypes on the sertraline concentration
The impact of the CYP2B6 variants on sertraline serum concentration was investigated using multivariate linear mixed‐model analysis, adjusting for CYP2C:TG, CYP2C19 genotype, age, sex, and time between last dose and blood sampling. Compared with the reference group (CYP2B6*1/*1, n = 454), patients carrying the CYP2B6*4 allele had a 17.4% (n = 37, p = 0.022) reduced serum concentration of sertraline (Table 2). On the contrary, compared to the reference group, patients homozygous or heterozygous for the CYP2B6*6 or *9 allele had a 25% (n = 40, p = 0.008) and 15% (n = 261, p < 0.001) increased serum concentration of sertraline, respectively (Table 2)
In addition, a significant impact of sex and age on sertraline serum concentration was found in the patient population, where a 11% (p < 0.001) and 18% (p < 0.001) increased serum concentration of sertraline was found for women and patients >65 years (n = 176), respectively.
Effect of genotype‐predicted phenotype subgroups on sertraline exposure
Patients were further divided into genotype‐predicted phenotype subgroups to investigate the combined effect of CYP2C19 and CYP2B6 phenotypes on the sertraline serum concentration which was investigated using a multivariate linear mixed model, adjusting for age, sex, and time between last dose and blood sampling. Patients with a normal metabolizer phenotype of both CYP2C19 and CYP2B6 were used as reference. Results of the mixed‐model analysis based on genotype‐predicted phenotype subgroups is presented in Table S1.
Predicted mean serum concentration of sertraline for the different combinations of CYP2C19 and CYP2B6 phenotypes were calculated using the estimates from the phenotype model, and the results are presented in Table 3 and Figure 2. Most notably, the predicted sertraline serum concentration in patients with a combined ultrarapid metabolizer (CYP2C19 UM+ CYP2B6 UM) phenotype was reduced by 35.4%, compared with the reference group (CYP2B6 NM and CYP2C19 NM). Patients with a combined poor metabolizer (CYP2C19 PM + CYP2B6 PM) phenotype had a 2.89‐fold increased predicted mean serum concentration, compared to the reference group.
TABLE 3.
Predicted mean serum concentration of sertraline in combined genotype‐predicted phenotype groups of CYP2B6 and CYP2C19
| Phenotype | Patients, n | Samples, n | Sertraline | ||
|---|---|---|---|---|---|
| CYP2B6 | CYP2C19 | Predicted mean serum concentration (95% CI) | Fold difference (95% CI) | ||
| UM | UM | 10 | 22 | 46.31 (35.0–61.3) | 0.65 (0.49–0.86) |
| UM | RM | 6 | 7 | 53.17 (40.9–69.1) | 0.74 (0.57–0.96) |
| UM | NM | 9 | 19 | 58.52 (49.6–69.1) | 0.82 (0.69–0.96) |
| UM | IM | 9 | 17 | 72.19 (55.3–94.2) | 1.01 (0.77–1.32) |
| UM | PM | 1 | 1 | 133.28 (92.9–191.2) | 1.86 (1.30–2.67) |
| NM | UM | 67 | 123 | 56.71 (50.5–63.6) | 0.79 (0.71–0.89) |
| NM | RM | 156 | 281 | 65.11 (59.1–71.7) | 0.91 (0.83–1.00) |
| NM | NM | 90 | 155 | 71.66 (62.4–82.3) | – |
| NM | IM | 119 | 224 | 88.4 (79.9–97.8) | 1.23 (1.12–1.36) |
| NM | PM | 14 | 52 | 163.21 (121.7–198.5) | 2.28 (1.87–2.77) |
| IM | UM | 40 | 60 | 65.64 (54.4–79.2) | 0.92 (0.76–1.11) |
| IM | RM | 93 | 144 | 75.37 (63.6–89.3) | 1.05 (0.89–1.25) |
| IM | NM | 43 | 72 | 82.95 (77.1–89.3) | 1.16 (1.08–1.25) |
| IM | IM | 73 | 110 | 102.33 (86.0–121.8) | 1.43 (1.20–1.70) |
| IM | PM | 10 | 15 | 188.92 (144.4–247.2) | 2.64 (2.01–3.45) |
| PM | UM | 6 | 7 | 71.99 (54.4–95.3) | 1.01 (0.76–1.33) |
| PM | RM | 14 | 18 | 82.65 (63.6–107.4) | 1.15 (0.89–1.50) |
| PM | NM | 5 | 6 | 90.96 (77.1–107.3) | 1.27 (1.08–1.50) |
| PM | IM | 10 | 10 | 112.22 (86.1–146.3) | 1.57 (1.20–2.04) |
| PM | PM | 2 | 2 | 207.18 (144.5–297.1) | 2.89 (2.02–4.15) |
Note: Patients with a CYP2B6 NM and CYP2C19 NM phenotype were set as the reference group. Fold difference represents the estimated relative impact of the occurrence of the combination of phenotype on sertraline serum concentration, as compared with the reference group.
Abbreviations: CI, confidence interval; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
FIGURE 2.
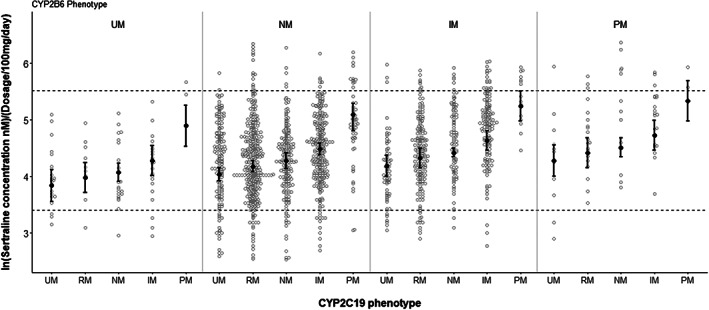
Sertraline serum concentration measurements in different phenotype subgroups of CYP2C19 and CYP2B6. The sertraline serum concentrations were normalized to the dosage of 100 mg/day and analyzed in the natural logarithmic scale. Predicted serum concentration is based on mixed‐model analyses of the different CYP2C19 and CYP2B6 phenotype subgroups and is presented as error bars for each phenotype combination. The dotted line indicates the therapeutic reference range (30–250 nM)
Formation of N‐desmethylsertraline in relation to CYP2C19 and CYP2B6 genotypes
In addition to sertraline exposure, the formation of the inactive metabolite N‐desmethylsertraline, in relation to CYP2C19 and CYP2B6 genotypes, was also analyzed (Table S2). The metabolite was previously reported to be formed by CYP2C19 and CYP2B6 to a limited extent, 3 , 23 which was supported by the only 1.2‐fold higher N‐desmethylsertraline‐to‐sertraline metabolic ratio (MR) in CYP2C19 PMs versus NMs (Table S3). For CYP2B6, IMs and PMs had similar MR as NMs (Table S3). The lack of effect of CYP2C19 and CYP2B6 genotypes on the rate of formation of N‐desmethylsertraline reflects the fact that the levels of this metabolite were directly proportional to sertraline exposure, that is, enzymatic access of the substrate sertraline in relation to the respective genotypes (Table S2), and indicates that enzymes other than CYP2C19 and CYP2B6 are responsible for the N‐desmethylsertraline formation, presumably CYP3A4. 7
DISCUSSION
In this study we have investigated the impact of CYP2C19 and CYP2B6 variants on sertraline serum levels, with the emphasis on the novel CYP2C:TG haplotype (rs2860840T and rs11188059G), in a large population of psychiatric patients. The results demonstrate that patients carrying the CYP2C:TG haplotype, which was previously suggested to encode increased CYP2C19 metabolic capacity, 18 had a lower serum concentration of sertraline, and that the decrease was similar as in the patients carrying the validated increased expression allele CYP2C19*17. These findings are in line with those recently reported based on another CYP2C19 substrate, escitalopram, 19 and hence these data together support the hypothesis that the CYP2C:TG haplotype is indeed associated with ultrarapid CYP2C19 metabolism. With a total population frequency of 10% for CYP2C:TG/CYP2C:TG and CYP2C:TG /CYP2C19*17, our discovery of CYP2C:TG may be of substantial clinical importance for the treatment of sertraline and potentially additional CYP2C19 substrates.
These results together imply that patients homozygous for either CYP2C19*17 or CYP2C:TG or a combination of both generally would require higher sertraline dosing than CYP2C19*1/*1 carriers to reach therapeutic concentrations. Furthermore, this study indicated that CYP2B6 genotype also significantly contributed to the individual variability in exposure and metabolism of sertraline, albeit at a lower extent as compared to variants encoding non‐functional CYP2C19 allelic variants. Regarding clinically personalized sertraline dosing, preemptive genotyping of CYP2C19, the CYP2C:TG haplotype, and CYP2B6 in sertraline‐treated patients may be more informative. Most notably, a group which may be of particular risk of sertraline underexposure and treatment failure are the patients with genotypes predicting UM phenotypes of both CYP2C19 and CYP2B6; 1.2% of the European population fall into this category associated with extremely fast sertraline clearance. Based on the results of the present study, such patients would require higher doses than recommended to ascertain optimal sertraline exposure. However, further studies are necessary to provide evidence for the clinical relevance of this pharmacokinetic/pharmacogenetic interpretation.
In a previous study, we observed a minor, but statistically significant lower serum concentration of sertraline of 8% in patients carrying the CYP2C19*17 allele compared with the reference group. 5 However, the reference group (CYP2C19*1/*1) in the previous study was in fact a mixture of NMs and the, at that time, unknown RMs (CYP2C19*1/CYP2C:TG) and UMs (CYP2C:TG/CYP2C:TG). Therefore, in the present study, where the CYP2C:TG haplotype was considered and when the reference group contained only prototypical CYP2C19*1/*1 NMs, the mentioned difference in sertraline exposure between CYP2C19 NM and UM increased to 22%. Accordingly, the CYP2C:TG haplotype identified among CYP2C19*1/*1 patients 19 may explain the previously observed underexposure of sertraline in CYP2C19 NMs, in a similar pattern as for the underexposure of escitalopram described for a substantial number of CYP2C19*1/*1 carriers reported by Jukic et al. 10
The molecular mechanism underlying the increased metabolism of CYP2C19 substrates caused by CYP2C:TG is unclear. Since CYP2C19 also metabolizes a multitude of other types of drugs such as proton pump inhibitors, diazepam, clopidogrel, voriconazole, diazepam, proguanil, and many others it appears of importance to consider the CYP2C:TG haplotype when conducting genotype phenotype linkage analyses for all these drugs.
In addition, the significant effect of CYP2B6 genotype on the serum concentration of sertraline shown in this study may have clinical relevance. Patients carrying the CYP2B6*4 allele exhibited a 17% reduced sertraline concentration, which is similar to the effect observed in CYP2C19 UM patients. There is a lack of in vivo data on the effect of CYP2B6*4 on sertraline metabolism, but studies investigating the effect of this variant on other CYP2B6 substrates have already indicated that it causes increased enzyme activity. 14 Regarding the effects of CYP2B6*6 or *9 alleles on sertraline metabolism, patients who were homo‐ or heterozygous carriers exhibited a 1.15–1.30‐fold increased serum concentration of sertraline, respectively. Regarding increased risk of overexposure and adverse effects, these differences alone are unlikely to be of major clinical relevance; however, combined with the presence of CYP2C19 PM genotypes, the impact of CYP2B6*6 or *9 may be relevant for sertraline dosing decisions. 5 For example, patients with a combined poor metabolizer (CYP2C19 PM + CYP2B6 PM) phenotype had a 2.89‐fold increased serum concentration, compared to the reference group. Overall, the present study confirms that CYP2B6 phenotype is an important determinant for the serum concentration and dose requirement of sertraline and that it should not be neglected as it currently is. Thus, individualized sertraline dosing based on preemptive genotyping of CYP2C19/CYP2B6 and/or TDM can be used to minimize the over‐ and under‐exposure of sertraline.
This study has some inherent limitations associated with the retrospective use of the TDM data. One is that information about comedication relies on the information provided by the physicians on the requisition form. The potential comedication with CYP2C19 inhibitors could be missed, although concurrent use of enzyme‐inducing anti‐epileptics, which were excluded from the study, is often specified on the requisition form. This represents a potential non‐pharmacogenetic source of variability in this patient population. Only common CYP2B6 allelic variants were investigated in this study; further studies investigating the effect of CYP2B6 variants on sertraline metabolism might also include other rare CYP2B6 variants in order to increase the resolution of the influence of CYP2B6 genotypes. This study did not include CYP3A4, whereas consideration of CYP3A4 may be relevant to sertraline dosing. Further, other factors possibly affecting the serum concentration of sertraline (e.g., body weight, renal function, liver function, inflammatory state, somatic diseases, and/or partial nonadherence) could not be controlled for. However, the potential interference of these confounding factors is likely outweighed by the large number of included patients.
CONCLUSIONS
Patients carrying the CYP2C:TG‐haplotype demonstrate similar reduced exposure of sertraline as the increased expression allele CYP2C19*17, that is, about 20% in homozygous carriers, respectively. These patients will generally require a 25% higher dose to obtain the full potential effect of sertraline. Furthermore, the study also reveals a significant impact of CYP2B6 genotype on sertraline exposure. Consequently, patients with a combined homozygous presence of CYP2C19*17 and CYP2C:TG haplotype as well as CYP2B6*4 are expected to require a 50% increased dose to reach target concentration levels. Based on the relative differences in serum concentrations compared to NMs, dose reductions of 65% and 30% should be considered in patients with a combined PM/PM and IM/IM phenotype, respectively, to reduce the risk of sertraline overexposure in these patients. The significant impact of combined genotype‐predicted phenotypes of CYP2B6 and CYP2C19, including the novel CYP2C‐haplotype, on sertraline concentration suggests that genotyping of both the CYP2C:TG and CYP2B6 (*4, *6, and *9) should be included in genotyping panels for sertraline metabolism, in addition to CYP2C19 genotyping. Further studies are necessary to elucidate the clinical implications of the novel CYP2C‐haplotype for the treatment of >20 drugs having pharmacogenomic labels for CYP2C19 variants, including substrates like omeprazole, voriconazole, and clopidogrel.
AUTHOR CONTRIBUTIONS
L.S.B., E.M., M.K.K., M.I.S., and M.M.J. wrote the manuscript. L.S.B, M.K.K., and E.M. designed the research. L.S.B. performed the research. L.S.B., E.M., M.I.S., M.M.J., and M.K.K. analyzed the data.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
Supporting information
Table S1
Table S2
Table S3
ACKNOWLEDGMENT
Open access funding enabled and organized by ProjektDEAL.
Bråten LS, Ingelman‐Sundberg M, Jukic MM, Molden E, Kringen MK. Impact of the novel CYP2C:TG haplotype and CYP2B6 variants on sertraline exposure in a large patient population. Clin Transl Sci. 2022;15:2135‐2145. doi: 10.1111/cts.13347
Founding information
The project was partly funded by the South‐Eastern Norway Regional Health Authority (to L.S.B), the European Union's Horizon 2020 research and innovation program U‐PGx (grant agreement number 668353 to M.I.‐S.), and The Swedish Research Council (grant 2021–02732 to M.I.‐S and E.M.).
REFERENCES
- 1. U.S. Food and Drug Administration . Zoloft. 2017; 12 December 2021, https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/019839s091lbl.pdf#page=27
- 2. Saiz‐Rodriguez M, Belmonte C, Roman M, et al. Effect of polymorphisms on the pharmacokinetics, pharmacodynamics and safety of sertraline in healthy volunteers. Basic Clin Pharmacol Toxicol. 2018;122:501‐511. [DOI] [PubMed] [Google Scholar]
- 3. Obach RS, Cox LM, Tremaine LM. Sertraline is metabolized by multiple cytochrome P450 enzymes, monoamine oxidases, and glucuronyl transferases in human: an in vitro study. Drug Metab Dispos. 2005;33:262‐270. [DOI] [PubMed] [Google Scholar]
- 4. Chen S, Wu Q, Li X, et al. The role of hepatic cytochrome P450s in the cytotoxicity of sertraline. Arch Toxicol. 2020;94:2401‐2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bråten LS, Haslemo T, Jukic MM, Ingelman‐Sundberg M, Molden E, Kringen MK. Impact of CYP2C19 genotype on sertraline exposure in 1200 Scandinavian patients. Neuropsychopharmacology. 2020;45:570‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuce‐Artun N, Baskak B, Ozel‐Kizil ET, et al. Influence of CYP2B6 and CYP2C19 polymorphisms on sertraline metabolism in major depression patients. Int J Clin Pharmacol. 2016;38:388‐394. [DOI] [PubMed] [Google Scholar]
- 7. Lee AJ, Chan WK, Harralson AF, Buffum J, Bui BCC. The effects of grapefruit juice on sertraline metabolism: an in vitro and in vivo study. Clin Ther. 1999;21:1890‐1899. [DOI] [PubMed] [Google Scholar]
- 8. Hodgson K, Tansey K, Dernovsek MZ, et al. Genetic differences in cytochrome P450 enzymes and antidepressant treatment response. J Psychopharmacol. 2014;28:133‐141. [DOI] [PubMed] [Google Scholar]
- 9. Tsai MH, Lin KM, Hsiao MC, et al. Genetic polymorphisms of cytochrome P450 enzymes influence metabolism of the antidepressant escitalopram and treatment response. Pharmacogenomics. 2010;11:537‐546. [DOI] [PubMed] [Google Scholar]
- 10. Jukic MM, Haslemo T, Molden E, et al. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a retrospective study based on 2,087 patients. Am J Psychiatry. 2018;175:463‐470. [DOI] [PubMed] [Google Scholar]
- 11. Sim SC, Risinger C, Dahl ML, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103‐113. [DOI] [PubMed] [Google Scholar]
- 12. Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102:37‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thorn CF, Lamba JK, Lamba V, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for CYP2B6. Pharmacogenet Genomics. 2010;20:520‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langmia IM, Just KS, Yamoune S, Brockmöller J, Masimirembwa C, Stingl JC. CYP2B6 functional variability in drug metabolism and exposure across populations—implication for drug safety, dosing, and individualized therapy. Front Genet. 2021;12. doi: 10.3389/fgene.2021.692234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desta Z, El‐Boraie A, Gong L, et al. PharmVar GeneFocus: CYP2B6. Clin Pharmacol Ther. 2021;110:82‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desta Z, Gammal RS, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2B6 and efavirenz‐containing antiretroviral therapy. Clin Pharmacol Ther. 2019;106:726‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whirl‐Carrillo M, Huddart R, Gong L, et al. An evidence‐based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2021;110:563‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. PharmGKB . Gene‐specific information tables for CYP2B6. https://www.pharmgkb.org/page/cyp2b6RefMaterials. 21 February.
- 19. Bråten LS, Haslemo T, Jukic MM, et al. A novel CYP2C‐haplotype associated with ultrarapid metabolism of escitalopram. Clin Pharmacol Ther. 2021;110:786‐793. [DOI] [PubMed] [Google Scholar]
- 20. RStudio Team . RStudio: Integrated Development Environment for R. RStudio, Inc.; 2019. http://www.rstudio.com/ [Google Scholar]
- 21. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2020. https://www.R‐project.org/ [Google Scholar]
- 22. Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kobayashi K, Ishizuka T, Shimada N, Yoshimura Y, Kamijima K, Chiba K. Sertraline N‐demethylation is catalyzed by multiple isoforms of human cytochrome P‐450 in vitro. Drug Metab Dispos. 1999;27:763‐766. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3


