Abstract
Objectives
Monkeypox outbreaks in nonendemic countries have been reported since early May 2022. The first case of monkeypox in the Republic of Korea was confirmed in a patient who traveled to Europe in June 2022, and an attempt was made to isolate and identify the monkeypox virus (MPXV) from the patient’s specimens.
Methods
Clinical specimens from the patient were inoculated in Vero E6 cells. The isolated virus was identified as MPXV by the observation of cytopathic effects on Vero E6 cells, transmission electron microscopy, conventional polymerase chain reaction (PCR), and sequencing of PCR products.
Results
Cytopathic effects were observed in Vero E6 cells that were inoculated with skin lesion swab eluates. After multiple passages from the primary culture, orthopoxvirus morphology was observed using transmission electron microscopy. In addition, both MPXV-specific (F3L and ATI) and orthopoxvirus-specific genes (A39R, B2R, and HA) were confirmed by conventional PCR and Sanger sequencing.
Conclusion
These results indicate the successful isolation and identification of MPXV from the first patient in the Republic of Korea. The isolated virus was named MPXV-ROK-P1-2022.
Keywords: Identification, Isolation, Monkeypox virus
Graphical abstract
Introduction
Monkeypox virus (MPXV), a DNA virus, belongs to the family Poxviridae, genus Orthopoxvirus [1,2]. MPXV shows a characteristically large brick or mulberry shape with a dumbbell-shaped DNA core. The diameters of the viral particles have been reported to be about 150 to 200 nm. MPXV is a zoonosis that is transmitted from animals to humans, and several rodent species and nonhuman primates are known to be susceptible hosts. Human-to-human transmission can also occur, mainly through close contact with the skin lesions or body fluids of an infected person [3]. In the past, MPXV was endemic in West and Central Africa, but it has recently spread in Europe and several countries, including the United States. As of August 11, 2022, more than 32,000 cases and 6 deaths across 74 countries have been reported since May 2022 [4]. On July 23, 2022, the MPXV outbreak was declared an international public health emergency by the World Health Organization [5]. In Korea, on June 22, 2022, the Korea Disease Control and Prevention Agency (KDCA) confirmed the first MPXV-infected case [6]. In this study, we report the isolation and identification of the first patient-derived MPXV. The virus was named MPXV-ROK-P1-2022.
Materials and Methods
Virus Isolation from Clinical Specimens
Clinical specimens, including skin lesion, nasopharyngeal, and oropharyngeal swabs, were collected at a medical center (where the patient was hospitalized) and sent to the KDCA. The specimens were diluted with antibiotics (penicillin/streptomycin and nystadine) at a 1:4 ratio, and then diluted with culture medium (2% fetal bovine serum [FBS]–Dulbecco’s modified Eagle’s Medium [DMEM]) at a 1:3 ratio and were incubated for 30 minutes at 4°C [7]. After centrifugation, the supernatant was inoculated into Vero E6 cells in 2% FBS-DMEM with antibiotics (penicillin-streptomycin) and was incubated at 37°C and 5% CO2. For virus isolation, inoculated cells were incubated with 1.5% carboxymethyl cellulose overlay medium, and plaque isolation was performed. All virus isolation procedures were performed according to laboratory biosafety guidelines for the MPXV by the KDCA in a biosafety level 3 facility.
Conventional PCR for the Detection of MPXV
Viral DNA was extracted from 200 μL of virus culture supernatants using a QIAamp blood mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Conventional polymerase chain reaction (PCR) was performed with a selected primer set. The sequences of the primer sets for conventional PCR were as follows: F3L-forward primer, 5'-CATCTATTATAGCATCAGCATCAGA-3', F3L-reverse primer, 5'-GATACTCCTCCTCGTTGGTCTAC-3' for the F3L gene; A39R-forward primer, 5'-TGGGATAACGAATCCAATGTCA-3', A39R-reverse primer, 5'-GCGTGCTTCCAGCAACACT-3' for the A39R gene [6]; 1995MPV-forward primer, 5'-CTGATAATGTAGAAGAC-3'; 1995MPV-reverse primer, 5'-TTGTATTTACGTGGGTG-3' for the B2R gene [8]; 1997MPV-forward primer, 5’-AATACAAGGAGGATCT-3’; 1997MPV-reverse primer, 5'-CTTAACTTTTTCTTTCTC-3' for the ATI gene [9]; HAOUT-forward primer, 5'-CCATTGGAAAAAACACAGTAC-3'; HAOUT-reverse primer, 5'-CCAAATATATTCCCATAGTC-3' for the HA gene [10]. The conventional PCR conditions were as follows: 94°C for 2 minutes, 35 cycles of 94°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds, followed by 72°C for 5 minutes.
Sequencing Analysis of the Conventional PCR Products
The PCR products were purified using a gel extraction kit (Qiagen). Next, sequence reactions using the F3L, A39R, 1995MPV, 1997MPV, and HAOUT primers with the BigDye Terminator Reaction Mix (Applied Biosystems, Foster City, CA, USA) were performed. The reaction products were purified using a BigDye Purification Kit (Applied Biosystems), and sequence analysis was performed with a 3500xL Dx Genetic Analyzer (Applied Biosystems) [11]. A nucleotide blast was carried out with the sequencing results to confirm whether the sequences matched those of MPXV.
Transmission Electron Microscopy
In order to observe virus particles, inoculated cells were fixed with 0.1 M phosphate buffer (pH 7.4) containing 2% paraformaldehyde and 2.5% glutaraldehyde. After the fixation, a 3-mL drop of the fixed sample was adsorbed to a glow-discharged formvar-coated 200-mesh copper grid, washed with 3 drops of deionized water 3 times, and stained with 2 drops of freshly prepared 0.5% uranyl acetate, and then washed with deionized water and dry at room temperature. The staining grid was observed with a transmission electron microscope (Libra120; Carl Zeiss, Oberkochen, Germany) at an acceleration voltage of 120 kV. To perform transmission electron microscopy with sectioning, prefixed cells were fixed sequentially with 0.1 M phosphate buffer (pH 7.4) containing 2% paraformaldehyde, 2.5% glutaraldehyde, and 1% osmium tetroxide before negative staining. The fixed samples were sectioned at 80 nm, followed by staining with 4% uranyl acetate and 1% lead citrate. The prepared samples were observed with a transmission electron microscope.
Results and Discussion
Virus Isolation
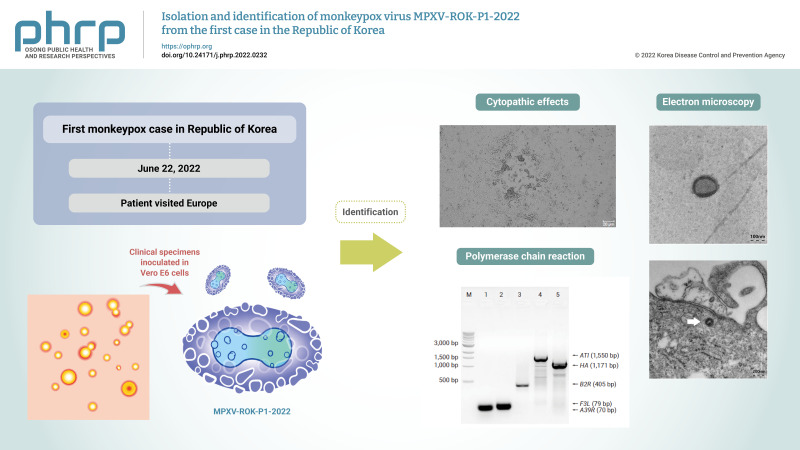
Of the specimens from the first patient, 2 specimens—the skin lesion swab and the oropharyngeal plus nasopharyngeal swabs—showed cytopathic effects (CPEs) on monolayers of Vero E6 cells (Figure 1). In the inoculation of the skin lesion swab, CPEs were observed after 36 hours on the monolayers. After 4 days, CPEs and detaching of cells were shown on the entire area of the monolayers. Through subsequent multiple passages and isolation of plaques, we obtained culture suspension and infected cells from a single plaque of the virus. The culture suspension and cells were used for conventional PCR and electron microscopy, as described in the following section. After confirmation of the virus as MPXV, we named the virus MPXV-ROK-P1-2022. Virus culture using the oropharyngeal plus nasopharyngeal swabs showed similar results (data not shown).
Figure 1.
Cytopathic effects in Vero E6 cells after 36 to 48 hours. (A) Mock infected cells (B) Skin swab specimen–infected cells.
Confirmation of MPXV by PCR
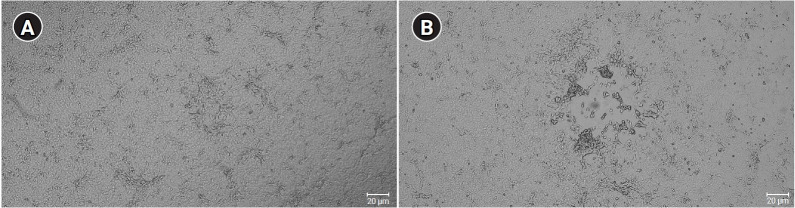
The isolated virus was confirmed as MPXV by PCR assays. The conventional PCR assays with F3L primers for the F3L gene, A39R primers for the A39R gene, 1995MPV primers for the B2R gene, 1997MPV primers for the ATI gene, and HAOUT primers for the HA gene were performed, followed by Sanger sequencing. Conventional PCR revealed PCR products of the expected sizes: 79 bp for the F3L gene, 70 bp for the A39R gene, 405 bp for the B2R gene, 1,550 bp for the ATI gene, and 1,171 bp for the HA gene, as shown in Figure 2. The Sanger sequencing results showed that sequences of those genes were >99% identical to previously reported human MPXVs (OP022170.1, OP022171.1, OP018607.1) (data not shown). Among the conventional PCR procedures, PCR of the ATI gene with 1997MPV primers was designed to distinguish 2 clades of MPXV—clade I (Congo-Basin clade, 450 bp) and clade II (West African, 1,550 bp) [8,12]—with different PCR product sizes. As shown in Figure 2 the PCR product of MPXV-ROK-P1-2022 showed a 1,550-bp product, indicating that the virus belonged to the West African clade (i.e., clade II).
Figure 2.
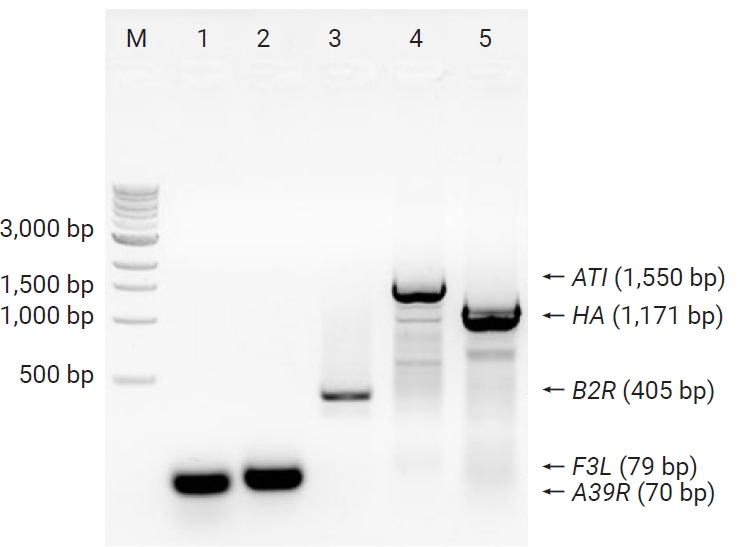
Conventional polymerase chain reaction products of MPXV-ROK-P1-2022.
M, size maker; Lane 1, A39R; lane 2, F3L; lane 3, B2R; lane 4, ATI; lane 5, HA; lane.
Transmission Electron Microscopy
The transmission electron microscopy experiment revealed MPXV-ROK-P1-2022 virus in Vero E6 cells. As shown in Figure 3, the virus itself or in the infected cells was visualized as a typical mulberry-shaped particle, which is characteristic of MPXV. In accordance with a previous report [13], the viral particle diameters were about 150 to 200 nm.
Figure 3.
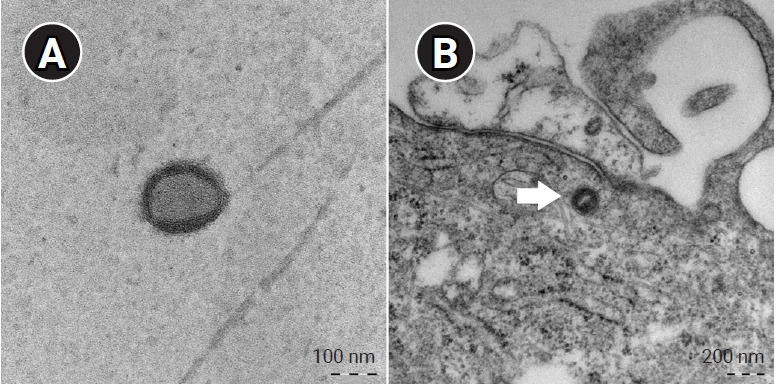
Electron microscopy of MPXV-ROK-P1-2022. (A) Mulberry-shaped virion from cell culture. (B) Virus particle (arrow) in an infected cell.
In summary, our results showed successful isolation and identification of MPXV-ROK-P1-2022 from the first monkeypox patient in Korea. The virus isolated using Vero E6 cells was identified as MPXV by electron microscopy, PCR, and sequencing. MPXV-ROK-P1-2022 will be used as a reference strain for diagnosis by the KDCA.
Footnotes
Ethics Approval
This study was approved by the Institutional Review Board of Korea Diseases Control and Prevention Agency (No: KDCA-IBC-2022-056).
Conflicts of Interest
The authors have no conflicts of interest to declare.
Funding
This work was supported by the Korea Diseases Control and Prevention Agency (No: 6331-301-210-13).
Availability of Data
The datasets are not publicly available but are available from the corresponding author upon reasonable request.
Authors’ Contributions
Conceptualization: GER, HY, JiWK, ML; Data curation: GER, HY, JiWK, ML, CHC; Formal analysis: HS, MMC; Funding acquisition: HY, GER; Investigation: HS, MMC, CHC; Methodology: HY, JiWK, ML, JeWK; Project administration: MMC, JiWK; Resources: JiWK, ML; Supervision: CKY, GER; Validation: HY, GER; Visualization: ML, JeWK; Writing–original draft: GER, HY, JiWK, ML; Writing–review & editing: all authors.
References
- 1.Marennikova SS, Seluhina EM, Mal'ceva NN, et al. Isolation and properties of the causal agent of a new variola-like disease (monkeypox) in man. Bull World Health Organ. 1972;46:599–611. [PMC free article] [PubMed] [Google Scholar]
- 2.Mukinda VB, Mwema G, Kilundu M, et al. Re-emergence of human monkeypox in Zaire in 1996. Monkeypox Epidemiologic Working Group. Lancet. 1997;349:1449–50. doi: 10.1016/S0140-6736(05)63725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthet N, Descorps-Declère S, Besombes C, et al. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Sci Rep. 2021;11:13085. doi: 10.1038/s41598-021-92315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johns Hopkins Center for Health Security. Monkeypox outbreak epi updates [Internet]. Baltimore: Johns Hopkins Bloomberg School of Public Health; 2022 [cited 2022 Aug 11]. Available from: https://www.centerforhealthsecurity.org/resources/monkeypox/index.html.
- 5. doi: 10.1016/S0140-6736(22)01437-4. Wenham C, Eccleston-Turner M. Monkeypox as a PHEIC: implications for global health governance. Lancet 2022:S0140-6736(22)01437-4. [DOI] [PMC free article] [PubMed]
- 6.Jang YR, Lee M, Shin H, et al. The first case of monkeypox in the Republic of Korea. J Korean Med Sci. 2022;37:e224. doi: 10.3346/jkms.2022.37.e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JM, Chung YS, Jo HJ, et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ropp SL, Jin Q, Knight JC, et al. PCR strategy for identification and differentiation of small pox and other orthopoxviruses. J Clin Microbiol. 1995;33:2069–76. doi: 10.1128/jcm.33.8.2069-2076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer H, Ropp SL, Esposito JJ. Gene for A-type inclusion body protein is useful for a polymerase chain reaction assay to differentiate orthopoxviruses. J Virol Methods. 1997;64:217–21. doi: 10.1016/S0166-0934(96)02155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damaso CR, Esposito JJ, Condit RC, et al. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology. 2000;277:439–49. doi: 10.1006/viro.2000.0603. [DOI] [PubMed] [Google Scholar]
- 11.Crossley BM, Bai J, Glaser A, et al. Guidelines for Sanger sequencing and molecular assay monitoring. J Vet Diagn Invest. 2020;32:767–75. doi: 10.1177/1040638720905833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saijo M, Ami Y, Suzaki Y, et al. Diagnosis and assessment of monkeypox virus (MPXV) infection by quantitative PCR assay: differentiation of Congo Basin and West African MPXV strains. Jpn J Infect Dis. 2008;61:140–2. [PubMed] [Google Scholar]
- 13.Reed KD, Melski JW, Graham MB, et al. The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med. 2004;350:342–50. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]