Abstract
To characterize the expression and possible regulation of reductive dehalogenation in halorespiring bacteria, a 11.5-kb genomic fragment containing the o-chlorophenol reductive dehalogenase-encoding cprBA genes of the gram-positive bacterium Desulfitobacterium dehalogenans was subjected to detailed molecular characterization. Sequence analysis revealed the presence of eight designated genes with the order cprTKZEBACD and with the same polarity except for cprT. The deduced cprC and cprK gene products belong to the NirI/NosR and CRP-FNR families of transcription regulatory proteins, respectively. CprD and CprE are predicted to be molecular chaperones of the GroEL type, whereas cprT may encode a homologue of the trigger factor folding catalysts. Northern blot analysis, reverse transcriptase PCR, and primer extension analysis were used to elucidate the transcriptional organization and regulation of the cpr gene cluster. Results indicated halorespiration-specific transcriptional induction of the monocistronic cprT gene and the biscistronic cprBA and cprZE genes. Occasional read-through at cprC gives rise to a tetracistronic cprBACD transcript. Transcription of cprBA was induced 15-fold upon addition of the o-chlorophenolic substrate 3-chloro-4-hydroxyphenylacetic acid within 30 min with concomitant induction of dehalogenation activity. Putative regulatory protein binding motifs that to some extent resemble the FNR box were identified in the cprT-cprK and cprK-cprZ intergenic regions and the promoter at cprB, suggesting a role for FNR-like CprK in the control of expression of the cprTKZEBACD genes.
Halorespiring bacteria have received increasing attention during the last decade, as they are able to couple the reductive dehalogenation of a large variety of halogenated aromatic and aliphatic hydrocarbons to energy conservation and hence to microbial growth. These compounds are present in the environment as a consequence of their past and present application in industry and agriculture and because of natural production, compromising environmental integrity and health (14, 15). Halorespiring bacteria are believed to play an important role in the in situ bioremediation of soil and groundwater polluted with halogenated hydrocarbons. The ability to perform halorespiration appears to be widespread throughout the Bacteria, as halorespiring bacteria have been found in the groups of low-G+C-content gram-positive bacteria, green nonsulfur bacteria, and δ and ɛ proteobacteria (9). Among these, the gram-positive genus Desulfitobacterium comprises a major group of isolates. The versatile Desulfitobacterium dehalogenans has been isolated because of its ability to use o-halogenated phenolic compounds as terminal electron acceptors in an anaerobic respiratory chain with lactate, pyruvate, formate, and molecular hydrogen as electron donors (38). Recently, the reductive dehalogenation of tetrachloroethene and hydroxylated polychlorinated biphenyls by the halorespirational system of D. dehalogenans has been reported (42).
In order to understand the molecular basis of this novel respiratory system, efforts have focused not only on the reductive dehalogenases as the central enzymes in halorespiration (16, 22, 40) but also on the identification of additional structural and regulatory components of the halorespiratory electron transport chain. An efficient conjugation system has been used for the integration of conjugative transposon Tn916 into the chromosome of D. dehalogenans, leading to the isolation of a number of halorespiration-deficient mutants, which were characterized at the physiological, biochemical, and genetic levels (31).
It is known from physiological experiments that halorespiration is induced by the presence of a halogenated substrate in most halorespiring bacteria described to date. For two halorespiring strains, Desulfomonile tiedjei and Desulfitobacterium frappieri TCE1, the influence of alternative electron acceptors on the activity of the dehalogenating system has been described, indicating that particularly sulfur oxyanions are potential inhibitors of halorespiration (12, 35). In contrast, expression of halorespiration by 3-chloro-4-hydroxyphenylacetic acid (Cl-OHPA) in nonacclimated cultures of D. dehalogenans was not affected by the presence of equimolar amounts of sulfite (20). However, the level at which regulation takes place, the control mechanisms involved, and the inducing signal remain to be elucidated.
This study addresses the molecular analysis of the regulation of reductive dehalogenation in a halorespiring bacterium. Chromosomal fragments flanking o-chlorophenol reductive dehalogenase-encoding gene cprA in D. dehalogenans were cloned and characterized, revealing the presence of open reading frames that encode polypeptides possibly involved in regulation and maturation of the dehalogenating system. The expression of the different genes identified in the cpr gene cluster was studied under various growth conditions and was found to be tightly controlled at the transcriptional level.
MATERIALS AND METHODS
Materials.
Cl-OHPA was purchased from Sigma-Aldrich Chemie (Zwijndrecht, The Netherlands) and filtered prior to use. All gases were obtained from Hoek Loos (Schiedam, The Netherlands). When appropriate, experiments were carried out in an anaerobic glove box (Coy Laboratories Products, Grass Lake, Mich.) under an atmosphere of 96% N2 and 4% H2. The oxygen concentration was kept low with the palladium catalyst RO-20, provided by BASF (Arnhem, The Netherlands).
Bacterial strains, plasmids, and growth and induction conditions.
D. dehalogenans strain JW/IU-DC1 (DSM 9161) (38) was routinely grown under anaerobic conditions (100% N2 gas phase) at 37°C in rubber-stoppered serum bottles containing basal mineral medium, as described by Neumann et al. (21), supplemented with 0.1% peptone, 30 mM NaHCO3, and trace elements and vitamin solution as recommended by the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). An electron donor and acceptor were added to a concentration of 20 mM from anaerobic stock solutions. Growth was monitored spectrophotometrically by determining the optical density at 600 nm (A600). The concentrations of Cl-OHPA and OHPA during growth in the presence of Cl-OHPA as the electron acceptor were determined by high-performance liquid chromatography analysis on a SpectraSystem high-performance liquid chromatograph, with a SpectraSystem P2000 pump, an AS3000 autosampler, and a UV1000 UV detector (ThermoQuest, Austin, Tex.). The sample (20 μl) was injected into a pesticide reversed-phase column (Chrompack, Middelburg, The Netherlands). The mobile phase was acetonitrile–0.01 M H3PO4 (10:90 [vol/vol]). A flow rate of 1 ml min−1 was applied, and Cl-OHPA and OHPA were quantified by their absorption at 206 nm. For the induction with Cl-OHPA, cells were grown with pyruvate to an A600 of 0.1 and supplemented with 5 mM Cl-OHPA. Samples were taken before and after induction and stored on ice prior to further processing.
Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) was used as a host for cloning vectors. The strain was grown in Luria-Bertani medium at 37°C (28), and ampicillin was added at 100 μg/ml when appropriate. The cloning vectors pUC18 and pUC19 were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden), and the PCR product cloning vectors pGEM-T and pMON38201 (3) were obtained from Promega (Madison, Wis.) and Monsanto (St. Louis, Mo.), respectively.
DNA isolation and manipulation.
Chromosomal DNA of D. dehalogenans was isolated as described previously (40). Plasmid DNA was isolated from E. coli by using the alkaline lysis method, and standard DNA manipulations were performed according to established procedures (28) and manufacturers' instructions. Enzymes were purchased from Life Technologies B.V. (Breda, The Netherlands), Roche Molecular Biochemicals (Mannheim, Germany), and New England Biolabs (Beverly, Mass.). Oligonucleotides were obtained from Eurogentec (Seraing, Belgium), Life Technologies B.V., and MWG Biotech (Ebersberg, Germany). PCR products were purified prior to subsequent manipulation using the QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany). A Hybond-N+ nylon transfer membrane (Amersham Pharmacia Biotech) was used for Southern blot analysis. Probes for hybridization experiments were labeled by nick translation in the presence of [α-32P]dATP (Amersham Pharmacia Biotech).
Sequence analysis of the cpr gene cluster.
In order to extend the sequence downstream of the cprA locus, as it was determined previously from pLUW910 and pLUW913 (40) (Fig. 1), a 0.9-kb HincII-EcoRI restriction fragment of pLUW910 was subcloned in pUC19, yielding pLUW910EH2, and sequenced. Subsequently, Southern blot analysis of HincII-HindIII-digested chromosomal DNA of D. dehalogenans revealed a 1.9-kb fragment that strongly hybridized with the aforementioned radiolabeled 0.9-kb fragment. The 1.9-kb fragment was cloned in E. coli using HincII-HindIII-digested pUC19, resulting in pLUW911. pLUW916 was obtained by inverse PCR (36), which was performed as described previously (40) from NcoI-digested and self-ligated chromosomal DNA of D. dehalogenans with the divergent primer pair BG580 and BG581 (BG580, positions 9023 to 9002, and BG581, positions 9671 to 9692, of the cpr gene cluster) (see below; Fig. 1). The resulting 1.8-kb PCR product was cloned in E. coli using XcmI-digested pMON38201. Subsequently, PCR was performed with chromosomal DNA of D. dehalogenans with primers BG581 and HS22 (positions 10260 to 10240) and HS23 and HS27 (HS23, positions 10237 to 10257, and HS27, positions 11117 to 11097). Both PCR products were cloned in E. coli using XcmI-digested pMON38201, yielding pLUW921 and pLUW922, respectively.
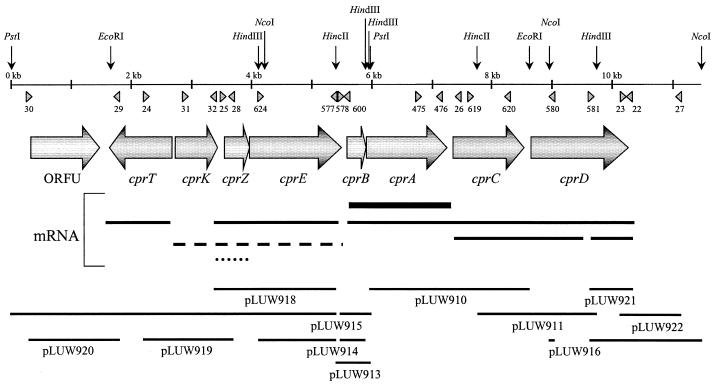
FIG. 1.
Physical map of the cpr gene locus in D. dehalogenans. Horizontal arrows, open reading frames; triangles, oligonucleotides used in this study; vertical arrows, DNA restriction sites which were relevant for the construction of clones (bars). mRNA: solid bars, apparent halorespiration-specific transcription products; dashed and dotted lines, apparent constitutive and halorespiration-repressed transcripts, respectively.
To elucidate the sequence upstream of cprB, inverse PCR products were obtained from HindIII- or PstI-digested and self-ligated chromosomal DNA using the divergent primer pair BG577 and BG578 (BG577, positions 5443 to 5423, and BG578, positions 5459 to 5485), resulting in pLUW914 and pLUW915, respectively. pLUW915EN was obtained by subcloning a 2.3-kb EcoRI-NcoI fragment of pLUW915 in E. coli using EcoRI-NcoI-digested pMON38201. Finally, pLUW918, pLUW919, and pLUW920 were obtained after PCR using chromosomal DNA as the template and primers BG577 and HS25, HS24 and HS28, and HS29 and HS30 (HS24, positions 2244 to 2267; HS25, positions 3521 to 3545; HS28, positions 3628 to 3605; HS29, positions 1768 to 1748; HS30, positions 266 to 284), respectively.
Using the above-mentioned set of clones, the almost-complete double-stranded cpr gene cluster nucleotide sequence could be elucidated. Where the sequence was only single stranded, sequence analysis of multiple, independently obtained PCR products was used to obtain an unambiguous result.
Amplification, cloning, and sequencing of rRNA genes.
The 16S rRNA-encoding gene was amplified from chromosomal DNA of D. dehalogenans with the universal primer pair 7f-1510r (19). Primers 1492f (19) and 23InsR (24) were used for the amplification of the 3′ and 5′ ends of the 16S and 23S rRNA genes, respectively, and the 16S-23S intergenic spacer. PCR products were cloned in the pGEM-T vector, yielding pLUW900 (16S) and pLUW901 (16S-23S), and their authenticity was verified by nucleotide sequence analysis.
DNA sequencing and DNA and protein sequence analysis.
DNA sequencing was performed using DNA sequencer 4000L (LiCor, Lincoln, Nebr.). Plasmid DNA used for sequencing reactions was purified with the QIAprep Spin Miniprep kit (Qiagen GmbH). Reactions were performed using the Thermo Sequenase fluorescently labeled primer cycle sequencing kit (Amersham Pharmacia Biotech). Fluorescently (IRD 800) labeled universal sequencing primers were purchased from MWG Biotech. Sequence similarity searches and alignments were performed using the BLAST, version 2.0, program (1) (National Center for Biotechnology Information, Bethesda, Md.) and the programs Clustal X, GeneDoc (34; K. B. Nicholas and H. B. J. Nicholas, GeneDoc: a tool for editing multiple sequence alignments, 1997), and the DNAstar package (DNASTAR Inc., Madison, Wis.). Protein secondary structure and helical transmembrane region predictions were made with the profile network systems PHDsec and PHDhtm (25–27), respectively. Prediction of helix-turn-helix (H-T-H) DNA-binding motifs was made using the method of Dodd and Egan (8).
Isolation of total RNA, Northern analysis, RT-PCR, and primer extension.
Total RNA was isolated from exponentially growing cultures of D. dehalogenans by the Macaloid method described by Kuipers et al. (18). For Northern blot analysis, 7 μg of RNA was separated on a formaldehyde–1% agarose gel, transferred to a Hybond-N+ nylon transfer membrane (Amersham Pharmacia Biotech) by downwards capillary transfer as described by Chomczynski (5) with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10 mM NaOH as the transfer liquid, and immobilized by UV cross-linking. Prehybridization and hybridization were performed in ULTRAhyb hybridization buffer (Ambion, Austin, Tex.) as recommended by the manufacturer.
A cprA-specific probe was generated by PCR amplification from chromosomal DNA of D. dehalogenans with the primer pair BG475 and BG476 (BG475, positions 6778 to 6797; BG476, positions 7213 to 7192). As probes specific for cprC, cprD, and cprE, PCR products that were obtained after PCR amplification with primer pairs BG619 and BG620 (BG619, positions 7632 to 7658, and BG620, positions 8282 to 8255), BG581 and HS22, and BG624 and BG577 (BG624, positions 4147 to 4171), respectively, were used. For the detection of transcription products of ORFU, cprT, cprK, and cprZE, probes were generated by PCR with primer pairs HS29 and HS30, HS24 and HS28, HS31 and HS32 (HS31, positions 2906 to 2927, and HS32, positions 3440 to 3419), and HS25 and BG577, respectively.
Reverse transcriptase PCR (RT-PCR) was performed to analyze the transcriptional organization of the genes in the cpr gene cluster. Five hundred nanograms of DNase-treated RNA (RQ1 RNase-free DNase; Promega) was used in a 25-μl reaction mixture containing the following: 25 pmol of each primer, 200 μM dATP, dCTP, dGTP, and dTTP, 1.7 mM MgSO4, 5 μl of avian myeloblastosis virus (AMV)–Tfl 5× reaction buffer, and 2.5 U of AMV RT and Tfl polymerase (Access RT-PCR system; Promega). In negative controls, AMV RT was omitted, and chromosomal DNA of D. dehalogenans was added to positive-control reaction mixtures. cDNA synthesis and subsequent PCR amplification were performed using the GeneAmp PCR system 9700 (Perkin-Elmer Cetus, Norwalk, Conn.). The reaction mixture was incubated at 48°C for 45 min. After the mixture was preheated to 94°C for 2 min, 40 amplification cycles, consisting of denaturation at 94°C for 20 s, primer annealing at 50°C for 30 s, and elongation at 68°C for 1 min 30 s, were performed. A final extension of 7 min at 68°C was performed.
Primer extension analysis was performed to determine the transcription start sites of the cprBA and cprCD transcripts. For this purpose, 10 or 30 μg of RNA and 4 pmol of the fluorescently (IRD 800) labeled oligonucleotides BG600 (positions 5670 to 5648) and HS26 (positions 7476 to 7456), respectively, were dissolved in 10 μl of 1× RT buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol), incubated at 70°C for 5 min, and slowly cooled to room temperature. After addition of 10 μl of 1× RT buffer containing 2 mM dATP, dCTP, dGTP, and dTTP, 10 U of RNasin, and 200 U of Superscript II RT (Life Technologies), the sample was incubated at 48°C for 30 min. RNase (0.2 mg/ml) was added, and the sample was precipitated with ethanol and washed once with 70% ethanol. The pellet was dried, dissolved in 2 μl of formamide loading buffer, and separated on a Li-Cor 4000L DNA sequencer.
Nucleotide sequence accession number.
The nucleotide sequence of the cpr gene cluster described here has been deposited in the GenBank database under accession no. AF115542.
RESULTS
Genetic organization of the cpr gene cluster.
Previously we analyzed the nucleotide sequence of the cprBA genes that encode the catalytic subunit and putative membrane anchor of the o-chlorophenol reductive dehalogenase in D. dehalogenans (40). Here we report the transcriptional organization of the cprBA genes. The chromosomal regions flanking the cluster were characterized by sequence and transcriptional analysis, revealing the presence of six additional transcribed genes, tentatively designated cprC, cprD, cprE, cprK, cprT, and cprZ, and one untranscribed open reading frame, ORFU (Fig. 1; see below). With the exception of cprT, all genes are transcribed in the same direction as cprBA. In front of each of the genes potential Shine-Dalgarno sequences are present; these sequences are complementary to the 3′ end of the D. dehalogenans 16S rRNA (3′-AGAAUCUUUCCUCCA-5′; see below). The cprC gene, previously designated orf1 (40), is located downstream of the structural gene for the o-chlorophenol reductive dehalogenase (cprA) and is predicted to encode a polypeptide of 395 amino acids with a molecular mass of 43,867 Da. Secondary structure prediction suggests the presence of six transmembrane helices (Fig. 2). Within the C-terminal cytoplasmic domains, two cysteine-rich signatures of the type CXXXCP were identified. For the C-terminal part of the predicted protein, CprC, significant similarity was observed with membrane-bound regulators of the NosR/NirI type, which have been shown to play a role in a signal transduction pathway that eventually controls the transcription of the nitrous oxide (nos) and nitrite reductase (nir) gene clusters of Pseudomonas stutzeri and Paracoccus denitrificans, respectively (7, 30). This similarity was most pronounced in the vicinity of the cysteine-rich motifs (Fig. 2). However, CprC is significantly smaller than known proteins of the NosR/NirI family, due to a much shorter N-terminal extracytoplasmic loop (approximately 170 amino acids compared to 350) and the lack of a C-terminal cytoplasmic domain containing two additional ferredoxin-like motifs binding either [2Fe-2S] or [4Fe-4S] clusters in NosR and NirI (4).
FIG. 2.
Amino acid sequence alignment of CprC from D. dehalogenans with NosR from Pseudomonas stutzeri (accession no. Q00790; PsNosR) and NirI from Paracoccus denitrificans (accession no. AJ001308; PdNirI). Residues conserved between two or all sequences are highlighted in gray and black, respectively. Horizontal bars, cysteine-rich motifs; boxes, predicted transmembrane helices; i and o, intra- and extracytoplasmic domains, respectively.
Downstream of cprC and upstream of cprB, two other genes, cprD and cprE (the latter was previously designated orfX [40]), were identified, both potentially coding for chaperonins of the GroEL type (Fig. 1). The predicted gene products of cprD and cprE are polypeptides of 537 and 516 amino acids with calculated molecular masses of 58,002 and 54,632 Da, respectively. The two proteins have a sequence identity of 34% at the amino acid level. Highest values of sequence similarity were observed with proteins from Thermus thermophilus (P45746; 45% identity at the protein level for CprD and 40% for CprE) and Clostridium thermocellum (P48212; 44% identity for CprD and 39% for CprE).
Upstream of cprE, the cprK, cprT, and cprZ genes and open reading frame ORFU were identified (Fig. 1). ORFU potentially encodes a polypeptide of 388 amino acids with a calculated molecular mass of 43,887 Da with no homologue in the databases. The predicted gene product of cprT is a polypeptide of 311 amino acids with a molecular mass of 35,667 Da. CprT exhibits significant similarity to the trigger factor, a peptidyl prolyl isomerase that is considered to act as a protein folding catalyst (11). Highest similarities were to RopA from Streptococcus pyogenes (AAC82391; 15% identity at the amino acid level) and the trigger factor from Bacillus subtilis (P80698; 14% identity). Relatively low values of sequence identity are mainly caused by the fact that CprT lacks an approximately 110-amino-acid N-terminal region which is present in known trigger factor homologues. CprT, however, still contains the complete FKBP domain, which is associated with the peptidyl prolyl isomerase activity of known Trigger factors (11). Downstream of cprT is the location of cprK, which could encode a polypeptide of 233 residues with a calculated molecular mass of 26,646 Da. CprK revealed low, but significant, sequence similarity to known members of the CRP-FNR family of transcriptional regulators (Fig. 3). Preliminary results indicate that CprK deeply branches within subclass III (NtcA) of the CPR-FNR family (41). By applying the method of Dodd and Egan (8), an H-T-H motif could be predicted with 71% probability, aligning with the H-T-H motif that is conserved among members of the CRP-FNR family. However, the sequence E--SR, which is conserved in the recognition helix of nearly all FNR-like proteins, is only partially conserved in CprK (Fig. 3). This suggests that the recognition motif for CprK binding might be different from the common FNR box TTGAT-N4-ATCAA (Fig. 3) (44). cprZ is located downstream of cprK, overlapping with cprE over four nucleotides, and may code for a polypeptide of 138 amino acids with a calculated molecular mass of 15,546 Da. Significant sequence similarity of 24% identity on the protein level was observed only with a hypothetical protein from Synechocystis sp. (BAA17004). Substantial conservation of the CprZ N terminus and a well-defined ribosome binding site suggested an alternative translation start codon (GTG) for cprZ.
FIG. 3.
Alignment of CprK with proteins of the CRP-FNR family of regulatory proteins (Lv-PrfA, Listeria ivanovii listeriolysin regulatory protein PrfA, accession no. CAA51231; Dr, Deinococcus radiodurans putative transcriptional regulator, accession no. AAF11910; Ec-FNR, E. coli FNR, accession no. P03019; Rm-FixK, Rhizobium meliloti FixK, accession no. S04122; Rs-NNR, Rhodobacter sphaeroides NNR-like protein, accession no. AAD27624; Pd-NNR, Paracoccus denitrificans Fnr-like transcriptional activator, accession no. AAA69977.1). Cysteine residues and residues conserved between three or more sequences are highlighted in black and gray, respectively. Asterisks, cysteine residues essential for activity of the E. coli FNR protein. Predicted H-T-H DNA-binding motifs are indicated.
Transcriptional analysis of the cpr gene cluster.
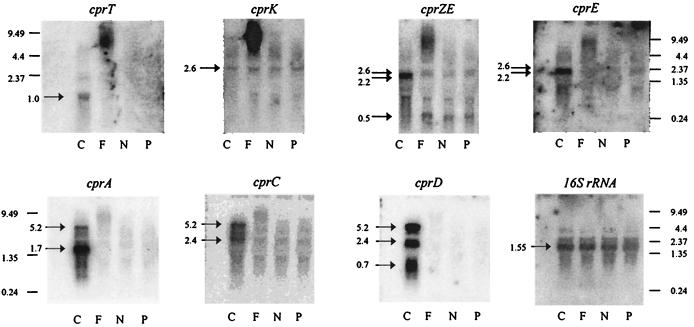
The observation that the cprBA genes for the o-chlorophenol reductive dehalogenase are flanked by five genes that could encode proteins which can be expected to play a role in regulation, maturation, or action of CprA prompted us to investigate the transcription of these genes under different conditions. Northern blot analysis was performed on total RNA isolated from cells grown with pyruvate as the electron donor and either Cl-OHPA, fumarate, nitrate, or pyruvate as the electron acceptor. The sizes of the transcripts were estimated by comparison with RNA molecular weight markers. Hybridization with a 32P-labeled cprA-specific probe revealed the presence of two transcripts of approximately 1.7 and 5.2 kb, which were solely detectable in RNA isolated from cells grown by halorespiration (Fig. 4). The major hybridizing transcript of approximately 1.7 kb indicated cotranscription of the structural gene cprA with cprB, as was anticipated because of the fact that both genes are only 12 nucleotides apart (40) (Fig. 4). Hybridization with probes specific for genes downstream of cprA unveiled transcripts of approximately 2.4 and 5.2 kb for cprC and 0.7, 2.4, and 5.2 kb for cprD. All transcripts were observed only in RNA obtained from cells grown with Cl-OHPA as the electron acceptor (Fig. 4). The presence of a large transcript of about 5.2 kb hybridizing with probes specific for cprA, cprC, and cprD and its concentration relative to that of the major 1.7-kb cprBA transcript, indicate occasional read-through after cprA. This would imply that the cprBACD genes are cotranscribed from the promoter preceding cprB (see below). The smaller transcripts (2.4 and 0.7 kb) detected with the cprC- and cprD-specific probes could be products of posttranscriptional processing of either the large 5.2-kb polycistronic cprBACD mRNA or a 3-kb biscistronic cprCD transcript transcribed from a promoter preceding cprC. Hybridization with probes specific for cprE, cprZE, and cprT indicated a bicistronic transcript of cprE and cprZ and monocistronic transcription of cprT specifically induced under halorespiring conditions (Fig. 4). A small 0.5-kb transcript that was detected with the cprZE-specific probe in RNA from cells grown on pyruvate, fumarate, or nitrate as the electron acceptor was absent from halorespiring cells, indicating that halorespiration induced read-through between cprZ and cprE. A transcript of approximately 2.6 kb could be detected with a probe specific for cprK, which was constitutively produced at very low levels. The same transcript was also detected with probes specific for cprE and cprZE, indicating constitutive transcription of a tricistronic cprKZE transcript (Fig. 4). Transcription of ORFU could not be detected by Northern blot analysis.
FIG. 4.
Northern blot analysis of total RNA extracted from cells of D. dehalogenans grown with pyruvate as the electron donor and various electron acceptors (C, Cl-OHPA; F, fumarate; N, nitrate; P, pyruvate). 32P-labeled probes that were specific for genes present at the cpr gene locus and the 16S rRNA-encoding gene of D. dehalogenans were applied. RNA size markers are in kilobases. Arrows, specific hybridizing signals that were obtained after 3- to 48-h exposures. The high-molecular-weight hybridization signals obtained with RNA isolated from fumarate-grown cells are due to residual amounts of chromosomal DNA.
To verify the transcriptional organization of the cpr gene cluster as proposed from the results of the Northern blot analysis, RT-PCR was performed using primer pairs that were designed to detect (i) transcription of each single gene and (ii) cotranscription of two neighboring genes. The results was in perfect agreement with the Northern blot analysis, i.e., cotranscription of cprB-cprA, cprA-cprC, cprC-cprD, and cprK-cprZ could be demonstrated, whereas no RT-PCR product was obtained for cprE-cprB (data not shown).
Transcription initiation from putative cprB and cprC promoters.
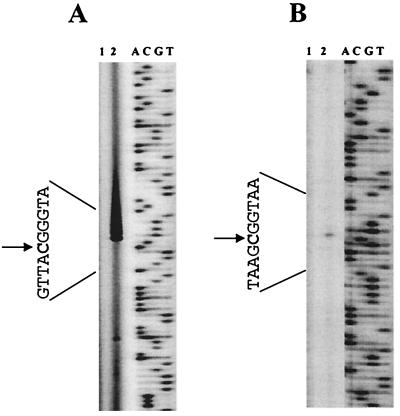
The results obtained by Northern blot analysis and RT-PCR indicated transcription initiation under halorespiring conditions from a promoter preceding cprB. The start site of Cl-OHPA-specific transcription from the cprB promoter could be identified 43 nucleotides upstream of the translation start site by primer extension using total RNA extracted from cells of D. dehalogenans grown by halorespiration with pyruvate as the electron donor and Cl-OHPA as the electron acceptor (Fig. 5A). No primer extension product was obtained with RNA isolated from cells grown by pyruvate fermentation. Upstream of the transcription initiation site, the consensus sequence for a −10 region could be detected, but no consensus −35 region was observed (Fig. 6A). Northern blot analysis also suggested posttranscriptional processing of a polycistronic mRNA or another transcription initiation at a site preceding cprC. A halorespiration-specific primer extension product indicated that this site is localized 58 nucleotides upstream of the translation start of cprC (Fig. 5B).
FIG. 5.
Analysis of the transcription initiation sites (arrows) at the cprB (A) and cprC (B) promoters by primer extension. Primer extension was performed with RNA isolated from cells grown on pyruvate (lane 1) or pyruvate and Cl-OHPA (lane 2). Primer extension products were electrophoresed in parallel with a sequence ladder (lanes A, C, G, and T) generated with the same primer on the noncoding strands of cprB and cprC, respectively.
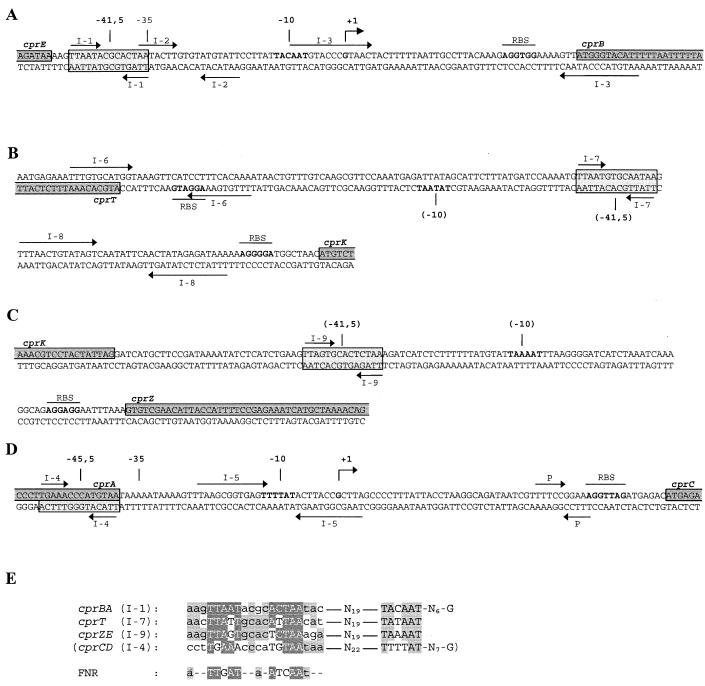
FIG. 6.
Detailed analysis of the cprE-cprB (A), cprT-cprK (B), cprK-cprZ (C), and cprA-cprC (D) intergenic regions. Bent arrows (+1), apparent transcription initiation sites; boldface, apparent and putative −10 regions and ribosome binding sites (RBS); horizontal arrows, palindromic sequences (P) and inverted repeats (I-1 to I-9); parentheses, hypothetical elements of putative promoters preceding the cprT and cprZ genes; dark and light gray boxes, protein-encoding sequences and FNR box-like motifs, respectively. (E) Alignment of putative CprK recognition motifs with the E. coli FNR recognition consensus motif (32). Conserved residues within the recognition motifs and at apparent and putative −10 consensus motifs are in gray.
Kinetics of induction of cprBA operon expression.
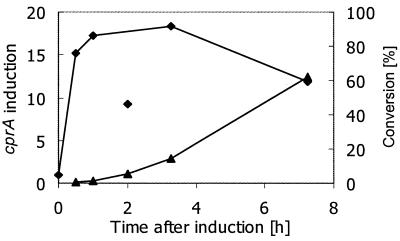
To investigate the induction kinetics of cprBA expression under halorespiring conditions, cells grown with pyruvate as the sole carbon and energy source were amended with Cl-OHPA during exponential growth and cprBA transcription and dechlorination of Cl-OHPA to OHPA were determined. Normalized by comparison to the 16S rRNA levels, transcription of cprBA was already induced 15-fold 30 min after induction, whereas significant amounts of the dechlorination product, OHPA (5.8% of Cl-OHPA added), were detected after 2 h (Fig. 7). Considering a specific growth rate of approximately 0.2 h−1 (generation time [tD] ≈ 3 h), the massive induction of halorespiration-specific transcription within 0.15 × tD is remarkably fast. Maximal induction of 18-fold was observed 3 h after addition of Cl-OHPA.
FIG. 7.
Kinetics of induction of cprBA transcription and dechlorination of Cl-OHPA. Northern blot analysis was performed with total RNA extracted from D. dehalogenans cells grown on pyruvate that were amended with Cl-OHPA during exponential growth. Relative transcription values were obtained after quantification of hybridization signals with 32P-labeled probes that were specific for cprA and the 16S rRNA-encoding gene of D. dehalogenans (⧫). The unusual ratio obtained after 2 h of induction may be due to degradation of the cprBA transcript compared to the 16S rRNA. Concentrations of Cl-OHPA and OHPA were determined by reverse-phase high-performance liquid chromatography analysis. The degree of conversion of Cl-OHPA to OHPA was calculated as [OHPA] × 100%/([Cl-OHPA] + [OHPA]) (▴).
DISCUSSION
Halorespiring bacteria have been demonstrated to be actively involved in the reductive dehalogenation of chlorinated aliphatic and aromatic compounds of natural and anthropogenic origin. Hence they contribute significantly to the detoxification of these contaminants in the environment (10). The reductively dehalogenating enzymes from a limited number of halorespiring bacteria have been characterized at the biochemical and genetic levels, indicating similar modes of catalytic action (16). However, insight into the regulatory circuits involved in the induction and repression of the halorespiration process is still very limited. For the first time, we here present a molecular analysis of the control of halorespiration-specific gene expression.
We have cloned and sequenced extended chromosomal fragments up- and downstream of the ortho-chlorophenol reductive dehalogenase-encoding cprBA gene cluster in halorespiring D. dehalogenans. Previously, we showed that cprA codes for a preprotein containing a twin-arginine (RR) signal sequence (40). This signal peptide is cleaved off in the mature protein and is thought to play a major role in the maturation and translocation of mainly periplasmic proteins binding different redox cofactors by the recently described twin-arginine translocation system (2). Putative functions of the newly detected open reading frames could in most cases be assigned by the similarity of the products to proteins present in the databases. CprC and CprK are potentially involved in regulation of transcription at different levels, whereas CprD, CprE, and CprT have significant similarity to molecular chaperones involved in the correct folding, processing, and assembly of proteins. However, no function could be assigned to the predicted gene products of ORFU and cprZ.
CprD and CprE belong to the family of GroEL chaperonins, which are found in prokaryotes, chloroplasts, and mitochondria. GroEL chaperonins are tetradecameric proteins that are involved in preventing protein aggregation and facilitating protein folding and assembly (11). In addition, it has been suggested that accessory proteins such as GroEL might play a role in correct assembly and cofactor insertion during the maturation of RR signal peptide-containing proteins, such as the reductive dehalogenases from halorespiring bacteria (2, 29). CprT has significant similarity to the trigger factor, a prolyl peptidyl isomerase that catalyzes proline cis-trans isomerization, a potential rate-limiting step in protein folding (11). Studies on the trigger factor from E. coli showed its association with nascent polypeptide chains and the 50S ribosome, suggesting a role in protein folding (33, 39). Interestingly, complexes between trigger factor and GroEL formed in vivo have been reported to have a much higher affinity for partially folded polypeptides than GroEL alone (17). Moreover, overexpression of trigger factor together with GroEL and GroES significantly improved the solubility of recombinant proteins, normally prone to aggregation into inclusion bodies (23). Unlike known trigger factors, CprT lacks part of the N-terminal domain, which is noncatalytic but which appears to be required for full activity in protein folding (43). Nonetheless, the specific coordinated expression of cprD, cprE, and cprT under halorespiring conditions suggests a synergistic role of these molecular chaperones in the maturation of the dehalogenating complex.
Northern blot analysis and RT-PCR revealed that the transcription of almost all genes identified in the cpr gene cluster is induced under halorespiring growth conditions, whereas no or significantly less-abundant transcription was observed under pyruvate-fermenting and fumarate- or nitrate-respiring conditions. We could reveal the transcriptional organization of the locus, i.e., two biscistronic units, cprZE and cprBA, with occasional read-through at cprC, yielding expression of the polycistronic cprBACD genes. Possibly, transcription of a third biscistronic unit, cprCD, might be initiated at a promoter preceding cprC. cprT, encoding a trigger factor, is obviously transcribed into a monocistronic mRNA (Fig. 4). Low-level constitutive transcription under all tested conditions was solely observed for tricistronic transcript cprKZE. Transcription from the cprB promoter was strongly induced within 30 min upon the addition of Cl-OHPA to cells growing by fermentation of pyruvate with concomitant dehalogenation of Cl-OHPA to OHPA, indicating o-chlorophenol reductive dehalogenase activity. This is in agreement with the earlier result that dehalogenation cannot be induced in the presence of chloramphenicol, showing that activation of dehalogenation requires de novo protein synthesis (37).
Sequence analysis revealed the presence of gene cprK, constitutively expressed at a low level, encoding a potential transcription regulatory protein. The observed tight control of the expression of the structural and putative accessory cpr genes might imply a direct involvement of CprK in the functionality of the D. dehalogenans halorespirational system. CprK has significant similarity to FNR- and FixK-like regulators, which are important trans-acting factors in regulatory networks of anaerobic assimilation and dissimilation. Like FixK, CprK lacks the N-terminal cysteine cluster, a characteristic of FNR, which is involved in the binding of an Fe/S center, and as such in redox sensing (44). However, CprK does show an unusually high content of five cysteine residues, among which is the conserved internal cysteine residue Cys105. In the E. coli FNR protein, the corresponding Cys122 has been shown to be essential for Fe binding, disulfide bond formation, and covalent modification (13). The FNR- and FixK-like regulatory proteins share a common conserved DNA-binding motif in their C-terminal recognition helices, which is complementary to a palindromic recognition motif, the so-called FNR or anaerobox, in the promoter of target genes (TTGAT-N4-ATCAA) (32). In positively regulated promoters, this FNR binding motif is preferentially centered at a distance of 41.5 nucleotides upstream of the transcription start (32). Inspection of the mapped halorespiration-inducible cprB promoter showed that it lacks the −35 consensus motif of strong constitutive promoters but does contain an anaerobox-like palindromic structure (I-1; TTAAT-N4-ACTAA). This putative regulatory protein binding motif is centered 41.5 nucleotides upstream of the apparent transcription start, suggesting positive regulation of transcription by an FNR-like factor (Fig. 6A). Another interesting feature of the cprB promoter is the presence and position of an additional long inverted repeat (I-3) that overlaps with the transcription start site, suggesting a function in control of transcription initiation. Northern blot analysis revealed that expression from putative promoters preceding cprT and cprZ was also stimulated under halorespiring conditions. FNR box-like motifs centered 87.5 and 77.5 bp upstream of cprT and cprZ, respectively (I-7 and I-9; Fig. 6B and C), could be identified. Moreover, in both cases conserved −10 consensus motifs were found at the same distance (19 bp) downstream of the FNR box-like motifs as in the mapped cprB promoter (Fig. 6B, C, and E). Both the lower degree of conservation of the proposed FNR box-like motif at cprC (Fig. 6D and E) and the small difference in spacing to the transcription start (45.5 bp instead of 41.5 bp) and a less well conserved −10 motif favor the idea that the mapped start site of the cprCD mRNA is the site of processing of the larger tetracistronic unit cprBACD rather than of transcription initiation.
In conclusion, it is tempting to speculate that the FNR homologue CprK is the factor binding to the different anaerobox-like motifs preceding at least the three halorespiration-inducible promoters present in the cpr gene cluster. An alignment of the identified motifs revealed a consensus sequence for all analyzed promoters that only slightly differs from that of the FNR box (Fig. 6E), which could reflect corresponding differences in the recognition helix of CprK (Fig. 3). CprK might be activated by the addition of a halogenated substrate, either directly or via an o-chlorophenol sensor, such as the two-component regulatory system that was previously detected from the detailed analysis of halorespiration-deficient mutants (31). If so, active CprK then induces transcription from the apparent cprB promoter and the putative cprT and cprZ promoters, generating the set of polypeptides required to obtain a functional dehalogenating complex. Such a model suggests a regulatory loop similar to that which has been proposed recently for nitrite reductase- and nitrous oxide reductase-encoding gene clusters from denitrifying bacteria. There, the expression of both the structural genes, nirS and nosZ, and of the membrane-bound regulator, encoded by nirI and nosR, is under the control of FNR-like regulatory proteins NNR and FnrD, respectively (6, 30). However, the exact function and mode of action of the NirI, NosR, and CprC regulatory proteins remain unknown. Interestingly, the analysis of the partially available genome sequence of the halorespiring Dehalococcoides ethenogenes (preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org) has revealed the occasionally close linkage of reductive dehalogenase-encoding genes with cprC and cprK homologues, as well as with genes potentially coding for two-component regulatory systems. This might serve as an additional, although only indirect, indication for the involvement of such regulatory proteins in the regulation of expression of reductive dehalogenases.
The molecular analysis of the cpr gene cluster reported here for the first time provides insight into the molecular bases of regulation and maturation of the halorespiratory system and suggests regulatory circuits which are similar to those proposed for respiratory complexes present in denitrifying bacteria. The remarkably fast induction of halorespiration-specific gene expression and its relative insensitivity to the presence of alternative electron acceptors (20; Smidt et al., unpublished results) indicate the potential of D. dehalogenans as a dedicated degrader in contaminated environments.
ACKNOWLEDGMENTS
This work was partly supported by a grant of the Studienstiftung des Deutschen Volkes and contract BIO4-98-0303 of the European Union.
Sequencing of Dehalococcoides ethenogenes was accomplished with support from the DOE Microbial Genome Program.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks B C, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 3.Borovkov A Y, Rivkin M I. XcmI-containing vector for direct cloning of PCR products. BioTechniques. 1997;22:812–814. doi: 10.2144/97225bm04. [DOI] [PubMed] [Google Scholar]
- 4.Bruschi M, Guerlesquin F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev. 1988;4:155–175. doi: 10.1111/j.1574-6968.1988.tb02741.x. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem. 1992;201:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- 6.Cuypers H, Berghoefer J, Zumft W G. Multiple nosZ promoters and anaerobic expression of nos genes necessary for Pseudomonas stutzerinitrous oxide reductase and assembly of its copper centers. Biochim Biophys Acta. 1995;1264:183–190. doi: 10.1016/0167-4781(95)00128-4. [DOI] [PubMed] [Google Scholar]
- 7.Cuypers H, Viebrock-Sambale A, Zumft W G. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J Bacteriol. 1992;174:5332–5339. doi: 10.1128/jb.174.16.5332-5339.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Fantroussi S, Naveau H, Agathos S N. Anaerobic dechlorinating bacteria. Biotechnol Prog. 1998;14:167–188. doi: 10.1021/bp980011k. [DOI] [PubMed] [Google Scholar]
- 10.Fetzner S. Bacterial dehalogenation. Appl Microbiol Biotechnol. 1998;50:633–657. doi: 10.1007/s002530051346. [DOI] [PubMed] [Google Scholar]
- 11.Fink A L. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 12.Gerritse J, Drzyzga O, Kloetstra G, Keijmel M, Wiersum L P, Hutson R, Collins M D, Gottschal J C. Influence of different electron donors and acceptors on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieriTCE1. Appl Environ Microbiol. 1999;65:5212–5221. doi: 10.1128/aem.65.12.5212-5221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green J, Sharrocks A D, Green B, Geisow M, Guest J R. Properties of FNR proteins substituted at each of the five cysteine residues. Mol Microbiol. 1993;8:61–68. doi: 10.1111/j.1365-2958.1993.tb01203.x. [DOI] [PubMed] [Google Scholar]
- 14.Gribble G W. Naturally occurring organohalogen compounds—a comprehensive survey. Fortschr Chem Org Naturst. 1996;68:1–423. [PubMed] [Google Scholar]
- 15.Hileman B. Concerns broaden over chlorine and chlorinated hydrocarbons. Chem Eng News. 1993;71:11–20. [Google Scholar]
- 16.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 17.Kandror O, Sherman M, Moerschell R, Goldberg A L. Trigger factor associates with GroEL in vivo and promotes its binding to certain polypeptides. J Biol Chem. 1997;272:1730–1734. doi: 10.1074/jbc.272.3.1730. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisIgenes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 19.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 20.Mackiewicz N, Wiegel J. Comparison of energy and growth yields for Desulfitobacterium dehalogenansduring utilization of chlorophenol and various traditional electron acceptors. Appl Environ Microbiol. 1998;64:352–355. doi: 10.1128/aem.64.1.352-355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann A, Scholz-Muramatsu H, Diekert G. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch Microbiol. 1994;162:295–301. doi: 10.1007/BF00301854. [DOI] [PubMed] [Google Scholar]
- 22.Neumann A, Wohlfarth G, Diekert G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J Bacteriol. 1998;180:4140–4145. doi: 10.1128/jb.180.16.4140-4145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishihara K, Kanemori M, Yanagi H, Yura T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl Environ Microbiol. 2000;66:884–889. doi: 10.1128/aem.66.3.884-889.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roller C, Ludwig W, Schleifer K H. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J Gen Microbiol. 1992;138:1167–1175. doi: 10.1099/00221287-138-6-1167. [DOI] [PubMed] [Google Scholar]
- 25.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 27.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Santini C L, Ize B, Chanal A, Muller M, Giordano G, Wu L F. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders N F W, Houben E N G, Koefoed S, de Weert S, Reijnders W N M, Westerhoff H V, de Boer A P N, van Spanning R J M. Transcription regulation of the nir gene cluster encoding nitrite reductase of Paracoccus denitrificansinvolves NNR and NirI, a novel type of membrane protein. Mol Microbiol. 1999;34:24–36. doi: 10.1046/j.1365-2958.1999.01563.x. [DOI] [PubMed] [Google Scholar]
- 31.Smidt H, Song D L, van der Oost J, de Vos W M. Random transposition by Tn916 in Desulfitobacterium dehalogenansallows for isolation and characterization of halorespiration-deficient mutants. J Bacteriol. 1999;181:6882–6888. doi: 10.1128/jb.181.22.6882-6888.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiro S, Guest J R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990;75:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 33.Stoller G, Rucknagel K P, Nierhaus K H, Schmid F X, Fischer G, Rahfeld J U. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 1995;14:4939–4948. doi: 10.1002/j.1460-2075.1995.tb00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend G T, Suflita J M. Influence of sulfur oxyanions on reductive dehalogenation activities in Desulfomonile tiedjei. Appl Environ Microbiol. 1997;63:3594–3599. doi: 10.1128/aem.63.9.3594-3599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Triglia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Utkin I, Dalton D D, Wiegel J. Specificity of reductive dehalogenation of substituted ortho-chlorophenols by Desulfitobacterium dehalogenansJW/IU-DC1. Appl Environ Microbiol. 1995;61:346–351. doi: 10.1128/aem.61.1.346-351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenansgen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 39.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Pas B A, Smidt H, Hagen W R, van der Oost J, Schraa G, Stams A J M, de Vos W M. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem. 1999;274:20287–20292. doi: 10.1074/jbc.274.29.20287. [DOI] [PubMed] [Google Scholar]
- 41.Vollack K U, Hartig E, Korner H, Zumft W G. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol Microbiol. 1999;31:1681–1694. doi: 10.1046/j.1365-2958.1999.01302.x. [DOI] [PubMed] [Google Scholar]
- 42.Wiegel J, Zhang X M, Wu Q Z. Anaerobic dehalogenation of hydroxylated polychlorinated biphenyls by Desulfitobacterium dehalogenans. Appl Environ Microbiol. 1999;65:2217–2221. doi: 10.1128/aem.65.5.2217-2221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarnt T, Tradler T, Stoller G, Scholz C, Schmid F X, Fischer G. Modular structure of the trigger factor required for high activity in protein folding. J Mol Biol. 1997;271:827–837. doi: 10.1006/jmbi.1997.1206. [DOI] [PubMed] [Google Scholar]
- 44.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]