Abstract
Three-dimensional (3D) bioprinting technology is an effective method for exploring the biological functions of hepatocytes by building biomimetic 3D microenvironments. Various hepatic tissue models have been developed for disease modeling, drug screening, and tissue regeneration using 3D bioprinting technology. Human-induced pluripotent stem cells (hiPSCs) are a promising cell source for the generation of functional hepatocytes for bioprinting. In this study, we introduced hiPSC-derived hepatocytes (hiPSC-Heps) as mature hepatocytes for the bioprinting of a 3D hepatic tissue model. The 3D-printed (3DP) model facilitated the formation of hiPSC-Hep spheroids with higher viability and proliferation than the commonly used non-printed sandwich-cultured model. hiPSC-Heps in the 3DP model exhibited higher mRNA expression of liver-specific functions than those in the two-dimensional-cultured model. Moreover, enhanced secretion of liver function-related proteins, including α-1-antitrypsin, albumin, and blood urea nitrogen, was observed in the 3DP model. For the evaluation of acetaminophen-induced hepatotoxicity, the 3DP model exhibited a favorable drug response with upregulation of the drug metabolism-related gene cytochrome P450-1A2 (CYP1A2). Overall, the bioprinted hepatic tissue model showed great biofunctional and drug-responsive performance, which could be potentially applied in in vitro toxicological studies.
Keywords: 3D bioprinting, Human-induced pluripotent stem cell, Hepatocytes, Sandwich culture, Hepatic model, Acetaminophen, Drug-induced hepatotoxicity
1. Introduction
Three-dimensional (3D) bioprinting is an emerging and promising technology for the construction of in vitro tissue and organ models. Incorporation of biocompatible biomaterials provides an effective method for reconstruction of the native cellular microenvironment for cell growth and biofunction maintenance[1]. Cells, biomaterials, and growth factors can be spatially arranged and assembled into complex heterogeneous architectures[2], and abundant cell-cell and cell-extracellular matrix (ECM) interactions have been established in bioprinted cell-laden structures. These characteristics could meet the demands of liver tissue engineering. Hepatocytes, as parenchymal cells of the liver, are attachment-dependent and require close interactions with other liver cells and within specific hepatic microenvironments[3]. The rapid loss of the normal phenotype and biological function of hepatocytes occurs in conventional two-dimensional (2D) cultures; however, hepatic tissue models created by 3D bioprinting have been shown to better facilitate the phenotype restoration and function preservation of hepatocytes, showing great superiority over 2D cultures[4].
To date, bioprinted hepatic tissue models have been widely used for disease modeling[5,6], drug screening[7], hepatotoxicity evaluation[8-10], and tissue regeneration[11,12]. Hepatocytes used in bioprinting mainly include hepatocarcinoma cell lines, primary human hepatocytes (PHHs)[11,13], and hepatocytes derived from human-induced pluripotent stem cells (hiPSCs)[14-16]. Among hepatocarcinoma cell lines, HepG2[8,17-19], Huh-7[20], and HepaRG[5,9,12] are commonly used in hepatic tissue models; however, these hepatoma cells cannot adequately represent typical hepatocytes because of a deficiency in liver-specific functions[21]. PHHs isolated from native liver tissue express excellent biofunctions but are difficult to culture and expand in vitro. hiPSC lines are capable of self-renewal. They can be expanded in vitro on a large scale and converted into hepatocyte-like cells through certain differentiation processes[22,23]. At present, most of the hepatocyte-like cells derived from hiPSCs retain a relatively immature phenotype and express limited metabolic functions compared to PHHs[24]. Nonetheless, hiPSCs could be a promising renewable and easily accessible cell source for the generation of functional hepatocytes which may obtain full maturity with the application of future biotechnologies. When applied in the bioprinting of hepatic tissue models using proper printing techniques and bioinks, hiPSC-derived hepatocytes (hiPSC-Heps) have demonstrated well-maintained cellular phenotypes and biofunctions[25]. Bioprinting of hiPSCs and human embryonic stem cells was first reported in 2015 by Faulkner-Jones et al.[14] The cells were bioprinted using RGD-coupled alginate bioink and then were further differentiated into hepatocyte-like cells. During the differentiation process after printing, the cells retained pluripotency with positive hepatocyte nuclear factor 4 alpha expressions, and they displayed biological functions and cellular morphology similar to functional hepatocytes. In another study by Ma et al.[15], hiPSC-derived hepatic progenitor cells (hiPSC-HPCs) were bioprinted into biomimetic hepatic lobule patterns using digital light processing-based 3D printing. In co-printing with human umbilical vein endothelial cells and adipose-derived stem cells, the phenotype and biofunction of hiPSC-HPCs were found to be enhanced in long-term culture. In addition, spheroid-based bioprinting has been shown to possess unique advantages in terms of biological characteristics of the spheroids[16]. Higher cell viability and better maintenance of hepatic function have been shown in bioprinted hepatoblast spheroids compared to bioprinted hepatoblasts using single-cell dispersion. However, most current studies use midterm-differentiated hepatoblasts or HPCs in bioprinting, which are involved in long-term, post-printing differentiation. Methods for in vitro differentiation into hepatocytes through bioprinting remain relatively underdeveloped. The bioprinting process and bioink components could potentially affect the differentiation efficiency of the cells, which needs to be optimized and standardized for successful bioprinting, and fully matured hepatocytes are required to construct hepatic tissue models for drug screening and toxicological studies[25]. In our previous study, we developed an effective differentiation system to generate large quantities of mature hepatocytes from hiPSCs[26]. Therefore, we considered maturating hiPSC-HPCs based on our well-developed differentiation protocol and then applied functional hepatocytes in the bioprinting of hepatic tissue models. hiPSC-Heps obtained using our optimized protocol showed excellent expression of liver-specific functions similar to PHHs, and they were successfully applied in bioartificial liver systems for acute liver failure treatment[26,27].
In this study, we bioprinted hiPSC-Heps using an alginate-gelatin bioink to construct a hepatic tissue model. We evaluated the success of cell growth, liver-specific function, and drug-induced hepatotoxicity of the 3D-printed (3DP) model compared with the conventional 2D-cultured (2D) and the non-printed sandwich-cultured (SW) models. The results of this study demonstrated the feasibility of hiPSC-Heps bioprinting using an alginate-gelatin bioink and confirmed that using the 3DP model showed biofunctional superiority, thus providing potential applications in the prediction of drug-induced liver injury.
2. Materials and methods
2.1. Cell culture and bioink preparation
The cell differentiation timeline from hiPSCs to hiPSC-Heps is shown in Figure 1A. hiPSC-Heps were obtained according to the manufacturer’s protocol[27], in which, hiPSCs were cultured in RPMI1640 medium with a combination of Activin A, BMP4, bFGF, B27, and Wnt3a for 1 day (24 h), and transferred to RPMI1640 medium with a combination of Activin A, BMP4, and bFGF for 3 days to induce differentiation into definitive endoderm cells. Subsequently, the endoderm cells were cultured in RPMI1640 medium with a combination of KGF, SB431542, and B27 for 2 days, then transferred to RPMI1640 medium with a combination of KGF, BMP4, BMP2, bFGF, and B27 for 3 days to produce hepatoblasts. Hepatoblasts were developed into hiPSC-HPCs in DMEM/F12 medium with a combination of B27, forskolin, SB431542, EGF, CHIR99021, LPA, Dex, and S1P for 6–8 days. Finally, hiPSC-HPCs were cultured in William’s E medium with a combination of B27, forskolin, and SB431542 for 21 days to completely mature into hiPSC-Heps. The final hiPSC-Heps were incubated at 37°C in a humidified 5% carbon dioxide (CO2) atmosphere and passaged every 3 days. To prepare the bioink, sodium alginate powder (A0682, Sigma-Aldrich, St Louis, MO, USA) and gelatin powder (G1890, Sigma-Aldrich) were dissolved in sterilized 0.9% NaCl (w/v) of 4% and 15%, respectively. Both solutions were sterilized in three rounds of repetitive heating (70°C) and cooling (25°C).
Figure 1.
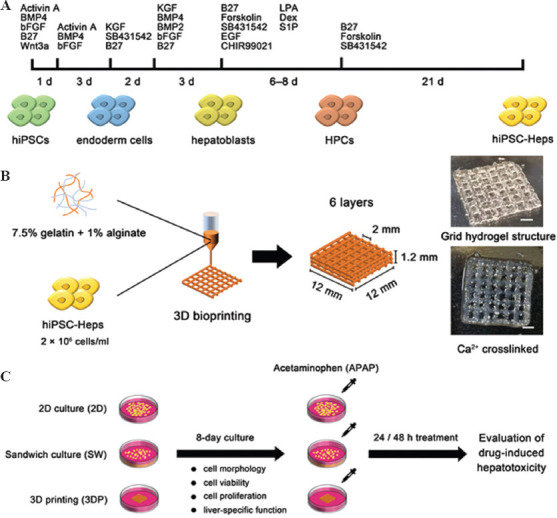
Schematic figure of cell differentiation, bioprinting process, and experimental timeline. (A) Cell differentiation timeline from hiPSCs to hiPSC-Heps. (B) Bioprinting process: hiPSC-Heps were bioprinted using alginate-gelatin bioink to form a grid hydrogel structure as a hepatic tissue model. (C) Experimental timeline: The 2D-cultured (2D), the sandwich-cultured (SW), and the 3DP hepatic tissue model were cultured for 8 days, then treated with acetaminophen (APAP) for 24 or 48 h to evaluate drug-induced hepatotoxicity (Scale bar: 200 μm). hiPSCs, human-induced pluripotent stem cells; hiPSC-Heps, human-induced pluripotent stem cell-derived hepatocytes; HPC, hepatic progenitor cells.
2.2. Bioprinting and culture of hepatic tissue models
hiPSC-Heps were harvested, and a cell suspension was formed at a density of 8 × 106 cells/mL. The cell suspension, 4% sodium alginate solution, and 15% gelatin solution were mixed at 37°C at a volume ratio of 1:1:2, resulting in a new solution of 7.5% gelatin and 1% sodium alginate with the suspended hiPSC-Heps at a density of 2 × 106 cells/mL. The cell-laden bioink was loaded into a syringe equipped with a 25 G printing nozzle. Grid-patterned hydrogel structures were bioprinted in a layer-by-layer fashion using an extrusive bioprinter (SUNP biomarker 2, SunP Biotech, Beijing, China). Briefly, a 3D cube model (12 × 12 × 1.2 mm) was uploaded and sliced using the bioprinter software. The sliced model consisted of six layers with a 0.2 mm layer height and a 2 mm line distance. The printer temperature was set at 21°C. Printing began 10 min after the syringe was loaded, to allow temperature equilibration. Before the start of printing, a 35 mm tissue culture plate was placed on the printing platform. The nozzle tip was manually moved to a height just touching the plate bottom. During printing, the nozzle moved along an automatically planned path, depositing bioink. After printing, the 3DP structures were ionically crosslinked for 3 min using a calcium dichloride (CaCl2) solution. A schematic of the bioprinting process is shown in Figure 1B. Moreover, non-printed sandwich cultures of hiPSC-Heps were performed by pipetting the bioink into 24-well culture plates, forming a hydrogel layer with the same height (1.2 mm) as the 3DP structures. In addition, hiPSC-Heps, at a density of 2 × 104 cells/cm2, were inoculated into 24-well culture plates for 2D cultures. An 8-day culture of the 2D, SW, and 3DP hepatic tissue models was implemented with culture medium refreshment occurring every other day. The experimental timeline for this study is shown in Figure 1C.
2.3. Spheroid morphology analysis and measurement of cell viability and proliferation
The morphology of the hiPSC-Heps spheroids was observed under a phase-contrast microscope (Eclipse TS100, Nikon, Tokyo, Japan). Cell viability was determined using the calcium acetoxymethyl ester and propidium iodide double staining method (C542, Dojindo, Kumamoto, Japan). The SW and 3DP models were incubated for 15 min in a staining solution containing 2 μg/mL calcium acetoxymethyl ester (green) and 3 μg/mL propidium iodide (red). The stained samples were observed under a confocal laser-scanning microscope (LSM710META, Zeiss, Oberkochen, Germany). The number of living and dead cells was counted to calculate the cell viability rates using ImageJ software. The cell viability data were analyzed using four randomly captured images from each sample. These fluorescent images were also used to measure the average diameter of hiPSC-Heps spheroids and the data were analyzed using ImageJ. Cell proliferation was measured using a Cell Counting Kit-8 (CCK8) (C10350, Dojindo), and finally, a standard curve was plotted using the known cell densities to convert the collected OD values to cell densities. All the experiments mentioned above were performed in triplicate.
2.4. Immunostaining
Samples from the 2D, SW, and 3DP models were collected for immunostaining after 8 days in culture. The samples were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100 for 20 min. Fluorescein isothiocyanate–conjugated phalloidin (ab176753, Abcam, Cambridge, UK) was used after permeabilization for cytoskeletal staining. The samples for ki-67 and albumin staining were subsequently blocked in 2% bovine albumin for another 2 h. Rabbit anti-human ki-67 (ab15580, Abcam) and rabbit anti-human albumin (ab207327, Abcam) were used as the primary antibodies. The blocked samples were incubated in the primary antibody solution overnight at 4°C and then treated with Alexa Fluor®488 (ab150077) - or Alexa Fluor®568 (ab175471) - conjugated goat anti-rabbit IgG (Abcam) as secondary antibodies. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole. The fluorescence images were captured using a confocal laser-scanning microscope (LSM710META, Zeiss, Oberkochen, Germany) by layer-by-layer scanning. All images were stacked and rebuilt to a single image. The mean fluorescence intensity of these images was analyzed using ImageJ to quantitatively determine the expression level of the biomarkers. Samples of each group were prepared in triplicate. Four randomly captured images from each sample were used for the analysis.
2.5. RNA extraction and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from hiPSC-Heps using TRIzol™ reagent (15596026, Invitrogen, Carlsbad, CA, USA). After RNA extraction, the concentration and purity of the RNA were measured using a spectrophotometer (Nanodrop 2000, Thermo Scientific, Waltham, MA, USA). Reverse transcription of RNA was performed using PrimeScript™ RT Master Mix (6210 A, TaKaRa, Kyoto, Japan), following the manufacturer’s protocol. The qRT-PCR reaction system was then prepared using Maxima SYBR Green qPCR Master Mix (RR420L, TaKaRa). Data collection was performed using a real-time PCR detection system (7500, Applied Biosystems, Carlsbad, CA, USA). The relative gene expression levels were calculated using the 2–DDCt method with b-actin selected as the housekeeping gene. Primers used for qRT-PCR are listed in Table A.1. Samples of each group were prepared in triplicate.
2.6. Enzyme-linked immunosorbent assay (ELISA) assay
The secretion of human albumin, a-1-antitrypsin (AAT), and blood urea nitrogen (BUN) was measured using a human albumin ELISA kit (EHALB, Invitrogen), a serpin A1 human ELISA kit (EHSERPINA1, Invitrogen), and a BUN colorimetric detection kit (EIABUN, Invitrogen), respectively. Culture supernatants were collected 24 h after the medium change on days 2, 4, 6, and 8. The number of cells at each time point for sample collection was measured using the CCK8 kit. The amounts of albumin, AAT, and BUN were calculated according to the standard curve, followed by data normalization per 1 million cells per day. A total of 12 individual samples (triplicates at each sampling time point) were used for data analysis.
2.7. Evaluation of drug-induced hepatotoxic response
In this study, acetaminophen (APAP) (A800441, Macklin, Shanghai, China) was used as a model drug for in vitro hepatotoxicity testing. Drug-induced hepatotoxicity in the 2D, SW, and 3DP models was evaluated after 8 days in culture. APAP stock solution was prepared by dissolving APAP powder in dimethyl sulfoxide, which made up 1% of the culture medium volume. Consecutive dilutions, ranging from 0 to 80 mM, were selected for drug testing. Cell viability was measured after 24 or 48 h of treatment using the CCK8 assay kit. Dose–response curves were obtained using the four-parameter variable slope-fitting method (GraphPad Prism 7.0). Internal controls were prepared with the same volume of culture medium without APAP. The half maximal inhibitory concentrations (IC50) were determined from respective dose-response curves. Samples of each group were prepared in triplicate.
2.8. Statistical analysis
Statistical analyses were performed using GraphPad Prism 7.0. All data are presented as the mean ± standard deviation (SD). Two-tailed Student’s t-test or one-way analysis of variance was used for two-group and multiple-group comparisons, respectively. Statistical significance is defined as *P < 0.05, **P < 0.01, and ***P < 0.001.
3. Results
3.1. Spheroid formation in the SW and the 3DP hepatic tissue model
In the 2D model, hiPSC-Heps attached to the bottom of culture plates and grew in a monolayer with no spheroid formed. In the SW and 3DP models, hiPSC-Heps were embedded in an alginate-gelatin hydrogel and surrounded by 3DP microenvironments. Cells proliferated and gradually formed spheroids during the culture period (Figure 2A). Figure 2B shows the average diameter of hiPSC-Heps spheroids in the SW and 3DP model. Interestingly, we observed that the spheroid growth was different between the central and edge regions of the 3DP model. Considering that the diffusion limit in avascular tissue is approximately 200 μm[28], we defined the region within 200 μm of the hydrogel edges as the edge region and that more than 200 μm from the edges as the central region. The average diameter of the hiPSC-Heps spheroids in both the central and edge regions of the 3DP model was significantly wider than that in the SW model from day 2 to day 4. However, the average diameter of the spheroids in the central region of the 3DP model was no longer significantly wider than that in the SW model from day 6 to day 8, while that in the edge region of the 3DP model was still found to be significantly wider. Notably, a significant difference in spheroid diameter between the central and edge regions of the 3DP model was observed on day 8, with 46.06 ± 6.38 μm and 70.25 ± 11.65 μm in the central and edge region, respectively. Figure 2C shows the live/dead fluorescent images of the SW and 3DP models on day 8. The average diameter of the spheroids located at different distances from the edge of the hydrogel structure was analyzed. We observed that most large spheroids were distributed toward the edge of the hydrogel structure in the SW and 3DP models. In the 3DP model, the spheroid diameter decreased rapidly as the distance to the edge of the hydrogel structure increased. However, this phenomenon was not seen in the SW model. These results suggest that the 3DP model is permissive for hiPSC-Heps to form larger spheroids, despite a concentrated distribution at the edge of the hydrogel fibers.
Figure 2.
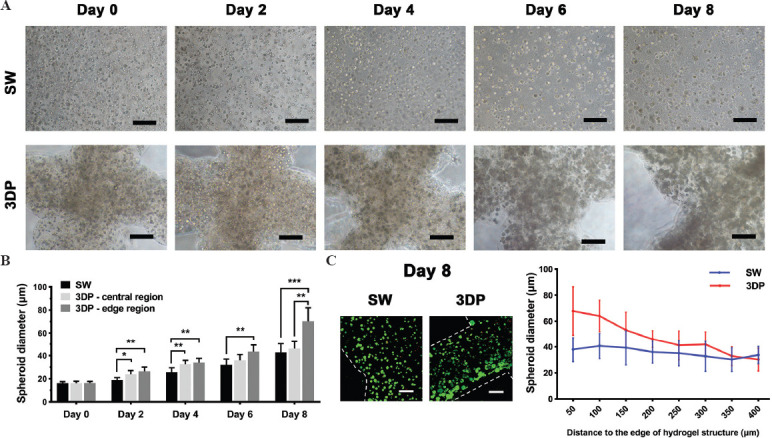
Formation of hiPSC-Heps spheroids in the SW and the 3DP hepatic tissue model. (A) Cellular morphology of hiPSC-Heps during 8 days of culture. (B) The average diameter of hiPSC-Heps spheroids in the SW and 3DP model. Central region: region more than 200 μm from the hydrogel edges; edge region: region within 200 μm of the hydrogel edges. (C) Variation of spheroid diameter in different regions of hydrogel structure on day 8. In the 3DP model, spheroid diameter rapidly decreased as the distance to the edge of hydrogel structure increased (Scale bar: 200 μm). Data are presented as means ± SD (n = 3). *P < 0.05, **P < 0.01.
3.2. Cell viability and proliferation in the SW and the 3DP hepatic tissue model
As mentioned above, cells in the two 3D-cultured models (SW and 3DP model) both formed spheroids after 8 days of culture. It is essential to compare the cell viability and proliferation to evaluate the cell growth during spheroid formation. As shown in Figure 3A, fluorescence images of living and dead cells on days 0, 2, 4, 6, and 8 were captured using the live/dead double staining method. The cell viability rates in the SW and 3DP models were calculated and are shown in Figure 3B. On day 0, cell viability in the 3DP model after the printing process was only 70.94% ± 3.44%, while hiPSC-Heps in the SW model maintained high viability close to 100%. However, after 2 days of culture, cell viability in the 3DP model significantly increased to 97.52% ± 0.80%, and a massive cell death occurred in the SW model with a sharp decrease in cell viability to 68.02% ± 5.09%. After 8 days of culture, cell viability in the 3DP model was significantly higher than that in the SW model. In both models, cell viability decreased moving from the peripheral to the central area of the hydrogel structure. According to the results in Figure 2C, it could be speculated that spheroid diameter may be a good proxy for cell viability.
Figure 3.
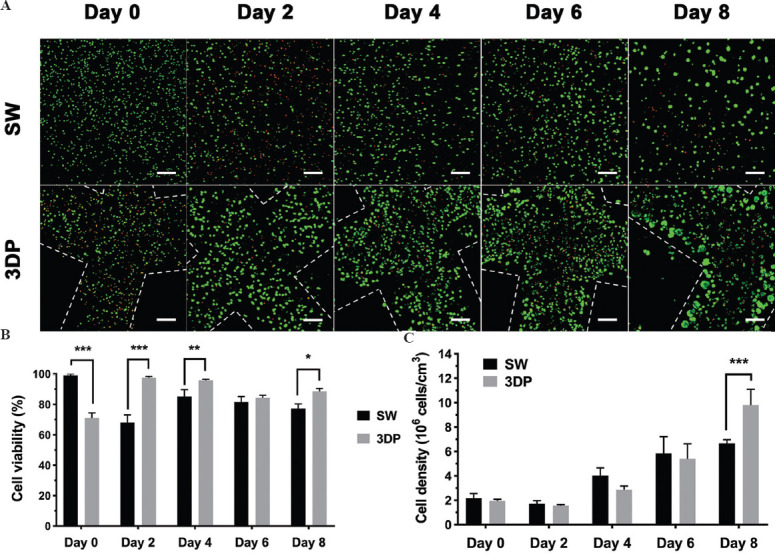
Cell viability and proliferation in the SW and the 3DP hepatic tissue model. (A) Images of live/dead double-staining during 8 days of culture. (B) Statistics of cell viability rate. (C) Statistics of cell density using CCK8 assay (Scale bar: 200 μm). Data are presented as means ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
Cell densities were calculated using the CCK8 assay to determine cell proliferation in the two models. As shown in Figure 3C, cell densities in the two models exhibited a slight decrease on day 2, followed by a continuous increase until day 8. On day 8, the cell density of the 3DP model reached at (9.79 ± 1.29) × 106 cells/cm3 and the SW model reached a density of (6.67 ± 0.30) × 106 cells/cm3, resulting in an approximately five- and three-fold increase from the initial seeding density (2 × 106 cells/cm3), respectively. On the basis of these results, it can be concluded that hiPSC-Heps exhibited higher viability and more robust proliferation in the 3DP model than hiPSC-Heps in the SW model.
3.3. Comparison of typical immunohistochemical characteristics
Immunostaining was performed to compare the typical immunohistochemical characteristics of the 2D, SW, and 3DP models. F-actin, ki67, and albumin were selected as biomarkers to assess cellular morphology, proliferation capability, and liver-specific function, respectively. Figure 4A shows the fluorescent images of the three models and Figure 4B shows the mean fluorescence intensity of these images. Widespread expression of F-actin and ki-67 was observed across the three models. As shown in Figure 4B, spheroid-formed hiPSC-Heps in the SW and the 3DP model exhibited stronger mean fluorescence intensity of F-actin and ki-67 than monolayer hiPSC-Heps in the 2D model. Despite the wide expression in the three models, stronger mean fluorescence intensity of F-actin and ki-67 in the SW and 3DP model revealed more integrated cellular morphology and active proliferation status of the hiPSC-Heps spheroids. Only a small fraction of hiPSC-Heps in the 2D model showed any expression of albumin, a marker of mature hepatocytes, whereas a significantly stronger mean fluorescence intensity with an extensive distribution throughout the spheroids indicated higher albumin expression in the SW and 3DP models. These results suggest the biofunctional superiority of hiPSC-Heps spheroids in 3D alginate-gelatin hydrogel cultures over monolayer cells in 2D cultures.
Figure 4.
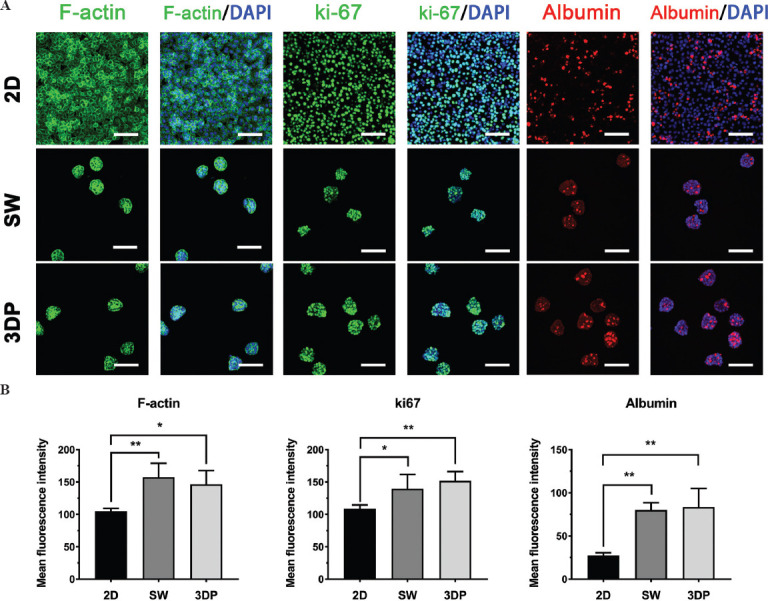
Immunohistochemical characteristics of the hepatic tissue models. Immunofluorescent images of F-actin, ki67, and albumin after 8 days of culture to evaluate cell morphology, proliferation ability, and liver-function expression, respectively (Scale bar: 100 μm). (A) F-actin: Green; ki67: Green; albumin: Red; DAPI: Blue. (B) Fluorescence intensity of F-actin, ki67, and albumin. Data are presented as means ± SD (n = 3). *P < 0.05, **P < 0.01.
3.4. Liver function-related mRNA and protein expression
We performed qRT-PCR and ELISA assays after 8 days of culture to determine the gene and protein expression of liver-specific functions in the three models. Biomarkers detected by qRT-PCR included cytochrome P450-1A2 (CYP1A2), cytochrome P450-3A4, AAT, tyrosine aminotransferase, albumin, transferrin, asialoglycoprotein receptor-1, and cytokeratin-18. As shown in Figure 5A, all biomarkers showed significantly higher expression levels in the 3DP model than those in the 2D model. Notably, high mean values and SDs of biomarker expression were observed in the SW model. However, no statistically significant difference was found between the SW model and the other two models. Figure 5B shows the secretion of AAT, albumin, and BUN. hiPSC-Heps in the 3DP model exhibited significantly higher levels of AAT, albumin, and BUN secretion than those in the other two models. The expressions of AAT and albumin were also found to be significantly higher in the SW model than those in the 2D model. However, no statistically significant difference was found in BUN secretion between the 2D and SW models. Interestingly, the results for gene expression and protein secretion of AAT and albumin were not consistent with each other. Despite the lower average level of gene expression in the 3DP model compared with that in the SW model, the higher quantitative secretion of these proteins in the 3DP model accurately reflected the liver-function activity of hiPSC-Heps.
Figure 5.
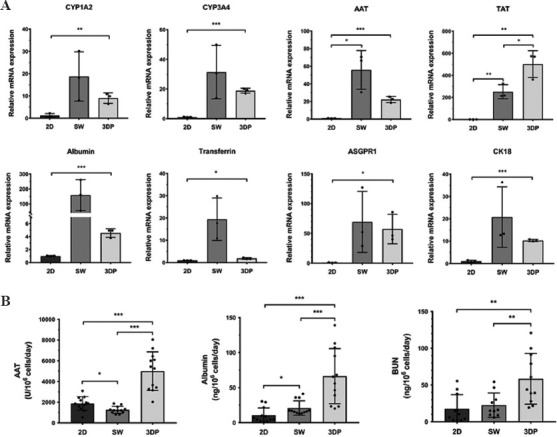
Liver function-related mRNA and protein expression in the 2D, SW, and 3DP hepatic tissue models. (A) Detection of liver function-related mRNA expression by qRT-PCR: cytochrome P450-1A2, cytochrome P450-3A4, α-1-Antitrypsin (AAT), tyrosine aminotransferase (TAT), albumin, transferrin, asialoglycoprotein receptor-1, and cytokeratin-18. Data are presented as means ± SD (n = 3). (B) Measurement of liver function-related proteins by ELISA: AAT, albumin, and blood urea nitrogen. Data are presented as means ± SD (n = 12). *P < 0.05, **P < 0.01, ***P < 0.001.
3.5. Assessment of drug-induced hepatotoxicity
After 8 days of culture, all three models were treated with APAP to evaluate drug-induced hepatotoxicity. Figure 6A shows the dose-response curves after 24 h of treatment with APAP. According to Table 1, the IC50 of the 2D, SW, and 3DP model after 24 h of drug treatment were 11.89, 26.48, and 49.54 mM, respectively. Therefore, we selected 10, 25, and 50 mM as the corresponding IC50 concentrations to further evaluate the metabolic activity of drug metabolism-related enzyme CYP1A2. The hiPSC-Heps in the 3DP model exhibited the strongest resistance to APAP, followed by those in the SW model. As shown in Figure 6B, the viability of hiPSC-Heps in all three models significantly decreased with prolonged drug exposure. The IC50 values after 48 h of treatment decreased to 0.89, 16.41, and 12.88 mM, respectively. At this point, the drug resistance of the 3DP model became slightly weaker than that of the SW model but still much stronger than that of the 2D model. Under the IC50 concentrations, the expression of the representative drug metabolic enzyme CYP1A2 was determined after 24 and 48 h of drug treatment. As shown in Figure 6C, CYP1A2 expression was significantly upregulated in all three models between 0 and 24 h. Between 24 h and 48 h, CYP1A2 expression was significantly upregulated in the SW and 3DP models, while no significant change was found in the 2D model. Moreover, spheroid-formed hiPSC-Heps in the SW and 3DP models presented higher levels of CYP1A2 expression than those in the 2D model.
Figure 6.
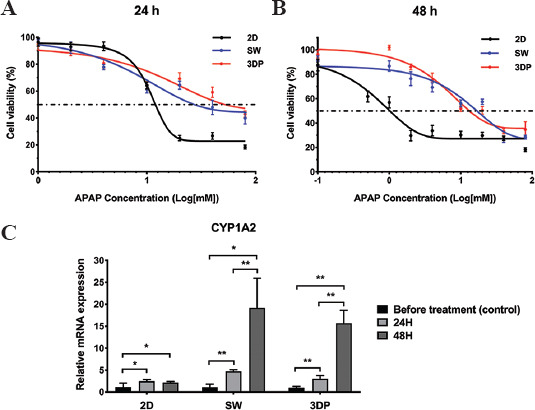
APAP-induced hepatotoxicity of the hepatic tissue models. (A) Dose-response curves after 24 h treatment of APAP. (B) Dose-response curves after 48 h of treatment with APAP. (C) Expression of cytochrome CYP1A2 measured at the 2 time points under the APAP exposure of IC50 concentrations. Data are presented as means ± SD (n = 3). *P < 0.05, **P < 0.01.
Table 1.
IC50 values (mM) of APAP-induced hepatotoxic response.
| 2D model | SW model | 3DP model | |
|---|---|---|---|
| 24 h treatment | 11.89 | 26.48 | 49.54 |
| 48 h treatment | 0.89 | 16.41 | 12.88 |
4. Discussion
With the development of 3D cell culture technologies, human hepatocytes have been used to construct 3D hepatic tissue models for a variety of biomedical applications. So far, methods of constructing 3D hepatic tissue models mainly include scaffold-free and scaffold-based approaches[6]. Scaffold-free cultures are aimed at generating spheroids without introducing external biomaterials. Cell suspensions self-aggregate into spheroids, avoiding cell adhesion onto substrates through gravitational, hydrodynamic, and electrostatic forces. Representative techniques are spinner flasks, rotary culture systems, non-adherent surfaces, hanging drop, and microwell arrays[29]. Among these techniques, arrayed platforms, including commercial ultra-low attachment culture plates[30], hanging drops[31], and microwells[32], have been applied widely for high-throughput drug screening. However, the absence of biomaterials in scaffold-free cultures leads to a lack of biochemical cues and inadequate recapitulation of actual hepatic microenvironments. Therefore, a myriad of natural and synthetic biomaterials with various chemical components and mechanical properties have been developed as porous bio-scaffolds to facilitate cell-cell and cell-ECM interactions in hepatic tissue models. At present, scaffold-based culture approaches mainly consist of cell microencapsulation[33], microfluidics[34,35], and 3D bioprinting[3,36]. Cell microencapsulation is a relatively easy way to generate liver microtissues, but this method does not always meet the standard of drug screening due to its low reproducibility. Artificial microstructures in microfluidic devices ensure the establishment of controllable microenvironments. However, high cost and specific requirements for fabrication technology limit the large-scale generation of hepatic tissues. Compared to cell microencapsulation and microfluidics, 3D bioprinting has unique benefits for the efficient, scalable, and reproducible fabrication of hepatic tissue models, which could easily meet the demands of high-throughput drug screening and toxicological tests.
Given these merits, we chose 3D bioprinting as the tissue model construction method for this study of hiPSCs, an easily accessible and highly expandable hepatic cell source. A standard type of bioink alginate-gelatin, which has been widely used for bioprinting various types of tissue models[37-39] including liver tissue[12,13,18], was employed to construct a simple and practical hepatic tissue model. Alginate, a natural polysaccharide with good biocompatibility, can be rapidly crosslinked by divalent ions, such as calcium ions (Ca2+). Despite their excellent formability, the lack of cell-binding domains leads to poor cell adhesion in pure alginate hydrogels[40]. Thus, peptide-containing gelatin is often added to enhance the cellular affinity of the bioink. By optimizing the bioink concentrations and printing parameters in our preliminary experiment, we successfully created a 12 × 12 × 1.2 mm grid hydrogel structure. Considering the high oxygen consumption of hepatocytes[41], the distance between adjacent hydrogel fibers was set to 2 mm to ensure sufficient oxygen supply to the medium.
In our study, we compared the cell growth, liver-specific function, and drug-induced hepatotoxicity of constructed 2D, SW, and 3DP models. In contrast to the 2D monolayer culture, cells are 3D-cultured in both the SW and 3DP models. Since the same material was used in the SW and 3DP models, topological structure is considered the major difference in microenvironment between the SW and 3DP models. In hydrogel structures, oxygen and nutrient supply to cells relies on diffusion through the culture medium. Cells in the peripheral region consume oxygen and nutrients and consequently hinder their penetration[42]. In the SW models, the cells in the inner region of a consolidated hydrogel layer experienced impaired medium supply. Compared to the consolidated hydrogel layer in the SW model, the interconnected microchannels of the 3DP grid structure allow greater inflow of culture medium, and thus may have facilitated mass transfer, supporting greater cell growth. In relevant studies of 3D bioprinting of cervical[43] and cholangiocarcinoma[44] tumor models, comparing 3DP models to SW models, it was found that cell spheroids in 3DP models were larger than those in SW models, indicating better cell growth in 3DP models. Similarly, in our study, hiPSC-Heps maintained active proliferation and formed larger spheroids in the 3DP model than those in the SW model. Nevertheless, diffusional gradients of oxygen and nutrients considerably limited the volume of spheroids formed in the inner region of the hydrogel fibers. We observed that a certain number of cell deaths occurred in the 3DP model immediately after the printing process, which could be attributed to shear stress during bioink extrusion. However, hiPSC-Heps rapidly recovered their high viability and started steady proliferation in the 3DP model. Conversely, in the SW model, the poor oxygen and nutrient supply in the inner region of the hydrogel layer could not support such a high density of cell culture, leading to substantial cell death. Therefore, 3D bioprinting plays a key role in efficient 3D culture of hiPSC-Heps by ameliorating oxygen and nutrient supply conditions.
Regarding biofunction in the 2D, SW, and 3DP models, spheroid-formed hiPSC-Heps in the SW and 3DP models showed significantly higher expression of albumin than did those in the 2D model according to the immunostaining results. Compared to monolayer cultures, the 3D spheroids in the hydrogel structures provide tight cell junctions for attachment-dependent hepatocytes, which promotes hepatocyte polarization and function maintenance[45]. In addition, the SW and 3DP models showed a higher mRNA expression of liver function-related biomarkers than did the 2D model. Notably, high SDs in the mRNA expression were observed in the SW model. On the one hand, the results were analyzed using only 3 data points, which could lead to large random errors. On the other hand, it could be also due to the rupture of the hydrogel structure in some samples, leading to unwanted changes in culture conditions. However, the secretion levels of AAT, albumin, and BUN were significantly higher in the 3DP model than those in either of the other two models, which clearly demonstrate the stronger liver functions of the 3DP model.
Finally, we conducted drug screening using APAP to evaluate drug-induced hepatotoxicity in the constructed hepatic tissue models. APAP overdose is the leading cause of acute liver failure in many countries. APAP induces liver impairment by causing mitochondrial damage and subsequent hepatocyte necrosis[46]. PHHs, HepaRG, and HepG2 cell lines are commonly used in APAP-induced hepatotoxic assays[47]. As an alternative hepatic cell source, the response behavior of hiPSC-Heps to APAP in the hepatic tissue models requires investigation. After 24 h of drug treatment, stronger drug resistance was observed in the 3DP model than in the SW model, possibly due to the larger and more viable spheroids formed in the 3DP model. However, cell viability in the 3DP model significantly decreased after 48 h of drug treatment. A pronounced upregulation of the drug metabolism-related enzyme CYP1A2 increased drug sensitivity, which is consistent with the findings of recent studies using other types of hepatocytes[8,48]. In addition, the hiPSC-Heps spheroids in our 3DP model displayed drug resistance comparable to that of hiPSC-derived hepatocyte spheroids generated by nanopillar plates[49] and 3D cellulosic scaffolds[50], suggesting good efficacy of the 3DP model in the assessment of APAP-induced hepatotoxicity. Overall, the 3DP model performed well in APAP drug test and proved its application value in in vitro hepatotoxicity evaluation.
Overall, 3D bioprinting of hiPSC-Heps proved to be an effective way to construct a functional hepatic tissue model. High cell viability, rapid cell proliferation, and enhanced liver-specific functions were observed in this 3DP model compared to other common models including SW and 2D models. This hepatic tissue model has potential applications in in vitro toxicological studies. However, further investigation into this methodology is needed. The mechanical strength of alginate-gelatin bioink is relatively weak and incapable of supporting long-term cultures; therefore, bioink components should be optimized to achieve better mechanical properties and favorable biofunctionality. In addition, the co-culture of hiPSC-Heps with non-parenchymal hepatic cells, such as mesenchymal stem cells[51], fibroblasts[52], vascular endothelial cells[53], stellate cells[54], and Kupffer cells[55], should be incorporated into the 3DP hepatic tissue model to more accurately mimic the cellular microenvironment of liver tissue. Furthermore, investigation of hepatotoxicity induced by other types of drugs, such as antituberculotic and antineoplastic drugs, should be considered in this 3DP model.
5. Conclusion
A hepatic tissue model was constructed by 3D bioprinting hiPSC-Heps using an alginate-gelatin bioink. Compared with the non-printed SW model, the hiPSC-Heps in the 3DP model formed 3D spheroids with higher overall viability and proliferation. The hiPSC-Heps in the SW and 3DP models showed upregulated mRNA expression of liver function-related biomarkers compared to those in the 2D model. The secretion of AAT, albumin, and BUN was found to be highest in the 3DP model of the three models, indicating the well-maintained biofunctions of hiPSC-Heps using this method. In the drug testing of APAP-induced hepatotoxicity, the 3DP model exhibited favorable performance with the upregulation of the drug metabolic enzyme CYP1A2. This 3DP model could be used as an advanced hepatic tissue model for potential applications in in vitro toxicological studies.
Acknowledgments
None.
Appendix
Table A.1.
Oligonucleotide primers used for qRT-PCR
| Primer | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| b-actin | TATTGGCAACGAGCGGTTC | ATGCCACAGGATTCCATACCC |
| CYP1A | CAATCAGGTGGTGGTGTCAG | GCTCCTGGACTGTTTTCTGC |
| CYP3A | AAGTCGCCTCGAAGATACACA | AAGGAGAGAACACTGCTCGTG |
| AAT | ATGCTGCCCAGAAGACAGATA | CTGAAGGCGAACTCAGCCA |
| TAT | TGCCGGGAAAAATGAAAGGC | CTCGGATGGGGTTGAAAGTTT |
| Albumin | GCACAGAATCCTTGGTGAACAG | ATGGAAGGTGAATGTTTTCAGCA |
| Transferrin | GTGTGCAGTGTCGGAGCAT | CATCGGATGGAATGACGCTTT |
| ASGPR1 | GAGAGAGACGTTCAGCAACTTC | GGGACTCTAGCGACTTCATCTT |
| CK-18 | GGCATCCAGAACGAGAAGGAG | ATTGTCCACAGTATTTGCGAAGA |
Funding
This work was supported by the National Natural Science Foundation of China (82100664, 52075285), the Key-Area Research and Development Program of Guangdong Province (2020B090923003), the National Key Research and Development Program of China (2018YFA0703004), Tsinghua University-Peking Union Medical College Hospital Initiative Scientific Research Program (20191080843) Tsinghua University Spring Breeze Fund (20201080760), the Natural Science Foundation of Jiangsu Province (BK20190114), Jiangsu Province Postdoctoral Research Funding Program (2021K116B), Key Project supported by Medical Science and technology development Foundation, Nanjing Department of Health (YKK19070), the Fundamental Research Funds for the Central Universities (0214-14380510), the Nanjing health science and technology development project for Distinguished Young Scholars (JQX19002), Project of Modern Hospital Management and Development Institute, Nanjing University and Aid project of Nanjing Drum Tower Hospital Health, Education & Research Foundation (NDYG2020047), fundings for Clinical Trials from the Affiliated Drum Tower Hospital, Medical School of Nanjing University (2021-LCYJ-PY-46), the Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province, China (CXPJJH121001—2021073).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
Research, writing, editing and formatting of the manuscript: JH, JW, and HR; Data collection and analysis: HY and KS; Administrative support and funding acquisition. YP, XQ, TX and HR. All authors contributed to the article and approved the submitted version.
References
- 1.Agarwal T, Banerjee D, Konwarh R, et al. Recent Advances in Bioprinting Technologies for Engineering Hepatic Tissue. Mater Sci Eng C Mater Biol Appl. 2021;123:112005. doi: 10.1016/j.msec.2021.112013. https://doi.org/10.1016/j.msec.2021.112013. [DOI] [PubMed] [Google Scholar]
- 2.Hwang DG, Choi YM, Jang J. 3D Bioprinting-Based Vascularized Tissue Models Mimicking Tissue-Specific Architecture and Pathophysiology for In Vitro Studies. Front Bioeng Biotechnol. 2021;9:685507. doi: 10.3389/fbioe.2021.685507. https://doi.org/10.3389/fbioe.2021.685507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma L, Wu YT, Li YT, et al. Current Advances on 3D-Bioprinted Liver Tissue Models. Adv HealthcMater. 2020;9:2001517. doi: 10.1002/adhm.202001517. https://doi.org/10.1002/adhm.202001517. [DOI] [PubMed] [Google Scholar]
- 4.Gough A, Soto-Gutierrez A, Vernetti L, et al. Human Biomimetic Liver Microphysiology Systems in Drug Development and Precision Medicine. Nat Rev Gastroenterol Hepatol. 2021;18:252–68. doi: 10.1038/s41575-020-00386-1. https://doi.org/10.1038/s41575-020-00386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiller T, Berg J, Elomaa L, et al. Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies. Int J Mol Sci. 2018;19:3129. doi: 10.3390/ijms19103129. https://doi.org/10.3390/ijms19103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang DT, Gibeley SB, Xu C, et al. Engineering Liver Microtissues for Disease Modeling and Regenerative Medicine. Adv Funct Mater. 2020;30:9553. doi: 10.1002/adfm.201909553. https://doi.org/10.1002/adfm.201909553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen DG, Funk J, Robbins JB, et al. Bioprinted 3D Primary Liver Tissues Allow Assessment of Organ-Level Response to Clinical Drug Induced Toxicity In Vitro. PLoS One. 2016;11:5867. doi: 10.1371/journal.pone.0158674. https://doi.org/10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gori M, Giannitelli SM, Torre M, et al. Biofabrication of hepatic constructs by 3D bioprinting of a cell-laden thermogel:An effective tool to assess drug-induced hepatotoxic response. Adv Healthc Mater. 2020;9:1163. doi: 10.1002/adhm.202001163. https://doi.org/10.1002/adhm.202001163. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt K, Berg J, Roehrs V, et al. 3D-bioprinted HepaRG Cultures as a Model for Testing Long Term Aflatoxin B1 Toxicity In Vitro. Toxicol Rep. 2020;7:1578–87. doi: 10.1016/j.toxrep.2020.11.003. https://doi.org/10.1016/j.toxrep.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu SM, Zhang JW, Lin S, et al. Recent Advances in the Development of In Vitro Liver Models for Hepatotoxicity Testing. Bio Des Manuf. 2021;4:717–34. https://doi.org/10.1007/s42242-021-00142-7. [Google Scholar]
- 11.Kryou C, Leva V, Chatzipetrou M, et al. Bioprinting for Liver Transplantation. Bioeng Basel. 2019;6:95. doi: 10.3390/bioengineering6040095. https://doi.org/10.3390/bioengineering6040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang HY, Sun LJ, Pang Y, et al. Three-dimensional Bioprinted Hepatorganoids Prolong Survival of Mice with Liver Failure. Gut. 2021;70:567–74. doi: 10.1136/gutjnl-2019-319960. https://doi.org/10.1136/gutjnl-2019-319960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie FH, Sun LJ, Pang Y, et al. Three-dimensional Bio-printing of Primary Human Hepatocellular Carcinoma for Personalized Medicine. Biomaterials. 2021;265:120416. doi: 10.1016/j.biomaterials.2020.120416. https://doi.org/10.1016/j.biomaterials.2020.120416. [DOI] [PubMed] [Google Scholar]
- 14.Faulkner-Jones A, Fyfe C, Cornelissen DJ, et al. Bioprinting of Human Pluripotent Stem Cells and their Directed Differentiation into Hepatocyte-like Cells for the Generation of Mini-livers in 3D. Biofabrication. 2015;7:044102. doi: 10.1088/1758-5090/7/4/044102. https://doi.org/10.1088/1758-5090/7/4/044102. [DOI] [PubMed] [Google Scholar]
- 15.Ma X Y, Qu X, Zhu W, et al. Deterministically Patterned Biomimetic Human iPSC-derived Hepatic Model Via Rapid 3D Bioprinting. Proc Natl Acad Sci U S A. 2016;113:2206–11. doi: 10.1073/pnas.1524510113. https://doi.org/10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulart E, de Caires LC, Telles-Silva KA, et al. 3D Bioprinting of Liver Spheroids Derived from Human Induced Pluripotent Stem Cells Sustain Liver Function and Viability In Vitro. Biofabrication. 2020;12:30. doi: 10.1088/1758-5090/ab4a30. https://doi.org/10.1088/1758-5090/ab4a30. [DOI] [PubMed] [Google Scholar]
- 17.Jeon H, Kang K, Park SA, et al. Generation of Multilayered 3D Structures of HepG2 Cells Using a Bio-printing Technique. Gut Liver. 2017;11:121–8. doi: 10.5009/gnl16010. https://doi.org/10.5009/gnl160https://doi.org/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun LJ, Yang HY, Wang YN, et al. Application of a 3D Bioprinted Hepatocellular Carcinoma Cell Model in Antitumor Drug Research. Front Oncol. 2020;10:878. doi: 10.3389/fonc.2020.00878. https://doi.org/10.3389/fonc.2020.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang HK, Sarsenova M, Kim DH, et al. Establishing a 3D In Vitro Hepatic Model Mimicking Physiologically Relevant to In Vivo State. Cells. 2021;10:1268. doi: 10.3390/cells10051268. https://doi.org/10.3390/cells10051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis PL, Green RM, Shah RN. 3D-printed Gelatin Scaffolds of Differing Pore Geometry Modulate Hepatocyte Function and Gene Expression. Acta Biomater. 2018;69:63–70. doi: 10.1016/j.actbio.2017.12.042. https://doi.org/10.1016/j.actbio.2017.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannoun Z, Steichen C, Dianat N, et al. The Potential of Induced Pluripotent Stem Cell Derived Hepatocytes. J Hepatol. 2016;65:182–99. doi: 10.1016/j.jhep.2016.02.025. https://doi.org/10.1016/j.jhep.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 22.Chen YF, Tseng CY, Wang HW, et al. Rapid Generation of Mature Hepatocyte-like Cells from Human Induced Pluripotent Stem Cells by an Efficient Three-step Protocol. Hepatology. 2012;55:1193–203. doi: 10.1002/hep.24790. https://doi.org/10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi F, Liu GH, Belmonte JC. Human Induced Pluripotent Stem Cells Derived Hepatocytes:Rising Promise for Disease Modeling, Drug Development and Cell Therapy. Protein Cell. 2012;3:246–50. doi: 10.1007/s13238-012-2918-4. https://doi.org/10.1007/s13238-012-2918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabar V, Studer L. Pluripotent Stem Cells in Regenerative Medicine:Challenges and Recent Progress. Nat Rev Genet. 2014;15:82–92. doi: 10.1038/nrg3563. https://doi.org/10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luce E, Messina A, Duclos-Vallee JC, et al. Advanced Techniques and Awaited Clinical Applications for Human Pluripotent Stem Cell Differentiation into Hepatocytes. Hepatology. 2021;74:1101–16. doi: 10.1002/hep.31705. https://doi.org/10.1002/hep.31705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ST, Wang JL, Ren HZ, et al. Hepatic Spheroids Derived from Human Induced Pluripotent Stem Cells in Bio-artificial Liver Rescue Porcine Acute Liver Failure. Cell Res. 2020;30:1–3. doi: 10.1038/s41422-019-0261-5. https://doi.org/10.1038/s41422-019-0261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Ren H, Liu Y, et al. Bioinspired Artificial Liver System with hiPSC-Derived Hepatocytes for Acute Liver Failure Treatment. Adv Healthc Mater. 2021;10:e2101580. doi: 10.1002/adhm.202101580. https://doi.org/10.1002/adhm.202101580. [DOI] [PubMed] [Google Scholar]
- 28.Lin RZ, Chang HY. Recent Advances in Three-dimensional Multicellular Spheroid Culture for Biomedical Research. Biotechnol J. 2008;3:1172–84. doi: 10.1002/biot.200700228. https://doi.org/10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- 29.Decarli MC, Amaral R, Dos Santos DP, et al. Cell Spheroids as a Versatile Research Platform:Formation Mechanisms, High Throughput Production, Characterization and Applications. Biofabrication. 2021;13:37. doi: 10.1088/1758-5090/abe6f2. https://doi.org/10.1088/1758-5090/abe6f2. [DOI] [PubMed] [Google Scholar]
- 30.Mandon M, Huet S, Dubreil E, et al. Three-dimensional HepaRG Spheroids as a Liver Model to Study Human Genotoxicity In Vitro with the Single Cell Gel Electrophoresis Assay. Sci Rep. 2019;9:1058. doi: 10.1038/s41598-019-47114-7. https://doi.org/10.1038/s41598-019-47114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurrell T, Ellero AA, Masso ZF, et al. Characterization and Reproducibility of HepG2 Hanging Drop Spheroids Toxicology In Vitro. Toxicol In Vitro. 2018;50:86–94. doi: 10.1016/j.tiv.2018.02.013. https://doi.org/10.1016/j.tiv.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Lee G, Lee J, Oh H, et al. Reproducible Construction of Surface Tension-Mediated Honeycomb Concave Microwell Arrays for Engineering of 3D Microtissues with Minimal Cell Loss. PLoS One. 2016;11:1026. doi: 10.1371/journal.pone.0161026. https://doi.org/10.1371/journal.pone.0161026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao YS, Xu YX, Zhang BF, et al. In Vivo Generation of Thick, Vascularized Hepatic Tissue from Collagen Hydrogel-Based Hepatic Units. Tissue Eng Part C Methods. 2010;16:653–9. doi: 10.1089/ten.TEC.2009.0053. https://doi.org/10.1089/ten.tec.2009.0053. [DOI] [PubMed] [Google Scholar]
- 34.Zuchowska A, Kwapiszewska K, Chudy M, et al. Studies of Anticancer Drug Cytotoxicity Based on Long-term HepG2 Spheroid Culture in a Microfluidic System. Electrophoresis. 2017;38:1206–16. doi: 10.1002/elps.201600417. https://doi.org/10.1002/elps.201600417. [DOI] [PubMed] [Google Scholar]
- 35.Ehrlich A, Duche D, Ouedraogo G, et al. Challenges and Opportunities in the Design of Liver-on-Chip Microdevices. Ann Rev Biomed Eng. 2019;21:219–39. doi: 10.1146/annurev-bioeng-060418-052305. https://doi.org/10.1146/annurev-bioeng-060418-052305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma XY, Liu J, Zhu W, et al. 3D Bioprinting of Functional Tissue Models for Personalized Drug Screening and In Vitro Disease Modeling. Adv Drug Deliv Rev. 2018;132:235–51. doi: 10.1016/j.addr.2018.06.011. https://doi.org/10.1016/j.addr.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XD, Wang X, Wang XZ, et al. 3D Bioprinted rat Schwann Cell-laden Structures with Shape Flexibility and Enhanced Nerve Growth Factor Expression. 3 Biotech. 2018;8:342. doi: 10.1007/s13205-018-1341-9. https://doi.org/10.1007/s13205-018-1341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu PC, Shen HZ, Zhi Y, et al. 3D Bioprinting and In Vitro Study of Bilayered Membranous Construct with Human Cells-laden Alginate/Gelatin Composite Hydrogels. Colloids Surfaces B Biointerfaces. 2019;181:1026–34. doi: 10.1016/j.colsurfb.2019.06.069. https://doi.org/10.1016/j.colsurfb.2019.06.069. [DOI] [PubMed] [Google Scholar]
- 39.Yu HY, Zhang XY, Song WJ, et al. Effects of 3-dimensional Bioprinting Alginate/Gelatin Hydrogel Scaffold Extract on Proliferation and Differentiation of Human Dental Pulp Stem Cells. J Endod. 2019;45:706–15. doi: 10.1016/j.joen.2019.03.004. https://doi.org/10.1016/j.joen.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Khoshnood N, Zamanian A, Abbasi M. The Potential Impact of Polyethylenimine on Biological Behavior of 3D-printed Alginate Scaffolds. Int J Biol Macromol. 2021;178:19–28. doi: 10.1016/j.ijbiomac.2021.02.152. https://doi.org/10.1016/j.ijbiomac.2021.02.152. [DOI] [PubMed] [Google Scholar]
- 41.Cho CH, Park J, Nagrath D, et al. Oxygen Uptake Rates and Liver-specific Functions of Hepatocyte and 3T3 Fibroblast Co-cultures. Biotechnol Bioeng. 2007;97:188–99. doi: 10.1002/bit.21225. https://doi.org/10.1002/bit.21225. [DOI] [PubMed] [Google Scholar]
- 42.Demol J, Lambrechts D, Geris L, et al. Towards a Quantitative Understanding of Oxygen Tension and Cell Density Evolution in Fibrin Hydrogels. Biomaterials. 2011;32:107–18. doi: 10.1016/j.biomaterials.2010.08.093. https://doi.org/10.1016/j.biomaterials.2010.08.093. [DOI] [PubMed] [Google Scholar]
- 43.Pang Y, Mao SS, Yao R, et al. TGF-beta Induced Epithelial-mesenchymal Transition in an Advanced Cervical Tumor Model by 3D Printing. Biofabrication. 2018;10:044102. doi: 10.1088/1758-5090/aadbde. https://doi.org/10.1088/1758-5090/aadbde. [DOI] [PubMed] [Google Scholar]
- 44.Mao SS, He JY, Zhao Y, et al. Bioprinting of Patient-derived In Vitro Intrahepatic Cholangiocarcinoma Tumor Model:Establishment, Evaluation and Anti-cancer Drug Testing. Biofabrication. 2020;12:045014. doi: 10.1088/1758-5090/aba0c3. https://doi.org/10.1088/1758-5090/aba0c3. [DOI] [PubMed] [Google Scholar]
- 45.Brophy CM, Luebke-Wheeler JL, Amiot BP, et al. Rat Hepatocyte Spheroids Formed by Rocked Technique Maintain Differentiated Hepatocyte Gene Expression and Function. Hepatology. 2009;49:578–86. doi: 10.1002/hep.22674. https://doi.org/10.1002/hep.22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni HM, Bockus A, Boggess N, et al. Activation of Autophagy Protects against Acetaminophen-induced Hepatotoxicity. Hepatology. 2012;55:222–31. doi: 10.1002/hep.24690. https://doi.org/10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigues RM, Heymans A, De Boe V, et al. Toxicogenomics-based Prediction of Acetaminophen-induced Liver Injury Using Human Hepatic Cell Systems. Toxicol Lett. 2016;240:50–9. doi: 10.1016/j.toxlet.2015.10.014. https://doi.org/10.1016/j.toxlet.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Gaskell H, Sharma P, Colley HE, et al. Characterization of a Functional C3A Liver Spheroid Model. Toxicol Res. 2016;5:1053–65. doi: 10.1039/c6tx00101g. https://doi.org/10.1039/c6tx00101g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takayama K, Kawabata K, Nagamoto Y, et al. 3D Spheroid Culture of hESC/hiPSC-derived Hepatocyte-like Cells for Drug Toxicity Testing. Biomaterials. 2013;34:1781–9. doi: 10.1016/j.biomaterials.2012.11.029. https://doi.org/10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 50.Tasnim F, Toh YC, Qu YH, et al. Functionally Enhanced Human Stem Cell Derived Hepatocytes in Galactosylated Cellulosic Sponges for Hepatotoxicity Testing. Mol Pharm. 2016;13:1947–57. doi: 10.1021/acs.molpharmaceut.6b00119. https://doi.org/10.1021/acs.molpharmaceut.6b00119. [DOI] [PubMed] [Google Scholar]
- 51.Gu JY, Shi XL, Zhang Y, et al. Study on the Effects and Mechanisms of Bone Marrow Mesenchymal Stem Cells on Porcine Primary Hepatocyte Culture In Vitro. Zhonghua Gan Zang Bing Za Zhi. 2009;17:867–71. [PubMed] [Google Scholar]
- 52.He JY, Pang Y, Yang HY, et al. Modular Assembly-based Approach of Loosely Packing Co-cultured Hepatic Tissue Elements with Endothelialization for Liver Tissue Engineering. Ann Transl Med. 2020;8:100. doi: 10.21037/atm-20-1598. https://doi.org/10.21037/atm-20-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salerno S, Campana C, Morelli S, et al. Human Hepatocytes and Endothelial Cells in Organotypic Membrane Systems. Biomaterials. 2011;32:8848–59. doi: 10.1016/j.biomaterials.2011.08.004. https://doi.org/10.1016/j.biomaterials.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Thomas RJ, Bhandari R, Barrett DA, et al. The Effect of Three-dimensional Co-culture of Hepatocytes and Hepatic Stellate Cells on Key Hepatocyte Functions In Vitro. Cells Tissues Organs. 2005;181:67–79. doi: 10.1159/000091096. https://doi.org/10.1159/000091096. [DOI] [PubMed] [Google Scholar]
- 55.Zinchenko YS, Schrum LW, Clemens M, et al. Hepatocyte and Kupffer Cells Co-cultured on Micropatterned Surfaces to Optimize Hepatocyte Function. Tissue Eng. 2006;12:751–61. doi: 10.1089/ten.2006.12.751. https://doi.org/10.1089/ten.2006.12.751. [DOI] [PubMed] [Google Scholar]


