Abstract
We have discovered that LE1, one of the plaque-forming phages previously described as lytic for the Leptospira biflexa saprophytic spirochete (I. Saint Girons, D. Margarita, P. Amouriaux, and G. Baranton, Res. Microbiol. 141:1131–1138, 1990), was indeed temperate. LE1 was found to be unusual, as Southern blot analysis indicated that it is one of the few phages to replicate in the prophage state as a circular plasmid. The unavailability of such small endogenous replicons has hindered genetic experimentation in Leptospira. We have developed a shuttle vector with DNA derived from LE1. Random LE1 DNA fragments were cloned into a pGEM 7Zf(+) derivative devoid of most of the bla gene but carrying a kanamycin resistance marker from the gram-positive bacterium Enterococcus (Streptococcus) faecalis. These constructs were transformed into L. biflexa strain Patoc 1 by electroporation, giving rise to kanamycin-resistant transformants. A 2.2-kb fragment from LE1 was responsible for replication of the vector in L. biflexa. However, a larger region including an intact parA gene homologue was necessary for the stability of the shuttle vector. Direct repeats and AT-rich regions characterized the LE1 origin of replication. Our data indicate that the replicon derived from the LE1 leptophage, together with the kanamycin resistance gene, is a promising tool with which to develop the genetics of Leptospira species.
Temperate bacteriophages, in contrast to virulent phages, can lysogenize their bacterial hosts. Most of these temperate phages integrate their DNA into host DNA. Some phages such as P1 and N15 are unusual in that they behave like a plasmid in the prophage state (12, 19).
The genus Leptospira belongs to the order Spirochaetales and is composed of both pathogenic and saprophytic species (10). 16S RNA analysis indicates that this phylum of eubacteria has a deep branching lineage (26). Few genetic tools are available for spirochetes (20); however, isolation of phage particles has been reported over the last 10 years. A transducing phage was recently used for developing the genetics of Brachyspira (Serpulina) hyodysenteriae (11). Numerous linear and circular plasmids are found in Borrelia burgdorferi, and one of them, a circular cp32 plasmid, was identified as a bacteriophage (8). An endogenous plasmid from Treponema denticola was engineered as a shuttle vector (5), while a broad-host-range plasmid was developed as a cloning vector for B. burgdorferi (23).
No plasmid had been found to be part of the Leptospira spp. genome, which is composed of two chromosomes (CI and CII) (27). The only available potential genetic tools consist of plaque-forming bacteriophages (LE1, LE3, and LE4) which were isolated from sewage water (21). These bacteriophages do not infect representative species of pathogenic Leptospira spp., and their host range is restricted to strain Patoc 1 (Pasteur Institute) of the Leptospira biflexa saprophytic species (21). Here we show that LE1 is a temperate phage which behaves like a plasmid within its host. We then cloned and characterized, at the nucleotide level, the region of the origin of replication of the LE1 genome. The L. biflexa-Escherichia coli shuttle vector thus constructed contains replication functions of the LE1 leptophage and a gene for antibiotic resistance to kanamycin from the gram-positive bacterium Enterococcus faecalis. It represents the first demonstration of DNA entry within Leptospira and is a stepping stone towards the development of the genetics of Leptospira species.
(This work represents a portion of a thesis submitted by Pascale Bourhy to the University of Paris VII for a Ph.D. degree.)
MATERIALS AND METHODS
Bacterial strains and media.
The Leptospira strains (Reference National Center, Institut Pasteur, Paris, France) used in this study are as follows: (i) L. biflexa serovar patoc strain Patoc 1 and Leptospira meyeri serovar semaranga strain Veldrat Semarang 173 (18), which belong to saprophytic species, and (ii) Leptospira interrogans serovar icterohaemorrhagiae strain Verdun, Leptospira inadai serovar lyme strain Lyme 10, and Leptospira fainei serovar hurstbridge strain BU T6, which belong to pathogenic species. The LE1 lysogens of L. biflexa strain Patoc 1 were called 3c and 6c. Leptospira strains were grown in EMJH medium (9, 13). Solid EMJH medium was prepared at a concentration of 1.1% agar. E. coli JM 109 (Promega) and XL10-Gold (Stratagene) competent strains were used for the propagation of plasmids and for cloning experiments, respectively. E. coli strains were grown in LB (22). Mitomycin C (Sigma) was used at a concentration from 1 to 100 ng/ml; isopropyl-β-d-thiogalactoside (IPTG) was used at a concentration of 0.5 mM; 3-indolyl-β-d-galactopyranoside (X-Gal) was used at 80 μg/ml; and ampicillin, tobramycin, and kanamycin were used at concentrations of 100, 20, and 50 μg/ml, respectively.
Phage purification and DNA extraction.
Large quantities of high-titer lysates of LE1 leptophage were produced by infection of exponentially growing L. biflexa strain Patoc 1 cells with a multiplicity of infection of 0.01 as described previously (21). Titer was determined by the soft agar overlay method (0.8% agar–0.23% EMJH base) (Difco) (21). Phage DNA was extracted three times with phenol, followed by ethanol precipitation.
Cloning of the phage origin of replication.
The pGEMK plasmid vector, used in this study, was constructed by replacing the ampicillin cassette with the kanamycin resistance cassette (in the same orientation) as follows. The kanamycin cassette (1,060 bp) derived from Enterococcus (Streptococcus) faecalis (24) was amplified with pAT21 as the template and the two primers 5′-ATCGGCTCCGTCGATACTAT-3′ and 5′-GTAGTCTCATACCTGTCAACGCCTACAT-3′ (Eurogentec). PfuTurbo DNA polymerase (Stratagene) was used according to the manufacturer's specifications. Most of the bla gene conferring ampicillin resistance (from the ScaI site to the BsaAI site) from pGEM 7Zf(+) (Promega) was thus replaced by the kanamycin cassette, giving rise to pGEMK.
Cloning of LE1 DNA fragments obtained by partial digestion with Sau3A into the pGEMK vector (linearized by BamHI) was performed according to the method of reference 22. Twenty-six recombinant plasmids, with a mean insert size of approximately 5 kb, were selected and used as a mixture for electroporation.
Electroporation.
A notable feature of the electroporation method for Leptospira is the temperature of 20°C for washing the cells (as with electroporation of Lactobacillus spp. [14]), instead of the usual 4°C and the use of water instead of an electroporation buffer. Briefly, 50 ml of a log-phase culture of L. biflexa strain Patoc 1 (2 × 108 to 5 × 108 cells per ml) was pelleted at 4,000 × g for 20 min at 20°C, washed twice with a half volume of sterile, deionized water (for an injectable preparation, pH 6, B. Braun), resuspended with 200 μl of water (final volume, 300 μl), and used immediately. Fifty microliters of cell suspension (4 × 109 cells) was mixed with 10 to 2,000 ng of plasmid DNA (in 5 μl of water), kept at 4°C for 10 min, and then transferred to a 0.2-cm-diameter ice-cold electroporation cuvette (Bio-Rad Laboratories, Richmond, Calif.) at 4°C. One pulse was delivered from a gene pulser with a Pulse controller (set at 1.8 [or 2.5] kV, 25 μF, and 200 Ω, producing a time constant of 4.5 ms; Bio-Rad). One milliliter of EMJH liquid medium was immediately added to the cuvette, and the cells were transferred to a 20-ml culture tube. Cultures were incubated at 30°C for 24 to 48 h with shaking in the absence of selection. Then 500 μl was plated (in duplicate), in solid EMJH medium with kanamycin (EMJK), with a soft agar overlay (0.8% agar–0.23% EMJH base). Plates were incubated for 4 to 6 days at 30°C. A survival curve was performed by plating the cells before and after electroporation on solid EMJH. A survival rate of 0.1 to 1% was obtained. Plasmid DNA was extracted from Kanr L. biflexa cultures by the alkaline lysis method using the Wizard Plus Maxipreps DNA purification system (Promega).
Stability of shuttle vectors within Leptospira.
Kanr Leptospira cells were grown in EMJH liquid medium with no selective pressure (dilution, 1/10 for 24 h at 30°C). Another 1/100 dilution was grown in fresh antibiotic-free EMJH medium for an additional 48 h. This last procedure was repeated twice. Cells were plated onto EMJH agar plates without antibiotic, and colonies were transferred onto plates with and without kanamycin. The percentage of resistant cells after 18 generations was determined.
PFGE and Southern blotting.
Previously described procedures were used for the preparation of high-molecular-weight genomic DNA, restriction endonuclease digestions in situ, pulsed-field gel electrophoresis (PFGE), Southern blotting, and DNA hybridization (6). PFGE was performed in a contour-clamped homogeneous electric field DRII apparatus (Bio-Rad Laboratories). A program with a ramping from 2 to 15 s for 20 h at 200 V was used for the separations. Fragment sizes were determined by comparison of band mobilities with those of bacteriophage λ DNA multimers (monomer, 44.3 kb) and Saccharomyces cerevisiae chromosomes (Bio-Rad Laboratories). The [α-33P]dATP labeling of the LE1 probe (digested by HindIII) was performed with a Megaprime kit (Amersham). Hybridizations were carried out for 18 h at 68°C.
DNA sequencing and sequence analysis.
DNA sequencing was performed as described previously (3). Nucleic acid and deduced amino acid sequences were analyzed by using BLAST software (National Center for Biotechnology Information) and programs from the Genetics Computer Group software package.
Nucleotide sequence accession number.
The GenBank accession number of the nucleotide sequence of the replication origin of LE1 is AF 261714.
RESULTS
LE1 lysogenizes L. biflexa strain Patoc 1.
The lysogenic states of the three previously isolated leptophages (LE1, LE3, and LE4) were reevaluated. Plaques obtained with LE3 and LE4 at days 3 and 4 after infection of L. biflexa strain Patoc 1 were clear, while those obtained with LE1 were slightly turbid. Clones isolated from the center of 14 turbid plaques obtained with LE1 were found to be resistant to superinfection by LE1 but were sensitive to LE3 and LE4. Lysogeny has the property of producing phage either spontaneously or in some cases after induction. In order to discriminate between phage-resistant clones and lysogens, we tested phage production by the 14 clones. All were found to spontaneously produce phage particles. Clone 6c was examined in detail. This clone produced phage at a level of approximately 1,000 PFU per ml after 3 days in culture. In addition, LE1 phage was found to be inducible, with a maximum yield of 108 PFU/ml from clone 6c using mitomycin C at 10 ng/ml. It was concluded that LE1 is a temperate phage but that LE3 and LE4 are lytic.
LE1 replicates as a circular plasmid.
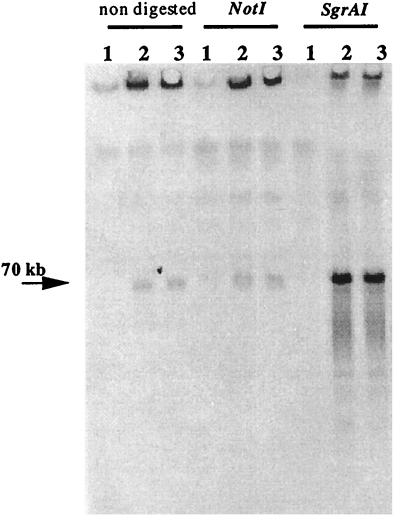
We determined whether LE1 leptophage DNA is inserted into the genome or whether it replicates as a plasmid. Restriction fragments produced following digestion of DNA from the lysogens were separated by PFGE and probed with labeled LE1 DNA. Results representative of three independent experiments are shown in Fig. 1. DNAs from the L. biflexa lysogens 3c and 6c, which either had not been digested or had been digested with NotI, gave a strong signal in the wells and a faint signal at 70 kb. DNAs from the L. biflexa lysogens 3c and 6c digested with SgrAI gave a faint signal in the wells and a strong signal at 70 kb, in agreement with the migration of the virion DNA, indicative of linearity (21). No signal was obtained with control leptospiral DNA. A likely explanation is that LE1 DNA, originating from the lysogen, is a supercoiled circular molecule which is trapped in the well of the agarose gel when it is unrestricted. One can conclude that LE1 is not integrated into the L. biflexa genome but that it behaves like an autonomous replicon of 70 kb.
FIG. 1.
Evidence for LE1 behaving like a plasmid. DNAs from the L. biflexa control strain Patoc 1 and lysogens 3c and 6c (lanes 1, 2, and 3, respectively) were not digested, restricted with NotI or SgrAI, and blotted on a membrane. [α-P33]dATP-labeled LE1 DNA was used as a probe.
DNA transfer into L. biflexa by electrotransformation.
Knowing that other spirochetes could be transformed by electroporation (for a review, see reference 20), we assumed that electroporation would be a likely technique for introducing DNA into Leptospira spp. The strategy consisted of two steps. First, we cloned LE1 DNA fragments into a plasmid vector with the aim of selecting the replication origin of the LE1 leptophage DNA. The vector plasmid chosen (pGEMK; see Materials and Methods) had an E. coli origin of replication and harbored a kanamycin resistance cassette from a gram-positive bacterium (24). Second, electroporation of L. biflexa strain Patoc 1 with selection for kanamycin resistance was performed. A set of 26 recombinant plasmids which represents twice the LE1 genome was thus constructed by cloning LE1 partial restriction fragments (mean size of 5 kb). This set of 26 recombinant plasmids was used as a mixture to transform L. biflexa strain Patoc 1 by electroporation. The transformants obtained were resistant to kanamycin and sensitive to tobramycin, indicating that they were not spontaneous mutants but bona fide transformants. Reelectrotranformation of L. biflexa with a plasmid extracted from Kanr L. biflexa cells also yielded Kanr transformants. Plasmid DNAs extracted from several Kanr L. biflexa transformants and analyzed after amplification in E. coli showed identical restriction profiles. One of the recombinant plasmids (pGKLep1) was further analyzed. Plasmid pGKLep1, isolated from the transformants, appeared to be identical by restriction analysis to one of the original 26 recombinant input plasmids that carried a 5.2-kb-long fragment from LE1 DNA. Electrotransformation of L. biflexa with pGKLep1 was quite efficient and gave reproducible results, yielding 100,000 transformants per μg of DNA. The number of transformants was found to be proportional to DNA concentrations up to 10 ng.
A shuttle vector with the LE1 origin of replication.
We showed that pGKLep1, a recombinant plasmid carrying 5.2 kb from the 70-kb LE1 leptophage DNA, could shuttle between L. biflexa and E. coli. This result indicates that pGKLep1 contains the origin replication of the LE1 leptophage, which allows replication of the plasmid within L. biflexa strain Patoc 1. The capacity of pGKLep1 to replicate in other species of Leptospira was analyzed. The strain Veldrat Semarang 173 serovar semaranga from L. meyeri, another saprophytic species, was found to be transformable by pGKLep1. However, attempts to transform three strains belonging to three different pathogenic species, L. interrogans, L. inadai, and L. fainei, were unsuccessful.
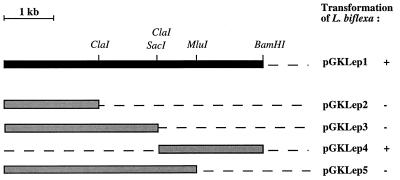
We determined whether a DNA fragment shorter than 5.2 kb was sufficient for replication within L. biflexa. A restriction endonuclease map of pGKLep1 allowed us to locate sites for ClaI, MluI, BamHI, and SacI. Restriction fragments were then subcloned and yielded pGKLep2 to pGKLep5 (Fig. 2). Electrotransformation of L. biflexa was performed with the latter recombinant plasmids. Only plasmid pGKLep4 with a SacI-BamHI 2.2-kb-long insert retained the ability to replicate in L. biflexa (Fig. 2). Southern blots of DNA extracted from L. biflexa transformed (either with pGKLep1 or with pGKLep4) with pGKLep1 as a probe confirm that both pGKLep1 and pGKLep4 replicate as autonomous plasmids (Fig. 3). Stability analyses of pGKLep1 and pGKLep4 were performed by diluting L. biflexa transformants in antibiotic-free medium and then testing for antibiotic resistance after 18 generations. While 99% of the leptospiral cells transformed with pGKLep1 were still Kanr after 18 generations, only 40% of the leptospiral cells transformed with pGKLep4 were still Kanr after 18 generations. The instability of pGKLep4 versus the stability of pGKLep1 was confirmed twice.
FIG. 2.
Subcloning of the LE1 origin of replication. Subcloning of plasmid pGKLep1 was performed to determine the minimum region required for replication in L. biflexa. The locations of some restriction sites used for subcloning are indicated. The ability (+) or the inability (−) of a given clone to replicate in L. biflexa is shown on the right.
FIG. 3.
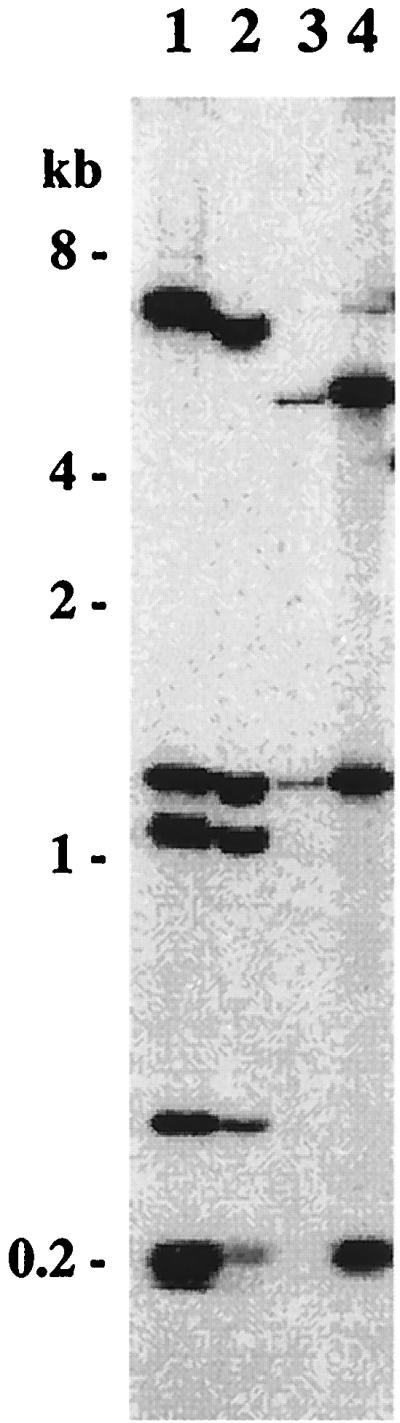
Evidence for the autonomous replication of pGKLep1 and pGKLep4 within L. biflexa. Shown are the results of a Southern blot analysis of DNA of L. biflexa transformed with pGKLep1 (lane 2) and pGKLep4 (lane 3). Lanes 1 and 4, original pGKLep1 and pGKLep4 plasmids, respectively. DNAs in all lanes were digested with EcoRI and probed with pGKLep1. Sizes of the fragments of the molecular mass markers are indicated on the left.
Sequence analysis of the LE1 origin of replication region.
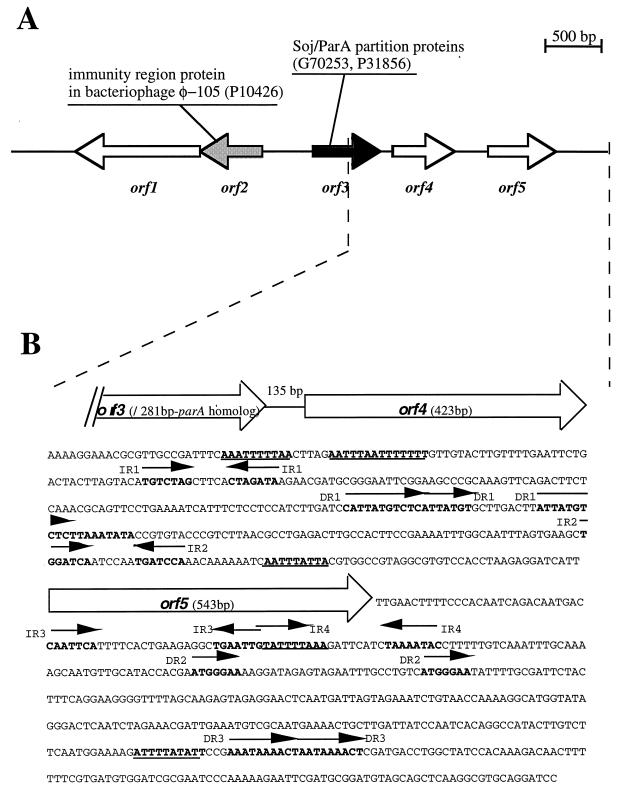
We have determined the sequence of the Sau3A 5.2-kb-long fragment from LE1 responsible for replication in L. biflexa (Fig. 4). The replication region of pGKLep1 contains five possible open reading frames (ORFs) (Fig. 4A) with no detectable homology at the nucleotide level with DNA sequences from the public databases. However, the amino acid sequence of Orf2 exhibits 31% identity with an immunity repressor protein from the Bacillus subtilis temperate bacteriophage φ 105. In addition, the amino acid sequence of Orf3 shows homologies with the ParA protein (up to 30% identity with ParAs from a wide variety of bacteria, including the spirochete B. burgdorferi), which is involved in the partitioning of low-copy-number plasmids (22).
FIG. 4.
Nucleotide sequence of the LE1 origin of replication. (A) Genetic organization of the replication region of pGKLep1. The putative genes and their transcription orientations are indicated by open arrows. Similarities with proteins in databases (accession numbers in brackets) are indicated. (B) Nucleotide sequence of the replication region of pGKLep4. Bold letters indicate repeats and runs of A and T (underlined). Arrows indicate inverted repeats (IR) and direct repeats (DR).
We also analyzed the 2.2-kb replication region of pGKLep4 for sequence patterns that have been described for plasmid origins. For example, the replication origins of many circular plasmids contain direct repeats which are the binding sites for the Rep protein and AT-rich regions, facilitating strand separation (7). Although the GC content of the 2.2-kb fragment is relatively low (37% GC), a region corresponding to the beginning of the 374-bp region between orf4 and orf5 exhibits an AT-rich content with runs of consecutive As and Ts (Fig. 4B). This intergenic region also contains two inverted repeats, IR1 and IR2, and a series of three 9-bp imperfect direct repeats, 5′-(T/C)ATTATGT(G/C)-3′. While the region between the parA homologue and orf4 does not contain repeats, the region downstream of orf5 contains two inverted repeats, IR3 and IR4; two direct repeats, DR2 and DR3; and two runs of As and Ts (Fig. 4B).
DISCUSSION
Genetic tools for spirochetes were scarce and nonexistent for the genus Leptospira. Until now, no small replicon had been isolated from Leptospira. However, LE1, one of the leptophages found at first to be lytic for L. biflexa (21), was shown on closer analysis to be temperate. This property led to our search for integration of the 70-kb-long LE1 DNA into the L. biflexa genome. No integration was found; rather, LE1 was found to replicate like a plasmid within L. biflexa. This property renders the LE1 leptophage similar to coliphages P1 and N15 (which is linear), both of which replicate as autonomous replicons (12, 19). Recently, a B. burgdorferi phage which also replicates as a circular plasmid has been described (8).
Since LE1 replicates as a plasmid, the next step was to isolate its origin of replication. Its isolation relied on the introduction of DNA into Leptospira. Previous attempts to electrotransform Leptospira strains from diverse pathogenic species with broad-host-range plasmids had failed (C. Baril, J. L. Herrmann, and I. Saint Girons, unpublished). To maximize our chances of detecting electrotransformation of Leptospira, we addressed two major issues. The first was the availability of a leptospiral replication origin, which was resolved by using the origin from leptophage LE1. The second was the choice of a selection marker. This was resolved by using an antibiotic resistance cassette from a gram-positive bacterium (Enterococcus faecalis). This marker was chosen for three reasons. First, the known sequences of Leptospira spp. genes are more similar to gene sequences from gram-positive than from gram-negative bacteria (2); second, the 30% GC content of Enterococcus faecalis is similar to that of Leptospira (34 to 39%); and third, the 3′5"-aminoglycoside phosphotransferase conferred resistance to kanamycin but not to tobramycin, a property which allowed distinction of transformants from spontaneous Kanr mutants which are also tobramycin resistant. We hypothesized that this kanamycin cassette would likely be expressed in Leptospira species. In addition, we had deleted the bla gene from the original plasmid pGEM 7Zf(+) (Promega), since penicillin derivatives are used to treat leptospirosis.
We thus reported for the first time the construction of an L. biflexa-E. coli shuttle vector with the following characteristics: (i) an E. coli origin of replication, (ii) a kanamycin cassette from Enterococcus faecalis, and (iii) a 2.2-kb fragment from LE1 leptophage DNA which contains its origin of replication. This shuttle vector, pGKLep4, could shuttle by electrotransformation between E. coli and L. biflexa and by regular transformation between L. biflexa and E. coli. The efficiency of electroporation of L. biflexa and L. meyeri (two saprophytic species) by pGKLep1, comprising the origin of replication region of the LE1 leptophage, is quite efficient when its electroporation efficiency is compared to the low electroporation efficiencies of other spirochetes with replicons derived from broad-host-range plasmids that need micrograms of DNA for electroporation (5, 23). However, no extrachromosomal replication of pGKLep1 has been demonstrated for representatives of three pathogenic Leptospira species. This may be due to several causes: (i) a replication origin unrecognized by these different species, (ii) a low electroporation efficiency, and (iii) the presence of restriction enzymes (D. E. A. Boursaux, unpublished data, 1996). Further studies will be needed to extend DNA transfer to all species of Leptospira.
The 2.2-kb fragment of LE1 necessary for replication has several features that are common to plasmid replication origins. These features include both AT-rich regions and direct repeats. In addition, the COOH terminus of a ParA homologue (Orf3) is also present in pGKLep4, further suggesting that this region functions as the LE1 replication origin. Indeed, in P1 and P7 bacteriophages, the partitioning operon, consisting of the parA and parB genes, is adjacent to the replication origin (1, 17) and a soj or parA homologue is found near the oriC region of many bacterial chromosomes (16). Both ParA and ParB are required for plasmid stability (25). All bacterial ParA-like proteins share a specific version of the Walker A motif for ATP binding (15), which facilitates their recognition. In contrast, sequence homology among ParB homologues can be more difficult to discern. In pGKLep1, we found Orf4, 134 bp downstream of the parA homologue and transcribed in the same direction. Orf4 has no significant similarity to genes of other bacteria but is located in an arrangement suggestive of the parB counterpart of a parA-parB operon (25). Along the same lines, such a genetic organization, with a parA homologue and an adjacent ORF of unknown function (both transcribed in the same direction and of sizes similar to those of Orf3 and Orf4 of pGKLep1), was also found in the numerous B. burgdorferi plasmids (4, 17a). The high stability of pGKLep1 suggests that the plasmid partition system is complete within the 5.2-kb fragment from LE1. In contrast, the low stability of pGKLep4 in L. biflexa may be due to an incomplete partitioning system, consisting of only the 3′-terminal fragment of the parA homologue.
The perspectives of the availability of a shuttle vector for the genus Leptospira supplies researchers with a genetic tool for mutagenesis, complementation of mutants, regulation of gene expression, and heterologous expression of genes from other spirochetes.
ACKNOWLEDGMENTS
The Institut Pasteur supported this work. Sabbatical leaves from West Virginia University, Morgantown, are acknowledged for N. Charon (supported by Public Health Service grant AI29743) in I. Saint Girons's and R. Hendrix's laboratories and D. Yelton (supported by USDA grant CRIS 0162600) in I. Saint Girons's laboratory. M. Picardeau thanks the Foundation de France for financial support (Prix Jacques Monod).
We thank F. Brossier for the gift of the pAT21 plasmid and the corresponding oligonucleotides; A. Labigne, C. Wandersman, A. Fouet, and P. Trieu-Cuot for helpful discussions; and G. Baranton, C. Werts, and A. Brenot for their constant interest in this work.
REFERENCES
- 1.Abeles A L, Friedman S A, Austin S J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985;185:261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- 2.Baril C, Richaud C, Fournié E, Baranton G, Saint Girons I. Cloning of dapD, aroD and asd of Leptospira interrogans serovar icterohaemorrhagiae, and nucleotide sequence of the asd gene. J Gen Microbiol. 1992;138:47–53. doi: 10.1099/00221287-138-1-47. [DOI] [PubMed] [Google Scholar]
- 3.Buchrieser C, Rusniok C, Frangeul L, Couve E, Billault A, Kunst F, Carniel E, Glaser P. The 102-kilobase pgm locus of Yersinia pestis: Sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect Immun. 1999;67:4851–4861. doi: 10.1128/iai.67.9.4851-4861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 5.Chi B, Chauhan S, Kuramitsu H K. Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infect Immun. 1999;67:3653–3656. doi: 10.1128/iai.67.7.3653-3656.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson B E, MacDougall J, Saint Girons I. Physical map of the linear chromosome of the bacterium Borrelia burgdorferi 212, a causative agent of Lyme disease, and localization of rRNA genes. J Bacteriol. 1992;174:3766–3774. doi: 10.1128/jb.174.11.3766-3774.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DelSolar G, Giraldo R, Ruiz-Echevarria M, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eggers C H, Samuels D S. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J Bacteriol. 1999;181:7308–7313. doi: 10.1128/jb.181.23.7308-7313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinghausen H C, McCullough W G. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and medium bovine albumin and polysorbate 80. Am J Vet Res. 1965;26:45–51. [PubMed] [Google Scholar]
- 10.Faine S, Adler B, Bolin C, Perolat P. Leptospira and leptospirosis. 2nd ed. Melbourne, Australia: Medisci; 1999. [Google Scholar]
- 11.Humphrey S B, Stanton T B, Jensen N S, Zuerner R L. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J Bacteriol. 1997;179:323–329. doi: 10.1128/jb.179.2.323-329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda H, Tomizawa J-I. Prophage P1, an extrachromosomal replication unit. Cold Spring Harbor Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- 13.Johnson R C, Harris V G. Differentiation of pathogenic and saprophytic Leptospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Josson K, Scheirlinck T, Michiels F, Platteeuw C, Stanssens P, Joos H, Dhaese P, Zabeau M, Mahillon J. Characterization of a gram-positive broad host range plasmid isolated from Lactobacillus hilgardii. Plasmid. 1989;21:9–20. doi: 10.1016/0147-619x(89)90082-6. [DOI] [PubMed] [Google Scholar]
- 15.Koonin E V. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 16.Lin D, Grossman A. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 17.Ludkte D N, Eichorn B G, Austin S J. Plasmid-partition functions of the P7 prophage. J Mol Biol. 1989;209:393–406. doi: 10.1016/0022-2836(89)90005-3. [DOI] [PubMed] [Google Scholar]
- 17a.Picardeau, M., J. R. Lobry, and B. J. Hinnebusch. Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 18.Postic D, Riquelme-Sertour N, Merien F, Perolat P, Baranton G. Interest of partial rDNA gene sequences to resolve heterogeneities between Leptospira collections: application to L. meyeri. Res Microbiol. 2000;151:333–341. doi: 10.1016/s0923-2508(00)00156-x. [DOI] [PubMed] [Google Scholar]
- 19.Ravin V K, Shulga M G. Evidence for extrachromosomal location of prophage N15. Virology. 1970;40:800–807. doi: 10.1016/0042-6822(70)90125-x. [DOI] [PubMed] [Google Scholar]
- 20.Rosa P, Stevenson B, Tilly K. Genetic methods in Borrelia and other spirochetes. Methods Microbiol. 1999;29:209–227. [Google Scholar]
- 21.Saint Girons I, Margarita D, Amouriaux P, Baranton G. First isolation of bacteriophages for a spirochaete: potential genetic tools for Leptospira. Res Microbiol. 1990;141:1131–1138. doi: 10.1016/0923-2508(90)90086-6. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sartakova M, Dobrikova E, Cabello F C. Development of an extrachromosomal cloning vector system for use in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2000;97:4850–4855. doi: 10.1073/pnas.080068797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 25.Williams D, Thomas C. Active partitioning of bacterial plasmids. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]
- 26.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuerner R L, Herrmann J L, Saint Girons I. Comparison of genetic maps for two Leptospira interrogans serovars provides evidence for two chromosomes and intraspecies heterogeneity. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]