Abstract
DNA polymerase I (PolI) functions both in nucleotide excision repair (NER) and in the processing of Okazaki fragments that are generated on the lagging strand during DNA replication. Escherichia coli cells completely lacking the PolI enzyme are viable as long as they are grown on minimal medium. Here we show that viability is fully dependent on the presence of functional UvrA, UvrB, and UvrD (helicase II) proteins but does not require UvrC. In contrast, ΔpolA cells grow even better when the uvrC gene has been deleted. Apparently UvrA, UvrB, and UvrD are needed in a replication backup system that replaces the PolI function, and UvrC interferes with this alternative replication pathway. With specific mutants of UvrC we could show that the inhibitory effect of this protein is related to its catalytic activity that on damaged DNA is responsible for the 3′ incision reaction. Specific mutants of UvrA and UvrB were also studied for their capacity to support the PolI-independent replication. Deletion of the UvrC-binding domain of UvrB resulted in a phenotype similar to that caused by deletion of the uvrC gene, showing that the inhibitory incision activity of UvrC is mediated via binding to UvrB. A mutation in the N-terminal zinc finger domain of UvrA does not affect NER in vivo or in vitro. The same mutation, however, does give inviability in combination with the ΔpolA mutation. Apparently the N-terminal zinc-binding domain of UvrA has specifically evolved for a function outside DNA repair. A model for the function of the UvrA, UvrB, and UvrD proteins in the alternative replication pathway is discussed.
In Escherichia coli, nucleotide excision repair (NER) is initiated by the action of the UvrA, UvrB, and UvrC proteins. The UvrA protein loads UvrB onto a damaged site, after which UvrC binds to UvrB, resulting in the UvrBC-DNA incision complex. In this complex, first an incision is made at the fourth or fifth phosphodiester bond on the 3′ side of the damage, followed by incision at the eighth phosphodiester bond on the 5′ side of the damage. Both incisions are catalyzed by the UvrC protein, which contains two distinct active sites, one for each incision (20, 45). UvrD (helicase II) subsequently removes the damaged strand, and DNA polymerase I (PolI) fills in the resulting gap. Finally, the remaining nick is closed by DNA ligase (for reviews, see references 8 and 36).
Besides its function in NER, it is generally believed that the major role of PolI in the cell is the processing of the lagging strand during DNA replication (16). In polA mutant strains the joining of Okazaki fragments is severely retarded (31, 32, 42). The protein possesses three enzymatic activities, a 5′-3′ exonuclease activity located in the N-terminal part of the protein (the small domain) and a DNA polymerase activity which, together with a 3′-5′ exonuclease activity, is located in the C-terminal part of the protein (the Klenow domain) (5, 15). The combination of the 5′-3′ exonuclease and the polymerase activities results in the so-called nick translation activity, which is responsible for the removal of the RNA primers and the resynthesis of DNA in the lagging strand (16).
More than 25 years ago it was proposed that UvrB and UvrD might also be involved in DNA replication, since in vivo in the absence of DNA damage-inducing treatments, uvrB or uvrD mutations were found to be lethal in combination with a mutation in the polA gene (11, 29, 38, 39). Combining a deletion of the uvrB gene with either a polA1 or a polA12 mutation leads to inviability (38). polA1 is an amber mutation introducing a stop codon at the position corresponding to residue 342, which results in a protein lacking the polymerase and proofreading activities but with a functional 5′-3′ exonuclease activity (14). polA12 is an undefined mutation, resulting in thermosensitivity for all three activities of the PolI enzyme (14). The inviability of the uvrB polA1 double mutant suggests that in the absence of the polymerase activity of PolI, DNA replication becomes dependent on the UvrB protein. Two different unidentified point mutations in uvrD are also lethal in combination with the polA1 mutation, indicating a role for UvrD in replication as well (11, 39). In contrast with uvrB and uvrD, strains with point mutations in uvrA or uvrC (18, 25, 29, 37) in a polA mutant background have been reported to be viable, although the plating efficiency of a uvrA6 polA12 strain was found to be reduced at 42°C (18, 37).
More recently it has been shown that E. coli cells in which the complete polA gene has been deleted are viable, although growth is restricted to synthetic media (13). Growth on rich media can be restored by introducing either the 5′-3′ exonuclease or the Klenow domain of PolI in this mutant strain. This implies that other enzymatic activities in the cell can substitute for the exonuclease and polymerase activities of PolI. To investigate the function of the Uvr proteins in these substituting activities, we have combined the polA deletion with defined deletions of the uvrA, uvrB, uvrC, and uvrD genes. We show that not only UvrB and UvrD but also UvrA are essential for the viability of a ΔpolA strain. Using defined mutations in uvrA and uvrB, we have analyzed the involvement of the different functional domains of the UvrA and UvrB proteins in the PolI-independent replication system. In contrast to UvrA, UvrB and UvrD the presence of the UvrC protein appear to have a negative effect on the viability of the ΔpolA cells, and we show that this negative effect is the result of its incision activity.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. For the construction of the chromosomal deletion of the uvrB gene, plasmid pNP12 was digested with EcoRI and StuI, thereby deleting the uvrB gene. The remaining flanking DNA was treated with Klenow polymerase, and XbaI linkers were ligated to the blunt ends. Next, an XbaI fragment containing the chloramphenicol resistance (Cmr) gene was ligated to the XbaI sites. The resulting plasmid was digested with PstI and BamHI, and the linear DNA containing the Cmr gene was introduced into JC7620. Homologous recombination with the chromosome of this strain resulted in a Cmr strain in which the uvrB gene has been deleted. In a similar way, a chromosomal deletion of the uvrC gene was constructed. Plasmid pCA32 was digested with BglII and ligated with a BglII-BamHI fragment containing the Cmr gene, thereby replacing the uvrC gene with the Cmr gene. The resulting plasmid was linearized with PstI and allowed to recombine with the chromosome of JC7620. The presence of the ΔuvrB::Cm and ΔuvrC::Cm mutations was confirmed both by Southern blotting and by PCR using oligonucleotides flanking the deleted and replaced region. The PCR product was analyzed on a gel for its size and restriction pattern (results not shown). Finally, it was shown that the Δuvr strains were UV sensitive and that this sensitivity could be complemented by the appropriate uvr gene located on a plasmid (not shown). The ΔuvrA::Cm, ΔuvrB::Cm, ΔuvrC::Cm, and ΔuvrD::Tc mutations were transferred to KMBL1001 by P1 transduction, and transductants were selected on Luria-Bertani (LB) medium containing 2.5 mM sodium citrate and the appropriate antibiotic. The presence of the mutation was verified by testing for UV sensitivity. For the construction of double mutants, the ΔuvrA::Cm and ΔuvrB::Cm mutations were transferred to KMBL1001 uvrC::Tn10, and the ΔuvrD::Tc mutation was transferred to KMBL1001 ΔuvrC::Cm.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or marker(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| JC7620 | recB21 recC22 sbcB12 | 17 |
| CS4985 | ΔuvrA::Cm | 4 |
| CS5316 | JC 7620 ΔuvrB::Cm | This paper |
| CS5388 | JC 7620 ΔuvrC::Cm | This paper |
| SY124 | AB1157 uvrC::Tn10 | P. Strike |
| GE1752 | ΔuvrD::Tc | 7 |
| S90C | Δ(lac-pro) | 33 |
| HP3430 | S90C Δ(bio-uvrB261) | 33 |
| CS5531 | S90C ΔuvrD::Tc | This paper |
| KMBL1001 | No known mutations (F− derivative of W1485) | R. Devoret |
| CS5428 | KMBL1001 ΔuvrA::Cm | This paper |
| CS5429 | KMBL1001 ΔuvrB::Cm | This paper |
| CS5430 | KMBL1001 ΔuvrC::Cm | This paper |
| CS5431 | KMBL1001 ΔuvrD::Tc | This paper |
| CS5432 | KMBL1001 ΔuvrC::Cm ΔuvrD::Tc | This paper |
| CS5458 | KMBL1001 uvrC::Tn10 ΔuvrA::Cm | This paper |
| CS5530 | KMBL1001 uvrC::Tn10 ΔuvrB::Cm | This paper |
| CJ225 | CM4722 Δpol::Km, pCJ100 (F′ polA+, Cm) | 13 |
| CJ231 | CM4722 Δpol::Km, pCJ102 (F′ 5′exo, Cm) | 13 |
| CJ233 | CM4722 Δpol::Km, pCJ103 (F′ Klenow, Cm) | 13 |
| Plasmids | ||
| pUvr-A7 | uvrA, Ap | 1 |
| pNP12 | uvrB, Km | 43 |
| pNP50 | uvrB, Km (pNP12 without dnaA box) | Our laboratory, unpublished |
| pCA32 | uvrC, Tc | 44 |
| pBL12 | uvrC, Ap | 49 |
| pSC101 | Tc | 3 |
| pCL1920 | Sm, ori pSC101 | 19 |
| pUC4-KSAC | Ap, Km | Pharmacia |
| pJA87-1 | uvrA with TAB linker in ATPase 1, Km | 4 |
| pJA87-6 | uvrA with TAB linker in ATPase 2, Km | 4 |
| pJA87-C253S | uvrA(C253S), mutation in Zn finger 1, Km | 46 |
| pJA87-C763S | uvrA(C763S), mutation in Zn finger 2, Km | 46 |
| pNP77-B430 | uvrB430, Km | 26 |
| pNP77-B(R544H) | uvrB(R544H), Km | 26 |
| pNP78-B(G509S) | uvrB(G509S), Km | 26 |
| pNP97 | uvrB630, Km | 27 |
| pDR3274 | uvrC(D466A), Tc | 20 |
| pCA161 | uvrC(R42A), Ap | 45 |
| pWU1 | Km Tc, ori pSC101 | This paper |
| pNP120 | uvrA, Tc, ori pSC101 | This paper |
| pNP121 | uvrB, Tc, ori pSC101 | This paper |
| pNP122 | uvrC, Tc, ori pSC101 | This paper |
| pNP136 | uvrA, Sm, ori pSC101 (Sm in pNP120) | This paper |
| pNP137 | pNP136, uvrA(ATP1) (from pJA87-1), Sm | This paper |
| pNP138 | pNP136, uvrA(ATP2) (from pJA87-6), Sm | This paper |
| pNP139 | pNP136, uvrA(Zn1) (from pJA87-C253S), Sm | This paper |
| pNP140 | pNP136, uvrA(Zn2) (from pJA87-C763S), Sm | This paper |
| pNP123 | pNP121, uvrB430 (from pNP77-B430), Tc | This paper |
| pNP129 | pNP121, uvrB630 (from pNP97), Tc | This paper |
| pNP132 | pNP121, uvrB(R544H) [from pNP77-B(R544H)], Tc | This paper |
| pNP133 | pNP121, uvrB(G509S) [from pNP77-B(G509S)], Tc | This paper |
| pCA154 | pNP122, uvrC(D466A) (from pDR3274), Tc | This paper |
| pCA179 | pNP122, uvrC(R42A) (from pCA161), Tc | This paper |
Media.
LB medium and plates were made as described previously (23). Minimal medium contained, per liter, 0.2 g of MgSO4 · 7H2O, 2 g of citric acid, 10 g of K2HPO4, 3.5 g of Na(NH4)HPO4 · 4H2O, 4 g of glucose, and 10 mg of thiamine. For derivatives of strain C90S, the minimal medium was supplemented with proline (50 μg/ml) and biotin (0.5 μg/ml). Strains containing the F plasmids with the polA gene or fragments thereof were plated on medium with 120 μg/ml IPTG (isopropyl-β-d-thiogalactopyranoside) to allow optimal expression of the (truncated) PolI proteins. Antibiotics were used in the following concentrations: chloramphenicol, 12.5 μg/ml; tetracycline, 25 μg/ml; streptomycin, 25 μg/ml; and kanamycin, 25 μg/ml.
Transduction of the ΔpolA mutation.
Strains were grown in LB medium containing the appropriate antibiotics at 37°C to an optical density at 600 nm of 0.4. Log-phase cells were spun down and resuspended in LB medium containing 2.5 mM CaCl2 and 5 mM MgSO4 at a concentration of 109 cells/ml. To 1 ml of cells, 10 μl of a P1 lysate from CJ225 was added (resulting in 0.1 phage per bacterial cell). The phage were allowed to infect the cells for 20 min at 37°C. The cells were centrifuged and resuspended in 2 ml of minimal medium, after which they were incubated for another hour at 37°C. The cells were washed with minimal medium, and finally 100 μl of the cells was plated on minimal medium plates or LB plates containing 2.5 mM sodium citrate and supplemented with the appropriate antibiotics.
Plasmids.
The plasmids used in this study are listed in Table 1. Plasmid pWU1 was used to clone the uvr genes on a vector containing a pSC101 origin (which does not require PolI for initiation of replication), and was constructed by insertion of the EcoRI fragment containing the kanamycin resistance (Kmr) gene from pUC4-KSAC (Pharmacia) into the EcoRI site of pSC101. Plasmid pNP120 was constructed by digestion of pUvr-A7 with HindIII and filling in of the ends with Klenow polymerase. Next, after digestion with PstI, the PstI-blunt-end fragment containing the uvrA gene was inserted into the PstI and PvuII sites of pWU1. Plasmid pNP121 was constructed by inserting the PstI-StuI fragment from pNP50 containing the uvrB gene into the PstI and PvuII sites of pWU1. Plasmid pNP122 was constructed by inserting the PvuII-PstI fragment from pCA32 containing the uvrC gene into the PvuII and PstI sites of pWU1. Plasmids expressing mutant uvr genes were constructed by restriction fragment exchange between previously isolated uvrA, uvrB, and uvrC mutants and plasmids pNP120, pNP121, and pNP122. Since overproduction of UvrA turned out to be lethal in a ΔpolA strain, the different uvrA plasmids had to be introduced into the double mutant KMBL1001 ΔuvrA::Cm uvrC::Tn10. To be able to do this, the tetracycline resistance (Tcr) genes of the pNP120 derivatives had to be replaced by another resistance gene. This was done by insertion of a HindIII fragment containing the streptomycin resistance (Smr) gene into the HindIII sites located in the Tcr genes of the pNP120 derivatives.
RESULTS
UvrA, UvrB, and UvrD proteins are essential in a ΔpolA background.
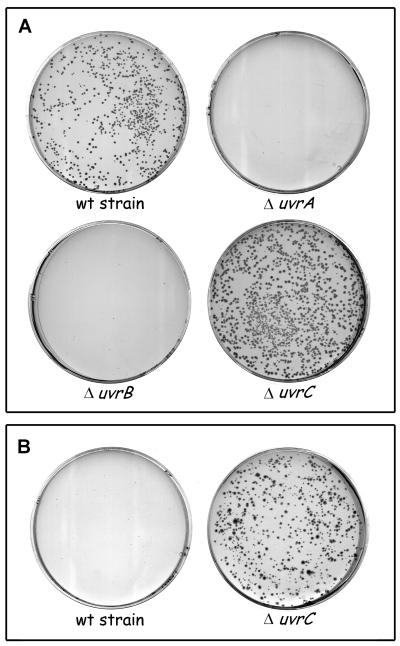
In the past, the viability of polA mutants has been tested with strains carrying a point mutation in the polA gene, which still might produce a partially functional PolI enzyme. To test the requirement for the different Uvr proteins in the complete absence of the PolI enzyme, we made use of a ΔpolA::Km mutation, in which the polA gene has been removed and replaced by a kanamycin resistance gene. First we constructed isogenic strains in which the uvrA, uvrB, or uvrC gene has been deleted and replaced by a chloramphenicol resistance gene and the uvrD gene has been replaced by a tetracycline resistance gene. These strains were infected with a P1 lysate that was made on the ΔpolA::Km strain, and transductants were selected on minimal medium with kanamycin at 30 and 37°C. The wild-type strain KMBL1001 yielded ΔpolA transductants at 30°C but not at 37°C (Fig. 1). Apparently, in our genetic background the ΔpolA strain is viable on minimal medium at low temperature only. The ΔuvrA, ΔuvrB, and ΔuvrD strains did not give rise to kanamycin-resistant transductants (Fig. 1 and Table 2), even after prolonged incubation at 30°C. Surprisingly, not only did the ΔuvrC strain produce ΔpolA transductants at 30°C, but these colonies were larger than the transductants of the isogenic wild-type strain (Fig. 1A), suggesting that the presence of the UvrC protein has a negative effect on the growth of a ΔpolA strain. This effect was even more clear at 37°C, where the wild-type strain yielded no ΔpolA transductants whereas the ΔuvrC strain did (Fig. 1B). Double mutants carrying ΔuvrA uvrC::Tn10, ΔuvrB uvrC::Tn10, or ΔuvrD ΔuvrC were also inviable in combination with the ΔpolA mutation (Table 2), showing that the requirement for the UvrA, UvrB, and UvrD proteins cannot be ascribed to a squelching of the negative effect of UvrC by these proteins. To demonstrate that the inability to obtain ΔpolA transductants in the ΔuvrA, ΔuvrB, and ΔuvrD strains was not due to a general transduction deficiency of these strains, we also did the reciprocal experiment by transferring the Δuvr mutations to KMBL1001 ΔpolA. As expected, the ΔuvrC::Cm mutation could be successfully introduced into the ΔpolA strain, whereas no transductants of the ΔuvrA::Cm, ΔuvrB::Cm, or ΔuvrD::Tc mutations were found. Taken together, the results show that the UvrA, UvrB, and UvrD proteins are essential for a process that substitutes for PolI function, whereas the UvrC protein seems to interfere with this process.
FIG. 1.
Transduction of the ΔpolA::Km mutation into different genetic backgrounds. After infection with a P1 lysate grown on CJ225, the cells were plated on minimal medium containing kanamycin and the plates were incubated for 60 h at 30°C (A) or 37°C (B). Shown are the results with KMBL1001 (wild-type [wt] strain) and the isogenic ΔuvrA, ΔuvrB, and ΔuvrC derivatives. The ΔuvrD mutant strain gave results identical to those with the ΔuvrA and ΔuvrB mutant strains (not shown).
TABLE 2.
Transduction of the Δpol::Km mutation to Δuvr strains
| Strain | Colonies formed on minimal medium ata:
|
|
|---|---|---|
| 30°C | 37°C | |
| KMBL1001 | + | − |
| KMBL1001 ΔuvrA | − | − |
| KMBL1001 ΔuvrB | − | − |
| KMBL1001 ΔuvrC | ++ | ++ |
| KMBL1001 ΔuvrD | − | − |
| KMBL1001 ΔuvrA uvrC::Tn10 | − | − |
| KMBL1001 ΔuvrB uvrC::Tn10 | − | − |
| KMBL1001 ΔuvrC ΔuvrD | − | − |
−, no colonies; +, small colonies; ++, larger colonies.
The 3′ incision activity of UvrC interferes with the PolI-independent replication process.
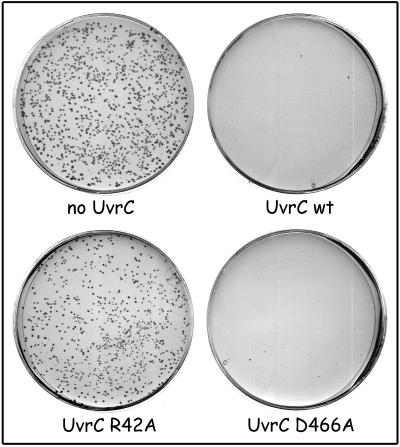
Plasmid pNP122 contains the wild-type uvrC gene inserted in a pSC101 derivative, a vector that does not require PolI for its replication initiation. Introduction of pNP122 into a wild-type strain (KMBL1001) or a ΔuvrC strain abolishes the generation of ΔpolA transductants, even at 30°C (Fig. 2 and Table 3). Apparently, a higher level of UvrC totally blocks the PolI-independent replication pathway. The UvrC protein has two catalytic sites for cleavage of the DNA during NER. The N-terminal half of the protein contains the active site for incision of the DNA at the 3′ side of the damage (45), and the C-terminal half contains the site for incision at the 5′ side of the damage (20). Mutations in uvrC that selectively inactivate one of the catalytic sites have been constructed. Mutant UvrC(R42A) is no longer capable of inducing the 3′ incision (45), and in UvrC(D466A) the 5′ incision is impaired (20). Each of the catalytic-site mutations was introduced in pNP122, and the resulting plasmids were tested for their capacity to allow transduction of the ΔpolA mutation (Fig. 2 and Table 3). Like wild-type UvrC, mutant UvrC(D466A) abolished the generation of ΔpolA transductants. Expression of the UvrC(R42A) mutant, however, was not lethal in combination with the ΔpolA mutation, although the colonies were somewhat smaller (Fig. 2; Table 3). Apparently it is the DNA incision activity by the 3′ catalytic site of UvrC that causes the lethality of the overproduction of the UvrC protein in a ΔpolA strain.
FIG. 2.
Effect of uvrC mutations on transduction of the ΔpolA::Km mutation. KMBL1001 ΔuvrC with pSC101 (no UvrC), pNP122 (wild-type [wt] UvrC), pCA154 (R42A), and pCA179 (D466A) was infected with a P1 lysate grown on CJ225. After infection, the cells were plated on minimal medium with kanamycin and the plates were incubated at 30°C for 60 h.
TABLE 3.
Effect of uvr mutation on the Δpol::Km transduction
| Mutation, strain, and plasmid | Colonies formed on minimal medium ata:
|
|
|---|---|---|
| 30°C | 37°C | |
| uvrC mutations | ||
| KMBL1001 + pNP122 (wtb UvrC) | − | − |
| KMBL1001 ΔuvrC + pSC101 (no UvrC) | ++ | ++ |
| KMBL1001 ΔuvrC + pNP122 (wt UvrC) | − | − |
| KMBL1001 ΔuvrC + pCA154 [UvrC(D466A)] | − | − |
| KMBL1001 ΔuvrC + pCA179 [UvrC(R42A)] | + | + |
| uvrB mutations | ||
| KMBL1001 + pNP121 (wt UvrB) | + | − |
| KMBL1001 ΔuvrB + pSC101 (no UvrB) | − | − |
| KMBL1001 ΔuvrB + pNP121 (wt UvrB) | + | − |
| KMBL1001 ΔuvrB + pNP129 (UvrB630) | ++ | ++ |
| KMBL1001 ΔuvrB + pNP123 (UvrB430) | − | − |
| KMBL1001 ΔuvrB + pNP133 [UvrB(G509S)] | − | − |
| KMBL1001 ΔuvrB + pNP132 [UvrB(R544H)] | − | − |
| uvrA mutations | ||
| KMBL1001 + pNP120 (wt UvrA) | − | − |
| KMBL1001 ΔuvrA + pNP120 (wt UvrA) | − | − |
| KMBL1001 ΔuvrC ΔuvrA + pCL1920 (no UvrA) | − | − |
| KMBL1001 ΔuvrC ΔuvrA + pNP136 (wt UvrA) | ++ | ++ |
| KMBL1001 ΔuvrC ΔuvrA + pNP137 [UvrA(ATP1)] | − | − |
| KMBL1001 ΔuvrC ΔuvrA + pNP138 [UvrA(ATP2)] | − | − |
| KMBL1001 ΔuvrC ΔuvrA + pNP139 [UvrA(Zn1)] | − | − |
| KMBL1001 ΔuvrC ΔuvrA + pNP140 [UvrA(Zn2)] | − | − |
−, no colonies; +, small colonies; ++, larger colonies.
wt, wild type.
The C-terminal domain of UvrB contains an important binding domain for UvrC (27, 40). A truncated UvrB protein lacking this domain (UvrB630) no longer stabilizes the binding of UvrC to the UvrB-DNA preincision complex during the repair reaction, and as a result the incision at the 3′ side of the damage is severely reduced (27). We have tested whether the same truncated UvrB protein does support DNA replication in the absence of a functional PolI enzyme. Table 3 shows that a ΔuvrB strain with a pSC101 plasmid that expresses either the wild-type UvrB protein (pNP121) or the truncated UvrB630 (pNP129) does allow the formation of ΔpolA transductants at 30°C. This means that the UvrC-binding domain of UvrB is dispensable for its role in PolI-independent replication. The pNP129-containing strain even allowed formation of ΔpolA transductants at 37°C, whereas the pNP121-containing strain did not (Table 3). This difference in growth is comparable to the difference found between the strain lacking the uvrC gene and the wild-type strain (KMBL1001) (Fig. 1; Table 2), suggesting that in the absence of the UvrC-binding domain of UvrB, the UvrC protein no longer exerts its negative effect in the PolI-independent replication pathway.
We have also inserted the uvrA gene in a pSC101 vector (pNP120). Surprisingly, when this plasmid was introduced in either a wild-type strain (KMBL1001) or a ΔuvrA strain, no ΔpolA transductants could be obtained (Table 3). Apparently, although the UvrA protein is essential, a higher level of the protein is unfavorable for E. coli lacking the PolI enzyme. The same plasmid, however, did allow deletion of the polA gene in a uvrC::Tn10 ΔuvrA double mutant (Table 3), indicating that a higher level of UvrA results in more deleterious incisions by the UvrC protein.
Role of functional domains of the UvrA and UvrB proteins in PolI-independent replication.
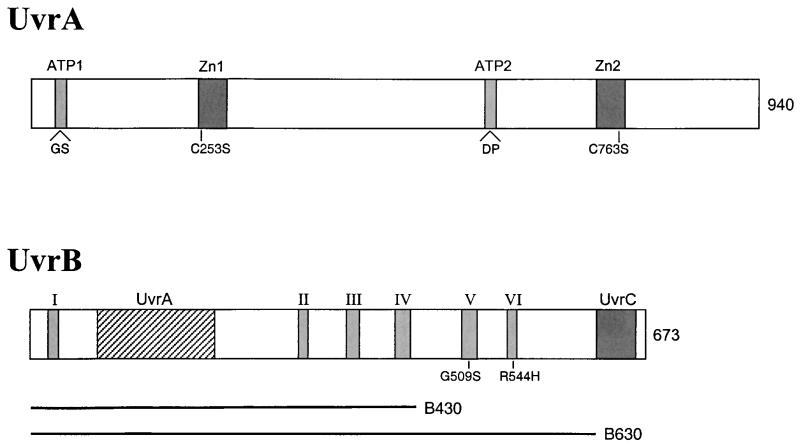
From structural and mutational studies (see reference 8 for a review), different functional domains in UvrA and UvrB can be indicated (Fig. 3). The UvrA protein contains two ATP-binding sites and two zinc-binding sites. The UvrB protein contains six so-called helicase motifs (I to VI) which are involved in ATPase and DNA-unwinding activity. In addition, an important UvrC-binding domain is located in the C-terminal part of the protein, and a putative UvrA-binding domain is present between motifs I and II.
FIG. 3.
Schematic representation of the UvrA and UvrB proteins. The UvrA protein contains two ATPase sites and two zinc-binding motifs, and the mutations in these domains used in this study are indicated. The UvrB protein contains UvrA- and UvrC-binding domains and six ATPase-helicase motifs (I to VI). The positions of the substitutions in motifs V and VI are shown. The lengths of the truncated UvrB proteins UvrB430 and UvrB630 are indicated.
As shown above, a UvrB protein lacking the C-binding domain still supports transduction of the ΔpolA mutation. We have also tested UvrB with a larger deletion (UvrB430), which lacks 243 amino acids from the C terminus, including helicase motifs V and VI. This truncated protein has been shown to form damage-specific UvrA2B-DNA complexes, but it can no longer form the UvrB-DNA preincision complex (our laboratory, unpublished data). Expression of the truncated UvrB protein in a ΔuvrB background does not allow formation of ΔpolA transductants (Table 3), suggesting that the helicase motifs are important for the activity of UvrB in PolI-independent replication. This was further tested with two UvrB mutant proteins having an amino acid substitution in helicase motif V (G509S) or VI (R544H). Both mutant proteins have been shown to bind UvrA and to bind to a damage site in the UvrA2B complex, but they are disturbed in ATPase and DNA-unwinding activity and as a result can no longer form the UvrB-DNA preincision complex (26). Like in the complete absence of the UvrB protein, no ΔpolA transductants were found upon expression of the point mutants, suggesting that the action that is required by the Uvr proteins for PolI-independent replication involves ATPase-induced conformational changes of the UvrB protein similar to those for formation of the preincision complex.
The two ATPase sites of UvrA have shown to be essential for the NER reaction (4, 41). The N-terminal ATPase site (ATP1) seems to be important for dimerization of UvrA (22, 30), and the C-terminal site (ATP2) is thought to be involved in the dissociation of UvrA from undamaged DNA (41). Mutant UvrA proteins with two amino acid insertions within ATP1 or ATP2 have been constructed and purified in the past (4). Both proteins displayed 50% of the ATPase activity of wild-type UvrA, and they were both defective in NER. We have inserted the uvrA genes with the corresponding mutations in pSC101 and introduced these plasmids in the double uvrC::Tn10 ΔuvrA double mutant. Table 3 shows that neither of the mutant UvrA proteins supported the PolI-independent replication, indicating that the two ATP sites are also important for this process.
Amino acid substitutions in the two zinc-binding domains have also been constructed. Substitution C763F (47) or C763S (46) in the C-terminal zinc-binding motif (Zn2), resulted in a UvrA protein that is completely defective in NER. In contrast, substitution C253S or C256S in the N-terminal zinc-binding domain (Zn1), although resulting in the loss of zinc coordination, did not lead to any defect in the repair reaction (46). We have tested the C763S and C253S mutations for their effect on the PolI-independent replication. In contrast to the differential effect on NER, both mutations now prevented the transduction of the ΔpolA mutation (Table 3), which means that both zinc-binding motifs are essential for the UvrA-mediated replication pathway. To test whether the C253S protein was properly expressed in the uvrC::Tn10 ΔuvrA strain, we also introduced pBL12, expressing the wild-type UvrC protein in the cells. The resulting strain containing both the uvrC and the uvrA(C253S) plasmids appeared to be UV resistant, whereas the same strain with only the uvrC or uvrA(C253S) plasmid was UV sensitive. This confirms not only that UvrA(C253S) is expressed but also that the mutant protein is indeed active in NER. The fact that the N-terminal zinc-binding domain is essential for PolI-independent replication but not for repair suggests that this domain has specifically evolved in UvrA for its function in replication.
Importance of the polymerase and exonuclease activities of PolI.
The PolI enzyme has two important enzymatic activities for the processing of the Okazaki fragments that are generated on the lagging strand during DNA replication. The polymerase activity (together with the 3′-5′ proofreading activity located in the Klenow fragment) extends the 3′ end of an Okazaki fragment, and the 5′-3′ exonuclease activity removes the RNA primers. E. coli strains with a deletion in the chromosomal polA gene are viable on minimal medium only, but expression of either the 5′-3′ exonuclease or the Klenow fragment portion of the enzyme is sufficient to allow growth on rich medium (13). This means that there must be alternative pathways for each of the two functions of PolI. To test which of these pathways involve the Uvr(A)B and UvrD enzymes we introduced F′ plasmids expressing either the complete PolI enzyme or only one of the functional domains in a strain lacking the uvrB (HP3430) or uvrD (CS5531) gene and repeated the ΔpolA transduction experiments.
As expected, the wild-type strain S90C gave rise to ΔpolA transductants only on minimal medium and not on LB medium (Table 4). In contrast to the case for KMBL1001, transductants were also found at 37°C, confirming that viability of the ΔpolA transductants is strongly dependent on the genetic background. In the presence of the F plasmid expressing either the complete PolI enzyme or the functional fragments, transductants were found on both LB and minimal media. The colonies on rich medium with the FExo plasmid were smaller than those observed with the other two plasmids. Possibly the expression or stability of the exonuclease part of the protein in our genetic background is somewhat reduced. The strain lacking the uvrB gene gave transductants on LB only in the presence of the complete PolI enzyme and not with either of the fragments. On minimal medium, however, normal transductants were found with both FPolA and FKlenow but not with FExo (Table 4). Apparently, under slow-growth conditions, UvrB is needed only when the polymerase activity is missing, but under fast-growth conditions, it is required to substitute for both the polymerase and the exonuclease activities.
TABLE 4.
Effect of the PolI domains on Δpol::Km transduction
| Strain and plasmid | Colonies formed ona:
|
|||
|---|---|---|---|---|
| LB medium
|
Minimal medium
|
|||
| 30°C | 37°C | 30°C | 37°C | |
| S90C | − | − | + | + |
| S90C + FPolA | ++ | ++ | ++ | ++ |
| S90C + FKlenow | ++ | ++ | ++ | ++ |
| S90C + FExo | + | + | ++ | ++ |
| S90C ΔuvrB261 | − | − | − | − |
| S90C ΔuvrB261 + FPolA | ++ | ++ | ++ | ++ |
| S90C ΔuvrB261 + FKlenow | − | − | ++ | ++ |
| S90C ΔuvrB261 + FExo | − | − | − | − |
| S90C ΔuvrD | − | − | − | − |
| S90C ΔuvrD + FPolA | ++ | ++ | ++ | ++ |
| S90C ΔuvrD + FKlenow | − | − | − | − |
| S90C ΔuvrD + FExo | − | − | − | − |
−, no colonies; +, small colonies; ++, larger colonies.
The isogenic strains lacking the uvrD gene gave a different result (Table 4). Now transductants could be found on both types of media only with the complete PolI enzyme and not with either of the truncations. This shows that the UvrD protein is essential for both alternative activities that replace polymerase and exonuclease activities.
DISCUSSION
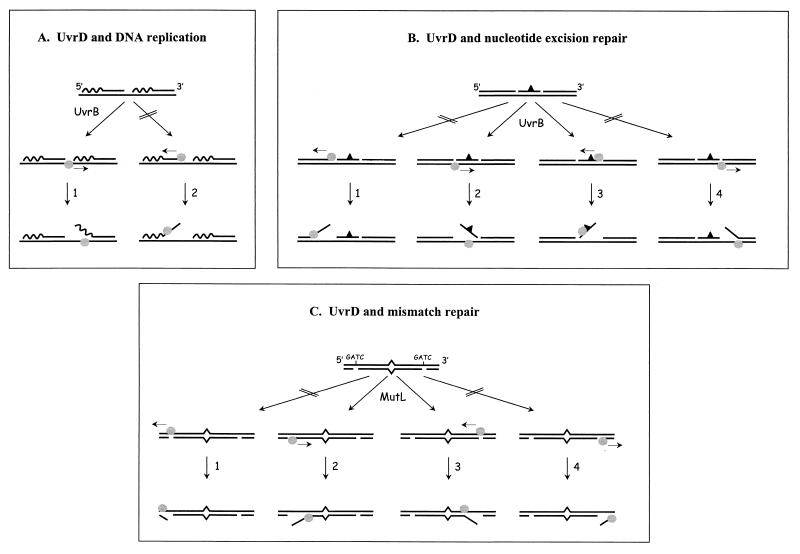
We have shown that the UvrA, UvrB, and UvrD (helicase II) proteins are essential for the viability of E. coli cells lacking the polA gene, indicating that they play a crucial role in alternative pathways that substitute for the polymerase and exonuclease functions of the PolI enzyme. The UvrD protein appears to be essential for both substituting activities, since expression of the polymerase or exonuclease activity alone is not sufficient for survival of a ΔuvrD strain. Being a very efficient DNA helicase, the most likely function of UvrD is to unwind the DNA-RNA hybrids in the Okazaki fragments. Not only would this unwinding facilitate the removal of the RNA primers by exonucleases or RNAses, but extension of the unwinding into the DNA-DNA hybrid would also result in larger gaps, which might facilitate the entry of an alternative polymerase like PolIII. The UvrD protein has been shown to be able to unwind DNA from a nick (34), but the initiation of this reaction requires very high UvrD concentrations (35). From the nick UvrD can unwind the DNA in both directions (34). The UvrD protein has a helicase activity with a 3′-to-5′ polarity (21), which means that the protein can be loaded on the nicked DNA in two different ways, either on the nicked strand or on the continuous strand. With respect to the processing of the lagging strand, loading of UvrD on the continuous strand would result in displacement of the RNA primer (Fig. 4A), which would account for the function of UvrD in the alternative replication pathway as described above. Loading of UvrD on the opposite strand, however, would displace the DNA end that needs to be elongated (Fig. 4A). Such a displacement is expected to interfere with the action of any alternative polymerase. It is therefore conceivable that in E. coli there is a mechanism to load UvrD onto the appropriate strand, so that it unwinds in the proper direction.
FIG. 4.
Models for protein-mediated orientation of the UvrD helicase (circles). (A) DNA replication. The lagging strand is shown. The wavy line represents the RNA. Loading of UvrD onto the continuous strand will unwind the RNA-DNA hybrid (1) and loading on the opposite strand will displace the 3′ end that needs to be elongated by DNA polymerase (2). An interaction of UvrD with UvrB orients UvrD in the proper direction (1). (B) Nucleotide excision repair. DNA with a damage (triangle) after incision by UvrC is shown. Loading of UvrD on the 3′ nick (3) or opposite the 5′ nick (2) will lead to removal of the damaged oligonucleotide. Loading of UvrD onto the 5′ nick (1) or opposite the 3′ nick (4) will lead to unwinding in the opposite direction. An interaction between UvrB and UvrD directs UvrD (2 or 3). (C) Mismatch repair. DNA with a mismatch and two nicked GATC sites is shown. For unwinding of the DNA by UvrD in the direction of the mismatch, the helicase needs to bind to the nicked strand when the GATC is located at the 5′ side of the mismatch (2) or to the continuous strand when the GATC is at the 3′ side (3). The other orientations will direct the helicase away from the mismatch (1 and 4). The interaction of UvrD with MutL orients the protein in the proper direction (2 and 3).
We would like to propose that the UvrA and UvrB proteins orient the UvrD protein, by binding at or near the entry site of the helicase. Several arguments for such a model can be given. (i) In NER the UvrD protein removes the damage-containing oligonucleotide that results from the two incisions made in the UvrBC-DNA complex. For this action the UvrD protein also needs to initiate unwinding from a nick. From each of the two nicks that are present, UvrD can potentially start to unwind the DNA in two directions: towards the DNA damage, thereby releasing the oligonucleotide, or in the opposite direction, which will not result in oligonucleotide removal (Fig. 4B). Possibly the UvrBC proteins shield one of the nicks, but efficient release of the damage-containing oligonucleotide still requires that the UvrD protein is properly oriented on the other nick. A possible physical interaction between UvrB and UvrD not only would account for such a directed DNA binding, but simultaneously it could stimulate the initiation of the helicase activity, which on a nicked DNA substrate in the absence of other proteins is very slow (35). The fact that the homologous Rep helicase can not substitute for UvrD in NER (12) supports the proposed specific interaction between UvrB and UvrD. For the PolI-independent replication, the same interaction between UvrB and UvrD on the lagging strand might stimulate and direct UvrD towards the unwinding of the DNA-RNA hybrid. (ii) In addition to its role in NER, UvrD is also an important factor in mismatch repair. In this process MutS and MutL bind to a mismatched base, and the MutH protein generates a nick at a nearby GATC sequence (for a review, see reference 24). UvrD, together with one of several nucleases, will remove the mismatch-containing strand, starting at the nicked GATC site. Since this nick can be located either 3′ or 5′ to the mismatch, the UvrD protein needs to be loaded onto the nicked or continuous strand, depending on the location of the GATC site (Fig. 4C). It has been shown that MutS and MutL not only activate the unwinding by UvrD but also bias the unwinding in the direction of the mismatch (6). A physical interaction between MutL and UvrD has been shown (10), and it is likely that this interaction serves to load the helicase on the proper strand. Such a MutL-mediated activation and orientation of UvrD is very similar to our proposed model for the UvrB-mediated activation and orientation of this helicase.
Our proposed model implies that UvrB specifically binds to the lagging strand at or near the junctions between the Okazaki fragments. The requirement for UvrA indicates that such a binding should be mediated via the same UvrA2B complex that in NER recognizes structural changes in the DNA as a result of a DNA damage. In the lagging strand, however, a different kind of DNA structure has to be recognized, since it is very unlikely that DNA damages play a role in the PolI-independent replication pathway. Possibly the UvrA2B complex is capable of recognizing the non-B conformation of RNA-DNA hybrids. The presence of nicks or small gaps might also be important for the recognition. The UvrA2B-DNA complex formed in the lagging strand could subsequently interact with UvrD, thereby directing its helicase activity. Alternatively, in analogy to the sequential reactions during NER, the UvrA protein could first dissociate from the complex and then UvrD could bind to the resulting UvrB-DNA complex. The requirements for functional ATPase sites in UvrA and ATPase-helicase motifs in UvrB indicate that for the proposed binding of UvrA2B or UvrB in the lagging strand, similar ATPase-induced conformational changes are required, which during NER lead to formation of the preincision complex.
Our finding that the N-terminal zinc-binding motif of UvrA is essential for the alternative replication pathway but not for NER suggests that this domain has specifically evolved for the role of UvrA2B in replication. In many cases zinc-binding domains have been found to participate in DNA interactions (2). Possibly the N-terminal zinc-binding domain is involved in the proposed specific binding of UvrA2B in the lagging strand, as discussed above. The C-terminal zinc-binding domain has been shown to be important for the binding of UvrA to damaged and nondamaged DNA (47). The fact that a mutation in the C-terminal zinc-binding domain of UvrA obstructs both DNA repair and PolI-independent replication suggests that this DNA-binding domain is important for both processes. On the other hand it cannot be excluded that the particular mutation not only affects the structure not only of the zinc-binding motif but also of other domains of the protein, thereby indirectly influencing the activity of UvrA in the two processes.
Unlike UvrD, the UvrB protein seems less important for the cells when the Klenow fragment of PolI is present. ΔpolA transductants of a uvrB strain expressing this polymerase domain can be found on minimal medium. If indeed the role of UvrB is to orient the UvrD protein, a possible explanation for the effect of the Klenow fragment could be that binding of the polymerase domain to the 3′ end of an Okazaki fragment prevents DNA unwinding from this 3′ end. As a consequence, the Klenow fragment itself would direct the helicase towards unwinding of the DNA-RNA hybrid. At higher growth rates (i.e., on LB medium) the amount of Klenow fragment probably becomes limiting, and therefore under these conditions, the UvrB protein is essential again.
A striking observation in this study is the fact that the UvrC protein has a strong negative effect on the alternative replication pathway. In the absence of UvrC, ΔpolA transductants grow much better, and overproduction of UvrC is lethal in a ΔpolA strain. UvrC contains two catalytic sites for incision of damaged DNA. The N-terminal part of the protein contains the active site for incision at the 3′ side of the damage, and the active site for 5′ incision is located in the C-terminal part. DNA incision by the N-terminal active site appeared to be mainly responsible for the negative effect of UvrC in a ΔpolA strain. A UvrB mutant lacking the UvrC-binding domain could counteract the negative effect of the presence of UvrC. This strongly suggests that UvrC induces strand incisions by binding to UvrB at a specific DNA target. In what way could such single-strand incisions influence the viability of a ΔpolA strain? As discussed above, UvrB or UvrA2B might bind specifically at or near the junction of an Okazaki fragment. A subsequent binding of UvrC, followed by a strand incision in the Okazaki fragment, would not obviously be deleterious. On the contrary, such an incision is expected to be advantageous, since it would help to remove the RNA primer. If, however, the orientation of the UvrA2B-DNA or UvrB-DNA complex in the lagging strand would lead to UvrC incision in the opposite (template) strand, a double-strand break would be generated, which, if not repaired, is lethal for the cell.
Expression of the UvrC mutant with a base substitution in the 3′ catalytic site (R42A) in a ΔpolA strain was not lethal, but the resulting colonies were clearly smaller than those of a ΔpolA strain without any UvrC. This could mean that the R42A mutant is somewhat leaky and that a limited number of incisions are still induced. Alternatively the R42A mutant could interfere with the PolI-independent replication just by binding to UvrB without inducing incisions, thereby hindering the proposed interaction of UvrD with UvrB.
Overexpression of the UvrA protein in a ΔpolA strain appeared to be lethal as well, whereas overexpression of the same protein in a ΔpolA ΔuvrC double mutant is not. Apparently a higher level of UvrA leads to more deleterious incisions by UvrC. A higher level of UvrB protein does not show this effect, suggesting that the UvrA concentration in the cell is limiting. Increasing the level of UvrA by the introduction of a multicopy plasmid will result in the formation of more UvrA2B complexes and subsequently the binding of more of these complexes to the proposed sites in the lagging strand. As a result, more targets for incision by UvrC are formed, finally leading to the death of the cells.
The viability of a ΔpolA strain in the presence of UvrA, UvrB, and UvrC strongly depends on the genetic background of the strain. Strain KMBL1001 (which does not have any known chromosomal mutations) with the ΔpolA mutation could survive on minimal medium only at 30°C, whereas strain S90C with the same mutation was viable on minimal medium at 30 and 37°C. The influence of the strain background on the severity of the ΔpolA mutation has been described before (14). A particular E. coli strain (SY203) carrying a polA deletion was shown to be nonviable on minimal medium at 37°C, although the authors did not report whether the strain could survive at lower temperatures. In this case also, the inviability could not be ascribed to a specific chromosomal mutation (14).
Additional deletion of the uvrC gene allowed KMBL1001 to grow at 37°C as well. Possibly the effect of the strain backgrounds is related to differences in uvrC expression in the different strains. A higher level of UvrC will lead to more harmful incisions, which need to be repaired for survival of the cell. At lower growth rates the cell has more time for repair, and therefore strains that contain more UvrC protein can survive at lower temperatures but not at higher temperatures.
The results in this paper show that the 3′ catalytic site of UvrC induces incisions in nondamaged DNA in vivo, causing a negative effect when the cell is dependent on the PolI-independent replication pathway. It is not clear from our experiments whether the same incision activity on nondamaged DNA has a function in other processes in the cell. DNA incision by a complex of UvrB and UvrC in the absence of DNA damage has also been shown in vitro (9, 28, 48). This incision, however, which takes place seven nucleotides from a single strand-double strand junction, is induced by the catalytic site that on damaged DNA makes the 5′ nick and is independent of UvrA (28). For this UvrC-induced incision also, a clear in vivo function has not yet been found. The determination of potential functions of the two types of damage-independent incision awaits a further characterization of substrates that are incised by Uvr(A)BC.
In summary, we have shown that UvrA, UvrB, and UvrD, together with other, as-yet-unidentified proteins like polymerase(s) and exonuclease(s), can take over the function of the PolI enzyme in DNA replication. The existence of such backup systems can be very important for the cell, since it provides flexibility, both on short- and long-term scales. On a short-term scale, backup systems can ensure the survival of cells in which, as a result of internal or external variations, the level of a specific protein drops below a critical level. On a long-term scale, backup systems allow proteins to evolve into having other functions, even if this results in the eventual loss of their original functions.
ACKNOWLEDGMENTS
We thank Catherine M. Joyce, Steven W. Matson, and Aziz Sancar for providing strains and plasmids and Esther Vogels for technical assistance.
This work was supported by the J. A. Cohen Institute for Radiopathology and Radiation Protection (IRS).
REFERENCES
- 1.Backendorf C, Brandsma J A, Kartasova T, van de Putte P. In vivo regulation of the uvrA gene: role of the −10 and −35 promoter regions. Nucleic Acids Res. 1983;11:5795–5810. doi: 10.1093/nar/11.17.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg J M, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 3.Bernardi A, Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984;12:9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandsma J A, de Ruijter M, Szpilewska H, Tasseron-de Jong J G, Brouwer J, van de Putte P. In: Mechanisms and consequences of DNA damage processing. Friedberg E C, Hanawalt P C, editors. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 95–103. [Google Scholar]
- 5.Brutlag D, Atkinson M R, Setlow P, Kornberg A. An active fragment of DNA polymerase produced by proteolytic cleavage. Biochem Biophys Res Commun. 1969;37:982–989. doi: 10.1016/0006-291x(69)90228-9. [DOI] [PubMed] [Google Scholar]
- 6.Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicaseII-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- 7.George J W, Brosh R M, Jr, Matson S W. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J Mol Biol. 1994;235:424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- 8.Goosen N, Moolenaar G F, Visse R, van de Putte P. Functional domains of the E. coli UvrABC proteins in nucleotide excision repair. In: Eckstein F, Lilley D M J, editors. Nucleic acids and molecular biology: DNA repair. Berlin, Germany: Springer Verlag; 1998. pp. 103–123. [Google Scholar]
- 9.Gordienko I, Rupp W D. A specific 3′ exonuclease activity of UvrABC. EMBO J. 1998;17:626–633. doi: 10.1093/emboj/17.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall M C, Jordan J R, Matson S W. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J. 1998;17:1535–1541. doi: 10.1093/emboj/17.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horiuchi T, Nagata T. Lethality of the Escherichia coli K12 cell doubly deficient in DNA polymerase I and DNA strand-joining activity. Mol Gen Genet. 1974;128:105–115. doi: 10.1007/BF02654484. [DOI] [PubMed] [Google Scholar]
- 12.Husain I, Van Houten B, Thomas D C, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci USA. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce C M, Grindley N D F. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce C M, Fujii D M, Laks H S, Hughes C M, Grindley N D. Genetic mapping and DNA sequence analysis of mutations in the polA gene of Escherichia coli. J Mol Biol. 1985;186:283–293. doi: 10.1016/0022-2836(85)90105-6. [DOI] [PubMed] [Google Scholar]
- 15.Klenow H, Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by a limited proteolysis. Proc Natl Acad Sci USA. 1970;65:168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman and Company; 1992. pp. 113–164. [Google Scholar]
- 17.Kushner S R, Nagagaishi H, Templin A, Clark A J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci USA. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert B, Roques B P, Le Pecq J-B. Induction of an abortive and futile DNA repair process in E. coli by the antitumor DNA bifunctional intercalator, ditercalinium: role in polA in death induction. Nucleic Acids Res. 1988;16:1063–1078. doi: 10.1093/nar/16.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J J, Sancar A. Active site of (A)BC excinuclease. I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J Biol Chem. 1992;267:17688–17692. [PubMed] [Google Scholar]
- 21.Matson S W. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3′ to 5′ direction. J Biol Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- 22.Mazur S J, Grossman L. Dimerization of Escherichia coli UvrA and its binding to undamaged and ultraviolet-light damaged DNA. Biochemistry. 1991;30:4432–4443. doi: 10.1021/bi00232a009. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 24.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 25.Monk M, Peacey M, Gross J D. Repair of damage induced by ultraviolet light in DNA polymerase-defective Escherichia coli cells. J Mol Biol. 1971;58:623–630. doi: 10.1016/0022-2836(71)90376-7. [DOI] [PubMed] [Google Scholar]
- 26.Moolenaar G F, Visse R, Ortiz-Buysse M, Goosen N, van de Putte P. Helicase motifs V and VI of the Escherichia coli UvrB protein of the UvrABC endonuclease are essential for the formation of the preincision complex. J Mol Biol. 1994;240:294–307. doi: 10.1006/jmbi.1994.1447. [DOI] [PubMed] [Google Scholar]
- 27.Moolenaar G F, Franken C L M C, Dijkstra D M, Thomas-Oates J E, Visse R, van de Putte P, Goosen N. The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex, but not the catalytic site for 3′ incision. J Biol Chem. 1995;270:30508–30515. doi: 10.1074/jbc.270.51.30508. [DOI] [PubMed] [Google Scholar]
- 28.Moolenaar G F, Bazuine M, van Knippenberg I C, Visse R, Goosen N. Characterization of the Escherichia coli damage-independent UvrBC endonuclease activity. J Biol Chem. 1998;273:34896–34903. doi: 10.1074/jbc.273.52.34896. [DOI] [PubMed] [Google Scholar]
- 29.Morimyo M, Shimazu Y. Evidence that the gene uvrB is indispensable for a polymerase I deficient strain of Escherichia coli K-12. Mol Gen Genet. 1976;147:243–250. doi: 10.1007/BF00582875. [DOI] [PubMed] [Google Scholar]
- 30.Myles G M, Sancar A. Isolation and characterization of functional domains of UvrA. Biochemistry. 1991;30:3834–3840. doi: 10.1021/bi00230a005. [DOI] [PubMed] [Google Scholar]
- 31.Okazaki R, Arisawa M, Sugino A. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc Natl Acad Sci USA. 1971;68:2954–2957. doi: 10.1073/pnas.68.12.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivera R M, Bonhoeffer E. Replication of Escherichia coli requires DNA polymerase I. Nature. 1974;250:513–514. doi: 10.1038/250513a0. [DOI] [PubMed] [Google Scholar]
- 33.Pannekoek H, Noordermeer I, van de Putte P. Expression of the cloned uvrB gene of Escherichia coli: dependency on nonsense suppressors. J Bacteriol. 1979;139:48–53. doi: 10.1128/jb.139.1.48-53.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Runyon G T, Bear D G, Lohman T M. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc Natl Acad Sci USA. 1990;87:6383–6387. doi: 10.1073/pnas.87.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Runyon G T, Lohman T M. Kinetics of Escherichia coli helicase II-catalyzed unwinding of fully duplex and nicked circular DNA. Biochemistry. 1993;32:4128–4138. doi: 10.1021/bi00066a039. [DOI] [PubMed] [Google Scholar]
- 36.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 37.Seeberg E, Rupp W D, Strike P. Impaired incision of ultraviolet-irradiated deoxyribonucleic acid in uvrC mutants of Escherichia coli. J Bacteriol. 1980;144:97–104. doi: 10.1128/jb.144.1.97-104.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shizuya H, Dykhuizen D. Conditional lethality of deletions which include uvrB in strains of Escherichia coli lacking deoxyribonucleic acid polymerase I. J Bacteriol. 1972;112:676–681. doi: 10.1128/jb.112.2.676-681.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel E C. Ultraviolet-sensitive mutator mutU4 of Escherichia coli inviable with polA. J Bacteriol. 1973;113:161–166. doi: 10.1128/jb.113.1.161-166.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohi M, Alexandrovich A, Moolenaar G F, Visse R, Goosen N, Vernede X, Fontecilla-Camps J, Champness J, Sanderson M R. Crystal structure of Escherichia coli UvrB C-terminal domain and a model for UvrB-UvrC interaction. FEBS Lett. 2000;465:161–164. doi: 10.1016/s0014-5793(99)01690-7. [DOI] [PubMed] [Google Scholar]
- 41.Thiagalingam S, Grossman L. The multiple roles for ATP in Escherichia coli UvrABC endonuclease-catalyzed incision reaction. J Biol Chem. 1993;268:18382–18389. [PubMed] [Google Scholar]
- 42.Uyemura D, Eichler D C, Lehman I R. Biochemical characterization of mutant forms of DNA polymerase I from Escherichia coli. II. The polAex1 mutation. J Biol Chem. 1976;251:4085–4089. [PubMed] [Google Scholar]
- 43.Van den Berg E, Zwetsloot J, Noordermeer I, Pannekoek H, Dekker B, Dijkema R, van Ormondt H. The structure and function of the regulatory elements of the Escherichia coli uvrB gene. Nucleic Acids Res. 1981;9:5623–5643. doi: 10.1093/nar/9.21.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Sluis C A, Brandsma J A. Plasmids carrying the uvrA and uvrC genes of Escherichia coli K12: construction and properties. In: Seeberg E, Kleppe K, editors. Chromosome damage and repair. New York, N.Y: Plenum Publishing Corporation; 1981. pp. 293–302. [Google Scholar]
- 45.Verhoeven E E A, van Kesteren M, Moolenaar G F, Visse R, Goosen N. Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J Biol Chem. 2000;275:5120–5123. doi: 10.1074/jbc.275.7.5120. [DOI] [PubMed] [Google Scholar]
- 46.Visse R, de Ruijter M, Ubbink M, Brandsma J A, van de Putte P. The first zinc-binding domain of UvrA is not essential for UvrABC-mediated DNA excision repair. Mutat Res. 1993;294:263–274. doi: 10.1016/0921-8777(93)90009-6. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Mueller K L, Grossman L. A mutational study of the C-terminal zinc-finger motif of the Escherichia coli UvrA protein. J Biol Chem. 1994;269:10771–10775. [PubMed] [Google Scholar]
- 48.Zou Y, Walker R, Bassett H, Geacintov N E, Van Houten B. Formation of DNA repair intermediates and incision by the ATP-dependent UvrB-UvrC endonuclease. J Biol Chem. 1997;272:4820–4827. doi: 10.1074/jbc.272.8.4820. [DOI] [PubMed] [Google Scholar]
- 49.Zwetsloot J C M, Barbeiro A P, Vermeulen W, Arthur H M, Hoeijmakers J H J, Backendorf C. Microinjection of Escherichia coli UvrA, B, C and D proteins into fibroblasts of xeroderma pigmentosum complementation groups A and C does not result in restoration of UV-induced unscheduled DNA synthesis. Mutat Res. 1985;166:89–98. doi: 10.1016/0167-8817(86)90044-1. [DOI] [PubMed] [Google Scholar]