Abstract
New approaches to target antibacterial agents toward Gram-negative bacteria are key, given the rise of antibiotic resistance. Since the discovery of polymyxin B nonapeptide as a potent Gram-negative outer membrane (OM)-permeabilizing synergist in the early 1980s, a vast amount of literature on such synergists has been published. This Review addresses a range of peptide-based and small organic compounds that disrupt the OM to elicit a synergistic effect with antibiotics that are otherwise inactive toward Gram-negative bacteria, with synergy defined as a fractional inhibitory concentration index (FICI) of <0.5. Another requirement for the inclusion of the synergists here covered is their potentiation of a specific set of clinically used antibiotics: erythromycin, rifampicin, novobiocin, or vancomycin. In addition, we have focused on those synergists with reported activity against Gram-negative members of the ESKAPE family of pathogens namely, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, and/or Acinetobacter baumannii. In cases where the FICI values were not directly reported in the primary literature but could be calculated from the published data, we have done so, allowing for more direct comparison of potency with other synergists. We also address the hemolytic activity of the various OM-disrupting synergists reported in the literature, an effect that is often downplayed but is of key importance in assessing the selectivity of such compounds for Gram-negative bacteria.
Keywords: Gram-negative bacteria, Gram-positive antibiotics, synergy, outer membrane, permeabilization, potentiators
The increasing occurrence of antibiotic resistance among Gram-negative pathogens highlights the need for novel antibacterial agents and therapeutic strategies. It is well established that Gram-negative bacteria are inherently harder to kill with antibiotics than Gram-positives, given the presence of the Gram-negative outer membrane (OM) as well as efflux pumps.1−4 Given the limited number of clinically effective anti-Gram-negative agents, there is an urgent need for new treatments against Gram-negative pathogens.5−7 This troubling reality is further exacerbated by increasing accounts of emerging resistance mechanisms against Gram-negative antibiotics, including extended spectrum β-lactamases (ESBLs) that can render even fifth-generation cephalosporins and carbapenems inactive,8−11 enzymes that structurally modify and deactivate aminoglycosides,12−15 and mcr-mediated polymyxin resistance.16−27 In this context, the World Health Organization (WHO) recently listed Acinetobacter baumannii (carbapenem-resistant), Pseudomonas aeruginosa (carbapenem-resistant), and the Enterobacteriaceae (carbapenem-resistant and ESBL-producing strains) as the bacterial pathogens of highest priority for the development of new antibiotics.28
The Gram-negative OM functions as a barrier that prevents many antibiotics, that are otherwise active against Gram-positive species, from reaching their targets.3,29 The OM itself consists of an asymmetrical lipid bilayer (see Figure 1A).30 The inner leaflet consist mostly of phospholipids and is similar to the cytoplasmic membrane.31 The outer leaflet is made up of an organized and fortified structure of densely packed lipopolysaccharides (LPSs) and Mg2+/Ca2+ cations that bridge the negatively charged phosphate groups of the lipid A component of LPS (see Figure 1B).3,32 Furthermore, the tightly packed saturated acyl chains result in a low level of membrane fluidity that limits the diffusion of hydrophobic compounds across the OM.2,3 The OM also contains porins, which function as size exclusion channels across the OM that mediate the diffusion of small hydrophilic molecules between the periplasm and the extracellular environment while keeping large, hydrophobic molecules, including many antibiotics, out.1,2,29 Additionally, when lipophilic or amphiphilic antibiotics do manage to cross the OM, multi-drug efflux pumps can transport these molecules back out.1−3,29 In many cases, the overexpression of efflux pumps provides an effective means for a Gram-negative pathogen to decrease its susceptibility to antibiotics.3,33 Taken together, their diverse resistance mechanisms and unique cellular features provide Gram-negative bacteria with a formidable range of defenses against antibacterial agents.
Figure 1.
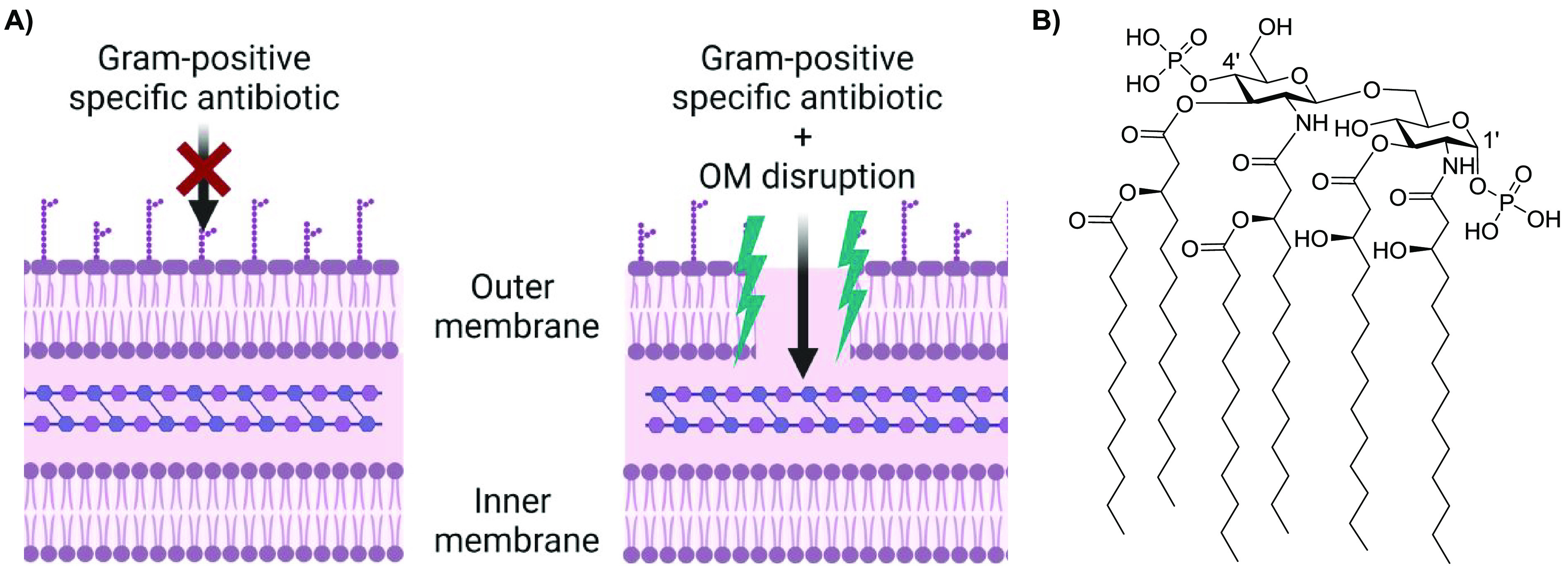
(A) Schematic depiction of the OM disruption required for potentiation of Gram-positive specific antibiotics (created with BioRender.com). (B) Lipid A (from Escherichia coli K-12), the hydrophobic anchor of LPS.
To address the specific challenges posed by Gram-negative bacteria, a number of new and innovative approaches are currently under investigation. Such strategies include interfering with LPS biosynthesis,34−37 targeting OM proteins such as the β-barrel assembly machine (BAM) complex,34,38,39 developing siderophore–antibiotic conjugates as Trojan horse agents, including the recently approved cefiderecol,40−42 co-administering different antibiotics to restrict or reverse antibiotic resistance,43,44 and blocking efflux pumps.45−48 In addition to these promising strategies, the development of agents that can selectively disrupt the OM offers the possibility of sensitizing Gram-negative bacteria to antibiotics that otherwise function only against Gram-positive bacteria.3,7,32 The pursuit of such synergists continues to be a very active field of research and is the basis for this Review.
The best-studied example of an OM-disrupting synergist is polymyxin B nonapeptide (PMBN), which is obtained by enzymatic degradation of the clinically used lipopeptide polymyxin B (PMB).7,32 The potentiating effects of PMBN were first reported in the 1980s, and in the decades since, a growing number of OM-disrupting synergists have been discovered.7,32,49 To date, a number of reviews have been published on the general topic of antibiotic synergy,50−57 including compounds that potentiate Gram-positive antibiotics through interactions with the OM58 and OM-disrupting synergists.32,59−63 However, a comprehensive overview of OM-disrupting synergists that also provides the reader with a direct comparison of both the potency and selectively of these compounds has, to date, been lacking. In this regard, the most widely accepted benchmark for synergistic activity is the so-called fractional inhibitory concentration index (FICI, Box 1).64 In this Review, we discuss only those synergists for which FICI values are reported or could be calculated from published data. The other criterion we have also chosen to emphasize is the selectivity of OM disruption associated with these synergists. In this regard, we pay special attention to the hemolytic activity reported for the various OM disrupters as a means of assessing their membrane specificity.
Box 1. An important formalism in the field of synergy is the fractional inhibitory concentration index (FICI).
The FICI is calculated from experimental minimum inhibitory concentration (MIC) data as shown in eq 1. A synergistic combination is generally defined as an FICI < 0.5. Additionally, it allows for a straightforward comparison of the potency of the synergistic combinations: the lower the FICI, the more potent the combination.
| 1 |
Among the Gram-negative bacteria for which OM-disrupting synergists have been reported, we have selected those pathogens noted on the WHO’s priority list: A. baumannii, Escherichia coli, Klebsiella pneumoniae, or P. aeruginosa.28 As for Gram-positive specific antibiotics whose activity is potentiated by OM-disrupting synergists, we have chosen to focus on clinically used agents that are most commonly evaluated for synergy with OM disrupters: erythromycin, rifampicin, vancomycin, and novobiocin.7,58 This criterion has, for example, led to the exclusion of OM-disrupting agents for which synergy was reported with macrolide antibiotics other than erythromycin.65−68 Also, while the specific media conditions used in antibacterial assays can strongly influence the outcome of synergy studies, for the sake of brevity, we do not include this level of detail here and instead provide clear referencing of the original studies wherein such information can be found. In addition, to further streamline the Review, synergists for which an OM-disrupting mechanism was not clearly demonstrated are not here discussed in detail.69−77 Furthermore, synergists that specifically engage with Gram-negative targets and subsequently cause OM disruption as a secondary effect are not discussed in this Review.78−86
The scope of the synergists included in this Review ranges from peptides to synthetic small molecules and small polymers of <1500 Da. In this regard, protein-based OM disrupters such as the membrane attack complex (MAC),87 lactoferrin,88 and the bactericidal/permeability-increasing protein (BPI)89 or larger polymers or polymer-like agents90−97 will not be discussed. This Review is further organized on the basis of the chemical families of the synergists covered. We begin with cyclic peptides based on PMBN, followed by linear peptides, cationic steroids, peptide–steroid hybrids, and small molecules. For each subgroup of synergists, a summary table has been assembled to provide a convenient comparative overview of FICI values. These tables also include the identity of the Gram-negative species and companion antibiotics employed in generating the FICIs. In addition, where possible, we have included the reported hemolytic activity of each synergist to provide an indication of its selectivity for Gram-negative cells.
1. Peptide-Based Potentiators
1.1. Polymyxin-Derived Synergists
Polymyxin-derived synergists have been extensively reviewed in the past, and therefore only a concise summary of these analogues is here included.7,32,63 PMBN is derived from the parent lipopeptide PMB (see Figure 2A). Unlike its parent compound, PMBN has no inherent antimicrobial activity, nor is it nephrotoxic.7,98 In their landmark 1983 paper, Martti and Timo Vaara demonstrated that the combination of PMBN with hydrophobic, generally Gram-positive-specific, antibiotics results in a potent synergistic effect (see Table 1).32,49 In this regard, PMBN is often used as a benchmark for synergistic activity.7 Apart from PMBN, other truncated derivatives of PMB, like deacylpolymyxin B (DAPB), polymyxin B octapeptide (PMBO), and polymyxin B heptapeptide (PMBH), also display synergistic activity (Figure 1A and Table 1).32 The peptide macrocycle is of key importance for these synergists, as linear PMBN variants lose their synergistic activity.99
Figure 2.
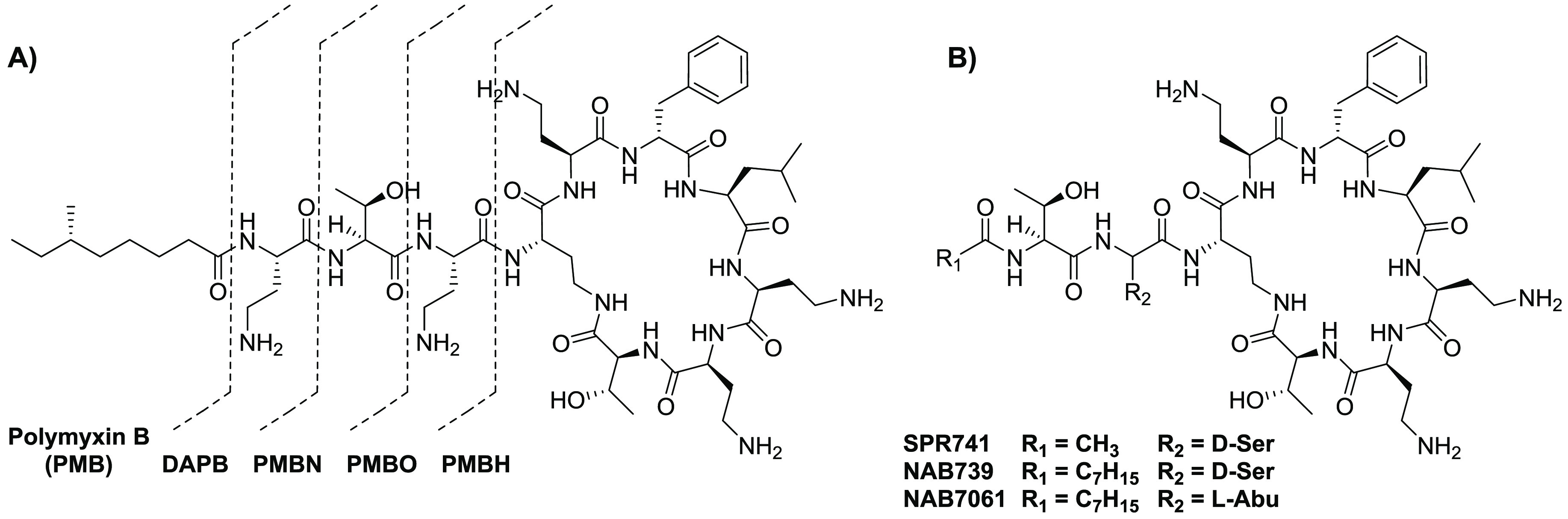
Molecular structures of (A) polymyxin B (PMB), deacylpolymyxin B (DAPB), polymyxin B nonapeptide (PMBN), polymyxin B octapeptide (PMBO), and polymyxin B heptapeptide (PMBH) and (B) PMBN analogues SPR741, NAB739, and NAB7061.
Table 1. Synergistic Activity of Polymyxin Analogues.
| name | ref | FICIa | pathogen | antibiotic |
|---|---|---|---|---|
| PMBN | (105) | 0.013a | E. coli | rifampicin |
| PMBO | (105) | 0.013a | E. coli | rifampicin |
| PMBH | (105) | 0.020a | E. coli | rifampicin |
| DAPB | (105) | 0.043a | E. coli | rifampicin |
| SPR741 | (106) | 0.06 | E. coli | rifampicin |
| NAB739 | (100) | 0.126 | A. baumannii | rifampicin |
| NAB7061 | (100) | 0.055 | E. coli | rifampicin |
A new generation of PMBN analogues containing only three positive charges was developed more recently.100,101 SPR741, previously named NAB741, has passed the Phase I clinical trials (see Figure 2B).7 Like PMBN, SPR741 has no lipophilic tail, resulting in improved renal clearance compared to PMB and other analogues that have a lipophilic tail, such as NAB739 and NAB7061.101 NAB7061 has little inherent antimicrobial activity but is a very potent synergist, while NAB739 has very potent antimicrobial activity (Table 1).102 Remarkably, this difference in activity between NAB739 and NAB7061 is attributed to the absence of one hydroxyl group in NAB7061 (see Figure 2B).100 NAB739 has been reported to exhibit generally moderate synergistic activity against wild-type strains, with the exception of the A. baumannii strain indicated in Table 1.100,103 Interestingly, against mcr-positive strains, the loss of antimicrobial activity for NAB739 is accompanied by a significant increase in its synergistic activity, an effect also noted for colistin.103,104
1.2. Dilipidated Polymyxins
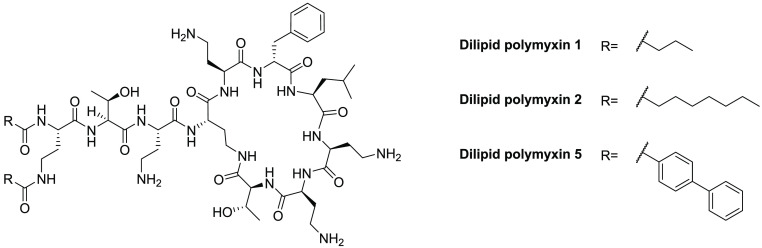
Polymyxin analogues bearing additional lipid tails have also been explored to test the hypothesis that additional hydrophobicity might enhance membrane interactions.107 To generate these variants, a variety of acyl tails were added to both amino groups of the N-terminal 2,4-diaminobutyric acid (Dab) residue of PMB (Figure 3).107,108 The introduction of simple propyl lipids, as in analogue 1, led to a complete loss of inherent activity (MIC ≥ 64 μg/mL), while the analogues 2 and 5, bearing larger, more hydrophobic groups, maintained moderate activity, with MICs of 4–64 μg/mL against most Gram-negative bacteria.107 Notably, the reduced inherent activity was accompanied by a higher synergistic potential (Table 2), indicating that these dilipidated analogues have an increased capacity to disrupt the OM.107 Also of note is the reported activity of analogues 2 and 5 against Gram-positive bacteria (MICs of 8–32 μg/mL) compared to colistin, which has no such activity (MIC > 128 μg/mL).107
Figure 3.
Molecular structures of the dilipidated polymyxin analogues.
Table 2. Synergistic Activities of Dilipidated Polymyxin Analogues.
| name | ref | FICI | pathogen | antibiotic | hemolytic activitya |
|---|---|---|---|---|---|
| dilipid polymyxin 1 | (107) | 0.02 | P. aeruginosa | rifampicin | <10% (1 h) |
| dilipid polymyxin 2 | (107) | 0.26 | P. aeruginosa | novobiocin | <10% (1 h) |
| dilipid polymyxin 5 | (107) | 0.31 | P. aeruginosa | rifampicin | <10% (1 h) |
Non-hemolytic is defined as <10% hemolysis compared to positive control, with incubation times denoted in parentheses.
1.3. Linear Peptide-Based Synergists
In most reviews published on the topic of OM-targeting synergists, relatively little attention has been paid to linear peptides. Peptides have several drawbacks, including poor metabolic stability, low bioavailability, potential immunogenicity, and high production costs.109−111 To improve their metabolic stability, the structures of peptides can be adapted by a number of approaches, including peptidomimetics, lipidation, head-to-tail cyclization, N- and C-terminus modifications, backbone stereochemistry changes, and incorporation of unnatural amino acids.109,110,112−116 Improvements to the bioavailability of peptides have also been explored by applying formulation techniques, adjusting the properties of peptides, or linking them to a moiety to improve passage over the blood–brain barrier.109−111 These advances, combined with the development of more economical methods for peptide synthesis, support a future role for peptide-based therapeutics, with a number of antimicrobial peptides (AMPs) already in (pre)clinical development.117−121
An increasing number of peptide synergists that function through OM disruption have been reported in the literature (see Table 3). In some studies, panels of structurally similar peptides are screened, resulting in the identification of multiple hits with FICI <0.5. In such cases, we have opted to select up to four of the most potent synergists to limit the number of peptides. Given that most peptide-based synergists are derived from specific lead proteins or AMPs, we have divided the linear peptide synergists accordingly, both in the discussion below and in the overview in Table 3.
Table 3. Overview of Linear Peptide-Based Synergists.
| namea | ref | peptide sequenceb | FICI | pathogen | antibiotic | hemolytic activityc |
|---|---|---|---|---|---|---|
| Cathelicidin-Derived Peptides | ||||||
| FK16 | (130) | FKRIVQRIKDFLRNLV | 0.25 | P. aeruginosa | vancomycin | <10% (1 h) |
| KR-12-a2 | (131, 214) | KRIVQRIKKWLR-NH2 | 0.156 | P. aeruginosa | erythromycin | <10% (1 h) |
| L-11 | (132) | RIVQRIKKWLR-NH2 | 0.070 | A. baumannii | vancomycin | NR |
| D-11 | (132, 133) | rivqrikkwlr-NH2 | 0.032 | A. baumannii | rifampicin | <10% (1 h) |
| novicidin | (134) | KNLRRIIRKGIHIIKKYF | 0.018 | E. coli | rifampicin | <10% (1 h) |
| G2 | (135) | RGARIVVIRVAR-NH2 | 0.38 | P. aeruginosa | erythromycin | NR |
| R2 | (135) | RRARIVVIRVAR-NH2 | 0.27 | P. aeruginosa | erythromycin | NR |
| DP7 | (138, 215) | VQWRIRVAVIRK | 0.25 | P. aeruginosa | vancomycin | <10% (1 h) |
| indopt 10 | (135) | ILKWKIFKWKWFR-NH2 | 0.38 | P. aeruginosa | erythromycin | NR |
| CLS001 | (138, 140) | ILRWPWWPWRRK-NH2 | 0.28 | P. aeruginosa | vancomycin | 10% (30 min) |
| Lactoferrin-Derived Peptides | ||||||
| P10 | (141) | FWQRNIRKVKKK-NH2 | 0.113 | P. aeruginosa | novobiocin | <10% (1 h) |
| P14 | (141) | FWQRNIRKVKKKI-NH2 | 0.113 | P. aeruginosa | novobiocin | <10% (1 h) |
| P22 | (141) | RFWQRNIRKYRR-NH2 | 0.431 | P. aeruginosa | novobiocin | <10% (1 h) |
| P2–16 | (142) | FWRNIRIWRR-NH2 | 0.116 | P. aeruginosa | novobiocin | NR |
| P12 | (145, 216) | RRWQWRMKKLGA | 0.43 | E. coli | erythromycin | <10% (2 h) |
| P15 | (145) | FK-Bip-RRWQWRMKKLGAd | 0.38 | E. coli | erythromycin | NR |
| P18 | (145) | PAWFKARRWAWRMLKKAA | 0.38 | E. coli | erythromycin | NR |
| Thrombin-Derived Peptides | ||||||
| peptide 6 | (148) | VFRLKKWIQKVI-NH2 | 0.094 | E. coli | rifampicin | <10% (20 h) |
| peptide 14 | (148) | VFRLKKAIQKVI-NH2 | 0.078 | E. coli | erythromycin | <10% (20 h) |
| peptide 19 | (148) | VFRLKKWIQKVA-NH2 | 0.078 | E. coli | rifampicin | <10% (20 h) |
| Histatin-Derived Peptides | ||||||
| Nal-P-113 | (153, 155) | Ac-AKR-Nal-Nal-GYKRKF-Nal-NH2e | 0.38 | E. coli | vancomycin | >10% (1 h) |
| Bip-P-113 | (153, 155) | Ac-AKR-Bip-Bip-GYKRKF-Bip-NH2d | 0.38 | E. coli | vancomycin | >10% (1 h) |
| Other Natural AMPs, Their Hybrids, and Derivatives | ||||||
| buforin II | (156, 217) | TRSSRAGLQFPVGRVHRLLRK | 0.312 | A. baumannii | rifampicin | <10% (1 h) |
| esculentin 1b | (157, 218) | GIFSKLAGKKLKNLLISG-NH2 | 0.36 | E. coli | erythromycin | >10% (1 h) |
| HE2α | (158, 162) | VHISHREARGPSFRICVGFLGPRWARGCSTGN | 0.3 | E.coli | rifampicin | <10% (1 h) |
| HE2β2 | (158, 162) | GDVPPGIRNTICRMQQGICRLFFCHSGTGQQHRQRCG | 0.2 | E.coli | rifampicin | <10% (1 h) |
| anoplin | (159) | GLLKRIKTLL | 0.3125 | P. aeruginosa | rifampicin | <10% (1 h) |
| magainin II | (160, 217) | GIGKFLHAAKKFAKAFVAEIMNS-NH2 | 0.312 | P. aeruginosa | rifampicin | >10% (1 h) |
| cecropin A | (160, 165) | KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK-NH2 | 0.312 | P. aeruginosa | rifampicin | <10% (1 h) |
| CAME | (219, 220) | KWKLFKKIGIGAVLKVLTTG-NH2 | 0.375 | A. baumannii | erythromycin | <10% (1 h) |
| CAMA | (219, 220) | KWKLFKKIGIGKFLHSAKKF-NH2 | 0.25 | A. baumannii | erythromycin | <10% (1 h) |
| HPMA | (219, 221) | AKKVFKRLGIGKFLHSAKKF-NH2 | 0.313 | A. baumannii | erythromcyin | <10% (1 h)n |
| H-TriA1 | (168, 169) | v-dab-GswS-Dab-dab-FEV-alle-Af,g | 0.002 | E. coli | rifampicin | <10% (30 min)n |
| SLAP-S25 | (173) | Ac-Dab-I-Dab-I-Dab-fL-Dab-vLA-NH2f | 0.031 | E. coli | rifampicin | <10% (1 h) |
| A13 | (159) | GWWKRIKTWW | 0.375 | K. pneumoniae | rifampicin | <10% (1 h) |
| A17 | (159) | KWWKRWKKWW | 0.3125 | P. aeruginosa | rifampicin | >10% (1 h) |
| A21 | (159) | KWWKKWKKWW | 0.3125 | K. pneumoniae | rifampicin | <10% (1 h) |
| L7A | (139) | LNLKALAAVAKKIL-NH2 | 0.31 | E. coli | rifampicin | <10% (1 h) |
| S1 | (181, 184) | Ac-KKWRKWLAKK-NH2 | 0.38 | A. baumannii | vancomycin | <10% (1 h)n |
| S1-Nal | (181, 184) | Ac-KKWRKWLAKK-Nal-NH2e | 0.27 | A. baumannii | vancomycin | <10% (1 h)n |
| S1-Nal-Nal | (181, 184) | Ac-KKWRKWLAKK-Nal-Nal-NH2e | 0.27 | A. baumannii | vancomycin | >10% (1 h) |
| Peptide Synergists via Library Screening | ||||||
| peptide 79 | (180, 185) | KKWRKWLKWLAKK-NH2 | 0.14 | E. coli | rifampicin | <10% (1 h) |
| peptide 1 | (71, 222) | KLWKKWKKWLK-NH2 | 0.02 | K. pneumoniae | rifampicin | <10% (1 h) |
| peptide 2 | (71, 188) | GKWKKILGKLIR-NH2 | 0.04 | K. pneumoniae | rifampicin | <10% (1 h) |
| peptide D1 | (71) | klwkkwkkwlk-NH2 | ≤0.03 | K. pneumoniae | rifampicin | NR |
| peptide D2 | (71) | gkwkkilgklir-NH2 | ≤0.04 | K. pneumoniae | rifampicin | NR |
| Peptide Synergists from Phage Display | ||||||
| EC5 | (131, 186) | RLLFRKIRRLKR | 0.266 | P. aeruginosa | erythromycin | <10% (24 h) |
| Designed Peptides | ||||||
| peptide 4 | (187) | KFFKFFKFF | 0.03 | E. coli | rifampicin | >10% (30 min) |
| peptide 5 | (187) | IKFLKFLKFL | 0.06 | E. coli | rifampicin | NR |
| peptide 7 | (187) | CKFKFKFKFC | 0.20 | E. coli | rifampicin | NR |
| ΔFm | (191) | Ac-GΔFRKΔFHKΔFWA-NH2h | 0.3 | E. coli | rifampicin | <10% (1 h) |
| ΔFmscr | (191) | Ac-GΔFRKΔFKAΔFWH-NH2h | 0.14 | E. coli | rifampicin | <10% (1 h) |
| LK-L8P | (223) | Ac-LKKLLKLPKKLLKL-NH2 | 0.18 | E. coli | erythromycin | <10% (4 h) |
| LK-L11P | (223) | Ac-LKKLLKLLKKPLKL-NH2 | 0.47 | E. coli | erythromycin | <10% (4 h) |
| KL-L6P | (223) | Ac-LKKLLPLLKKLLKL-NH2 | 0.33 | E. coli | erythromycin | >10% (4 h) |
| KL-L9P | (223) | Ac-LKKLLKLLPKLLKL-NH2 | 0.12 | E. coli | erythromycin | <10% (4 h) |
| zp12 | (196) | GIKRGIIKIIKRIKRI-NH2 | 0.25 | K. pneumoniae | vancomycin | NR |
| zp16 | (196) | GIKRGIIKIIRRIKRI-NH2 | 0.06 | K. pneumoniae | vancomycin | <10% (1 h) |
| K4 | (197, 198) | WRKWRKWRKWRK-NH2 | 0.2 | K. pneumoniae | rifampicin | <10% (1 h) |
| K5 | (197, 198) | WRKWRKWRKWRKWRK-NH2 | 0.2 | E. coli | rifampicin | <10% (1 h) |
| Lipopeptide Synergists | ||||||
| paenipeptin 1 | (199, 200) | C6-Dab-I-Dab-fL-Dab-vLS-NH2f,i | 0.125o | E. coli | rifampicin | <10% (30 min) |
| paenipeptin 9 | (199) | C8-Dab-I-Dab-fL-Dab-vL-Dab-NH2f,j | ≤0.03o | K. pneumoniae | rifampicin | <10% (30 min) |
| paenipeptin 15 | (199) | Cbz-Dab-I-Dab-fL-Dab-vLS-NH2f,k | ≤0.03o | K. pneumoniae | rifampicin | <10% (30 min) |
| paenipeptin 16 | (199) | Cha-Dab-I-Dab-fL-Dab-vLS-NH2f,l | 0.06o | K. pneumoniae | rifampicin | <10% (30 min) |
| dUSCL 2 | (201) | C10-K(C10)KKK-NH2m (Figure 4A) | 0.07 | P. aeruginosa | rifampicin | <10% (1 h) |
| dUSCL 6 | (201) | C10-K(C10)KGK-NH2m (Figure 4A) | 0.25 | P. aeruginosa | rifampicin | <10% (1 h) |
| UTBLP 5 | (202) | C8-K(C8)KKKK-NH2j (Figure 4B) | ≥0.016 | P. aeruginosa | novobiocin | NR |
| UTBLP 6 | (202) | C8-K(C8)K(Me)K(Me)K(Me)K(Me)-NH2j (Figure 4B) | 0.047 | A. baumannii | rifampicin | NR |
| Lipopeptidomimetic Synergists | ||||||
| dUSTBP 2 | (206) | Figure 4C | ≥0.250 | P. aeruginosa | rifampicin | <10% (1 h) |
| dUSTBP 5 | (206) | Figure 4C | ≥0.125 | P. aeruginosa | rifampicin | <10% (1 h) |
| dUSTBP 8 | (206) | Figure 4C | ≥0.002 | A. baumannii | novobiocin | <10% (1 h) |
| OAK C12(ω7) | (212) | Figure 4D | ≤0.073o | E. coli | rifampicin | >10% (3 h) |
| OAK C12 | (212) | Figure 4D | ≤0.211o | E. coli | rifampicin | >10% (3 h) |
| OAK C10 | (212) | Figure 4D | ≤0.036o | E. coli | rifampicin | <10% (3 h)n |
| OAK C8 | (212) | Figure 4D | ≤0.078o | E. coli | rifampicin | <10% (3 h)n |
| OAK C14(ω5)OOc10O | (213) | Figure 4D | 0.20o | K. pneumoniae | rifampicin | <10% (3 h)n |
Compound names are provided as given in the cited literature references.
Lowercase letters indicate d-amino acids.
Non-hemolytic is defined as <10% hemolysis compared to positive control, with incubation times denoted in parentheses; NR denotes no data reported.
Bip = biphenylalanine.
Nal = β-naphthylalanine.
Dab = 2,4-diaminobutyric acid.
alle = d-allo-isoleucine.
ΔF = α,β-didehydrophenylalanine.
C6 = hexanoyl.
C8 = octanoyl.
Cbz = benzyloxycarbonyl.
Cha = cyclohexylalanyl.
C10 = decanoyl.
Concentration tested was lower than 100 μg/mL.
FICI calculated from MIC values reported in the cited literature references.
1.3.1. Cathelicidin Antimicrobial Peptides
The cathelicidins are AMPs that play an important role in the innate immune defense system of mammals and function by binding to bacterial membranes, resulting in their destabilization and lysis.122−125 In addition to their direct antibacterial activity, cathelicidins have also been found to play a role in recruiting immune cells to the site of infection as well as in LPS neutralization.56,122,126 The sole human cathelicidin-AMP gene encodes for hCAP-18, which is cleaved by proteases into the active LL-37.123−125 The mature LL-37 peptide forms an amphipathic α-helix that, upon interaction with bacterial cell surfaces, is associated with a detergent-like antimicrobial activity.127−129 Recently, a truncated version of LL-37, termed FK16, was reported to potentiate the activity of vancomycin against P. aeruginosa (Table 3).130 Similarly, the Kuipers group showed that another LL-37-derived sequence, termed KR-12-2, is able to synergize with azithromycin (and erythromycin, Table 3).131 Further optimization of the peptide sequence resulted in peptide L11, which was also synthesized as the d-amino acid variant (D11) as a means of improving serum stability (Table 3).131,132 These peptides were screened in combination with multiple antibiotics against different Gram-negative strains, and OM disruption assays verified their mode of action.131−133
In addition to the human cathelicidins, derivatives of cathelicidins from other mammals have also been screened for synergistic activity, including novicidin (sheep), bactenectin (bovine), and indolicidine (bovine).122,134,135 Among these, only novicidin was reported to display potent synergy (Table 3).134 In the case of bactenectin, which normally contains a disulfide bridge, a number of linear analogues have been prepared, including peptides G2, R2, and DP7, which were found to exhibit OM disruption and moderate synergy (Table 3).135−138 In the case of indolicidin, structure–activity relationship (SAR) studies have led to the discovery of the synergists Indopt 10 and CLS001 (Table 3). CLS001 is particularly effective and displays synergy with both vancomycin and azithromycin against multiple Gram-negative pathogens.135,138 Marketed under the name Omiganan, CLS001 is also much less hemolytic than indolicidin and is currently in clinical trials for the treatment of skin-related infections.102,139,140
1.3.2. Lactoferrin-Derived Peptides
Lactoferrin is a multifunctional protein found in mammals and plays key roles in the human immune system. Lactoferrin has inherent activity against a range of bacterial, fungal, and viral pathogens, and in the case of Gram-negative bacteria, it can disrupt the OM.88 Based on the LPS-binding region of lactoferrin, known as LF11, the Martínez-de-Tejada group synthesized a series of LF11 homologues (Table 3) that were screened in combination with novobiocin for synergistic activity.141 Based on these findings, a new generation of peptide synergists was designed using PEptide DEscriptors from Sequence (PEDES) software to predict OM-permeabilizing sequences.142 The peptides thus obtained (i.e., peptide P2-16, Table 3) generally showed synergistic activity on par with that of the original series.142 Given the abundance of lactoferrins in other mammals, Svendsen and co-workers also investigated a series of peptides derived from bovine lactoferrin for both antimicrobial activity and synergistic activity.143−146 This led to the identification of a 12-mer peptide termed P12, along with P15, a 15-mer containing biphenylalanine (Bip), and a longer 18-mer termed P18, all of which were found to exhibit moderate synergy with erythromycin when tested against E. coli (Table 3).
1.3.3. Thrombin-Derived Peptides
Thrombin is an enzyme that plays a critical role in coagulation, and recent studies have also shown that certain thrombin-derived C-terminal peptides are capable of binding to LPS and neutralizing its toxic and inflammatory effects.147 Given the capacity of PMB to also bind and neutralize LPS, our group was interested in assessing whether these thrombin-derived peptides might also exhibit the synergistic behavior of PMBN. To this end, we prepared a series of 12-mer thrombin-derived peptides and showed that a number of them are, indeed, potent synergists.148 The most active synergist thus identified (peptide 6, Table 3) was further investigated by means of an alanine scan, leading to the discovery of more potent variants (peptides 14 and 19, Table 3). Notably, these peptides were found to be non-hemolytic, and their synergistic activity was shown to extend to rifampicin, erythromycin, and novobiocin against multiple Gram-negative strains, including those with mcr-mediated resistance.148
1.3.4. Histatins
The histatins are a unique group of histidine-rich peptides found in human saliva that play roles in defending against infection as well as in aiding wound-healing.149 Among the most common histatins, the 24 amino acid histatin 5 has been shown to bind Lipid A and has endotoxin-neutralizing properties.150 SAR studies with histatin 5 led to the identification of a 12-mer sub-region termed P-113 that exhibits antimicrobial activity against Gram-positive and Gram-negative bacteria.149,151−153 Further structural optimization to enhance the stability of P-113 led to analogues incorporating β-naphthylalanine (Nal) and Bip residues to yield Nal-P-113 and Bip-P-113 and wherein the 4th, 5th, and 12th histidine resides were replaced by Nal or Bip, respectively (Table 3).153 Bip-P-113 and Nal-P-113 exhibit antimicrobial activity and improved serum proteolytic stability, and they were also found to permeabilize LPSs containing large unilamellar vesicles used to model the Gram-negative OM.153,154 These findings prompted investigation of vancomycin potentiation by Bip-P-113 and Nal-P-113, revealing both to exhibit moderate synergy.155 However, a notable drawback of Bip-P-113 and Nal-P-113 is their significantly increased hemolytic activity relative to that of P-113.153
1.3.5. Other Natural AMPs, Their Hybrids, and Derivatives
A number of other naturally occurring AMPs have been reported to potentiate antibiotics that are otherwise excluded by the OM. These AMPs are all polycationic and include buforrin II, esculentin 1b, sphistin, HE2α, HE2β2, anoplin, magainin II, and cecropin A (Table 3).156−160 The sources of these AMPs are diverse and include toads, wasp venom, or even the human male reproductive tract.158,159,161 The AMPs here discussed have all been reported to disrupt the OM,157,159,162−164 bind to LPS, and/or show endotoxin-neutralizing activity.156,160,165,166 In general, these AMPs exhibit modest FICIs (0.2–0.36), which has also led to interest in hybrids and derivatives with enhanced synergistic activity. For example, Park and co-workers developed a series of hybrid peptide synergists, termed CAME, CAMA, and HPMA, containing sequences derived from crecopin A, magainin II, and melittin (Table 3).165,167 Other approaches include truncation, as in the case of the lipopeptide AMPs tridecaptin A1 and B1 (TriA1 and TriB1), which themselves exhibit potent inherent anti-Gram-negative activity and, when truncated, were found to be effective synergists.168−171 Specifically, removal of the TriA1 N-terminal lipid yielded H-TriA1, which was found to be much less active as an antibiotic but exhibited very potent synergism when combined with rifampicin, resulting in an FICI of 0.002 against E. coli (Table 3).168,169 Like the tridecaptins, the recently discovered paenipeptins contain a number of Dab residues and have been the subject of SAR studies.172 These efforts led to the discovery of a potent paenipeptin-inspired synergist termed SLAP-S25, which effectively potentiates the activity of rifampicin and vancomycin against E. coli (Table 3).173 In addition to OM disruption, the binding of SLAP-S25 to LPS and phosphatidylglycerol (PG) was established, suggesting that SLAP-S25 is also an inner membrane disrupter.173 This was confirmed by dose-dependent uptake of propidium iodide and release of cellular contents in cells treated with SLAP-S25.173 Notably, SLAP-S25 was also demonstrated to effectively enhance the in vivo activity of colistin against a colistin-resistant strain of E. coli in both Galleria mellonella and mouse infection models.173
Originally isolated from wasp venom, anoplin is one of the smallest known amphipathic, α-helical AMPs.159,161 Multiple SAR investigations have been performed to improve its antimicrobial activity and stability.174−178 A recent study with anoplin reported the systematic introduction of tryptophan and lysine residues to determine the optimal hydrophobicity, amphipathicity, and number of positive charges required for antibacterial activity and minimal cytotoxicity.159 A number of these analogues were also found to be synergistic when combined with rifampicin (see peptides A13, A17, and A21 in Table 3) via a mechanism involving OM disruption.159 A similar study with mastoparan-C, a peptide found in the venom of the European hornet, led to the identification of an analogue termed L7A (Table 3), which also displays synergy via OM perturbation.139 Another example of a synergist derived from a toxic peptide is myotoxin II, which is isolated from certain snake venoms. Studies with peptide sequences based on the C-terminus of myotoxin II resulted in peptide S1 (Table 3), which showed a good balance of synergy with vancomycin and low hemolytic activity.179,180 Attempts at further improving the S1 peptide involved the introduction of Nal residues at the C-terminus to generate S1-Nal, which exhibited enhanced synergistic activity, and S1-Nal-Nal, which also exhibited enhanced synergistic activity but at the expense of increased hemolytic activity (Table 3).181−184
1.3.6. Peptide Synergists Discovered via Library Screening
Guardabassi and co-workers recently reported the development and validation of an assay meant to enable high-throughput screens for identifying OM disruption agents.185 To this end, they applied a whole-cell screening platform that allows for detection of OM permeabilization in E. coli based on the signal generated by a chromogenic substrate reporter for a cytoplasmic β-galactosidase. To validate the assay, a library of peptides and peptidomimetics was screened, which generated a notable hit termed peptide 79 that showed potentiation of various antibiotics at therapeutically relevant levels (Table 3).185 In a follow-up study, the same group went on to develop two improved synergists, termed peptides 1 and 2, along with the all d-amino acid variants, which were also found to effectively potentiate rifampicin against K. pneumoniae (Table 3).71,185
1.3.7. Peptide Synergists from Phage Display
Phage display techniques have also been applied to identify novel peptides capable of interaction with the OM. In one such investigation, a phage library displaying random 12-mer peptides was screened for the ability to bind to the cell surface of Gram-negative bacteria.186 Specificity for the Gram-negative OM was ensured by removal of peptides binding to Gram-positive bacteria by pre-incubation of the library with Staphylococcus aureus.186 This approach led to the identification of a peptide termed EC5 that exhibits moderate antibacterial activity against E. coli and P. aeruginosa, with MICs in the range of 8–16 μg/mL against both.186 The EC5 peptide was shown to cause OM disruption and cytoplasmic membrane depolarization while exhibiting very little hemolytic activity.186 Subsequent synergy studies showed that the peptide was also capable of potentiating the activity of erythromycin, clarithromycin, and telithromycin against P. aeruginosa.131
1.3.8. Rationally Designed Peptide Synergists
Inspired by the structure of DAPB (see Figure 2), Vaara and co-workers designed a series of linear and cyclic peptides for evaluation as synergists.187 The sequences of these peptides were based on an ABBn motif, in which A is a basic amino acid and B a hydrophobic residue (see peptides 4 and 5, Table 3).187 Cyclic peptides were also prepared bearing a similar ABn motif (see peptide 7, Table 3).187 All peptides were screened for synergistic activity with erythromycin, rifampicin, novobiocin, and fusidic acid, with the rifampicin combinations being the most potent (Table 3).187 While the synergistic activity of these peptides could be correlated to their OM-disrupting activity, the effect was not specific, given their high hemolytic activity.187
De novo-designed peptides have also been explored as a means of generating novel synergists. To this end, the Sahal group developed a number of peptides incorporating key elements found in AMPs and synergists, including amphipathicity, positive charge, and helical conformation.188,189 Of note was the introduction of α,β-didehydrophenylalanine (ΔF) into the peptides as a means of constraining the helical conformation of the peptides.190−192 Using this approach, two peptides termed ΔFm and ΔFmscr were identified as effective synergists with low toxicity toward mammalian cells (Table 3).
In another recent approach to identifying novel peptide synergists, Yu and colleagues reported the construction of a small library wherein amphipathic peptides where subjected to a proline-scanning strategy to generate novel hinged peptides.193 Such proline-hinged peptides are reported to have lower toxicity toward mammalian cells, given that their membrane binding is reduced compared to that of conventional AMPs with a high α-helical conformation.194 Proline scanning of two model peptides, LK (LKKLLKLLKKLLKL) and KL (KLLKLLKKLLKLLK), provided a set of peptides that were screened for synergistic activity, with the four most potent peptides displayed in Table 3. The peptides were also screened for hemolysis, which led to identification of peptide KL-L9P as the most promising hit. This peptide was subsequently shown to permeabilize the OM, as evidenced by uptake of N-phenylnaphthalen-1-amine (NPN), and was also found to bind LPS without disturbing the inner membrane.193 Mouse sepsis studies were also performed to evaluate the in vivo synergistic effect of KL-L9P, which displayed a significant potentiation of a number of clinically used antibiotics and resulted in improved overall survival.193
In another recently reported study, Zeng et al. described the application of rational design approaches to generate novel helix-forming AMPs based on cytolytic peptide toxins produced by highly virulent strains of S. aureus.195,196 The peptides thus obtained were shown to have improved physicochemical properties and antibacterial activity, while maintaining low hemolytic activity and cytotoxicity. Among the 16-mers thus generated, two peptides, termed zp12 and zp16, were also found to exhibit potent synergy (Table 3). Notable in this regard is the finding that peptide zp16 specifically potentiates the effect of the glycopeptide antibiotics vancomycin and teicoplanin against highly pathogenic K. pneumoniae.196 The vancomycin-zp16 combination exhibits negligible toxicity in vitro and in vivo, and mechanistic studies indicate that zp16 enhances vancomycin’s cell permeability, leading to markedly reduced biofilm formation and rapid bactericidal effect.196
In 2022, the group of Ni reported the potentiation of multiple antibiotics, including rifampicin, by two rationally designed peptides named K4 and K5 (Table 3).197 These peptides were selected from a library of variants all containing a repeating motif, (WRX)n, wherein X represents I, K, L, F, and W.198 Hemolysis and cytotoxicity assays led to the selection of peptides K4 and K5 as leads.198 The finding that these peptides permeabilize the OM resulted in follow-up studies on the potentiation of antibiotics against Gram-negative bacteria.197 Apart from synergy, a 15-day resistance assay was also performed for the K4 and K5 peptides, with or without antibiotics, showing no significant resistance development.197,198 Also of note, while the inherent activity of K4 was found to be comparable to that of PMB, K4 was reported to display no in vivo toxicity when tested as high as 40 mg/kg, while all mice dosed with PMB at the same concentration died within 24 h.198
1.4. Lipopeptide Synergists
In addition to the exclusively peptide-based synergists described above, lipopeptides have also been explored as synergists. We here cover examples of lipopeptides that do not possess potent inherent antibacterial activity but rather have the capacity to effectively potentiate the activity of other antibiotics. A recent example includes the synthetic paenipeptins developed by Huang and co-workers.199 The design of these lipopeptides is based on peptides produced by Paenibacillus sp. strain OSY-N that contain a number of unnatural and d-amino acids. Using low hemolytic activity as a selection criterion, a subset of these lipopeptides were selected and screened for synergistic activity. This led to the identification of paenipeptins 1, 9, 15, and 16, which exhibit potent synergy (Table 3).199,200 These lipopeptides were further shown to have OM-disrupting activity, as indicated by the NPN assay. Furthermore, in a murine thigh infection model, paenipeptin 1 was shown to effectively potentiate the in vivo activity of both clarithromycin and rifampin against polymyxin-resistant E. coli.200
Small cationic lipopeptides have also been explored as synergists, with the aim of identifying smaller, less hemolytic agents. To this end, Schweizer and co-workers recently reported a series of dilipid ultrashort cationic lipopeptides (dUSCLs) capable of enhancing the activity of clinically used antibiotics against Gram-negative bacteria.201 The design of these dUSCLs consists of lysine-rich tetrapeptides bearing various lipids at the N-terminal residue, as illustrated in Figure 4A. It was found that dUSCLs bearing lipids of ≥11 carbon atoms caused significant hemolysis. However, analogues with slightly shorter lipids were found to achieve an acceptable balance of low hemolytic activity and synergistic activity. This led to the identification of dUSCLs 2 and 6 as the most promising synergists (Table 3) capable of sensitizing a range of Gram-negative strains to various antibiotics. The authors also noted that, in addition to permeabilizing the OM, the dUSCLs may also function by indirectly disrupting antibiotic efflux.201
Figure 4.
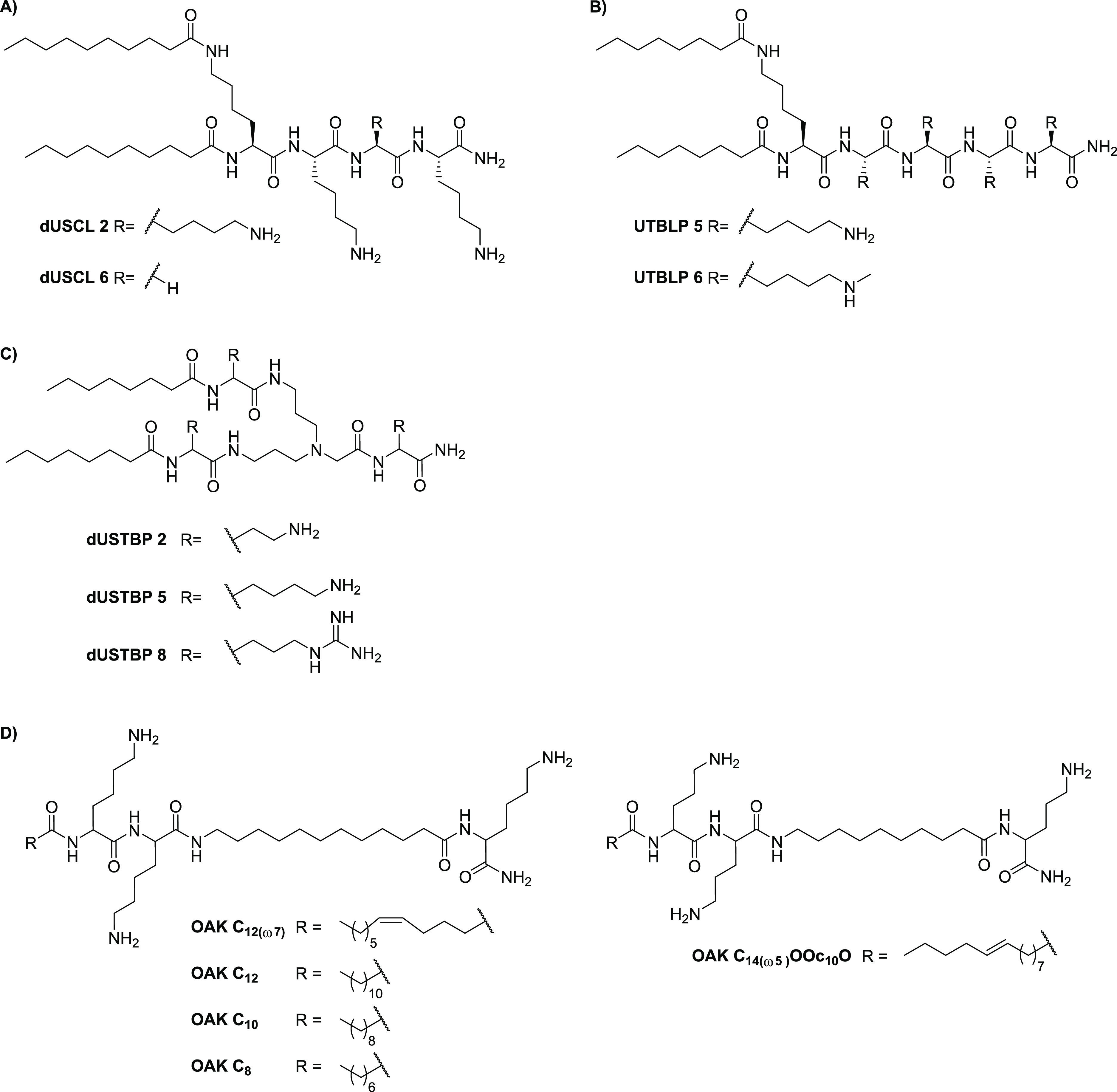
Lipopeptide and lipopeptidomimetic synergists. Representative structures of (A) dilipid ultrashort cationic lipopeptides (dUSCLs), (B) ultrashort tetrabasic lipopeptides (UTBLPs), (C) dilipid ultrashort tetrabasic peptidomimetics (dUSTBPs), and (D) oligo-acyl-lysyls (OAKs).
The Schweizer group also recently reported a series of ultrashort tetrabasic lipopeptides (UTBLPs) synergists.202 These compounds were specifically prepared to assess the effect of lysine N-ζ-methylation on the potentiation of antibiotics, inspired by reports suggesting that N-methylation can lead to reduced hemolysis, increased proteolytic stability, and improved antibacterial activity.203−205 Compared to the dUSCLs, UTBLPs 5 and 6 contain an extra lysine, while an octanoyl group was employed as the lipophilic moiety (Figure 4B).201,202 Methylation of the lysine side chain resulted in a reduction of potentiation for rifampicin and novobiocin in both wild-type and resistant Gram-negative strains.202 A correlation between the number of methyl groups and loss of activity was seen, while the increase in NPN fluorescence of the trimethylated UTBLPs was on par with that of their un- or monomethylated analogues.202
1.5. Lipopeptidomimetic Synergists
The Schweizer group also expanded the scope of their dUSCLs by exploring a series of dilipid ultrashort tetrabasic peptidomimetics (dUSTBPs) as proteolytically stable alternatives.206 In a focused SAR study, they prepared dUSTBPs consisting of three basic amino acids separated by a molecular scaffold, bis(3-aminopropyl)glycine, along with ligation to simple fatty acids (see Figure 4C).206 This led to identification of a number of dUSTBPs capable of potentiating the activity of several antibiotics against pathogenic Gram-negative bacteria while exhibiting low hemolytic activity (Table 3). In particular, dUSTBP 8, consisting of three l-arginine units and a dilipid eight carbons long, was found to potentiate novobiocin and rifampicin against multi-drug-resistant (MDR) clinical isolates of P. aeruginosa, A. baumannii, and Enterobacteriaceae species.206
In 2007, Mor and co-workers introduced the oligo-acyl-lysyls (OAKs) as peptidomimetics of the antimalarial peptide dermseptin S3 (Figure 4D) that were initially evaluated primarily for antimicrobial activity.207−209 Among the first series of analogues prepared, OAK C12(ω7) was found to adhere to the OM with minimal insertion, and its antibacterial activity against Gram-negative bacteria improved in combination with ethylenediaminetetraacetate (EDTA).209−211 The introduction of a double bond in OAK C12(ω7) resulted in a significant reduction of hemolytic activity compared to that of OAK C12, while the slightly less hydrophobic OAK C10 and OAK C8 analogues also showed no hemolytic activity.209,212 In 2013, these four OAKs, as well as the more recently described OAK C14(ω5)OOc10O, containing ornithine instead of lysine (Figure 4D), were reported to potentiate rifampicin against Gram-negative bacteria (Table 3).212,213 Interestingly, the synergistic activity of the OAKs was maintained in human plasma but was suppressed by addition of anti-complement antibodies, suggesting that these compounds sensitize Gram-negative bacteria to the action of antibacterial innate immune mechanisms.213
2. Cationic Steroids
In 1993, the isolation of squalamine from tissues of the dogfish shark Squalus acanthias was reported.224 Squalamine consists of a steroid core linked to a spermidine moiety (Figure 5A) and was found to exhibit broad antimicrobial activity.224 Later, it was established that squalamine disrupts membranes and is also hemolytic. Notably, investigations into its synergistic activity showed that it was unable to potentiate erythromycin against wild-type strains, showing an effect only against a P. aeruginosa strain overproducing MexAB-OprM efflux pumps (see Table 4).225,226 A few years after its discovery, novel squalamine mimics (SMs) were synthesized in an attempt to enhance antibacterial activities (Figure 5B).227 These synthetic analogues consist of cholic and deoxycholic acid as the steroid backbone to which a spermidine chain is appended. This approach resulted in the identification of analogue SM-7, which was found to potentiate rifampicin against multiple Gram-negative bacteria (Table 4).227 However, like squalamine, SM-7 also possesses significant hemolytic activity, limiting its potential for systemic use.227
Figure 5.
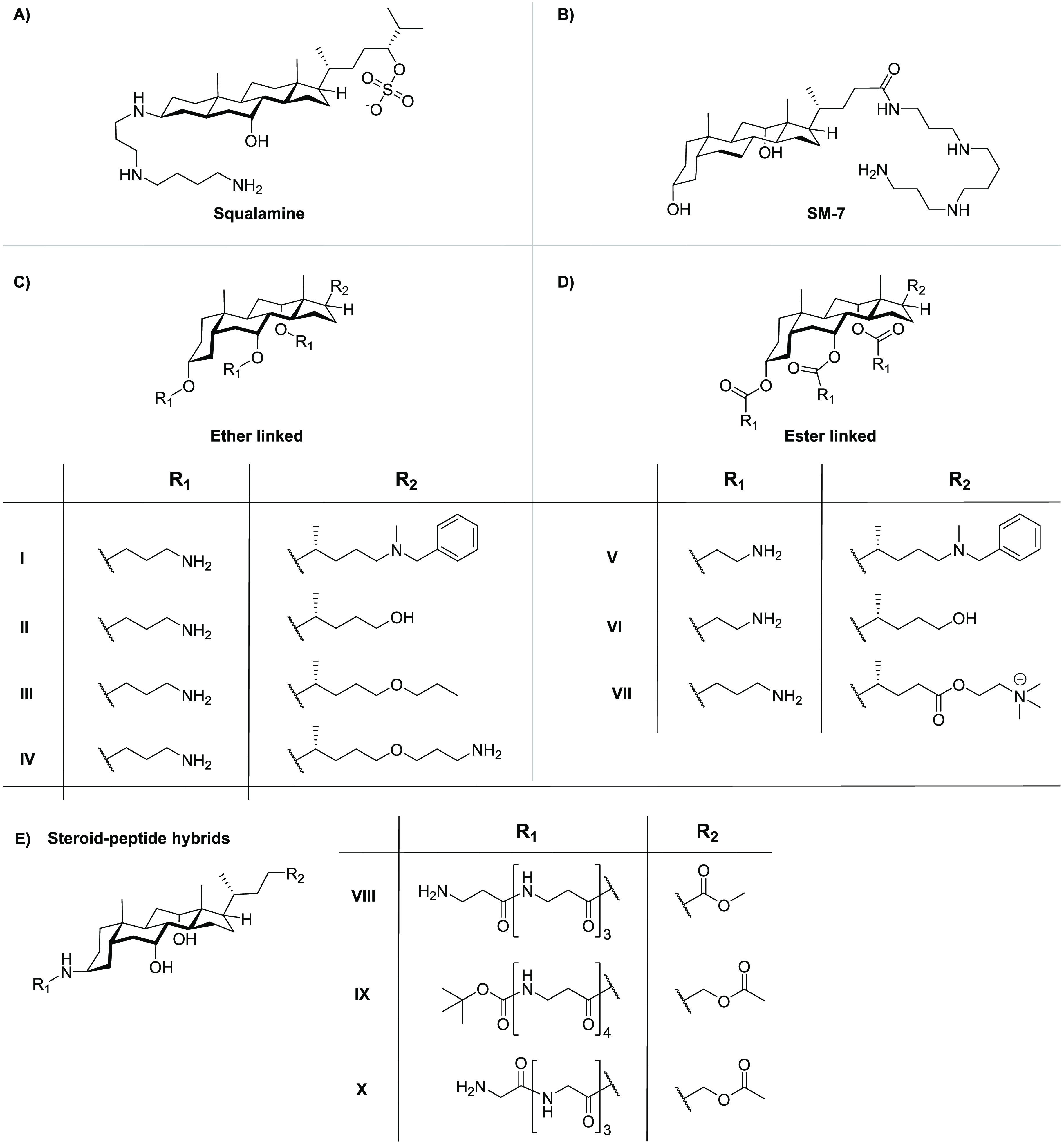
Overview of the synergistic steroids (A) squalamine, (B) squalamine mimic SM-7, (C) polycationic cholic acid ether-linked steroid synergists, (D) polycationic cholic acid ester-linked steroid synergists, and (E) steroid–peptide hybrids.
Table 4. Overview of Synergists Based on Cationic Steroids.
| name | ref | FICI | pathogen | antibiotic | hemolytic activitya |
|---|---|---|---|---|---|
| squalamine | (224, 226) | 0.35b | P. aeruginosa | erythromycin | >10% (10 min) |
| SM-7 | (227) | 0.063 | K. pneumoniae | rifampicin | <10% (24 h) |
| Polycationic Cholic Acid Analogues | |||||
| Ether-linked | |||||
| I | (229, 230) | 0.035 | K. pneumoniae | rifampicin | >10% (24 h) |
| II | (230) | 0.029 | K. pneumoniae | novobiocin | <10% (24 h) |
| III | (230) | 0.022 | K. pneumoniae | novobiocin | >10% (24 h) |
| IV | (232) | 0.13 | K. pneumoniae | rifampicin | <10% (24 h) |
| Ester-Linked | |||||
| V | (233) | 0.057b | E. coli | erythromycin | NR |
| VI | (233) | 0.064b | E. coli | erythromycin | NR |
| VII | (234) | 0.176b | E. coli | erythromycin | <10% (24 h) |
| Steroid–Peptide Hybrids | |||||
| VIII | (239) | 0.099 | E. coli | erythromycin | NR |
| IX | (239) | 0.093 | E. coli | erythromycin | NR |
| X | (239) | 0.078 | E. coli | erythromycin | NR |
Non-hemolytic is defined as <10% hemolysis compared to positive control, with incubation times denoted in parentheses; NR denotes no data reported.
FICI calculated from MIC values reported in the cited literature references.
In another approach, the Savage group also employed the cholic acid backbone but with the aim of mimicking polymyxins through the amphiphilic positioning of positive charges (Figure 5C,D).228,229 In doing so, a variety of cationic steroids were developed and screened for inherent antimicrobial activity as well as the capacity to potentiate antibiotics against Gram-negative bacteria.229−237 The hydroxyl groups on the cholic acid backbone provide convenient functionalities for the incorporation of positively charged moieties via formation of ether (Figure 5C) or ester (Figure 5D) linkages. Among the ether-linked series, an analogue bearing three carbon atom spacers between the steroid and the primary amine groups, along with an N-benzylated tertiary amino group at the C24 position (analogue I, Figure 5C), was found to exhibit both inherent antimicrobial activity and synergistic activity.229 Interestingly, replacement of the lipophilic N-benzyl moiety with a hydroxyl group led to analogue II, which showed a significant reduction of inherent activity while maintaining a strong ability to potentiate the activity of erythromycin against E. coli.228,229 The decreased lipophilicity of analogue II also reduced the hemolytic activity seen with analogue I (Table 4). Follow-up studies revealed that conversion of the free hydroxyl group at the C24 position to the propyl ether, as in analogue III, significantly increased the hemolytic activity.230,231 Notably, addition of a terminal amino group to the propyl ether moiety provided analogue IV, which exhibited significantly reduced hemolysis relative to that of analogue III while maintaining effective synergistic activity (Table 4).232 A series of ester-linked analogues were also prepared by the Savage group (Figure 5D), wherein compounds V, VI, and VII exhibited synergistic activity comparable to that of the corresponding ether variants (Table 4).233,234 Amide analogues were also explored; however, they exhibited a significant lower potentiation of erythromycin, presumably due to conformational constraints, relative to the more active esters.233
In addition to the polycationic steroids described above, steroid–peptide hybrids have also been explored as synergists.237−239 In one case, Bavikar et al. reported a series of hybrids wherein simple tetrapeptides were coupled to cholic acid in an attempt to mimic the squalamine tail (Figure 5E).239 As indicated in Table 4, these steroid–peptide hybrids exhibit potent synergy with erythromycin against E. coli. While the hemolytic activity of these compounds was not reported, they were described as having low cytotoxicity toward HEK293 and MCF-7 cells.239
3. Non-steroid Small-Molecule Synergists
3.1. Synergists Based on Approved Drugs
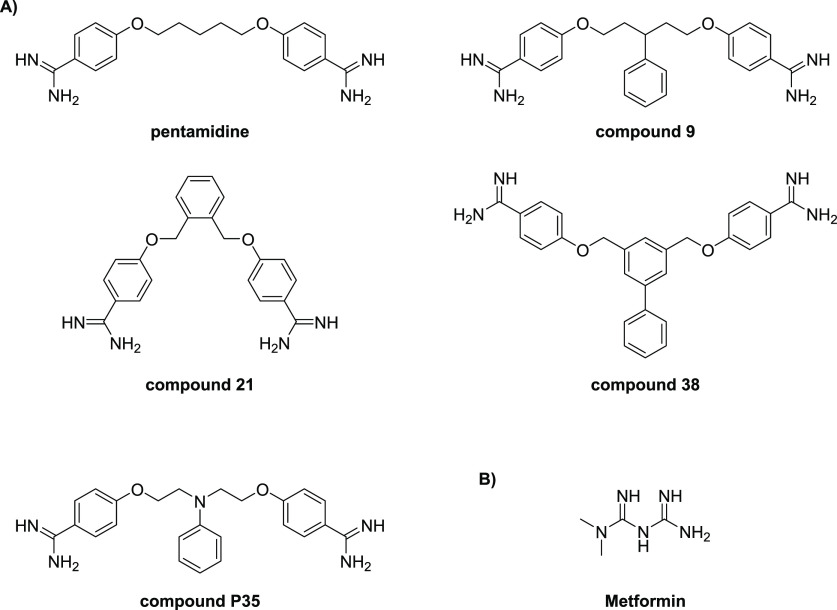
Recently, Brown and co-workers reported an innovative screening platform for the identification of non-lethal, OM-active compounds with potential as adjuvants for conventional antibiotics.240 They applied their screen to a library of 1440 previously approved drugs, which resulted in the identification of three hits. Among the three hits identified, the anti-protozoal agent pentamidine (Figure 6A) was subsequently found to display the highest synergistic potency (Table 5).240 Notably, while pentamidine’s OM-targeting mechanism was found to be driven by interaction with LPS, mcr-resistance did not affect its synergistic potential.240 The potentiation of novobiocin by pentamidine was also established in vivo against wild-type and resistant A. baumannii.240 Subsequently, a focused SAR study using commercially available bis-amidines similar in structure to pentamidine led to the identification of compound 9 as an even more potent synergist (Figure 6a and Table 5).240
Figure 6.
Representative structures of recently reported (A) bis-amidine synergists and (B) metformin.
Table 5. Overview of Non-steroid Small-Molecule Synergists.
| namea | ref | FICI | pathogen | antibiotic | hemolytic activityb |
|---|---|---|---|---|---|
| Synergists Based on Approved Drugs | |||||
| pentamidine | (240, 241) | 0.25 | E. coli | rifampicin | <10% (20 h) |
| compound 9 | (240, 241) | <0.047 | E. coli | rifampicin | >10% (20 h) |
| compound 21 | (241) | ≤0.094 | E. coli | rifampicin | <10% (20 h) |
| compound 38 | (241) | ≤0.039 | E. coli | rifampicin | >10% (20 h) |
| compound P35 | (242) | 0.094 | A. baumannii | novobiocin | <10% (45 min)c |
| metformin | (245) | 0.375 | E. coli | vancomycin | <10% (1 h) |
| High-Throughput Screening Hits | |||||
| MAC-0568743 | (246) | ≤0.16 | E. coli | rifampicin | NR |
| liproxstatin-1 | (246) | 0.25d | E. coli | rifampicin | NR |
| BWC-Aza1 | (247) | 0.258 | E. coli | rifampicin | <10% (45 min) |
| BWC-Aza2 | (247) | 0.06 | A. baumannii | rifampicin | <10% (45 min) |
| Peptidomimetics | |||||
| OAK C12(ω7) | (212) | ≤0.073d | E. coli | rifampicin | >10% (3 h) |
| OAK C12 | (212) | ≤0.211d | E. coli | rifampicin | >10% (3 h) |
| OAK C10 | (212) | ≤0.036d | E. coli | rifampicin | <10% (3 h)c |
| OAK C8 | (212) | ≤0.078d | E. coli | rifampicin | <10% (3 h)c |
| C14(ω5)OOC10O | (213) | 0.20d | K. pneumoniae | rifampicin | <10% (3 h)c |
| dUSTBP 2 | (206) | ≥0.250 | P. aeruginosa | rifampicin | <10% (1 h) |
| dUSTBP 5 | (206) | ≥0.125 | P. aeruginosa | rifampicin | <10% (1 h) |
| dUSTBP 8 | (206) | ≥0.002 | A. baumannii | novobiocin | <10% (1 h) |
| Synergists with a Polyamine Motif | |||||
| d-LANA-14 | (249, 250) | 0.09 | P. aeruginosa | rifampicin | <10% (1 h) |
| naphthylacetylspermine | (251) | 0.125d | E. coli | novobiocin | nr |
| bisacyl-homospermine 8a | (253) | 0.304d | E. coli | rifampicin | <10% (30 min) |
| bisacyl-homospermine 8b | (253) | 0.297d | E. coli | rifampicin | >10% (30 min) |
| spermidine analogue 14 | (258) | 0.255d | E. coli | erythromycin | <10% (1 h)c |
| spermidine analogue 17 | (258) | 0.255d | P. aeruginosa | erythromycin | <10% (1 h)c |
| 600-Da BPEI | (261, 275) | 0.26 | P. aeruginosa | erythromycin | <10% (1 h) |
| Plant-Derived Synergists | |||||
| eugenol | (262, 276) | ≤0.2d | P. aeruginosa | rifampicin | <10% (24 h) |
| linalool | (263, 277) | 0.37 | E. coli | erythromycin | <10% (4 h) |
| thymol | (271, 278) | 0.25 | E. coli | erythromycin | <10% (1 h) |
| cinnamaldehyde | (271, 279) | 0.24 | E. coli | erythromycin | <10% (48 h) |
| trans-cinnamic acid | (272, 280) | 0.36 | E. coli | erythromycin | <50% (1 h) |
| ferulic acid | (272, 280) | 0.48 | E. coli | erythromycin | <50% (1 h) |
| 3,4-dimethoxycinnamic acid | (272, 280) | 0.42 | E. coli | erythromycin | <50% (1 h) |
| 2,4,5-trimethoxycinnamic acid | (272, 280) | 0.22 | E. coli | erythromycin | <50% (1 h) |
Compound names are provided as given in the cited literature references.
Non-hemolytic is defined as <10% hemolysis compared to positive control, with incubation times denoted in parentheses; NR denotes no data reported.
Concentration tested was lower than 100 μg/mL.
FICI calculated from MIC values reported in the cited literature references.
Inspired by these findings, our group recently undertook a broad SAR investigation wherein a number of structurally unique bis-amidines were synthesized and evaluated as synergists.241 Specifically, we focused our attention on the length and rigidity of the linker motif as well as the geometry of the amidine groups on the aromatic rings. In addition to assessing the synergistic activity of the new bis-amidines prepared, we also performed hemolysis assays with each compound to ascertain OM selectivity. Given the potent synergy previously reported for bis-amidine 9,240 we also synthesized it to use as a benchmark. Among the compounds prepared in our study, bis-amidine 21, containing an ortho-substituted benzene linker, was found to be significantly more synergistic than pentamidine and displayed no hemolytic activity (Figure 6A and Table 5).241 We also found that the introduction of additional aromatic groups to the linker, such as in compound 38, led to further enhancement of synergy; however, this came at the cost of increased hemolytic activity (Table 5). Interestingly, our studies also revealed benchmark bis-amidine 9 to be hemolytic. These findings further highlight the importance of assessing OM selectivity when pursuing synergists.241
The Brown group also recently reported a follow-up SAR study aimed at further enhancing the therapeutic potential of bis-amidine synergists.242 Similar to our own SAR study, the rigidity, conformational flexibility, and lipophilicity were further explored. In addition, the roles of chirality and charge were also investigated.242 A key focus of this study was to identify bis-amidine synergists with improved off-target effects relative to those of pentamidine, especially the QT prolongation resulting from its effect on the hERG ion channel.242−244 This led to compound P35, which was shown to have the same synergistic mode of action as pentamidine; it displayed a strong potentiation of novobiocin and no hemolytic activity (Table 5). Furthermore, compound P35 outperformed pentamidine on multiple levels: an improvement in cytoxicity, a higher efficacy in a mouse infection model, and reduced hERG inhibition.242
Wang and co-workers also recently reported a study wherein the Prestwick Chemical Library, comprising 158 FDA-approved drugs, was assessed for compounds exhibiting synergy with doxycycline.245 This led to the finding that metformin, a commonly prescribed anti-diabetic agent (Figure 6B), effectively potentiates vancomycin as well as tetracycline antibiotics, particularly doxycycline and minocycline, against MDR S. aureus, Enterococcus faecalis, E. coli, and Salmonella enteritidis.245 The capacity for metformin to disturb the OM was assessed using the NPN assay, revealing an increase in E. coli OM permeability in a dose-dependent manner. Of particular note was the finding that metformin was also able to fully restore the activity of doxycycline in animal infection models.245
3.2. Small-Molecule Synergists via High-Throughput Screening
Following the success in applying their OM perturbation reporter assay to identify pentamidine as a potent synergist, the Brown group applied the same approach in a much larger high-throughput screening (HTS) campaign with a library of ca. 140 000 synthetic compounds.240,246 This, in turn, led to the identification of 39 hits that were subsequently screened for synergistic activity with rifampicin.246 Among these hits, MAC-0568743 and liproxstatin-1 (Figure 7A) were found to be particularly active synergists (Table 5).246 Both compounds were found to potentiate the activity of the Gram-positive-targeting antibiotics rifampicin, novobiocin, erythromycin, and linezolid. This potentiation was further shown to be due to selective disruption of the OM, driven by interactions with LPS, and neither compound impacted the inner membrane.246
Figure 7.
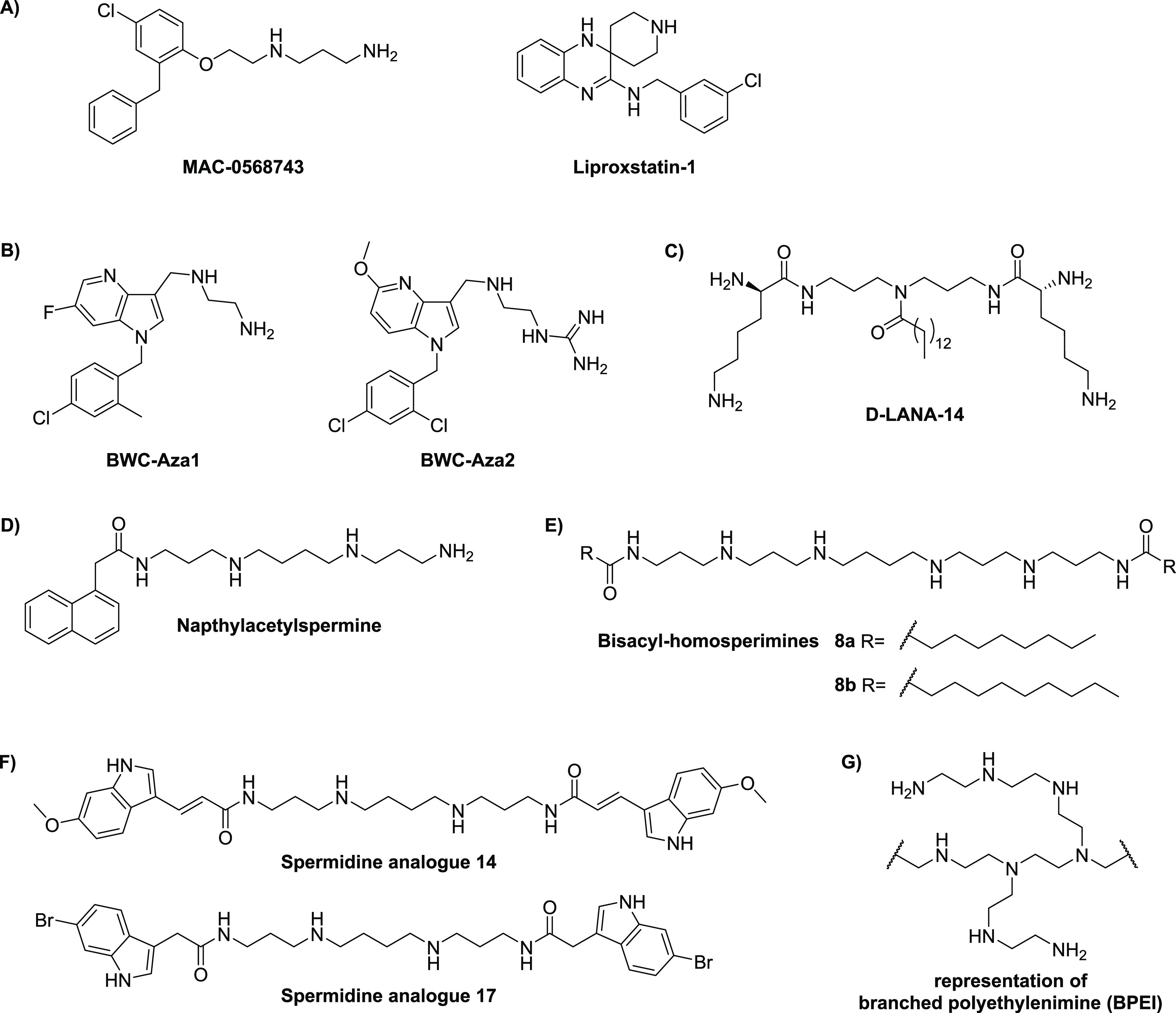
Non-steroid small-molecule synergists: (A) synergists identified via HTS, (B) azaindole synergists, (C) d-LANA-14 based on a norspermidine core linked to two d-lysine residues and a central tetradecanoyl moiety, (D) joro spider toxin-inspired naphthylacetylspermine, (E) bisacyl-homospermines, (F) indole-3-acrylamidospermine conjugates, and (G) representation of 600 Da branched polyethylenimine (BPEI).
In another recently reported campaign, Datta and co-workers screened a focused library of 3000 drug-like compounds for antibiotic synergy using a whole-cell-based phenotypic assay.247 This led to the identification of a series of azaindoles that potentiate the MICs of novobiocin and rifampicin by 100–1000-fold vs Gram-negative bacteria. Optimization studies led to compounds BWC-Aza1 and BWC-Aza2 (see Figure 7B), both of which were screened for synergistic activity with an extensive panel of antibiotics against E. coli (Table 5). The OM-permeabilizing activity of the azaindoles was also probed using the NPN assay, revealing dose-dependent disruption.247
3.3. Small-Molecule Polyamine Synergists
In recent years, the polyamines norspermine and norspermidine have been explored as starting points for the development of antibacterial and antibiofilm agents.248,249 Building on this work, the Haldar group recently reported the development of d-LANA-14, composed of a norspermidine core linked to two d-lysines, along with conjugation to a tetradecanoyl chain at the central secondary amine (Figure 7C).250d-LANA-14 showed potent synergy with tetracycline or rifampicin against meropenem-resistant A. baumannii and P. aeruginosa clinical isolates (Table 5) and, importantly, was also found to disrupt established biofilms formed by these pathogens.250d-LANA-14 was shown to perturb the OM by means of the NPN assay and, importantly, was also found to exhibit potent in vivo activity when combined with rifampicin, resulting in a significant reduction of bacterial burden in a mouse model of burn-wound infection.250
In another study involving small-molecule polyamines, Katsu and co-workers investigated synthetic analogues of the joro spider toxin as OM-disrupting agents, leading to the identification of naphthylacetylspermine (Figure 7D), which was found to potentiate the activity of novobiocin against E. coli (Table 5).251 Mechanistic studies revealed that administration of naphthylacetylspermine causes OM disruption, which was attributed to displacement of LPS-associated Ca2+. In addition, naphthylacetylspermine was found to promote cellular uptake of the tetraphenylphosphonium (TPP+), indicating membrane permeabilization, a finding similar to that obtained with PMBN.251,252 Interestingly, spermidine and spermine were also found to induce loss of Ca2+ but did not cause uptake of TPP+, pointing to the importance of the naphthyl moiety for membrane permeabilization.252 Given that no hemolysis data was reported for naphthylacetylspermine, it is not possible to assess the selectively of its OM activity.
The David group also reported the development of acylated polyamines as LPS neutralizing agents capable of functioning as OM-disrupting synergists.253−255 A series of monoacyl- and bisacyl-homospermines were prepared and evaluated as potentiators of rifampicin, resulting in the identification of two potent synergists, compounds 8a and 8b (see Figure 7E and Table 5).253 A clear correlation between the length of the lipophilic tail and hemolytic activity was seen, with compound 8a appearing to strike an optimal balance.253 Using a similar approach, Copp and co-workers introduced the indole-3-acrylamido-spermine conjugates inspired by a class of indole-spermidine alkaloid natural products.256,257 An SAR study led to the development of spermidine analogues like 14 and 17, which exhibited effective synergy with various antibiotics (Figure 7F and Table 5).256,258 These compounds affect bacterial membrane integrity and show low cytotoxicity and hemolytic activity. Interestingly, compound 14 was also found to inhibit bacterial efflux pumps, suggesting that the potentiation of antibiotics by these compounds may be attributed to a dual mechanism of action.256,258
Given the inclusion criteria noted in the introduction, only small-molecule synergists (MW under 1500 kDa) are included in this Review, and as such we do not discuss larger polycationic polymers even though some have been shown to exhibit synergistic activity.90−96,259,260 It is noteworthy, however, that branched polyethylenimine (BPEI) with a MW of 600 Da shows synergistic activity (Figure 7G, Table 5) and can also eradicate biofilms when co-administered with a variety of antibiotics.261 Mechanistic studies using isothermal titration calorimetry and fluorescence spectroscopy indicate that, at the concentration required for antibiotic potentiation, 600 Da BPEI reduces diffusion barriers from LPS without disrupting the OM itself.261
3.4. Plant-Derived Synergists
A number of plant-derived compounds have also been reported to potentiate the activity of antibiotics against Gram-negative bacteria (Table 5). These include natural products like eugenol, a major component of clove oil; linalool, which can be isolated from coriander; thymol, which is extracted from thyme; and cinnamaldehyde and cinnamic acid, which are found in the bark and leaves of the cinnamon tree (Figure 8).262−268 Important to note is that only pure compounds derived from plants are included in our assessment. We refer the reader to other reviews on the synergistic activity of essential oils or crude extracts.269,270 Notably, most plant-derived compounds reported to potentiate antibiotics against Gram-negative bacteria are not cationic, setting them apart from most other synergists. Despite their lack of positive charge, a number of investigations have shown that the synergy associated with these compounds is a function of their ability to induce OM permeabilization (Table 6).262,263,271−273 The broad range of biological activities associated with cinnamic acid and its derivatives, including ferulic acid, 3,4-dimethoxycinnamic acid, and 2,4,5-trimethoxycinnamic acid (Figure 8), has been recently reviewed including synergistic effects associated with OM disruption.274 Interestingly, despite its clear structural similarities with cinnamic acid, studies with cinnamaldehyde suggest that it may operate via a different synergistic mechanism. Unlike cinnamic acid, cinnamaldehyde does not increase OM permeabilization based on the NPN assay, but it does exhibit synergistic effects with erythromycin and novobiocin (Table 5).271,273
Figure 8.
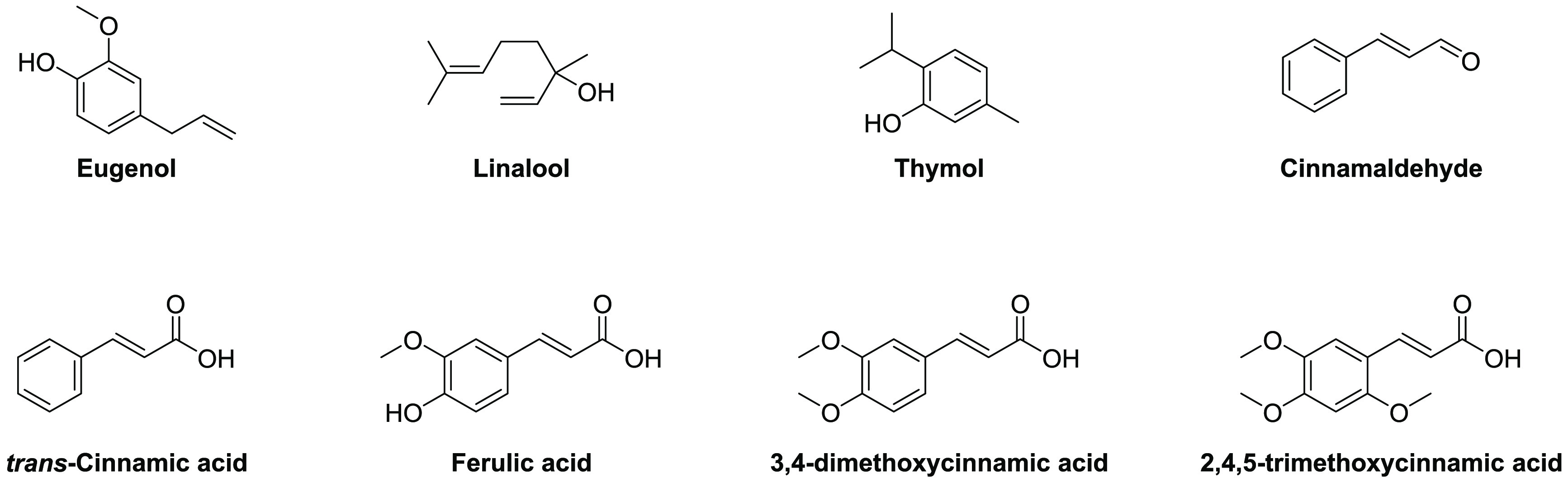
Plant-derived natural products reported to potentiate the activity of antibiotics against Gram-negative bacteria.
Table 6. Overview of Synergists Based on Clinically Used Antibiotics.
| namea | ref | FICI | pathogen | antibiotic | hemolytic activityb |
|---|---|---|---|---|---|
| Tobramycin Derivatives | |||||
| TOB-MOX 1 | (291) | 0.125 | P. aeruginosa | novobiocin | <10% (30 min) |
| tobramycin-ciprofloxacin 1e | (292) | <0.04 | P. aeruginosa | rifampicin | <10% (30 min) |
| tobramycin-rifampicin 1 | (293) | 0.28 | P. aeruginosa | rifampicin | <10% (1 h) |
| tobramycin-rifampicin 2 | (293) | 0.15 | P. aeruginosa | erythromycin | <10% (1 h) |
| tobramycin-rifampicin 3 | (293) | 0.06 | P. aeruginosa | erythromycin | <10% (1 h) |
| tobramycin-lysine 3 | (294) | 0.008 | P. aeruginosa | novobiocin | <10% (1 h) |
| TOB-NMP 1 | (296) | ≥0.008 | P. aeruginosa | rifampicin | <10% (30 min) |
| TOB-PAR 2 | (296) | ≥0.008 | P. aeruginosa | rifampicin | <10% (30 min) |
| tobramycin homodimer 1 | (297) | 0.07 | P. aeruginosa | novobiocin | <10% (1 h) |
| tobramycin homodimer 2 | (297) | 0.08 | P. aeruginosa | novobiocin | <10% (1 h) |
| tobramycin homodimer 3 | (297) | 0.05 | P. aeruginosa | novobiocin | <10% (1 h) |
| tobramycin-cyclam 1 | (298) | 0.13 | P. aeruginosa | novobiocin | <10% (30 min) |
| tobramycin-cyclam 2 | (298) | 0.13 | P. aeruginosa | novobiocin | <10% (30 min) |
| tobramycin-cyclam 3 | (298) | 0.08 | P. aeruginosa | novobiocin | <10% (30 min) |
| Nebramine Derivatives | |||||
| NEB-MOX 1a | (299) | ≥0.002 | K. pneumoniae | rifampicin | NR |
| NEB-CIP 1b | (299) | ≥0.008 | P. aeruginosa | rifampicin | <10% (1 h) |
| NEB-NMP 2 | (299) | ≥0.004 | P. aeruginosa | rifampicin | NR |
| nebramine-cyclam | (300) | 0.25 | P. aeruginosa | rifampicin | <10% (1 h) |
| Levofloxacin–Polybasic Peptide Conjugates | |||||
| levofloxacin conjugate 10 | (301) | 0.10 | P. aeruginosa | rifampicin | <10% (1 h) |
| levofloxacin conjugate 11 | (301) | 0.10 | P. aeruginosa | novobiocin | <10% (1 h) |
| levofloxacin conjugate 12 | (301) | 0.08 | P. aeruginosa | novobiocin | <10% (1 h) |
Compound names are provided as given in the cited literature references.
Non-hemolytic is defined as <10% hemolysis compared to positive control, with incubation times denoted in parentheses; NR denotes no data reported.
4. Antibiotic-Derived Synergists
In general, the antibiotic potentiators discussed above show little to no inherent antibacterial activity. There are, however, a number of reports describing antibacterial compounds that also exhibit OM-disrupting effects and, in doing so, synergize with antibiotics that are otherwise inactive toward Gram-negative bacteria. The synergists described in this section are specifically included on the basis of their OM-disrupting activity rather than a contribution of their inherent activity to synergy. We therefore do not include the combination of rifampicin with imipenem or trimethoprim, which is solely based on functional synergy.281,282 In addition, we also do not cover reports describing systems where an OM-perturbing motif like PMBN is covalently linked to another antibiotic as a means of enhancing anti-Gram-negative activity.39,283−285
4.1. Tobramycin-Derived Synergists
Tobramycin (Figure 9A) belongs to the aminoglycoside class of antibiotics that function by inhibiting ribosomal protein synthesis in bacteria. Recent studies have also revealed that aminoglycosides like tobramycin also interact with bacterial membranes by specifically binding to LPS and, in doing so, cause membrane depolarization.286−290 Building on these insights, Schweizer and co-workers prepared and assessed a number of conjugates wherein one tobramycin molecule is linked to a second antibiotic, providing hybrid systems that possess both inherent antibacterial activity and potent synergy with other antibiotics (Figure 9A).291−294,283,295−301 Among the first hybrids prepared was a series tobramycin–fluoroquinolone conjugates.291,292 Both the optimal sites of conjugation and linker lengths between the two antibiotics were investigated, revealing TOB-MOX, a tobramycin–moxifloxacin hybrid, and tobramycin–ciprofloxacin conjugate 1e to be potent synergists (Table 6).292 OM disruption was confirmed for both hybrids using the NPN assay, and both were found to potentiate multiple antibiotics, including rifampicin, erythromycin, novobiocin, and vancomycin.291,292 Also of note was the finding that these hybrids exhibited a significantly reduced capacity to inhibit protein translation compared to that of tobramycin.291,292 Conversely, the hybrids were found to maintain, and in some cases exceed, the gyrase-inhibiting activity of the parent fluoroquinolones.291,292 Another series of hybrids was prepared by coupling tobramycin with rifampicin, which targets the bacterial RNA polymerase.293 As for the fluoroquinolone conjugates, the inherent activity of the tobramycin–rifampicin conjugates was significantly reduced compared to that of the parent antibiotics. Again, however, some hybrids were found to exhibit synergy via an OM-disrupting mechanism (see tobramycin–rifampicins 1–3, Figure 9A).292−294,302
Figure 9.
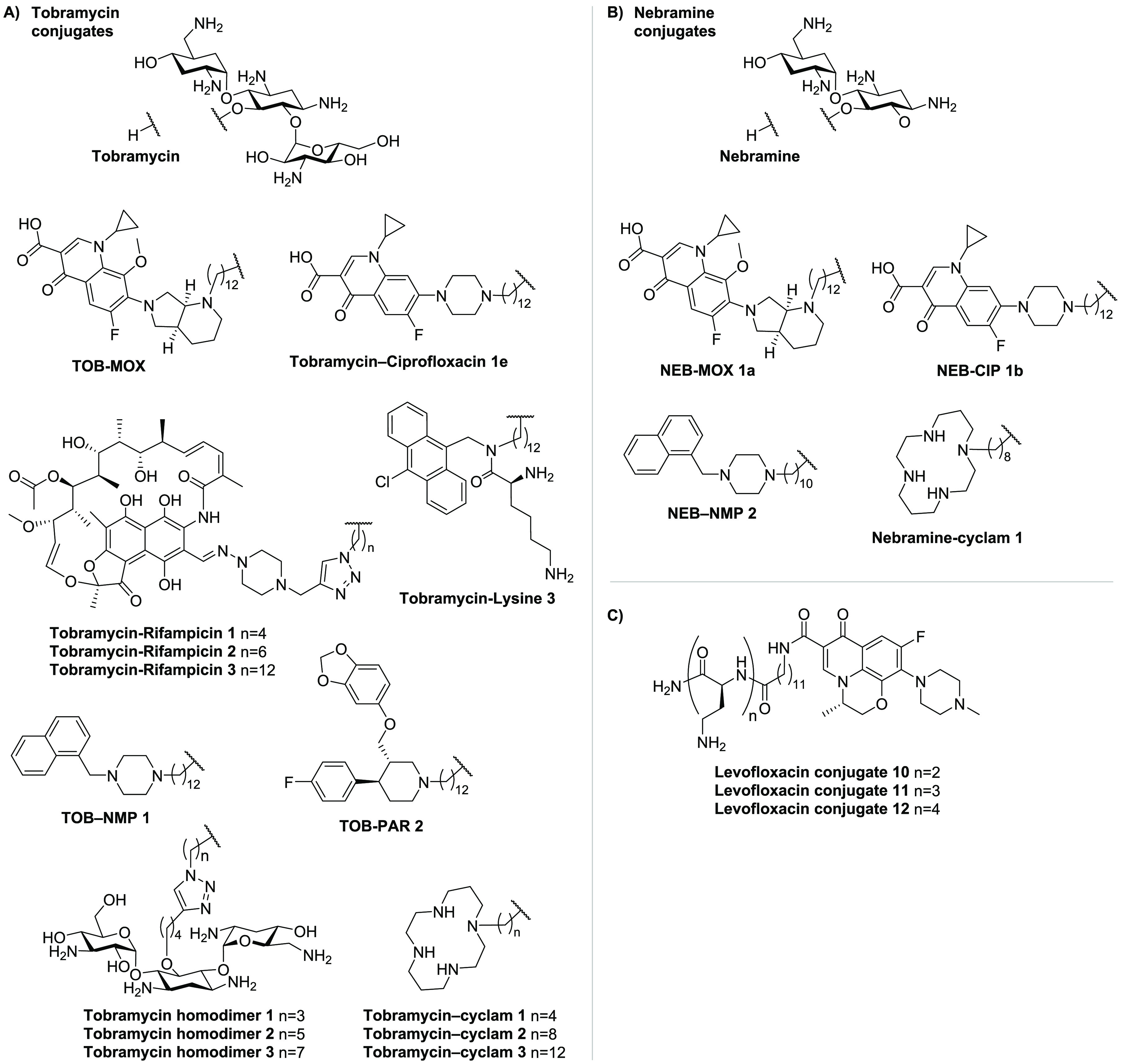
Synergists based on clinically used antibiotics: (A) tobramycin (TOB) conjugates, (B) nebramine (NEB) analogues, and (C) polybasic conjugated levofloxacin hybrids.
A number of other hybrids have also been reported by the Schweizer group wherein tobramycin was coupled to various other small molecules known to engage with different bacterial targets. In one case, tobramycin was coupled to a lysine-based amphiphile known to function as a membrane permeabilizer (see tobramycin–lysine 3, Figure 9A).294,303 This conjugate was found to effectively potentiate the activity of novobiocin, erythromycin, and vancomycin (Table 6).294,304 The same group also explored hybrids wherein tobramycin was coupled to small-molecule efflux pump inhibitors such as 1-(1-naphthylmethyl)piperazine (NMP) and paroxetine (PAR) (Figure 9A).45,295,305−307 Along with potent synergy against P. aeruginosa (Table 6), these hybrids were also found to cause OM disruption and inner membrane depolarization.295,296 Two additional generations of tobramycin conjugates were also reported: tobramycin homodimers and tobramycin coupled to chelating cyclams (Figure 9A).297,298 The dimerization of tobramycin was conveniently achieved by means of copper-catalyzed azide–alkyne click chemistry, resulting in potent synergists that also exhibit enhanced OM disruption relative to tobramycin itself (Table 6).297 A combination of novobiocin and tobramycin homodimer 1 (both administered at 50 μg/mL) was further shown to have in vivo efficacy against A. baumannii in a wax worm larvae model.297 Studies with the corresponding monomeric tobramycin azide and alkyne precursors revealed neither to be synergistic, underscoring the need for dimerization to achieve synergy.297 In the case of the tobramycin–cyclam conjugates, the introduction of the cyclam chelating group was hypothesized to aid in the OM permeabilization by sequestration of divalent cations bridging the Lipid A phosphate groups.298,308−310 While tobramycin–cyclam hybrids 1–3 effectively potentiated novobiocin, rifampicin, vancomycin, and erythromycin (Table 6), it is also particularly noteworthy that they also enhanced the activity of meropenem against both carbapenem-resistant and -sensitive strains.298 This effect was abrogated by the addition of excess MgCl2, further supporting a mode of action driven by OM disruption.298
4.2. Nebramine-Derived Synergists
Following on their work with tobramycin hybrids, the Schweizer group also prepared a number of analogous nebramine conjugates (Figure 9B). Nebramine (NEB) is a disaccharide sub-unit of tobramycin that interestingly displays activity against tobramycin-resistant strains and also interacts with the OM.287,311−317 The NEB hybrids synthesized included conjugates with moxifloxacin (MOX), ciprofloxacin (CIP), NMP, and cyclam (Figure 9B).299,300 These hybrids were all found to effectively potentiate the activity of multiple classes of antibiotics against a range of Gram-negative bacteria (Table 6). Furthermore, NEB-MOX 1a, NEB-CIP 1b, and NEB-NMP 2 were also reported to dissipate proton motive force and proposed to cause OM disruption, as for the corresponding tobramycin conjugates.291,294,295,299,300
4.3. Levofloxacin–Polybasic Peptide Conjugates as Synergists
Schweizer and co-workers also recently reported another class of antibiotic-based synergists, consisting of levofloxacin conjugated to polybasic peptides of varying lengths (Figure 9C).301 While these levofloxacin–peptide hybrids were found to be non-hemolytic, they were also shown to be essentially devoid of inherent antimicrobial activity (MICs typically >128 μg/mL). They did, however, exhibit strong potentiation of numerous antibiotics against MDR clinical isolates of P. aeruginosa, E. coli, K. pneumoniae, and, to a lesser extent, A. baumannii (Table 6).301 Preliminary mechanistic studies indicate that these conjugates potentiate other antibiotics both by blocking active efflux and by permeabilization of the OM.301
5. Chelating Agents as OM-Disrupting Synergists
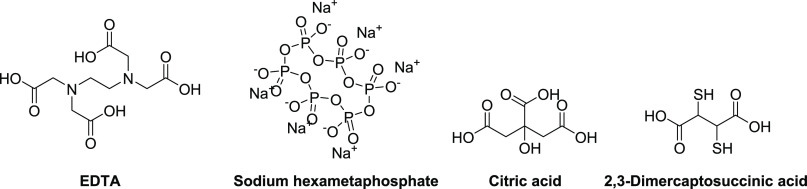
The activity of antibiotics can also be potentiated by chelating agents that disturb the integrity of the OM by sequestering the divalent cations Mg2+ or Ca2+ coordinated by the phosphate groups of the lipid A core of LPS (Figure 1B).32 The pre-eminent chelating agent, EDTA (Figure 10), is a well-described synergist, and its reported ability to potentiate antibiotics actually pre-dates the reported synergistic activity of PMBN.49,318−321 Exposure of Gram-negative bacteria to EDTA is accompanied by the significant release of LPS and, as for treatment with PMBN, also results in the increased uptake of NPN.322−324 While the potentiating effects of EDTA on antibiotics such as novobiocin and rifampicin are well documented, FICI values have not been reported in literature and cannot be readily calculated from published data.320,321,323,325 Similarly, for the other chelating agents here discussed, no FICI values could be found in the literature, and, as such, we do not provide a summary table as was done for the other synergists discussed in this Review.
Figure 10.
Chelating agents with demonstrated synergistic activity.
In additional to his seminal work with PMBN, Vaara also reported the potentiation of hydrophobic antibiotics by sodium hexametaphosphate (HMP, Figure 10) against Gram-negative bacteria as well as the increase in NPN uptake in cells treated with this potent Ca2+-binding agent.326 In a similar study, Ayres and Russell also described sodium polyphosphates as potent synergists with several antibiotics (structures not shown).327 In the same study, citric acid (Figure 10) was also demonstrated to exhibit synergistic activity with erythromycin, novobiocin, rifampicin, methicillin, and gentamicin.327 In addition, 2,3-dimercaptosuccinic acid (Figure 10), clinically used in the treatment of lead intoxication, was also found to potentiate the activity of hydrophobic antibiotics.323 The synergistic activity of 2,3-dimercaptosuccinic acid was attributed to an OM-permeabilizing mechanism, as evidenced by increased NPN uptake in bacterial cells treated with the compound.323
Concluding Remarks
New strategies are required to address the growing threat posed by MDR Gram-negative pathogens. To this end, a large and growing number of synergists capable of potentiating Gram-positive-specific antibiotics against Gram-negative bacteria have been described in the literature to date. Within this Review, we provide the reader with a comprehensive and up-to-date overview of those synergists reported to have a demonstrated OM-targeting mechanism. We also draw attention to the importance of selective OM disruption, a factor that has often been overlooked by researchers when characterizing their synergists. In this regard, and based on our assessment of the literature, the majority of hemolysis studies reported for such synergists use relatively short incubation times compared to the incubation times actually used in assessing synergy (i.e., in checkerboard assays). Based on our own experience, not only is the concentration at which hemolysis is assessed relevant, but incubation time can also make a significant difference in describing a compound as hemolytic or not. For example, in cases where 5% hemolysis is reported after 1 h, it is our experience that such compounds are often much more hemolytic after overnight incubation. For this reason, we have included both the concentrations and incubation times of the synergists described in this Review. Doing so provides for a more honest and accurate assessment of the OM specificity of these synergists.
To provide a means of comparing the relative activity of the synergists here summarized, we have emphasized their FICI values, a descriptor broadly applied as a scale to quantify synergistic potency. However, another important consideration that is not directly revealed by the FICI is, of course, the concentration at which a synergist actually potentiates the companion antibiotic. As for the concentrations of the antibiotics being potentiated, we suggest using the corresponding Gram-positive breakpoints as a guide for assessing whether the synergistic MICs determined against Gram-negative bacteria (for which Gram-positive antibiotics have no breakpoint) are within therapeutically relevant concentrations. Also related to this is the importance of the pharmacokinetic/pharmacodynamic (PK/PD) profile of the synergist and how well it matches that of the antibiotic it potentiates. Given that the vast majority of antibiotic synergists reported to date have only been characterized using cell-based in vitro and biochemical assays, we have not touched on this. Clearly, significant in vivo studies are needed to establish and optimize such parameters and will be essential to the (pre)clinical development of any such synergist. It is also notable that OM-perturbing synergists have been investigated as a means of enhancing the effect of other multi-drug cocktails, further underscoring the importance of such PK/PD considerations. Specifically, addition of the polymyxin-derived SPR741 has recently been studied as a means of enhancing the activity of β-lactam/β-lactamase inhibitor combinations such as piperacillin–tazobactam.328 Given the challenges associated with developing anti-Gram-negative agents and therapeutic strategies, the pursuit of antibiotic synergists is likely to remain an active field of research for the years to come.
Acknowledgments
Financial support provided by the European Research Council (ERC consolidator grant to N.I.M., grant agreement no. 725523).
Glossary
Abbreviations
- ΔF
α,β-didehydrophenylalanine
- alle
d-allo-isoleucine
- AMP
antimicrobial peptide
- BAM
β-barrel assembly machine
- Bip
biphenylalanine
- BPEI
branched polyethylenimine
- BPI
bactericidal/permeability-increasing protein
- C6
hexanoyl
- C8
octanoyl
- C10
decanoyl
- Cbz
benzyloxycarbonyl
- Cha
cyclohexylalanyl
- Dab
2,4-diaminobutyric acid
- DAPB
deacylpolymyxin B
- dUSCL
dilipid ultrashort cationic lipopeptide
- dUSTBP
dilipid ultrashort tetrabasic peptidomimetic
- EDTA
ethylenediaminetetraacetate
- ESBL
extended spectrum β-lactamase
- FICI
fractional inhibitory concentration index
- HMP
hexametaphosphate
- HTS
high-throughput screening
- LPS
lipopolysaccharide
- MAC
membrane attack complex
- MDR
multi-drug-resistant
- MIC
minimum inhibitory concentration
- MOX
moxifloxacin
- Nal
β-naphthylalanine
- NEB
nebramine
- NMP
1-(1-naphthylmethyl)piperazine
- NPN
N-phenylnaphthalen-1-amine
- NR
no data reported
- OAK
oligo-acyl-lysyl
- OM
outer membrane
- PAR
paroxetine
- PEDES
PEptide DEscriptors from Sequence
- PG
phosphatidylglycerol
- PK/PD
pharmacokinetic/pharmacodynamic
- PMB
polymyxin B
- PMBH
polymyxin B heptapeptide
- PMBN
polymyxin B nonapeptide
- PMBO
polymyxin B octapeptide
- SAR
structure–activity relationship
- SM
squalamine mimic
- TOB
tobramycin
- TPP
tetraphenylphosphonium
- TriA1
tridecaptin A1
- UTBLP
ultrashort tetrabasic lipopeptide
- WHO
World Health Organization
The authors declare no competing financial interest.
References
- Delcour A. H. Outer Membrane Permeability and Antibiotic Resistance. Biochim. Biophys. Acta BBA - Proteins Proteomics 2009, 1794 (5), 808–816. 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67 (4), 593–656. 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. The Role of Outer Membrane and Efflux Pumps in the Resistance of Gram-Negative Bacteria. Can We Improve Drug Access?. Drug Resist. Updat. 1998, 1 (2), 93–98. 10.1016/S1368-7646(98)80023-X. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J.; Kahne D.; Walker S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2 (5), a000414. 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Moran L.; Aronsson B.; Manz C.; Gyssens I. C.; So A. D.; Monnet D. L.; Cars O. Critical Shortage of New Antibiotics in Development against Multidrug-Resistant Bacteria—Time to React Is Now. Drug Resist. Updat. 2011, 14 (2), 118–124. 10.1016/j.drup.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Nicolau D. P. Carbapenems: A Potent Class of Antibiotics. Expert Opin. Pharmacother. 2008, 9 (1), 23–37. 10.1517/14656566.9.1.23. [DOI] [PubMed] [Google Scholar]
- Vaara M. Polymyxin Derivatives That Sensitize Gram-Negative Bacteria to Other Antibiotics. Molecules 2019, 24 (2), 249. 10.3390/molecules24020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos C.; Pozo J. L. D. Emerging Agents to Combat Complicated and Resistant Infections: Focus on Ceftobiprole. Infect. Drug Resist. 2010, 3, 5–14. 10.2147/IDR.S3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccobene T. A.; Su S. F.; Rank D. Single- and Multiple-Dose Study To Determine the Safety, Tolerability, and Pharmacokinetics of Ceftaroline Fosamil in Combination with Avibactam in Healthy Subjects. Antimicrob. Agents Chemother. 2013, 57 (3), 1496–1504. 10.1128/AAC.02134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisgen J.; Whitney D. Ceftobiprole, a Broad-Spectrum Cephalosporin With Activity against Methicillin-Resistant Staphylococcus Aureus (MRSA). Pharm. Ther. 2008, 33 (11), 631–641. [PMC free article] [PubMed] [Google Scholar]
- Chaudhary U.; Aggarwal R. Extended Spectrum β-Lactamases (ESBL) – an Emerging Threat to Clinical Therapeutics. Indian J. Med. Microbiol. 2004, 22 (2), 75–80. 10.1016/S0255-0857(21)02884-X. [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq M.-P.; Glupczynski Y.; Tulkens P. M. Aminoglycosides: Activity and Resistance. Antimicrob. Agents Chemother. 1999, 43 (4), 727–737. 10.1128/AAC.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. J.; Rather P. N.; Hare R. S.; Miller G. H. Molecular Genetics of Aminoglycoside Resistance Genes and Familial Relationships of the Aminoglycoside-Modifying Enzymes. Microbiol. Rev. 1993, 57 (1), 138–163. 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M. S.; Tolmasky M. E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat. 2010, 13 (6), 151–171. 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Aminoglycoside Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2005, 49 (2), 479–487. 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-Y.; Wang Y.; Walsh T. R.; Yi L.-X.; Zhang R.; Spencer J.; Doi Y.; Tian G.; Dong B.; Huang X.; Yu L.-F.; Gu D.; Ren H.; Chen X.; Lv L.; He D.; Zhou H.; Liang Z.; Liu J.-H.; Shen J. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16 (2), 161–168. 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- Partridge S. R.; Di Pilato V.; Doi Y.; Feldgarden M.; Haft D. H.; Klimke W.; Kumar-Singh S.; Liu J.-H.; Malhotra-Kumar S.; Prasad A.; Rossolini G. M.; Schwarz S.; Shen J.; Walsh T.; Wang Y.; Xavier B. B. Proposal for Assignment of Allele Numbers for Mobile Colistin Resistance (Mcr) Genes. J. Antimicrob. Chemother. 2018, 73 (10), 2625–2630. 10.1093/jac/dky262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier B. B.; Lammens C.; Ruhal R.; Kumar-Singh S.; Butaye P.; Goossens H.; Malhotra-Kumar S. Identification of a Novel Plasmid-Mediated Colistin-Resistance Gene, Mcr-2, in Escherichia Coli, Belgium, June 2016. Eurosurveillance 2016, 21 (27), 30280. 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- Yin W.; Li H.; Shen Y.; Liu Z.; Wang S.; Shen Z.; Zhang R.; Walsh T. R.; Shen J.; Wang Y. Novel Plasmid-Mediated Colistin Resistance Gene Mcr-3 in Escherichia Coli. mBio 2017, 8 (3), e00543–17. 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A.; Villa L.; Feudi C.; Curcio L.; Orsini S.; Luppi A.; Pezzotti G.; Magistrali C. F. Novel Plasmid-Mediated Colistin Resistance Mcr-4 Gene in Salmonella and Escherichia Coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22 (31), 30589. 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M.; Fischer J.; Hammerl J. A.; Hendriksen R. S.; Szabo I.; Malorny B. Identification of a Novel Transposon-Associated Phosphoethanolamine Transferase Gene, Mcr-5, Conferring Colistin Resistance in d-Tartrate Fermenting Salmonella Enterica Subsp. Enterica Serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72 (12), 3317–3324. 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- AbuOun M.; Stubberfield E. J.; Duggett N. A.; Kirchner M.; Dormer L.; Nunez-Garcia J.; Randall L. P.; Lemma F.; Crook D. W.; Teale C.; Smith R. P.; Anjum M. F. Mcr-1 and Mcr-2 (Mcr-6.1) Variant Genes Identified in Moraxella Species Isolated from Pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2017, 72 (10), 2745–2749. 10.1093/jac/dkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-Q.; Li Y.-X.; Lei C.-W.; Zhang A.-Y.; Wang H.-N. Novel Plasmid-Mediated Colistin Resistance Gene Mcr-7.1 in Klebsiella Pneumoniae. J. Antimicrob. Chemother. 2018, 73 (7), 1791–1795. 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- Wang X.; Wang Y.; Zhou Y.; Li J.; Yin W.; Wang S.; Zhang S.; Shen J.; Shen Z.; Wang Y. Emergence of a Novel Mobile Colistin Resistance Gene, Mcr-8, in NDM-Producing Klebsiella Pneumoniae. Emerg. Microbes Infect. 2018, 7 (1), 1–9. 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. M.; Gaballa A.; Guldimann C.; Sullivan G.; Henderson L. O.; Wiedmann M. Identification of Novel Mobilized Colistin Resistance Gene Mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella Enterica Serotype Typhimurium Isolate. mBio 2019, 10 (3), e00853-19. 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Feng Y.; Liu L.; Wei L.; Kang M.; Zong Z. Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerg. Microbes Infect. 2020, 9 (1), 508–516. 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein N. H.; AL-Kadmy I. M. S.; Taha B. M.; Hussein J. D. Mobilized Colistin Resistance (Mcr) Genes from 1 to 10: A Comprehensive Review. Mol. Biol. Rep. 2021, 48 (3), 2897–2907. 10.1007/s11033-021-06307-y. [DOI] [PubMed] [Google Scholar]
- Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis, WHO/EMP/IAU/2017.12; World Health Organization, 2017. https://www.who.int/publications/i/item/WHO-EMP-IAU-2017.12
- Ghai I.; Ghai S. Understanding Antibiotic Resistance via Outer Membrane Permeability. Infect. Drug Resist. 2018, 11, 523–530. 10.2147/IDR.S156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamio Y.; Nikaido H. Outer Membrane of Salmonella Typhimurium: Accessibility of Phospholipid Head Groups to Phospholipase C and Cyanogen Bromide Activated Dextran in the External Medium. Biochemistry 1976, 15 (12), 2561–2570. 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- Kadner R. J.Cytoplasmic: Membrane. Cytoplasmic membrane. In Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed.; Neidhardt F. C., Curtiss R. III, Ingraham J. L.; Lin E. C. C.; Low K. B.; Magasanik B.; Reznikoff W. S.; Riley M.; Schaechter M.; Umbarger H. E., Eds.; American Society for Microbiology Press: Washington, DC, 1996; Vol. I, pp 58–87. [Google Scholar]
- Vaara M. Agents That Increase the Permeability of the Outer Membrane. Microbiol. Rev. 1992, 56 (3), 395–411. 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Efflux-Mediated Antimicrobial Resistance. Efflux-Mediat. Antimicrob. Resist. 2005, 56 (1), 20–51. 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- Robinson J. A. Folded Synthetic Peptides and Other Molecules Targeting Outer Membrane Protein Complexes in Gram-Negative Bacteria. Front. Chem. 2019, 7, 45. 10.3389/fchem.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högenauer G.; Woisetschläger M. A Diazaborine Derivative Inhibits Lipopolysaccharide Biosynthesis. Nature 1981, 293 (5834), 662–664. 10.1038/293662a0. [DOI] [PubMed] [Google Scholar]
- Onishi H. R.; Pelak B. A.; Gerckens L. S.; Silver L. L.; Kahan F. M.; Chen M.-H.; Patchett A. A.; Galloway S. M.; Hyland S. A.; Anderson M. S.; Raetz C. R. H. Antibacterial Agents That Inhibit Lipid a Biosynthesis. Science 1996, 274 (5289), 980–982. 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- Hammond S. M.; Claesson A.; Jansson A. M.; Larsson L.-G.; Pring B. G.; Town C. M.; Ekström B. A New Class of Synthetic Antibacterials Acting on Lipopolysaccharide Biosynthesis. Nature 1987, 327 (6124), 730–732. 10.1038/327730a0. [DOI] [PubMed] [Google Scholar]
- Imai Y.; Meyer K. J.; Iinishi A.; Favre-Godal Q.; Green R.; Manuse S.; Caboni M.; Mori M.; Niles S.; Ghiglieri M.; Honrao C.; Ma X.; Guo J. J.; Makriyannis A.; Linares-Otoya L.; Böhringer N.; Wuisan Z. G.; Kaur H.; Wu R.; Mateus A.; Typas A.; Savitski M. M.; Espinoza J. L.; O’Rourke A.; Nelson K. E.; Hiller S.; Noinaj N.; Schäberle T. F.; D’Onofrio A.; Lewis K. A New Antibiotic Selectively Kills Gram-Negative Pathogens. Nature 2019, 576 (7787), 459–464. 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther A.; Urfer M.; Zahn M.; Müller M.; Wang S.-Y.; Mondal M.; Vitale A.; Hartmann J.-B.; Sharpe T.; Monte F. L.; Kocherla H.; Cline E.; Pessi G.; Rath P.; Modaresi S. M.; Chiquet P.; Stiegeler S.; Verbree C.; Remus T.; Schmitt M.; Kolopp C.; Westwood M.-A.; Desjonquères N.; Brabet E.; Hell S.; LePoupon K.; Vermeulen A.; Jaisson R.; Rithié V.; Upert G.; Lederer A.; Zbinden P.; Wach A.; Moehle K.; Zerbe K.; Locher H. H.; Bernardini F.; Dale G. E.; Eberl L.; Wollscheid B.; Hiller S.; Robinson J. A.; Obrecht D. Chimeric Peptidomimetic Antibiotics against Gram-Negative Bacteria. Nature 2019, 576 (7787), 452–458. 10.1038/s41586-019-1665-6. [DOI] [PubMed] [Google Scholar]
- Zähner H.; Diddens H.; Keller-Schierlein W.; Nägeli H. U. Some Experiments with Semisynthetic Sideromycins. Jpn. J. Antibiot. 1977, 30, 201–206. [PubMed] [Google Scholar]
- Arisawa M.; Sekine Y.; Shimizu S.; Takano H.; Angehrn P.; Then R. L. In Vitro and in Vivo Evaluation of Ro 09-1428, a New Parenteral Cephalosporin with High Antipseudomonal Activity. Antimicrob. Agents Chemother. 1991, 35 (4), 653–659. 10.1128/AAC.35.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möllmann U.; Heinisch L.; Bauernfeind A.; Köhler T.; Ankel-Fuchs D. Siderophores as Drug Delivery Agents: Application of the “Trojan Horse” Strategy. BioMetals 2009, 22 (4), 615–624. 10.1007/s10534-009-9219-2. [DOI] [PubMed] [Google Scholar]
- Baym M.; Stone L. K.; Kishony R. Multidrug Evolutionary Strategies to Reverse Antibiotic Resistance. Science 2016, 351 (6268), aad3292. 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. D.; Wright G. D. Antibacterial Drug Discovery in the Resistance Era. Nature 2016, 529 (7586), 336–343. 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- Bambeke F.; Pagès J.-M.; Lee V. J. Inhibitors of Bacterial Efflux Pumps as Adjuvants in Antibiotic Treatments and Diagnostic Tools for Detection of Resistance by Efflux. Recent Patents Anti-Infect. Drug Disc. 2006, 1 (2), 157–175. 10.2174/157489106777452692. [DOI] [PubMed] [Google Scholar]
- Pagès J.-M.; Amaral L. Mechanisms of Drug Efflux and Strategies to Combat Them: Challenging the Efflux Pump of Gram-Negative Bacteria. Biochim. Biophys. Acta BBA - Proteins Proteomics 2009, 1794 (5), 826–833. 10.1016/j.bbapap.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Blanco P.; Sanz-García F.; Hernando-Amado S.; Martínez J. L.; Alcalde-Rico M. The Development of Efflux Pump Inhibitors to Treat Gram-Negative Infections. Expert Opin. Drug Discovery 2018, 13 (10), 919–931. 10.1080/17460441.2018.1514386. [DOI] [PubMed] [Google Scholar]
- Cox G.; Wright G. D. Intrinsic Antibiotic Resistance: Mechanisms, Origins, Challenges and Solutions. Int. J. Med. Microbiol. 2013, 303 (6), 287–292. 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Vaara M.; Vaara T. Sensitization of Gram-Negative Bacteria to Antibiotics and Complement by a Nontoxic Oligopeptide. Nature 1983, 303 (5917), 526–528. 10.1038/303526a0. [DOI] [PubMed] [Google Scholar]
- Gadelii A.; Hassan K.-O.; Hakansson A. P.. Sensitizing Agents to Restore Antibiotic Resistance. In Antibiotic Drug Resistance; Capelo-Martínez J.-L.; Igrejas G., Eds.; John Wiley & Sons, Ltd, 2019; pp 429–452. 10.1002/9781119282549.ch17. [DOI] [Google Scholar]
- Schweizer F.Repurposing Antibiotics to Treat Resistant Gram-Negative Pathogens. In Antibiotic Drug Resistance; Capelo-Martínez J.-L.; Igrejas G., Eds.; John Wiley & Sons, Ltd., 2019; pp 453–476. 10.1002/9781119282549.ch18. [DOI] [Google Scholar]
- Bernal P.; Molina-Santiago C.; Daddaoua A.; Llamas M. A. Antibiotic Adjuvants: Identification and Clinical Use. Microb. Biotechnol. 2013, 6 (5), 445–449. 10.1111/1751-7915.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington R. J.; Melander C. Combination Approaches to Combat Multidrug-Resistant Bacteria. Trends Biotechnol. 2013, 31 (3), 177–184. 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander R. J.; Melander C.. Antibiotic Adjuvants. In Antibacterials, Vol. I; Fisher J. F., Mobashery S., Miller M. J., Eds.; Topics in Medicinal Chemistry; Springer International Publishing: Cham, 2018; pp 89–118. 10.1007/7355_2017_10. [DOI] [Google Scholar]
- Tyers M.; Wright G. D. Drug Combinations: A Strategy to Extend the Life of Antibiotics in the 21st Century. Nat. Rev. Microbiol. 2019, 17 (3), 141–155. 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- Wright G. D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016, 24 (11), 862–871. 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Li R.; Xiao X.; Wang Z. Antibiotic Adjuvants: An Alternative Approach to Overcome Multi-Drug Resistant Gram-Negative Bacteria. Crit. Rev. Microbiol. 2019, 45 (3), 301–314. 10.1080/1040841X.2019.1599813. [DOI] [PubMed] [Google Scholar]
- Klobucar K.; Brown E. D. New Potentiators of Ineffective Antibiotics: Targeting the Gram-Negative Outer Membrane to Overcome Intrinsic Resistance. Curr. Opin. Chem. Biol. 2022, 66, 102099. 10.1016/j.cbpa.2021.102099. [DOI] [PubMed] [Google Scholar]
- Savage P. B. Multidrug-Resistant Bacteria: Overcoming Antibiotic Permeability Barriers of Gram-Negative Bacteria. Ann. Med. 2001, 33 (3), 167–171. 10.3109/07853890109002073. [DOI] [PubMed] [Google Scholar]
- Nikaido H.; Vaara M. Molecular Basis of Bacterial Outer Membrane Permeability. Microbiol. Rev. 1985, 49 (1), 1–32. 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E. Alterations in Outer Membrane Permeability. Annu. Rev. Microbiol. 1984, 38, 237–264. 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- Douafer H.; Andrieu V.; Phanstiel O.; Brunel J. M. Antibiotic Adjuvants: Make Antibiotics Great Again!. J. Med. Chem. 2019, 62 (19), 8665–8681. 10.1021/acs.jmedchem.8b01781. [DOI] [PubMed] [Google Scholar]
- Zabawa T. P.; Pucci M. J.; Parr T. R.; Lister T. Treatment of Gram-Negative Bacterial Infections by Potentiation of Antibiotics. Curr. Opin. Microbiol. 2016, 33, 7–12. 10.1016/j.mib.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Odds F. C. Synergy, Antagonism, and What the Chequerboard Puts between Them. J. Antimicrob. Chemother. 2003, 52 (1), 1–1. 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- Blankson G.; Parhi A. K.; Kaul M.; Pilch D. S.; LaVoie E. J. Structure-Activity Relationships of Potentiators of the Antibiotic Activity of Clarithromycin against Escherichia Coli. Eur. J. Med. Chem. 2019, 178, 30–38. 10.1016/j.ejmech.2019.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti A.; Cirioni O.; Del Prete M. S.; Paggi A. M.; D’Errico M. M.; Scalise G. Combination Studies between Polycationic Peptides and Clinically Used Antibiotics against Gram-Positive and Gram-Negative Bacteria. Peptides 2000, 21 (8), 1155–1160. 10.1016/S0196-9781(00)00254-0. [DOI] [PubMed] [Google Scholar]
- Belanger C. R.; Lee A. H.-Y.; Pletzer D.; Dhillon B. K.; Falsafi R.; Hancock R. E. W. Identification of Novel Targets of Azithromycin Activity against Pseudomonas Aeruginosa Grown in Physiologically Relevant Media. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (52), 33519–33529. 10.1073/pnas.2007626117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisuria V. B.; Okshevsky M.; Déziel E.; Tufenkji N. Proanthocyanidin Interferes with Intrinsic Antibiotic Resistance Mechanisms of Gram-Negative Bacteria. Adv. Sci. Weinh. Baden-Wurtt. Ger. 2019, 6 (15), 1802333. 10.1002/advs.201802333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Md. S.; Choi Y. H.; Choi Y. S.; Yoo J. C. Glycin-Rich Antimicrobial Peptide YD1 from B. Amyloliquefaciens, Induced Morphological Alteration in and Showed Affinity for Plasmid DNA of E. Coli. AMB Express 2017, 7 (1), 8. 10.1186/s13568-016-0315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subratti A.; Ramkissoon A.; Lalgee L. J.; Jalsa N. K. Synthesis and Evaluation of the Antibiotic-Adjuvant Activity of Carbohydrate-Based Phosphoramidate Derivatives. Carbohydr. Res. 2021, 500, 108216. 10.1016/j.carres.2020.108216. [DOI] [PubMed] [Google Scholar]
- Baker K. R.; Jana B.; Hansen A. M.; Vissing K. J.; Nielsen H. M.; Franzyk H.; Guardabassi L. Repurposing Azithromycin and Rifampicin against Gram-Negative Pathogens by Combination with Peptide Potentiators. Int. J. Antimicrob. Agents 2019, 53 (6), 868–872. 10.1016/j.ijantimicag.2018.10.025. [DOI] [PubMed] [Google Scholar]
- Faure M.-E.Engineering Therapies against Pseudomonas Aeruginosa Based on Iron Chelation. Ph.D. Thesis, King’s College London, 2021. [Google Scholar]
- Lee H.; Hwang J. S.; Lee D. G. Periplanetasin-2 Enhances the Antibacterial Properties of Vancomycin or Chloramphenicol in Escherichia Coli. J. Microbiol. Biotechnol. 2021, 31 (2), 189–196. 10.4014/jmb.2010.10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R.; Sanchak Y.; Koskei L. C.; Lyu Y.; Zhanel G. G.; Arthur G.; Schweizer F. Short Proline-Rich Lipopeptide Potentiates Minocycline and Rifampin against Multidrug- and Extensively Drug-Resistant Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2018, 62 (4), e02374–17. 10.1128/AAC.02374-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangoni M. L.; Rinaldi A. C.; Di Giulio A.; Mignogna G.; Bozzi A.; Barra D.; Simmaco M. Structure-Function Relationships of Temporins, Small Antimicrobial Peptides from Amphibian Skin. Eur. J. Biochem. 2000, 267, 1447–1454. 10.1046/j.1432-1327.2000.01143.x. [DOI] [PubMed] [Google Scholar]
- Liu J.; Chen F.; Wang X.; Peng H.; Zhang H.; Wang K.-J. The Synergistic Effect of Mud Crab Antimicrobial Peptides Sphistin and Sph12–38 With Antibiotics Azithromycin and Rifampicin Enhances Bactericidal Activity Against Pseudomonas Aeruginosa. Front. Cell. Infect. Microbiol. 2020, 10, 572849. 10.3389/fcimb.2020.572849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaaytah A.; Qaoud M. T.; Abualhaijaa A.; Al-Balas Q.; Alzoubi K. H. Hybridization and Antibiotic Synergism as a Tool for Reducing the Cytotoxicity of Antimicrobial Peptides. Infect. Drug Resist. 2018, 11, 835–847. 10.2147/IDR.S166236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P. L.; Rossi L.; De Pascale G.; Wright G. D. A Forward Chemical Screen Identifies Antibiotic Adjuvants in Escherichia Coli. ACS Chem. Biol. 2012, 7 (9), 1547–1555. 10.1021/cb300269g. [DOI] [PubMed] [Google Scholar]
- Wang G.; Brunel J.-M.; Bolla J.-M.; Van Bambeke F. The Polyaminoisoprenyl Potentiator NV716 Revives Old Disused Antibiotics against Intracellular Forms of Infection by Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2021, 65 (3), e02028-20. 10.1128/AAC.02028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borselli D.; Blanchet M.; Bolla J.; Muth A.; Skruber K.; Phanstiel O.; Brunel J. M. Motuporamine Derivatives as Antimicrobial Agents and Antibiotic Enhancers against Resistant Gram-Negative Bacteria. Chembiochem 2017, 18 (3), 276–283. 10.1002/cbic.201600532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers R. P.; Cavallari J. F.; Burrows L. L. The Efflux Inhibitor Phenylalanine-Arginine Beta-Naphthylamide (PAβN) Permeabilizes the Outer Membrane of Gram-Negative Bacteria. PLoS One 2013, 8 (3), e60666. 10.1371/journal.pone.0060666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur U. J.; Chopra A.; Preet S.; Raj K.; Kondepudi K. K.; Gupta V.; Rishi P. Potential of 1-(1-Napthylmethyl)-Piperazine, an Efflux Pump Inhibitor against Cadmium-Induced Multidrug Resistance in Salmonella Enterica Serovar Typhi as an Adjunct to Antibiotics. Braz. J. Microbiol. 2021, 52 (3), 1303–1313. 10.1007/s42770-021-00492-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankson G. A.; Parhi A. K.; Kaul M.; Pilch D. S.; LaVoie E. J. Advances in the Structural Studies of Antibiotic Potentiators against Escherichia Coli. Bioorg. Med. Chem. 2019, 27 (15), 3254–3278. 10.1016/j.bmc.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart E. M.; Mitchell A. M.; Konovalova A.; Grabowicz M.; Sheng J.; Han X.; Rodriguez-Rivera F. P.; Schwaid A. G.; Malinverni J. C.; Balibar C. J.; Bodea S.; Si Q.; Wang H.; Homsher M. F.; Painter R. E.; Ogawa A. K.; Sutterlin H.; Roemer T.; Black T. A.; Rothman D. M.; Walker S. S.; Silhavy T. J. A Small-Molecule Inhibitor of BamA Impervious to Efflux and the Outer Membrane Permeability Barrier. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (43), 21748–21757. 10.1073/pnas.1912345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muheim C.; Götzke H.; Eriksson A. U.; Lindberg S.; Lauritsen I.; Nørholm M. H. H.; Daley D. O. Increasing the Permeability of Escherichia Coli Using MAC13243. Sci. Rep. 2017, 7 (1), 17629. 10.1038/s41598-017-17772-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzialampou A.; Protonotariou E.; Skoura L.; Sivropoulou A. Synergistic Antibacterial and Antibiofilm Activity of the MreB Inhibitor A22 Hydrochloride in Combination with Conventional Antibiotics against Pseudomonas Aeruginosa and Escherichia Coli Clinical Isolates. Int. J. Microbiol. 2021, 2021, e3057754. 10.1155/2021/3057754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesterbeek D. A. C.; Martin N. I.; Velthuizen A.; Duijst M.; Ruyken M.; Wubbolts R.; Rooijakkers S. H. M.; Bardoel B. W. Complement-Dependent Outer Membrane Perturbation Sensitizes Gram-Negative Bacteria to Gram-Positive Specific Antibiotics. Sci. Rep. 2019, 9 (1), 3074. 10.1038/s41598-019-38577-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison R. T.; Giehl T. J.; LaForce F. M. Damage of the Outer Membrane of Enteric Gram-Negative Bacteria by Lactoferrin and Transferrin. Infect. Immun. 1988, 56 (11), 2774–2781. 10.1128/iai.56.11.2774-2781.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.; Victor M.; Elsbach P. Role of Charge and Hydrophobic Interactions in the Action of Bactericidal/Permeability-Increasing Protein of Neutrophils on Gram-Negative Bacteria. J. Clin. Invest. 1983, 71 (3), 540–549. 10.1172/JCI110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S.; Mukherjee S.; Ghosh S.; Haldar J. Amino-Acid-Conjugated Polymer-Rifampicin Combination: Effective at Tackling Drug-Resistant Gram-Negative Clinical Isolates. ACS Appl. Bio Mater. 2019, 2 (12), 5404–5414. 10.1021/acsabm.9b00732. [DOI] [PubMed] [Google Scholar]
- Kim J.-H.; Yu D.; Eom S.-H.; Kim S.-H.; Oh J.; Jung W.-K.; Kim Y.-M. Synergistic Antibacterial Effects of Chitosan-Caffeic Acid Conjugate against Antibiotic-Resistant Acne-Related Bacteria. Mar. Drugs 2017, 15 (6), 167. 10.3390/md15060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je J.-Y.; Kim S.-K. Chitosan Derivatives Killed Bacteria by Disrupting the Outer and Inner Membrane. J. Agric. Food Chem. 2006, 54 (18), 6629–6633. 10.1021/jf061310p. [DOI] [PubMed] [Google Scholar]
- Qiao J.; Purro M.; Liu Z.; Xiong M. P. Effects of Polyethyelene Glycol-Desferrioxamine:Gallium Conjugates on Pseudomonas Aeruginosa Outer Membrane Permeability and Vancomycin Potentiation. Mol. Pharmaceutics 2021, 18 (2), 735–742. 10.1021/acs.molpharmaceut.0c00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J.; Liu Z.; Purro M.; Xiong M. P. Antibacterial and Potentiation Properties of Charge-Optimized Polyrotaxanes for Combating Opportunistic Bacteria. J. Mater. Chem. B 2018, 6 (33), 5353–5361. 10.1039/C8TB01610K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantisuwanno C.; Dang F.; Bender K.; Spencer J. D.; Jennings M. E.; Barton H. A.; Joy A. Synergism between Rifampicin and Cationic Polyurethanes Overcomes Intrinsic Resistance of Escherichia Coli. Biomacromolecules 2021, 22 (7), 2910–2920. 10.1021/acs.biomac.1c00306. [DOI] [PubMed] [Google Scholar]
- Livne L.; Epand R. F.; Papahadjopoulos-Sternberg B.; Epand R. M.; Mor A. OAK-Based Cochleates as a Novel Approach to Overcome Multidrug Resistance in Bacteria. FASEB J. 2010, 24 (12), 5092–5101. 10.1096/fj.10.167809. [DOI] [PubMed] [Google Scholar]
- Si Z.; Hou Z.; Vikhe Y. S.; Thappeta K. R. V.; Marimuthu K.; De P. P.; Ng O. T.; Li P.; Zhu Y.; Pethe K.; Chan-Park M. B. Antimicrobial Effect of a Novel Chitosan Derivative and Its Synergistic Effect with Antibiotics. ACS Appl. Mater. Interfaces 2021, 13 (2), 3237–3245. 10.1021/acsami.0c20881. [DOI] [PubMed] [Google Scholar]
- Danner R. L.; Joiner K. A.; Rubin M.; Patterson W. H.; Johnson N.; Ayers K. M.; Parrillo J. E. Purification, Toxicity, and Antiendotoxin Activity of Polymyxin B Nonapeptide. Antimicrob. Agents Chemother. 1989, 33 (9), 1428–1434. 10.1128/AAC.33.9.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. The Outer Membrane Permeability-Increasing Action of Linear Analogues of Polymyxin B Nonapeptide. Drugs Exp. Clin. Res. 1991, 17 (9), 437–443. [PubMed] [Google Scholar]
- Vaara M.; Fox J.; Loidl G.; Siikanen O.; Apajalahti J.; Hansen F.; Frimodt-Møller N.; Nagai J.; Takano M.; Vaara T. Novel Polymyxin Derivatives Carrying Only Three Positive Charges Are Effective Antibacterial Agents. Antimicrob. Agents Chemother. 2008, 52 (9), 3229–3236. 10.1128/AAC.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M.; Siikanen O.; Apajalahti J.; Fox J.; Frimodt-Møller N.; He H.; Poudyal A.; Li J.; Nation R. L.; Vaara T. A Novel Polymyxin Derivative That Lacks the Fatty Acid Tail and Carries Only Three Positive Charges Has Strong Synergism with Agents Excluded by the Intact Outer Membrane. Antimicrob. Agents Chemother. 2010, 54 (8), 3341–3346. 10.1128/AAC.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. New Approaches in Peptide Antibiotics. Curr. Opin. Pharmacol. 2009, 9 (5), 571–576. 10.1016/j.coph.2009.08.002. [DOI] [PubMed] [Google Scholar]
- MacNair C. R.; Stokes J. M.; Carfrae L. A.; Fiebig-Comyn A. A.; Coombes B. K.; Mulvey M. R.; Brown E. D. Overcoming Mcr-1 Mediated Colistin Resistance with Colistin in Combination with Other Antibiotics. Nat. Commun. 2018, 9 (1), 458. 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell J. M.; Aboklaish A. F.; Walsh T. R.; Vaara T.; Vaara M. The Polymyxin Derivative NAB739 Is Synergistic with Several Antibiotics against Polymyxin-Resistant Strains of Escherichia Coli, Klebsiella Pneumoniae and Acinetobacter Baumannii. Peptides 2019, 112, 149–153. 10.1016/j.peptides.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Kimura Y.; Matsunaga H.; Vaara M. Polymyxin B Octapeptide and Polymyxin B Heptapeptide Are Potent Outer Membrane Permeability-Increasing Agents. J. Antibiot. (Tokyo) 1992, 45 (5), 742–749. 10.7164/antibiotics.45.742. [DOI] [PubMed] [Google Scholar]
- Corbett D.; Wise A.; Langley T.; Skinner K.; Trimby E.; Birchall S.; Dorali A.; Sandiford S.; Williams J.; Warn P.; Vaara M.; Lister T. Potentiation of Antibiotic Activity by a Novel Cationic Peptide: Potency and Spectrum of Activity of SPR741. Antimicrob. Agents Chemother. 2017, 61 (8), e00200–17. 10.1128/AAC.00200-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R.; Berry L.; Tays Q.; Zhanel G. G.; Schweizer F. Development of Dilipid Polymyxins: Investigation on the Effect of Hydrophobicity through Its Fatty Acyl Component. Bioorganic Chem. 2018, 80, 639–648. 10.1016/j.bioorg.2018.07.018. [DOI] [PubMed] [Google Scholar]
- Kanazawa K.; Sato Y.; Ohki K.; Okimura K.; Uchida Y.; Shindo M.; Sakura N. Contribution of Each Amino Acid Residue in Polymyxin B3 to Antimicrobial and Lipopolysaccharide Binding Activity. Chem. Pharm. Bull. (Tokyo) 2009, 57 (3), 240–244. 10.1248/cpb.57.240. [DOI] [PubMed] [Google Scholar]
- Seo M.-D.; Won H.-S.; Kim J.-H.; Mishig-Ochir T.; Lee B.-J. Antimicrobial Peptides for Therapeutic Applications: A Review. Molecules 2012, 17 (10), 12276–12286. 10.3390/molecules171012276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adessi C.; Soto C. Converting a Peptide into a Drug: Strategies to Improve Stability and Bioavailability. Curr. Med. Chem. 2002, 9 (9), 963–978. 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- Bruckdorfer T.; Marder O.; Albericio F. From Production of Peptides in Milligram Amounts for Research to Multi-Tons Quantities for Drugs of the Future. Curr. Pharm. Biotechnol. 2004, 5 (1), 29–43. 10.2174/1389201043489620. [DOI] [PubMed] [Google Scholar]
- Ovadia O.; Greenberg S.; Laufer B.; Gilon C.; Hoffman A.; Kessler H. Improvement of Drug-like Properties of Peptides: The Somatostatin Paradigm. Expert Opin. Drug Discovery 2010, 5 (7), 655–671. 10.1517/17460441.2010.493935. [DOI] [PubMed] [Google Scholar]
- Godballe T.; Nilsson L. L.; Petersen P. D.; Jenssen H. Antimicrobial β-Peptides and α-Peptoids. Chem. Biol. Drug Des. 2011, 77 (2), 107–116. 10.1111/j.1747-0285.2010.01067.x. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Bulaj G. Converting Peptides into Drug Leads by Lipidation. Curr. Med. Chem. 2012, 19 (11), 1602–1618. 10.2174/092986712799945003. [DOI] [PubMed] [Google Scholar]
- Gentilucci L.; De Marco R.; Cerisoli L. Chemical Modifications Designed to Improve Peptide Stability: Incorporation of Non-Natural Amino Acids, Pseudo-Peptide Bonds, and Cyclization. Curr. Pharm. Des. 2010, 16 (28), 3185–3203. 10.2174/138161210793292555. [DOI] [PubMed] [Google Scholar]
- deGruyter J. N.; Malins L. R.; Baran P. S. Residue-Specific Peptide Modification: A Chemist’s Guide. Biochemistry 2017, 56 (30), 3863–3873. 10.1021/acs.biochem.7b00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. L. Antimicrobial Peptides Stage a Comeback. Nat. Biotechnol. 2013, 31 (5), 379–382. 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Hao W.; Wang X.; Ouyang J.; Deng X.; Yu H.; Wang Y. Antimicrobial Peptides, Conventional Antibiotics, and Their Synergistic Utility for the Treatment of Drug-Resistant Infections. Med. Res. Rev. 2022, 42 (4), 1377–1422. 10.1002/med.21879. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Sahl H.-G. Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies. Nat. Biotechnol. 2006, 24 (12), 1551–1557. 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Zompra A. A.; Galanis A. S.; Werbitzky O.; Albericio F. Manufacturing Peptides as Active Pharmaceutical Ingredients. Future Med. Chem. 2009, 1 (2), 361–377. 10.4155/fmc.09.23. [DOI] [PubMed] [Google Scholar]
- Albericio F.; Kruger H. G. Therapeutic Peptides. Future Med. Chem. 2012, 4 (12), 1527–1531. 10.4155/fmc.12.94. [DOI] [PubMed] [Google Scholar]
- Bals R.; Wilson J. M. Cathelicidins - a Family of Multifunctional Antimicrobial Peptides. Cell. Mol. Life Sci. CMLS 2003, 60 (4), 711–720. 10.1007/s00018-003-2186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A.; Hancock R. E. The Roles of Cathelicidin LL-37 in Immune Defences and Novel Clinical Applications. Curr. Opin. Hematol. 2009, 16 (1), 41–47. 10.1097/MOH.0b013e32831ac517. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Haney E. F.; Gill E. E. The Immunology of Host Defence Peptides: Beyond Antimicrobial Activity. Nat. Rev. Immunol. 2016, 16 (5), 321–334. 10.1038/nri.2016.29. [DOI] [PubMed] [Google Scholar]
- Bowdish D. M. E.; Davidson D. J.; Hancock R. E. W.. Immunomodulatory Properties of Defensins and Cathelicidins. In Antimicrobial Peptides and Human Disease; Shafer W. M., Ed.; Current Topics in Microbiology and Immunology; Springer: Berlin, Heidelberg, 2006; pp 27–66. 10.1007/3-540-29916-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. G.; Davidson D. J.; Gold M. R.; Bowdish D.; Hancock R. E. W. The Human Antimicrobial Peptide LL-37 Is a Multifunctional Modulator of Innate Immune Responses. J. Immunol. 2002, 169 (7), 3883–3891. 10.4049/jimmunol.169.7.3883. [DOI] [PubMed] [Google Scholar]
- Gudmundsson G. H.; Agerberth B.; Odeberg J.; Bergman T.; Olsson B.; Salcedo R. The Human Gene FALL39 and Processing of the Cathelin Precursor to the Antibacterial Peptide LL-37 in Granulocytes. Eur. J. Biochem. 1996, 238 (2), 325–332. 10.1111/j.1432-1033.1996.0325z.x. [DOI] [PubMed] [Google Scholar]
- Oren Z.; Lerman J. C.; Gudmundsson G. H.; Agerberth B.; Shai Y. Structure and Organization of the Human Antimicrobial Peptide LL-37 in Phospholipid Membranes: Relevance to the Molecular Basis for Its Non-Cell-Selective Activity. Biochem. J. 1999, 341 (3), 501–513. 10.1042/bj3410501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerberth B.; Gunne H.; Odeberg J.; Kogner P.; Boman H. G.; Gudmundsson G. H. FALL-39, a Putative Human Peptide Antibiotic, Is Cysteine-Free and Expressed in Bone Marrow and Testis. Proc. Natl. Acad. Sci. U. S. A. 1995, 92 (1), 195–199. 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed I.; Said D. G.; Nubile M.; Mastropasqua L.; Dua H. S. Cathelicidin-Derived Synthetic Peptide Improves Therapeutic Potential of Vancomycin Against Pseudomonas Aeruginosa. Front. Microbiol. 2019, 10, 2190. 10.3389/fmicb.2019.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y.; Cebrián R.; Xu C.; Jong A. de; Wu W.; Kuipers O. P. Elucidating the Mechanism by Which Synthetic Helper Peptides Sensitize Pseudomonas Aeruginosa to Multiple Antibiotics. PLOS Pathog. 2021, 17 (9), e1009909. 10.1371/journal.ppat.1009909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Cebrián R.; Montalbán-López M.; Ren H.; Wu W.; Kuipers O. P. Outer-Membrane-Acting Peptides and Lipid II-Targeting Antibiotics Cooperatively Kill Gram-Negative Pathogens. Commun. Biol. 2021, 4 (1), 31. 10.1038/s42003-020-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrián R.; Xu C.; Xia Y.; Wu W.; Kuipers O. P. The Cathelicidin-Derived Close-to-Nature Peptide D-11 Sensitises Klebsiella Pneumoniae to a Range of Antibiotics in Vitro, Ex Vivo and in Vivo. Int. J. Antimicrob. Agents 2021, 58 (5), 106434. 10.1016/j.ijantimicag.2021.106434. [DOI] [PubMed] [Google Scholar]
- Soren O.; Brinch K. S.; Patel D.; Liu Y.; Liu A.; Coates A.; Hu Y. Antimicrobial Peptide Novicidin Synergizes with Rifampin, Ceftriaxone, and Ceftazidime against Antibiotic-Resistant Enterobacteriaceae In Vitro. Antimicrob. Agents Chemother. 2015, 59 (10), 6233–6240. 10.1128/AAC.01245-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden S.; Rieder A.; Chis Ster I.; Schwartz T.; Mikut R.; Hilpert K. Synergy Pattern of Short Cationic Antimicrobial Peptides Against Multidrug-Resistant Pseudomonas Aeruginosa. Front. Microbiol. 2019, 10, 2740. 10.3389/fmicb.2019.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Wang H.; Cao S.; Jiang C.; Hou J. Characterization of Antibacterial Activity and Mechanisms of Two Linear Derivatives of Bactenecin. LWT 2019, 107, 89–97. 10.1016/j.lwt.2019.02.073. [DOI] [Google Scholar]
- Hilpert K.; Volkmer-Engert R.; Walter T.; Hancock R. E. W. High-Throughput Generation of Small Antibacterial Peptides with Improved Activity. Nat. Biotechnol. 2005, 23 (8), 1008–1012. 10.1038/nbt1113. [DOI] [PubMed] [Google Scholar]
- Wu X.; Li Z.; Li X.; Tian Y.; Fan Y.; Yu C.; Zhou B.; Liu Y.; Xiang R.; Yang L. Synergistic Effects of Antimicrobial Peptide DP7 Combined with Antibiotics against Multidrug-Resistant Bacteria. Drug Des. Devel. Ther. 2017, 11, 939–946. 10.2147/DDDT.S107195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N.; Zhong C.; Liu T.; Zhu Y.; Gou S.; Bao H.; Yao J.; Ni J. Newly Designed Antimicrobial Peptides with Potent Bioactivity and Enhanced Cell Selectivity Prevent and Reverse Rifampin Resistance in Gram-Negative Bacteria. Eur. J. Pharm. Sci. 2021, 158, 105665. 10.1016/j.ejps.2020.105665. [DOI] [PubMed] [Google Scholar]
- Faccone D.; Veliz O.; Corso A.; Noguera M.; Martínez M.; Payes C.; Semorile L.; Maffía P. C. Antimicrobial Activity of de Novo Designed Cationic Peptides against Multi-Resistant Clinical Isolates. Eur. J. Med. Chem. 2014, 71, 31–35. 10.1016/j.ejmech.2013.10.065. [DOI] [PubMed] [Google Scholar]
- Sánchez-Gómez S.; Lamata M.; Leiva J.; Blondelle S. E.; Jerala R.; Andrä J.; Brandenburg K.; Lohner K.; Moriyón I.; Martínez-de-Tejada G. Comparative Analysis of Selected Methods for the Assessment of Antimicrobial and Membrane-Permeabilizing Activity: A Case Study for Lactoferricin Derived Peptides. BMC Microbiol. 2008, 8 (1), 196. 10.1186/1471-2180-8-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gómez S.; Japelj B.; Jerala R.; Moriyón I.; Fernández Alonso M.; Leiva J.; Blondelle S. E.; Andrä J.; Brandenburg K.; Lohner K.; Martínez de Tejada G. Structural Features Governing the Activity of Lactoferricin-Derived Peptides That Act in Synergy with Antibiotics against Pseudomonas Aeruginosa In Vitro and In Vivo. Antimicrob. Agents Chemother. 2011, 55 (1), 218–228. 10.1128/AAC.00904-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strøm M. B.; Haug B. E.; Rekdal Ø.; Skar M. L.; Stensen W.; Svendsen J. S. Important Structural Features of 15-Residue Lactoferricin Derivatives and Methods for Improvement of Antimicrobial Activity. Biochem. Cell Biol. 2002, 80 (1), 65–74. 10.1139/o01-236. [DOI] [PubMed] [Google Scholar]
- Strøm M. B.; Svendsen J. S.; Rekdal Ø. Antibacterial Activity of 15-Residue Lactoferricin Derivatives. J. Pept. Res. 2000, 56 (5), 265–274. 10.1034/j.1399-3011.2000.00770.x. [DOI] [PubMed] [Google Scholar]
- Ulvatne H.; Karoliussen S.; Stiberg T.; Rekdal Ø.; Svendsen J. S. Short Antibacterial Peptides and Erythromycin Act Synergically against Escherichia Coli. J. Antimicrob. Chemother. 2001, 48 (2), 203–208. 10.1093/jac/48.2.203. [DOI] [PubMed] [Google Scholar]
- Rekdal Ø.; Andersen J.; Vorland L. H.; Svendsen J. S. Construction and Synthesis of Lactoferricin Derivatives with Enhanced Antibacterial Activity. J. Pept. Sci. 1999, 5 (1), 32–45. . [DOI] [Google Scholar]
- Saravanan R.; Holdbrook D. A.; Petrlova J.; Singh S.; Berglund N. A.; Choong Y. K.; Kjellström S.; Bond P. J.; Malmsten M.; Schmidtchen A. Structural Basis for Endotoxin Neutralisation and Anti-Inflammatory Activity of Thrombin-Derived C-Terminal Peptides. Nat. Commun. 2018, 9 (1), 2762. 10.1038/s41467-018-05242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling C. M. J.; Wood T. M.; Slingerland C. J.; Bertheussen K.; Lok S.; Martin N. I. Thrombin-Derived Peptides Potentiate the Activity of Gram-Positive-Specific Antibiotics against Gram-Negative Bacteria. Molecules 2021, 26 (7), 1954–1954. 10.3390/molecules26071954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti A.; Cirioni O.; Kamysz W.; D’Amato G.; Silvestri C.; Prete M. S. D.; Licci A.; Riva A.; Łukasiak J.; Scalise G. In Vitro Activity of the Histatin Derivative P-113 against Multidrug-Resistant Pathogens Responsible for Pneumonia in Immunocompromised Patients. Antimicrob. Agents Chemother. 2005, 49 (3), 1249–1252. 10.1128/AAC.49.3.1249-1252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama K. Anti-Lipopolysaccharide Activity of Histatins, Peptides from Human Saliva. Experientia 1993, 49 (12), 1095–1097. 10.1007/BF01929920. [DOI] [PubMed] [Google Scholar]
- Sajjan U. S.; Tran L. T.; Sole N.; Rovaldi C.; Akiyama A.; Friden P. M.; Forstner J. F.; Rothstein D. M. P-113D, an Antimicrobial Peptide Active against Pseudomonas Aeruginosa, Retains Activity in the Presence of Sputum from Cystic Fibrosis Patients. Antimicrob. Agents Chemother. 2001, 45 (12), 3437–3444. 10.1128/AAC.45.12.3437-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirioni O.; Giacometti A.; Ghiselli R.; Orlando F.; Kamysz W.; D’Amato G.; Mocchegiani F.; Lukasiak J.; Silvestri C.; Saba V.; Scalise G. Potential Therapeutic Role of Histatin Derivative P-113d in Experimental Rat Models of Pseudomonas Aeruginosa Sepsis. J. Infect. Dis. 2004, 190 (2), 356–364. 10.1086/421913. [DOI] [PubMed] [Google Scholar]
- Yu H.-Y.; Tu C.-H.; Yip B.-S.; Chen H.-L.; Cheng H.-T.; Huang K.-C.; Lo H.-J.; Cheng J.-W. Easy Strategy To Increase Salt Resistance of Antimicrobial Peptides. Antimicrob. Agents Chemother. 2011, 55, 4918. 10.1128/AAC.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih Y.-H.; Wang S.-Y.; Yip B.-S.; Cheng K.-T.; Hsu S.-Y.; Wu C.-L.; Yu H.-Y.; Cheng J.-W. Dependence on Size and Shape of Non-Nature Amino Acids in the Enhancement of Lipopolysaccharide (LPS) Neutralizing Activities of Antimicrobial Peptides. J. Colloid Interface Sci. 2019, 533, 492–502. 10.1016/j.jcis.2018.08.042. [DOI] [PubMed] [Google Scholar]
- Wu C.-L.; Hsueh J.-Y.; Yip B.-S.; Chih Y.-H.; Peng K.-L.; Cheng J.-W. Antimicrobial Peptides Display Strong Synergy with Vancomycin Against Vancomycin-Resistant E. Faecium, S. Aureus, and Wild-Type E. Coli. Int. J. Mol. Sci. 2020, 21 (13), 4578. 10.3390/ijms21134578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirioni O.; Silvestri C.; Ghiselli R.; Orlando F.; Riva A.; Gabrielli E.; Mocchegiani F.; Cianforlini N.; Trombettoni M. M. C.; Saba V.; Scalise G.; Giacometti A. Therapeutic Efficacy of Buforin II and Rifampin in a Rat Model of Acinetobacter Baumannii Sepsis. Crit. Care Med. 2009, 37 (4), 1403–1407. 10.1097/CCM.0b013e31819c3e22. [DOI] [PubMed] [Google Scholar]
- Marcellini L.; Borro M.; Gentile G.; Rinaldi A.; Stella L.; Aimola P.; Barra D.; Mangoni M. Esculentin-1b(1–18) - A Membrane-Active Antimicrobial Peptide That Synergizes with Antibiotics and Modifies the Expression Level of a Limited Number of Proteins in Escherichia Coli. FEBS J. 2009, 276, 5647–5664. 10.1111/j.1742-4658.2009.07257.x. [DOI] [PubMed] [Google Scholar]
- Yenugu S.; Narmadha G. The Human Male Reproductive Tract Antimicrobial Peptides of the HE2 Family Exhibit Potent Synergy with Standard Antibiotics. J. Pept. Sci. 2010, 16 (7), 337–341. 10.1002/psc.1246. [DOI] [PubMed] [Google Scholar]
- Gou S.; Li B.; Ouyang X.; Ba Z.; Zhong C.; Zhang T.; Chang L.; Zhu Y.; Zhang J.; Zhu N.; Zhang Y.; Liu H.; Ni J. Novel Broad-Spectrum Antimicrobial Peptide Derived from Anoplin and Its Activity on Bacterial Pneumonia in Mice. J. Med. Chem. 2021, 64 (15), 11247–11266. 10.1021/acs.jmedchem.1c00614. [DOI] [PubMed] [Google Scholar]
- Cirioni O.; Silvestri C.; Ghiselli R.; Orlando F.; Riva A.; Mocchegiani F.; Chiodi L.; Castelletti S.; Gabrielli E.; Saba V.; Scalise G.; Giacometti A. Protective Effects of the Combination of α-Helical Antimicrobial Peptides and Rifampicin in Three Rat Models of Pseudomonas Aeruginosa Infection. J. Antimicrob. Chemother. 2008, 62 (6), 1332–1338. 10.1093/jac/dkn393. [DOI] [PubMed] [Google Scholar]
- Konno K.; Hisada M.; Fontana R.; Lorenzi C. C. B.; Naoki H.; Itagaki Y.; Miwa A.; Kawai N.; Nakata Y.; Yasuhara T.; Ruggiero Neto J.; de Azevedo W. F.; Palma M. S.; Nakajima T. Anoplin, a Novel Antimicrobial Peptide from the Venom of the Solitary Wasp Anoplius Samariensis. Biochim. Biophys. Acta BBA - Protein Struct. Mol. Enzymol. 2001, 1550 (1), 70–80. 10.1016/S0167-4838(01)00271-0. [DOI] [PubMed] [Google Scholar]
- Yenugu S.; Hamil K. G.; Birse C. E.; Ruben S. M.; French F. S.; Hall S. H. Antibacterial Properties of the Sperm-Binding Proteins and Peptides of Human Epididymis 2 (HE2) Family; Salt Sensitivity, Structural Dependence and Their Interaction with Outer and Cytoplasmic Membranes of Escherichia Coli. Biochem. J. 2003, 372 (2), 473–483. 10.1042/bj20030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajish C.; Yang S.; Kumar S. D.; Shin S. Y. Proadrenomedullin N-Terminal 20 Peptide (PAMP) and Its C-Terminal 12-Residue Peptide, PAMP(9–20): Cell Selectivity and Antimicrobial Mechanism. Biochem. Biophys. Res. Commun. 2020, 527 (3), 744–750. 10.1016/j.bbrc.2020.04.063. [DOI] [PubMed] [Google Scholar]
- Kim M. K.; Kang N. H.; Ko S. J.; Park J.; Park E.; Shin D. W.; Kim S. H.; Lee S. A.; Lee J. I.; Lee S. H.; Ha E. G.; Jeon S. H.; Park Y. Antibacterial and Antibiofilm Activity and Mode of Action of Magainin 2 against Drug-Resistant Acinetobacter Baumannii. Int. J. Mol. Sci. 2018, 19 (10), 3041. 10.3390/ijms19103041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.; Shin A.; Kim Y. Anti-Inflammatory Activities of Cecropin a and Its Mechanism of Action. Arch. Insect Biochem. Physiol. 2015, 88 (1), 31–44. 10.1002/arch.21193. [DOI] [PubMed] [Google Scholar]
- Cochrane S. A.; Findlay B.; Bakhtiary A.; Acedo J. Z.; Rodriguez-Lopez E. M.; Mercier P.; Vederas J. C. Antimicrobial Lipopeptide Tridecaptin A1 Selectively Binds to Gram-Negative Lipid II. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (41), 11561–11566. 10.1073/pnas.1608623113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. G.; Yan H.; Hancock R. E. W. Biological Properties of Structurally Related α-Helical Cationic Antimicrobial Peptides. Infect. Immun. 1999, 67, 2005. 10.1128/IAI.67.4.2005-2009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane S. A.; Vederas J. C. Unacylated Tridecaptin A1 Acts as an Effective Sensitiser of Gram-Negative Bacteria to Other Antibiotics. Int. J. Antimicrob. Agents 2014, 44 (6), 493–499. 10.1016/j.ijantimicag.2014.08.008. [DOI] [PubMed] [Google Scholar]
- Cochrane S. A.; Lohans C. T.; Brandelli J. R.; Mulvey G.; Armstrong G. D.; Vederas J. C. Synthesis and Structure–Activity Relationship Studies of N-Terminal Analogues of the Antimicrobial Peptide Tridecaptin A1. J. Med. Chem. 2014, 57 (3), 1127–1131. 10.1021/jm401779d. [DOI] [PubMed] [Google Scholar]
- Cochrane S. A.; Lohans C. T.; van Belkum M. J.; Bels M. A.; Vederas J. C. Studies on Tridecaptin B1, a Lipopeptide with Activity against Multidrug Resistant Gram-Negative Bacteria. Org. Biomol. Chem. 2015, 13 (21), 6073–6081. 10.1039/C5OB00780A. [DOI] [PubMed] [Google Scholar]
- Chiorean S.; Antwi I.; Carney D. W.; Kotsogianni I.; Giltrap A. M.; Alexander F. M.; Cochrane S. A.; Payne R. J.; Martin N. I.; Henninot A.; Vederas J. C. Dissecting the Binding Interactions of Teixobactin with the Bacterial Cell-Wall Precursor Lipid II. ChemBioChem. 2020, 21 (6), 789–792. 10.1002/cbic.201900504. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Ding S.; Shen J.; Zhu K. Nonribosomal Antibacterial Peptides That Target Multidrug-Resistant Bacteria. Nat. Prod. Rep. 2019, 36 (4), 573–592. 10.1039/C8NP00031J. [DOI] [PubMed] [Google Scholar]
- Song M.; Liu Y.; Huang X.; Ding S.; Wang Y.; Shen J.; Zhu K. A Broad-Spectrum Antibiotic Adjuvant Reverses Multidrug-Resistant Gram-Negative Pathogens. Nat. Microbiol. 2020, 5 (8), 1040–1050. 10.1038/s41564-020-0723-z. [DOI] [PubMed] [Google Scholar]
- Slootweg J. C.; van Schaik T. B.; Quarles van Ufford H. C.; Breukink E.; Liskamp R. M. J.; Rijkers D. T. S. Improving the Biological Activity of the Antimicrobial Peptide Anoplin by Membrane Anchoring through a Lipophilic Amino Acid Derivative. Bioorg. Med. Chem. Lett. 2013, 23 (13), 3749–3752. 10.1016/j.bmcl.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Chen J.; Zheng X.; Yang X.; Ma P.; Cai Y.; Zhang B.; Chen Y. Design of Novel Analogues of Short Antimicrobial Peptide Anoplin with Improved Antimicrobial Activity. J. Pept. Sci. 2014, 20 (12), 945–951. 10.1002/psc.2705. [DOI] [PubMed] [Google Scholar]
- Munk J. K.; Ritz C.; Fliedner F. P.; Frimodt-Møller N.; Hansen P. R. Novel Method To Identify the Optimal Antimicrobial Peptide in a Combination Matrix, Using Anoplin as an Example. Antimicrob. Agents Chemother. 2014, 58 (2), 1063–1070. 10.1128/AAC.02369-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libardo M. D. J.; Nagella S.; Lugo A.; Pierce S.; Angeles-Boza A. M. Copper-Binding Tripeptide Motif Increases Potency of the Antimicrobial Peptide Anoplin via Reactive Oxygen Species Generation. Biochem. Biophys. Res. Commun. 2015, 456 (1), 446–451. 10.1016/j.bbrc.2014.11.104. [DOI] [PubMed] [Google Scholar]
- Zhong C.; Liu T.; Gou S.; He Y.; Zhu N.; Zhu Y.; Wang L.; Liu H.; Zhang Y.; Yao J.; Ni J. Design and Synthesis of New N-Terminal Fatty Acid Modified-Antimicrobial Peptide Analogues with Potent in Vitro Biological Activity. Eur. J. Med. Chem. 2019, 182, 111636. 10.1016/j.ejmech.2019.111636. [DOI] [PubMed] [Google Scholar]
- Santamaría C.; Larios S.; Angulo Y.; Pizarro-Cerda J.; Gorvel J.-P.; Moreno E.; Lomonte B. Antimicrobial Activity of Myotoxic Phospholipases A2 from Crotalid Snake Venoms and Synthetic Peptide Variants Derived from Their C-Terminal Region. Toxicon 2005, 45 (7), 807–815. 10.1016/j.toxicon.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Yu H.-Y.; Huang K.-C.; Yip B.-S.; Tu C.-H.; Chen H.-L.; Cheng H.-T.; Cheng J.-W. Rational Design of Tryptophan-Rich Antimicrobial Peptides with Enhanced Antimicrobial Activities and Specificities. ChemBioChem. 2010, 11 (16), 2273–2282. 10.1002/cbic.201000372. [DOI] [PubMed] [Google Scholar]
- Chu H.-L.; Yu H.-Y.; Yip B.-S.; Chih Y.-H.; Liang C.-W.; Cheng H.-T.; Cheng J.-W. Boosting Salt Resistance of Short Antimicrobial Peptides. Antimicrob. Agents Chemother. 2013, 57, 4050–4052. 10.1128/AAC.00252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih Y.-H.; Lin Y.-S.; Yip B.-S.; Wei H.-J.; Chu H.-L.; Yu H.-Y.; Cheng H.-T.; Chou Y.-T.; Cheng J.-W. Ultrashort Antimicrobial Peptides with Antiendotoxin Properties. Antimicrob. Agents Chemother. 2015, 59, 5052–5056. 10.1128/AAC.00519-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.-Y.; Chen Y.-A.; Yip B.-S.; Wang S.-Y.; Wei H.-J.; Chih Y.-H.; Chen K.-H.; Cheng J.-W. Role of β-Naphthylalanine End-Tags in the Enhancement of Antiendotoxin Activities: Solution Structure of the Antimicrobial Peptide S1-Nal-Nal in Complex with Lipopolysaccharide. Biochim. Biophys. Acta BBA - Biomembr. 2017, 1859 (6), 1114–1123. 10.1016/j.bbamem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Wu C.-L.; Peng K.-L.; Yip B.-S.; Chih Y.-H.; Cheng J.-W. Boosting Synergistic Effects of Short Antimicrobial Peptides With Conventional Antibiotics Against Resistant Bacteria. Front. Microbiol. 2021, 12, 3145. 10.3389/fmicb.2021.747760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. R.; Jana B.; Franzyk H.; Guardabassi L. A High-Throughput Approach To Identify Compounds That Impair Envelope Integrity in Escherichia Coli. Antimicrob. Agents Chemother. 2016, 60 (10), 5995–6002. 10.1128/AAC.00537-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S.; Mohan K. V. K.; Atreya C. D. A Peptide Derived from Phage Display Library Exhibits Antibacterial Activity against E. Coli and Pseudomonas Aeruginosa. PLoS One 2013, 8 (2), e56081. 10.1371/journal.pone.0056081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M.; Porro M. Group of Peptides That Act Synergistically with Hydrophobic Antibiotics against Gram-Negative Enteric Bacteria. Antimicrob. Agents Chemother. 1996, 40 (8), 1801–1805. 10.1128/AAC.40.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monincová L.; Buděšínský M.; Slaninová J.; Hovorka O.; Cvačka J.; Voburka Z.; Fučík V.; Borovičková L.; Bednárová L.; Straka J.; Čeřovský V. Novel Antimicrobial Peptides from the Venom of the Eusocial Bee Halictus Sexcinctus (Hymenoptera: Halictidae) and Their Analogs. Amino Acids 2010, 39 (3), 763–775. 10.1007/s00726-010-0519-1. [DOI] [PubMed] [Google Scholar]
- Dewan P. C.; Anantharaman A.; Chauhan V. S.; Sahal D. Antimicrobial Action of Prototypic Amphipathic Cationic Decapeptides and Their Branched Dimers. Biochemistry 2009, 48 (24), 5642–5657. 10.1021/bi900272r. [DOI] [PubMed] [Google Scholar]
- Ramagopal U. A.; Ramakumar S.; Sahal D.; Chauhan V. S. De Novo Design and Characterization of an Apolar Helical Hairpin Peptide at Atomic Resolution: Compaction Mediated by Weak Interactions. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (3), 870–874. 10.1073/pnas.98.3.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman A.; Rizvi M. S.; Sahal D. Synergy with Rifampin and Kanamycin Enhances Potency, Kill Kinetics, and Selectivity of DeNovo-Designed Antimicrobial Peptides. Antimicrob. Agents Chemother. 2010, 54 (5), 1693–1699. 10.1128/AAC.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman M. R.; Yount N. Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55 (1), 27–55. 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Hyun S.; Choi Y.; Jo D.; Choo S.; Park T. W.; Park S.-J.; Kim S.; Lee S.; Park S.; Jin S. M.; Cheon D. H.; Yoo W.; Arya R.; Chong Y. P.; Kim K. K.; Kim Y. S.; Lee Y.; Yu J. Proline Hinged Amphipathic α-Helical Peptide Sensitizes Gram-Negative Bacteria to Various Gram-Positive Antibiotics. J. Med. Chem. 2020, 63 (23), 14937–14950. 10.1021/acs.jmedchem.0c01506. [DOI] [PubMed] [Google Scholar]
- Fernandez D. I.; Lee T.-H.; Sani M.-A.; Aguilar M.-I.; Separovic F. Proline Facilitates Membrane Insertion of the Antimicrobial Peptide Maculatin 1.1 via Surface Indentation and Subsequent Lipid Disordering. Biophys. J. 2013, 104 (7), 1495–1507. 10.1016/j.bpj.2013.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P.; Xu C.; Cheng Q.; Liu J.; Gao W.; Yang X.; Wong K.-Y.; Chen S.; Chan K.-F. Phenol-Soluble-Modulin-Inspired Amphipathic Peptides Have Bactericidal Activity against Multidrug-Resistant Bacteria. ChemMedChem. 2019, 14 (16), 1547–1559. 10.1002/cmdc.201900364. [DOI] [PubMed] [Google Scholar]
- Zeng P.; Xu C.; Liu C.; Liu J.; Cheng Q.; Gao W.; Yang X.; Chen S.; Chan K. F.; Wong K.-Y. De Novo Designed Hexadecapeptides Synergize Glycopeptide Antibiotics Vancomycin and Teicoplanin against Pathogenic Klebsiella Pneumoniae via Disruption of Cell Permeability and Potential. ACS Appl. Bio Mater. 2020, 3 (3), 1738–1752. 10.1021/acsabm.0c00044. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Zhong C.; Yao J.; Zhang J.; Zhang T.; Li B.; Gou S.; Ni J. Antimicrobial Peptides–Antibiotics Combination: An Effective Strategy Targeting Drug-Resistant Gram-Negative Bacteria. Pept. Sci. 2022, e24261. 10.1002/pep2.24261. [DOI] [Google Scholar]
- Zhong C.; Zhang F.; Yao J.; Zhu Y.; Zhu N.; Zhang J.; Ouyang X.; Zhang T.; Li B.; Xie J.; Ni J. New Antimicrobial Peptides with Repeating Unit against Multidrug-Resistant Bacteria. ACS Infect. Dis. 2021, 7 (6), 1619–1637. 10.1021/acsinfecdis.0c00797. [DOI] [PubMed] [Google Scholar]
- Moon S. H.; Zhang X.; Zheng G.; Meeker D. G.; Smeltzer M. S.; Huang E. Novel Linear Lipopeptide Paenipeptins with Potential for Eradicating Biofilms and Sensitizing Gram-Negative Bacteria to Rifampicin and Clarithromycin. J. Med. Chem. 2017, 60 (23), 9630–9640. 10.1021/acs.jmedchem.7b01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S. H.; Kaufmann Y.; Huang E. Paenipeptin Analogues Potentiate Clarithromycin and Rifampin against Mcr-1-Mediated Polymyxin-Resistant Escherichia Coli In Vivo. Antimicrob. Agents Chemother. 2020, 64 (4), e02045–19. 10.1128/AAC.02045-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R.; Brizuela M.; Eisner B.; Findlay B.; Zhanel G. G.; Schweizer F. Dilipid Ultrashort Cationic Lipopeptides as Adjuvants for Chloramphenicol and Other Conventional Antibiotics against Gram-Negative Bacteria. Amino Acids 2019, 51 (3), 383–393. 10.1007/s00726-018-2673-9. [DOI] [PubMed] [Google Scholar]
- Schweizer L.; Ramirez D.; Schweizer F. Effects of Lysine N-ζ-Methylation in Ultrashort Tetrabasic Lipopeptides (UTBLPs) on the Potentiation of Rifampicin, Novobiocin, and Niclosamide in Gram-Negative Bacteria. Antibiotics 2022, 11 (3), 335. 10.3390/antibiotics11030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y.; Deng S.; Cong Z.; Zhang H.; Lu Z.; Shao N.; Bhatti S. A.; Zhou C.; Cheng J.; Gellman S. H.; Liu R. Secondary Amine Pendant β-Peptide Polymers Displaying Potent Antibacterial Activity and Promising Therapeutic Potential in Treating MRSA-Induced Wound Infections and Keratitis. J. Am. Chem. Soc. 2022, 144 (4), 1690–1699. 10.1021/jacs.1c10659. [DOI] [PubMed] [Google Scholar]
- Li H.; Hu Y.; Pu Q.; He T.; Zhang Q.; Wu W.; Xia X.; Zhang J. Novel Stapling by Lysine Tethering Provides Stable and Low Hemolytic Cationic Antimicrobial Peptides. J. Med. Chem. 2020, 63 (8), 4081–4089. 10.1021/acs.jmedchem.9b02025. [DOI] [PubMed] [Google Scholar]
- Fernández-Reyes M.; Díaz D.; de la Torre B. G.; Cabrales-Rico A.; Vallès-Miret M.; Jiménez-Barbero J.; Andreu D.; Rivas L. Lysine N(Epsilon)-Trimethylation, a Tool for Improving the Selectivity of Antimicrobial Peptides. J. Med. Chem. 2010, 53 (15), 5587–5596. 10.1021/jm100261r. [DOI] [PubMed] [Google Scholar]
- Ramirez D.; Berry L.; Domalaon R.; Brizuela M.; Schweizer F. Dilipid Ultrashort Tetrabasic Peptidomimetics Potentiate Novobiocin and Rifampicin Against Multidrug-Resistant Gram-Negative Bacteria. ACS Infect. Dis. 2020, 6 (6), 1413–1426. 10.1021/acsinfecdis.0c00017. [DOI] [PubMed] [Google Scholar]
- Radzishevsky I. S.; Rotem S.; Bourdetsky D.; Navon-Venezia S.; Carmeli Y.; Mor A. Improved Antimicrobial Peptides Based on Acyl-Lysine Oligomers. Nat. Biotechnol. 2007, 25 (6), 657–659. 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- Radzishevsky I.; Krugliak M.; Ginsburg H.; Mor A. Antiplasmodial Activity of Lauryl-Lysine Oligomers. Antimicrob. Agents Chemother. 2007, 51 (5), 1753–1759. 10.1128/AAC.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarig H.; Livne L.; Held-Kuznetsov V.; Zaknoon F.; Ivankin A.; Gidalevitz D.; Mor A. A Miniature Mimic of Host Defense Peptides with Systemic Antibacterial Efficacy. FASEB J. 2010, 24 (6), 1904–1913. 10.1096/fj.09-149427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem S.; Radzishevsky I. S.; Bourdetsky D.; Navon-Venezia S.; Carmeli Y.; Mor A. Analogous Oligo-Acyl-Lysines with Distinct Antibacterial Mechanisms. FASEB J. 2008, 22 (8), 2652–2661. 10.1096/fj.07-105015. [DOI] [PubMed] [Google Scholar]
- Epand R. F.; Sarig H.; Mor A.; Epand R. M. Cell-Wall Interactions and the Selective Bacteriostatic Activity of a Miniature Oligo-Acyl-Lysyl. Biophys. J. 2009, 97 (8), 2250–2257. 10.1016/j.bpj.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammal J.; Zaknoon F.; Kaneti G.; Goldberg K.; Mor A. Sensitization of Gram-Negative Bacteria to Rifampin and OAK Combinations. Sci. Rep. 2015, 5 (1), 9216. 10.1038/srep09216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaknoon F.; Meir O.; Mor A. Mechanistic Studies of Antibiotic Adjuvants Reducing Kidney’s Bacterial Loads upon Systemic Monotherapy. Pharmaceutics 2021, 13 (11), 1947. 10.3390/pharmaceutics13111947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob B.; Park I.-S.; Bang J.-K.; Shin S. Y. Short KR-12 Analogs Designed from Human Cathelicidin LL-37 Possessing Both Antimicrobial and Antiendotoxic Activities without Mammalian Cell Toxicity. J. Pept. Sci. 2013, 19 (11), 700–707. 10.1002/psc.2552. [DOI] [PubMed] [Google Scholar]
- Wu X.; Wang Z.; Li X.; Fan Y.; He G.; Wan Y.; Yu C.; Tang J.; Li M.; Zhang X.; Zhang H.; Xiang R.; Pan Y.; Liu Y.; Lu L.; Yang L. In Vitro and In Vivo Activities of Antimicrobial Peptides Developed Using an Amino Acid-Based Activity Prediction Method. Antimicrob. Agents Chemother. 2014, 58, 5342–5349. 10.1128/AAC.02823-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas N. d. J.; Rivera Monroy Z. J.; Fierro Medina R.; García Castañeda J. E. Antimicrobial Activity of Truncated and Polyvalent Peptides Derived from the FKCRRWQWRMKKGLA Sequence against Escherichia Coli ATCC 25922 and Staphylococcus Aureus ATCC 25923. Mol. J. Synth. Chem. Nat. Prod. Chem. 2017, 22 (6), 987. 10.3390/molecules22060987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H.; Kumar K. Antimicrobial Activity and Protease Stability of Peptides Containing Fluorinated Amino Acids. J. Am. Chem. Soc. 2007, 129 (50), 15615–15622. 10.1021/ja075373f. [DOI] [PubMed] [Google Scholar]
- Mangoni M. L.; Fiocco D.; Mignogna G.; Barra D.; Simmaco M. Functional Characterisation of the 1–18 Fragment of Esculentin-1b, an Antimicrobial Peptide from Rana Esculenta. Peptides 2003, 24 (11), 1771–1777. 10.1016/j.peptides.2003.07.029. [DOI] [PubMed] [Google Scholar]
- Gopal R.; Kim Y. G.; Lee J. H.; Lee S. K.; Chae J. D.; Son B. K.; Seo C. H.; Park Y. Synergistic Effects and Antibiofilm Properties of Chimeric Peptides against Multidrug-Resistant Acinetobacter Baumannii Strains. Antimicrob. Agents Chemother. 2014, 58 (3), 1622–1629. 10.1128/AAC.02473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. Y.; Kang J. H.; Lee M. K.; Kim S. Y.; Kim Y.; Hahm K.-S. Cecropin a - Magainin 2 Hybrid Peptides Having Potent Antimicrobial Activity with Low Hemolytic Effect. IUBMB Life 1998, 44 (6), 1119–1126. 10.1080/15216549800202192. [DOI] [PubMed] [Google Scholar]
- Keun Kim H.; Gun Lee D.; Park Y.; Nam Kim H.; Hwa Choi B.; Choi C.-H.; Hahm K.-S. Antibacterial Activities of Peptides Designed as Hybrids of Antimicrobial Peptides. Biotechnol. Lett. 2002, 24 (5), 347–353. 10.1023/A:1014573005866. [DOI] [Google Scholar]
- Park K. H.; Nan Y. H.; Park Y.; Kim J. I.; Park I.-S.; Hahm K.-S.; Shin S. Y. Cell Specificity, Anti-Inflammatory Activity, and Plausible Bactericidal Mechanism of Designed Trp-Rich Model Antimicrobial Peptides. Biochim. Biophys. Acta BBA - Biomembr. 2009, 1788 (5), 1193–1203. 10.1016/j.bbamem.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Yunhwa C.Proline Hinged Amphipathic α-Helical Peptide Enhances Synergistic Antimicrobial Activity with Various Antibiotics by Perturbing Outer Membrane of Gram-Negative Bacteria. Master’s Thesis, Seoul National University, 2017. [Google Scholar]
- Moore K. S.; Wehrli S.; Roder H.; Rogers M.; Forrest J. N.; McCrimmon D.; Zasloff M. Squalamine: An Aminosterol Antibiotic from the Shark. Proc. Natl. Acad. Sci. U. S. A. 1993, 90 (4), 1354–1358. 10.1073/pnas.90.4.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmi C.; Loncle C.; Vidal N.; Letourneux Y.; Fantini J.; Maresca M.; Taïeb N.; Pagès J.-M.; Brunel J. M. Squalamine: An Appropriate Strategy against the Emergence of Multidrug Resistant Gram-Negative Bacteria?. PLoS One 2008, 3 (7), e2765. 10.1371/journal.pone.0002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne J.-P.; Brunel J.-M.; Chevalier J.; Pagès J.-M. Squalamine, an Original Chemosensitizer to Combat Antibiotic-Resistant Gram-Negative Bacteria. J. Antimicrob. Chemother. 2010, 65 (4), 799–801. 10.1093/jac/dkq031. [DOI] [PubMed] [Google Scholar]
- Kikuchi K.; Bernard E. M.; Sadownik A.; Regen S. L.; Armstrong D. Antimicrobial Activities of Squalamine Mimics. Antimicrob. Agents Chemother. 1997, 41 (7), 1433–1438. 10.1128/AAC.41.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage P. B.; Li C. Cholic Acid Derivatives: Novel Antimicrobials. Expert Opin. Investig. Drugs 2000, 9 (2), 263–272. 10.1517/13543784.9.2.263. [DOI] [PubMed] [Google Scholar]
- Li C.; Peters A. S.; Meredith E. L.; Allman G. W.; Savage P. B. Design and Synthesis of Potent Sensitizers of Gram-Negative Bacteria Based on a Cholic Acid Scaffolding. J. Am. Chem. Soc. 1998, 120 (12), 2961–2962. 10.1021/ja973881r. [DOI] [Google Scholar]
- Li C.; Lewis M. R.; Gilbert A. B.; Noel M. D.; Scoville D. H.; Allman G. W.; Savage P. B. Antimicrobial Activities of Amine- and Guanidine-Functionalized Cholic Acid Derivatives. Antimicrob. Agents Chemother. 1999, 43 (6), 1347–1349. 10.1128/AAC.43.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Budge L. P.; Driscoll C. D.; Willardson B. M.; Allman G. W.; Savage P. B. Incremental Conversion of Outer-Membrane Permeabilizers into Potent Antibiotics for Gram-Negative Bacteria. J. Am. Chem. Soc. 1999, 121 (5), 931–940. 10.1021/ja982938m. [DOI] [Google Scholar]
- Schmidt E. J.; Boswell J. S.; Walsh J. P.; Schellenberg M. M.; Winter T. W.; Li C.; Allman G. W.; Savage P. B. Activities of Cholic Acid-Derived Antimicrobial Agents against Multidrug-Resistant Bacteria. J. Antimicrob. Chemother. 2001, 47 (5), 671–674. 10.1093/jac/47.5.671. [DOI] [PubMed] [Google Scholar]
- Atiq-ur-Rehman; Li C.; Budge L. P.; Street S. E.; Savage P. B. Preparation of Amino Acid-Appended Cholic Acid Derivatives as Sensitizers of Gram-Negative Bacteria. Tetrahedron Lett. 1999, 40 (10), 1865–1868. 10.1016/S0040-4039(99)00075-1. [DOI] [Google Scholar]
- Guan Q.; Li C.; Schmidt E. J.; Boswell J. S.; Walsh J. P.; Allman G. W.; Savage P. B. Preparation and Characterization of Cholic Acid-Derived Antimicrobial Agents with Controlled Stabilities. Org. Lett. 2000, 2 (18), 2837–2840. 10.1021/ol0062704. [DOI] [PubMed] [Google Scholar]
- Savage P. B.; Li C.; Taotafa U.; Ding B.; Guan Q. Antibacterial Properties of Cationic Steroid Antibiotics. FEMS Microbiol. Lett. 2002, 217 (1), 1–7. 10.1111/j.1574-6968.2002.tb11448.x. [DOI] [PubMed] [Google Scholar]
- Ding B.; Taotofa U.; Orsak T.; Chadwell M.; Savage P. B. Synthesis and Characterization of Peptide–Cationic Steroid Antibiotic Conjugates. Org. Lett. 2004, 6 (20), 3433–3436. 10.1021/ol048845t. [DOI] [PubMed] [Google Scholar]
- Lai X.-Z.; Feng Y.; Pollard J.; Chin J. N.; Rybak M. J.; Bucki R.; Epand R. F.; Epand R. M.; Savage P. B. Ceragenins: Cholic Acid-Based Mimics of Antimicrobial Peptides. Acc. Chem. Res. 2008, 41 (10), 1233–1240. 10.1021/ar700270t. [DOI] [PubMed] [Google Scholar]
- Saha S.; Savage P. b.; Bal M. Enhancement of the Efficacy of Erythromycin in Multiple Antibiotic-Resistant Gram-Negative Bacterial Pathogens. J. Appl. Microbiol. 2008, 105 (3), 822–828. 10.1111/j.1365-2672.2008.03820.x. [DOI] [PubMed] [Google Scholar]
- Bavikar S. N.; Salunke D. B.; Hazra B. G.; Pore V. S.; Dodd R. H.; Thierry J.; Shirazi F.; Deshpande M. V.; Kadreppa S.; Chattopadhyay S. Synthesis of Chimeric Tetrapeptide-Linked Cholic Acid Derivatives: Impending Synergistic Agents. Bioorg. Med. Chem. Lett. 2008, 18 (20), 5512–5517. 10.1016/j.bmcl.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Stokes J. M.; MacNair C. R.; Ilyas B.; French S.; Côté J.-P.; Bouwman C.; Farha M. A.; Sieron A. O.; Whitfield C.; Coombes B. K.; Brown E. D. Pentamidine Sensitizes Gram-Negative Pathogens to Antibiotics and Overcomes Acquired Colistin Resistance. Nat. Microbiol. 2017, 2 (5), 1–8. 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling C. M. J.; Slingerland C. J.; Veraar S.; Lok S.; Martin N. I. Structure–Activity Studies with Bis-Amidines That Potentiate Gram-Positive Specific Antibiotics against Gram-Negative Pathogens. ACS Infect. Dis. 2021, 7 (12), 3314–3335. 10.1021/acsinfecdis.1c00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNair C. R.; Farha M. A.; Serrano-Wu M. H.; Lee K. K.; Hubbard B.; Côté J.-P.; Carfrae L. A.; Tu M. M.; Gaulin J. L.; Hunt D. K.; Hung D. T.; Brown E. D. Preclinical Development of Pentamidine Analogs Identifies a Potent and Nontoxic Antibiotic Adjuvant. ACS Infect. Dis. 2022, 8, 768. 10.1021/acsinfecdis.1c00482. [DOI] [PubMed] [Google Scholar]
- Sands M.; Kron M. A.; Brown R. B. Pentamidine: A Review. Rev. Infect. Dis. 1985, 7 (5), 625–634. 10.1093/clinids/7.5.625. [DOI] [PubMed] [Google Scholar]
- Kuryshev Y. A.; Ficker E.; Wang L.; Hawryluk P.; Dennis A. T.; Wible B. A.; Brown A. M.; Kang J.; Chen X.-L.; Sawamura K.; Reynolds W.; Rampe D. Pentamidine-Induced Long QT Syndrome and Block of HERG Trafficking. J. Pharmacol. Exp. Ther. 2005, 312 (1), 316–323. 10.1124/jpet.104.073692. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Jia Y.; Yang K.; Li R.; Xiao X.; Zhu K.; Wang Z. Metformin Restores Tetracyclines Susceptibility against Multidrug Resistant Bacteria. Adv. Sci. 2020, 7 (12), 1902227. 10.1002/advs.201902227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobucar K.; Côté J.-P.; French S.; Borrillo L.; Guo A. B. Y.; Serrano-Wu M. H.; Lee K. K.; Hubbard B.; Johnson J. W.; Gaulin J. L.; Magolan J.; Hung D. T.; Brown E. D. Chemical Screen for Vancomycin Antagonism Uncovers Probes of the Gram-Negative Outer Membrane. ACS Chem. Biol. 2021, 16 (5), 929–942. 10.1021/acschembio.1c00179. [DOI] [PubMed] [Google Scholar]
- Sharma S.; Rao R.; Reeve S. M.; Phelps G. A.; Bharatham N.; Katagihallimath N.; Ramachandran V.; Raveendran S.; Sarma M.; Nath A.; Thomas T.; Manickam D.; Nagaraj S.; Balasubramanian V.; Lee R. E.; Hameed P S.; Datta S. Azaindole Based Potentiator of Antibiotics against Gram-Negative Bacteria. ACS Infect. Dis. 2021, 7 (11), 3009–3024. 10.1021/acsinfecdis.1c00171. [DOI] [PubMed] [Google Scholar]
- Böttcher T.; Kolodkin-Gal I.; Kolter R.; Losick R.; Clardy J. Synthesis and Activity of Biomimetic Biofilm Disruptors. J. Am. Chem. Soc. 2013, 135 (8), 2927–2930. 10.1021/ja3120955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konai M. M.; Haldar J. Lysine-Based Small Molecules That Disrupt Biofilms and Kill Both Actively Growing Planktonic and Nondividing Stationary Phase Bacteria. ACS Infect. Dis. 2015, 1 (10), 469–478. 10.1021/acsinfecdis.5b00056. [DOI] [PubMed] [Google Scholar]
- Konai M. M.; Haldar J. Lysine-Based Small Molecule Sensitizes Rifampicin and Tetracycline against Multidrug-Resistant Acinetobacter Baumannii and Pseudomonas Aeruginosa. ACS Infect. Dis. 2020, 6 (1), 91–99. 10.1021/acsinfecdis.9b00221. [DOI] [PubMed] [Google Scholar]
- Yasuda K.; Ohmizo C.; Katsu T. Mode of Action of Novel Polyamines Increasing the Permeability of Bacterial Outer Membrane. Int. J. Antimicrob. Agents 2004, 24 (1), 67–71. 10.1016/j.ijantimicag.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Katsu T.; Nakagawa H.; Yasuda K. Interaction between Polyamines and Bacterial Outer Membranes as Investigated with Ion-Selective Electrodes. Antimicrob. Agents Chemother. 2002, 46 (4), 1073–1079. 10.1128/AAC.46.4.1073-1079.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishna R.; Wood S. J.; Nguyen T. B.; Miller K. A.; Suresh Kumar E. V. K.; Datta A.; David S. A. Structural Correlates of Antibacterial and Membrane-Permeabilizing Activities in Acylpolyamines. Antimicrob. Agents Chemother. 2006, 50 (3), 852–861. 10.1128/AAC.50.3.852-861.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S. A. Towards a Rational Development of Anti-Endotoxin Agents: Novel Approaches to Sequestration of Bacterial Endotoxins with Small Molecules. J. Mol. Recognit. 2001, 14 (6), 370–387. 10.1002/jmr.549. [DOI] [PubMed] [Google Scholar]
- David S. A.; Silverstein R.; Amura C. R.; Kielian T.; Morrison D. C. Lipopolyamines: Novel Antiendotoxin Compounds That Reduce Mortality in Experimental Sepsis Caused by Gram-Negative Bacteria. Antimicrob. Agents Chemother. 1999, 43 (4), 912–919. 10.1128/AAC.43.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. A.; Cadelis M. M.; Sue K.; Blanchet M.; Vidal N.; Brunel J. M.; Bourguet-Kondracki M.-L.; Copp B. R. 6-Bromoindolglyoxylamido Derivatives as Antimicrobial Agents and Antibiotic Enhancers. Bioorg. Med. Chem. 2019, 27 (10), 2090–2099. 10.1016/j.bmc.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Finlayson R.; Pearce A. N.; Page M. J.; Kaiser M.; Bourguet-Kondracki M.-L.; Harper J. L.; Webb V. L.; Copp B. R. Didemnidines A and B, Indole Spermidine Alkaloids from the New Zealand Ascidian Didemnum Sp. J. Nat. Prod. 2011, 74 (4), 888–892. 10.1021/np1008619. [DOI] [PubMed] [Google Scholar]
- Cadelis M. M.; Li S. A.; Bourguet-Kondracki M.-L.; Blanchet M.; Douafer H.; Brunel J. M.; Copp B. R. Spermine Derivatives of Indole-3-Carboxylic Acid, Indole-3-Acetic Acid and Indole-3-Acrylic Acid as Gram-Negative Antibiotic Adjuvants. ChemMedChem. 2021, 16 (3), 513–523. 10.1002/cmdc.202000359. [DOI] [PubMed] [Google Scholar]
- Helander I. M.; Nurmiaho-Lassila E.-L.; Ahvenainen R.; Rhoades J.; Roller S. Chitosan Disrupts the Barrier Properties of the Outer Membrane of Gram-Negative Bacteria. Int. J. Food Microbiol. 2001, 71 (2), 235–244. 10.1016/S0168-1605(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Helander I. M.; Latva-Kala K.; Lounatmaa K. Permeabilizing Action of Polyethyleneimine on Salmonella Typhimurium Involves Disruption of the Outer Membrane and Interactions with Lipopolysaccharide. Microbiology 1998, 144 (2), 385–390. 10.1099/00221287-144-2-385. [DOI] [PubMed] [Google Scholar]
- Lam A. K.; Panlilio H.; Pusavat J.; Wouters C. L.; Moen E. L.; Rice C. V. Overcoming Multidrug Resistance and Biofilms of Pseudomonas Aeruginosa with a Single Dual-Function Potentiator of β-Lactams. ACS Infect. Dis. 2020, 6 (5), 1085–1097. 10.1021/acsinfecdis.9b00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemaiswarya S.; Doble M. Synergistic Interaction of Eugenol with Antibiotics against Gram Negative Bacteria. Phytomedicine 2009, 16 (11), 997–1005. 10.1016/j.phymed.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Aelenei P.; Rimbu C. M.; Guguianu E.; Dimitriu G.; Aprotosoaie A. C.; Brebu M.; Horhogea C. E.; Miron A. Coriander Essential Oil and Linalool - Interactions with Antibiotics against Gram-Positive and Gram-Negative Bacteria. Lett. Appl. Microbiol. 2019, 68 (2), 156–164. 10.1111/lam.13100. [DOI] [PubMed] [Google Scholar]
- Farag R. S.; Daw Z. Y.; Hewedi F. M.; El-Baroty G. S. A. Antimicrobial Activity of Some Egyptian Spice Essential Oils. J. Food Prot. 1989, 52 (9), 665–667. 10.4315/0362-028X-52.9.665. [DOI] [PubMed] [Google Scholar]
- Bauer K.; Garbe D.; Surburg H.. Common Fragrance and Flavor Materials: Preparation, Properties and Uses; John Wiley & Sons, 2008. [Google Scholar]
- Burt S. Essential Oils: Their Antibacterial Properties and Potential Applications in Foods—a Review. Int. J. Food Microbiol. 2004, 94 (3), 223–253. 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Wijesekera R. O. B.; Chichester C. O. The Chemistry and Technology of Cinnamon. C R C Crit. Rev. Food Sci. Nutr. 1978, 10 (1), 1–30. 10.1080/10408397809527243. [DOI] [PubMed] [Google Scholar]
- ter Heide R. Qualitative Analysis of the Essential Oil of Cassia (Cinnamomum Cassia Blume). J. Agric. Food Chem. 1972, 20 (4), 747–751. 10.1021/jf60182a023. [DOI] [Google Scholar]
- Langeveld W. T.; Veldhuizen E. J. A.; Burt S. A. Synergy between Essential Oil Components and Antibiotics: A Review. Crit. Rev. Microbiol. 2014, 40 (1), 76–94. 10.3109/1040841X.2013.763219. [DOI] [PubMed] [Google Scholar]
- Aelenei P.; Miron A.; Trifan A.; Bujor A.; Gille E.; Aprotosoaie A. C. Essential Oils and Their Components as Modulators of Antibiotic Activity against Gram-Negative Bacteria. Medicines 2016, 3 (3), 19. 10.3390/medicines3030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan K.; Holley R. A. Use of Natural Antimicrobials to Increase Antibiotic Susceptibility of Drug Resistant Bacteria. Int. J. Food Microbiol. 2010, 140 (2), 164–168. 10.1016/j.ijfoodmicro.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Hemaiswarya S.; Doble M. Synergistic Interaction of Phenylpropanoids with Antibiotics against Bacteria. J. Med. Microbiol. 2010, 59, 1469–1476. 10.1099/jmm.0.022426-0. [DOI] [PubMed] [Google Scholar]
- Helander I. M.; Alakomi H.-L.; Latva-Kala K.; Mattila-Sandholm T.; Pol I.; Smid E. J.; Gorris L. G. M.; von Wright A. Characterization of the Action of Selected Essential Oil Components on Gram-Negative Bacteria. J. Agric. Food Chem. 1998, 46 (9), 3590–3595. 10.1021/jf980154m. [DOI] [Google Scholar]
- Ruwizhi N.; Aderibigbe B. A. Cinnamic Acid Derivatives and Their Biological Efficacy. Int. J. Mol. Sci. 2020, 21 (16), 5712. 10.3390/ijms21165712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney K.; Sovadinova I.; Lopez A. I.; Urban M.; Ridgway Z.; Caputo G. A.; Kuroda K. Poly(Ethylene Imine)s as Antimicrobial Agents with Selective Activity. Macromol. Biosci. 2012, 12 (9), 1279–1289. 10.1002/mabi.201200052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes K. A. O.; Silva L. S.; Santos E. C.; Pinheiro A. S.; Teixeira D. E.; Peruchetti D. B.; Silva-Aguiar R. P.; Wendt C. H. C.; Miranda K. R.; Coelho-de-Souza A. N.; Leal-Cardoso J. H.; Caruso-Neves C.; Pinheiro A. A. S. Eugenol Disrupts Plasmodium Falciparum Intracellular Development during the Erythrocytic Cycle and Protects against Cerebral Malaria. Biochim. Biophys. Acta BBA - Gen. Subj. 2021, 1865 (3), 129813. 10.1016/j.bbagen.2020.129813. [DOI] [PubMed] [Google Scholar]
- Togashi N.; Hamashima H.; Shiraishi A.; Inoue Y.; Takano A. Antibacterial Activities Against Staphylococcus Aureus of Terpene Alcohols With Aliphatic Carbon Chains. J. Essent. Oil Res. 2010, 22 (3), 263–269. 10.1080/10412905.2010.9700321. [DOI] [Google Scholar]
- Ahmad A.; Khan A.; Akhtar F.; Yousuf S.; Xess I.; Khan L. A.; Manzoor N. Fungicidal Activity of Thymol and Carvacrol by Disrupting Ergosterol Biosynthesis and Membrane Integrity against Candida. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30 (1), 41–50. 10.1007/s10096-010-1050-8. [DOI] [PubMed] [Google Scholar]
- Theurer M.; Shaik N.; Lang F. Stimulation of Suicidal Erythrocyte Death by Trans-Cinnamaldehyde. Phytomedicine 2013, 20 (12), 1119–1123. 10.1016/j.phymed.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Hemaiswarya S.; Doble M. Combination of Phenylpropanoids with 5-Fluorouracil as Anti-Cancer Agents against Human Cervical Cancer (HeLa) Cell Line. Phytomedicine 2013, 20 (2), 151–158. 10.1016/j.phymed.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Farrell W.; Wilks M.; Drasar F. A. The Action of Trimethoprim and Rifampicin in Combination against Gram-Negative Rods Resistant to Gentamicin. J. Antimicrob. Chemother. 1977, 3 (5), 459–462. 10.1093/jac/3.5.459. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Bao W.; Guo N.; Chen H.; Cheng W.; Jin K.; Shen F.; Xu J.; Zhang Q.; Wang C.; An Y.; Zhang K.; Wang F.; Yu L. Antimicrobial Activity of the Imipenem/Rifampicin Combination against Clinical Isolates of Acinetobacter Baumannii Grown in Planktonic and Biofilm Cultures. World J. Microbiol. Biotechnol. 2014, 30 (12), 3015–3025. 10.1007/s11274-014-1728-7. [DOI] [PubMed] [Google Scholar]
- Domalaon R.; Yang X.; Lyu Y.; Zhanel G. G.; Schweizer F. Polymyxin B3–Tobramycin Hybrids with Pseudomonas Aeruginosa-Selective Antibacterial Activity and Strong Potentiation of Rifampicin, Minocycline, and Vancomycin. ACS Infect. Dis. 2017, 3 (12), 941–954. 10.1021/acsinfecdis.7b00145. [DOI] [PubMed] [Google Scholar]
- Wood T. M.; Slingerland C. J.; Martin N. I. A Convenient Chemoenzymatic Preparation of Chimeric Macrocyclic Peptide Antibiotics with Potent Activity against Gram-Negative Pathogens. J. Med. Chem. 2021, 64 (15), 10890–10899. 10.1021/acs.jmedchem.1c00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Groesen E.; Slingerland C. J.; Innocenti P.; Mihajlovic M.; Masereeuw R.; Martin N. I. Vancomyxins: Vancomycin-Polymyxin Nonapeptide Conjugates That Retain Anti-Gram-Positive Activity with Enhanced Potency against Gram-Negative Strains. ACS Infect. Dis. 2021, 7 (9), 2746–2754. 10.1021/acsinfecdis.1c00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog I. M.; Green K. D.; Berkov-Zrihen Y.; Feldman M.; Vidavski R. R.; Eldar-Boock A.; Satchi-Fainaro R.; Eldar A.; Garneau-Tsodikova S.; Fridman M. 6″-Thioether Tobramycin Analogues: Towards Selective Targeting of Bacterial Membranes. Angew. Chem. 2012, 124 (23), 5750–5754. 10.1002/ange.201200761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouberai M.; El Garch F.; Bussiere A.; Riou M.; Alsteens D.; Lins L.; Baussanne I.; Dufrêne Y. F.; Brasseur R.; Decout J.-L.; Mingeot-Leclercq M.-P. The Pseudomonas Aeruginosa Membranes: A Target for a New Amphiphilic Aminoglycoside Derivative?. Biochim. Biophys. Acta BBA - Biomembr. 2011, 1808 (6), 1716–1727. 10.1016/j.bbamem.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Guchhait G.; Altieri A.; Gorityala B.; Yang X.; Findlay B.; Zhanel G. G.; Mookherjee N.; Schweizer F. Amphiphilic Tobramycins with Immunomodulatory Properties. Angew. Chem., Int. Ed. 2015, 54 (21), 6278–6282. 10.1002/anie.201500598. [DOI] [PubMed] [Google Scholar]
- Loh B.; Grant C.; Hancock R. E. Use of the Fluorescent Probe 1-N-Phenylnaphthylamine to Study the Interactions of Aminoglycoside Antibiotics with the Outer Membrane of Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 1984, 26 (4), 546–551. 10.1128/AAC.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulitta J. B.; Ly N. S.; Landersdorfer C. B.; Wanigaratne N. A.; Velkov T.; Yadav R.; Oliver A.; Martin L.; Shin B. S.; Forrest A.; Tsuji B. T. Two Mechanisms of Killing of Pseudomonas Aeruginosa by Tobramycin Assessed at Multiple Inocula via Mechanism-Based Modeling. Antimicrob. Agents Chemother. 2015, 59 (4), 2315–2327. 10.1128/AAC.04099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorityala B. K.; Guchhait G.; Goswami S.; Fernando D. M.; Kumar A.; Zhanel G. G.; Schweizer F. Hybrid Antibiotic Overcomes Resistance in P. Aeruginosa by Enhancing Outer Membrane Penetration and Reducing Efflux. J. Med. Chem. 2016, 59 (18), 8441–8455. 10.1021/acs.jmedchem.6b00867. [DOI] [PubMed] [Google Scholar]
- Gorityala B. K.; Guchhait G.; Fernando D. M.; Deo S.; McKenna S. A.; Zhanel G. G.; Kumar A.; Schweizer F. Adjuvants Based on Hybrid Antibiotics Overcome Resistance in Pseudomonas Aeruginosa and Enhance Fluoroquinolone Efficacy. Angew. Chem., Int. Ed. 2016, 55 (2), 555–559. 10.1002/anie.201508330. [DOI] [PubMed] [Google Scholar]
- Idowu T.; Arthur G.; Zhanel G. G.; Schweizer F. Heterodimeric Rifampicin–Tobramycin Conjugates Break Intrinsic Resistance of Pseudomonas Aeruginosa to Doxycycline and Chloramphenicol in Vitro and in a Galleria Mellonella in Vivo Model. Eur. J. Med. Chem. 2019, 174, 16–32. 10.1016/j.ejmech.2019.04.034. [DOI] [PubMed] [Google Scholar]
- Lyu Y.; Yang X.; Goswami S.; Gorityala B. K.; Idowu T.; Domalaon R.; Zhanel G. G.; Shan A.; Schweizer F. Amphiphilic Tobramycin–Lysine Conjugates Sensitize Multidrug Resistant Gram-Negative Bacteria to Rifampicin and Minocycline. J. Med. Chem. 2017, 60 (9), 3684–3702. 10.1021/acs.jmedchem.6b01742. [DOI] [PubMed] [Google Scholar]
- Yang X.; Goswami S.; Gorityala B. K.; Domalaon R.; Lyu Y.; Kumar A.; Zhanel G. G.; Schweizer F. A Tobramycin Vector Enhances Synergy and Efficacy of Efflux Pump Inhibitors against Multidrug-Resistant Gram-Negative Bacteria. J. Med. Chem. 2017, 60 (9), 3913–3932. 10.1021/acs.jmedchem.7b00156. [DOI] [PubMed] [Google Scholar]
- Yang X.; Domalaon R.; Lyu Y.; Zhanel G. G.; Schweizer F. Tobramycin-Linked Efflux Pump Inhibitor Conjugates Synergize Fluoroquinolones, Rifampicin and Fosfomycin against Multidrug-Resistant Pseudomonas Aeruginosa. J. Clin. Med. 2018, 7 (7), 158. 10.3390/jcm7070158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idowu T.; Ammeter D.; Rossong H.; Zhanel G. G.; Schweizer F. Homodimeric Tobramycin Adjuvant Repurposes Novobiocin as an Effective Antibacterial Agent against Gram-Negative Bacteria. J. Med. Chem. 2019, 62 (20), 9103–9115. 10.1021/acs.jmedchem.9b00876. [DOI] [PubMed] [Google Scholar]
- Idowu T.; Ammeter D.; Arthur G.; Zhanel G. G.; Schweizer F. Potentiation of β-Lactam Antibiotics and β-Lactam/β-Lactamase Inhibitor Combinations against MDR and XDR Pseudomonas Aeruginosa Using Non-Ribosomal Tobramycin–Cyclam Conjugates. J. Antimicrob. Chemother. 2019, 74 (9), 2640–2648. 10.1093/jac/dkz228. [DOI] [PubMed] [Google Scholar]
- Yang X.; Ammeter D.; Idowu T.; Domalaon R.; Brizuela M.; Okunnu O.; Bi L.; Guerrero Y. A.; Zhanel G. G.; Kumar A.; Schweizer F. Amphiphilic Nebramine-Based Hybrids Rescue Legacy Antibiotics from Intrinsic Resistance in Multidrug-Resistant Gram-Negative Bacilli. Eur. J. Med. Chem. 2019, 175, 187–200. 10.1016/j.ejmech.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Ammeter D.; Idowu T.; Zhanel G. G.; Schweizer F. Development of a Nebramine-Cyclam Conjugate as an Antibacterial Adjuvant to Potentiate β-Lactam Antibiotics against Multidrug-Resistant P. Aeruginosa. J. Antibiot. (Tokyo) 2019, 72 (11), 816–826. 10.1038/s41429-019-0221-9. [DOI] [PubMed] [Google Scholar]
- Berry L.; Domalaon R.; Brizuela M.; Zhanel G. G.; Schweizer F. Polybasic Peptide–Levofloxacin Conjugates Potentiate Fluoroquinolones and Other Classes of Antibiotics against Multidrug-Resistant Gram-Negative Bacteria. MedChemComm 2019, 10 (4), 517–527. 10.1039/C9MD00051H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R.; Idowu T.; Zhanel G. G.; Schweizer F. Antibiotic Hybrids: The Next Generation of Agents and Adjuvants against Gram-Negative Pathogens?. Clin. Microbiol. Rev. 2018, 31 (2), 17. 10.1128/CMR.00077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh C.; Manjunath G. B.; Akkapeddi P.; Yarlagadda V.; Hoque J.; Uppu D. S. S. M.; Konai M. M.; Haldar J. Small Molecular Antibacterial Peptoid Mimics: The Simpler the Better!. J. Med. Chem. 2014, 57 (4), 1428–1436. 10.1021/jm401680a. [DOI] [PubMed] [Google Scholar]
- Lyu Y.; Domalaon R.; Yang X.; Schweizer F. Amphiphilic Lysine Conjugated to Tobramycin Synergizes Legacy Antibiotics against Wild-Type and Multidrug-Resistant Pseudomonas Aeruginosa. Pept. Sci. 2019, 111 (1), e23091. 10.1002/bip.23091. [DOI] [PubMed] [Google Scholar]
- Bohnert J. A.; Kern W. V. Selected Arylpiperazines Are Capable of Reversing Multidrug Resistance in Escherichia Coli Overexpressing RND Efflux Pumps. Antimicrob. Agents Chemother. 2005, 49 (2), 849–852. 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaatz G. W.; Moudgal V. V.; Seo S. M.; Hansen J. B.; Kristiansen J. E. Phenylpiperidine Selective Serotonin Reuptake Inhibitors Interfere with Multidrug Efflux Pump Activity in Staphylococcus Aureus. Int. J. Antimicrob. Agents 2003, 22 (3), 254–261. 10.1016/S0924-8579(03)00220-6. [DOI] [PubMed] [Google Scholar]
- Kriengkauykiat J.; Porter E.; Lomovskaya O.; Wong-Beringer A. Use of an Efflux Pump Inhibitor To Determine the Prevalence of Efflux Pump-Mediated Fluoroquinolone Resistance and Multidrug Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2005, 49 (2), 565–570. 10.1128/AAC.49.2.565-570.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conejo M. C.; García I.; Martínez-Martínez L.; Picabea L.; Pascual Á. Zinc Eluted from Siliconized Latex Urinary Catheters Decreases OprD Expression, Causing Carbapenem Resistance in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2003, 47 (7), 2313–2315. 10.1128/AAC.47.7.2313-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron K.; Caille O.; Rossier C.; Van Delden C.; Dumas J.-L.; Köhler T. CzcR-CzcS, a Two-Component System Involved in Heavy Metal and Carbapenem Resistance in Pseudomonas Aeruginosa. J. Biol. Chem. 2004, 279 (10), 8761–8768. 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- Caille O.; Rossier C.; Perron K. A Copper-Activated Two-Component System Interacts with Zinc and Imipenem Resistance in Pseudomonas Aeruginosa. J. Bacteriol. 2007, 189 (13), 4561–4568. 10.1128/JB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L.; Kempf J.; Briée F.; Swain J.; Mingeot-Leclercq M.-P.; Décout J.-L. Broad-Spectrum Antibacterial Amphiphilic Aminoglycosides: A New Focus on the Structure of the Lipophilic Groups Extends the Series of Active Dialkyl Neamines. Eur. J. Med. Chem. 2018, 157, 1512–1525. 10.1016/j.ejmech.2018.08.022. [DOI] [PubMed] [Google Scholar]
- Zimmermann L.; Das I.; Désiré J.; Sautrey G.; Barros R. S V.; El Khoury M.; Mingeot-Leclercq M.-P.; Décout J.-L. New Broad-Spectrum Antibacterial Amphiphilic Aminoglycosides Active against Resistant Bacteria: From Neamine Derivatives to Smaller Neosamine Analogues. J. Med. Chem. 2016, 59 (20), 9350–9369. 10.1021/acs.jmedchem.6b00818. [DOI] [PubMed] [Google Scholar]
- Fourmy D.; Recht M. I.; Puglisi J. D. Binding of Neomycin-Class Aminoglycoside Antibiotics to the A-Site of 16 s RRNA11Edited by I. Tinoco. J. Mol. Biol. 1998, 277 (2), 347–362. 10.1006/jmbi.1997.1552. [DOI] [PubMed] [Google Scholar]
- Fourmy D.; Recht M. I.; Blanchard S. C.; Puglisi J. D. Structure of the A Site of Escherichia Coli 16S Ribosomal RNA Complexed with an Aminoglycoside Antibiotic. Science 1996, 274 (5291), 1367–1371. 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Agnelli F.; Sucheck S. J.; Marby K. A.; Rabuka D.; Yao S.-L.; Sears P. S.; Liang F.-S.; Wong C.-H. Dimeric Aminoglycosides as Antibiotics. Angew. Chem., Int. Ed. 2004, 43 (12), 1562–1566. 10.1002/anie.200353225. [DOI] [PubMed] [Google Scholar]
- Vicens Q.; Westhof E. Crystal Structure of a Complex between the Aminoglycoside Tobramycin and an Oligonucleotide Containing the Ribosomal Decoding A Site. Chem. Biol. 2002, 9 (6), 747–755. 10.1016/S1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- Lynch S. R.; Gonzalez R. L.; Puglisi J. D. Comparison of X-Ray Crystal Structure of the 30S Subunit-Antibiotic Complex with NMR Structure of Decoding Site Oligonucleotide-Paromomycin Complex. Structure 2003, 11 (1), 43–53. 10.1016/S0969-2126(02)00934-6. [DOI] [PubMed] [Google Scholar]
- Russell A. D. Effect of Magnesium Ions and Ethylenediamine Tetra-Acetic Acid on the Activity of Vancomycin against Escherichia Coli and Staphylococcus Aureus. J. Appl. Bacteriol. 1967, 30 (2), 395–401. 10.1111/j.1365-2672.1967.tb00314.x. [DOI] [PubMed] [Google Scholar]
- Russell A. D.Chapter 3G - Ethylenediaminetetra-Acetic Acid. In Inhibition and Destruction of the Microbial Cell; Hugo W. B., Ed.; Academic Press, 1971; pp 209–224. 10.1016/B978-0-12-361150-5.50013-4. [DOI] [Google Scholar]
- Leive L. The Barrier Function of the Gram-Negative Envelope. Ann. N.Y. Acad. Sci. 1974, 235 (1), 109–129. 10.1111/j.1749-6632.1974.tb43261.x. [DOI] [PubMed] [Google Scholar]
- Mosteller R. D.; Yanofsky C. Transcription of the Tryptophan Operon in Escherichia Coli: Rifampicin as an Inhibitor of Initiation. J. Mol. Biol. 1970, 48 (3), 525–531. 10.1016/0022-2836(70)90064-1. [DOI] [PubMed] [Google Scholar]
- Hancock R. E.; Wong P. G. Compounds Which Increase the Permeability of the Pseudomonas Aeruginosa Outer Membrane. Antimicrob. Agents Chemother. 1984, 26 (1), 48–52. 10.1128/AAC.26.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakomi H.-L.; Paananen A.; Suihko M.-L.; Helander I. M.; Saarela M. Weakening Effect of Cell Permeabilizers on Gram-Negative Bacteria Causing Biodeterioration. Appl. Environ. Microbiol. 2006, 72 (7), 4695–4703. 10.1128/AEM.00142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leive L. Release of Lipopolysaccharide by EDTA Treatment of E., Coli. Biochem. Biophys. Res. Commun. 1965, 21 (4), 290–296. 10.1016/0006-291X(65)90191-9. [DOI] [PubMed] [Google Scholar]
- Scudamore R. A.; Beveridge T. J.; Goldner M. Outer-Membrane Penetration Barriers as Components of Intrinsic Resistance to Beta-Lactam and Other Antibiotics in Escherichia Coli K-12. Antimicrob. Agents Chemother. 1979, 15, 182. 10.1128/AAC.15.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M.; Jaakkola J. Sodium Hexametaphosphate Sensitizes Pseudomonas Aeruginosa, Several Other Species of Pseudomonas, and Escherichia Coli to Hydrophobic Drugs. Antimicrob. Agents Chemother. 1989, 33 (10), 1741–1747. 10.1128/AAC.33.10.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres H. M.; Furr J. R.; Russell A. D. Effect of Permeabilizers on Antibiotic Sensitivity of Pseudomonas Aeruginosa. Lett. Appl. Microbiol. 1999, 28 (1), 13–16. 10.1046/j.1365-2672.1999.00486.x. [DOI] [PubMed] [Google Scholar]
- Eckburg P. B.; Lister T.; Walpole S.; Keutzer T.; Utley L.; Tomayko J.; Kopp E.; Farinola N.; Coleman S. Safety, Tolerability, Pharmacokinetics, and Drug Interaction Potential of SPR741, an Intravenous Potentiator, after Single and Multiple Ascending Doses and When Combined with β-Lactam Antibiotics in Healthy Subjects. Antimicrob. Agents Chemother. 2019, 63 (9), e00892–19. 10.1128/AAC.00892-19. [DOI] [PMC free article] [PubMed] [Google Scholar]