Abstract
Mycobacterium tuberculosis possesses a homologue of glnE, potentially encoding a regulator of glutamine synthetase activity. We attempted to construct glnE-disrupted mutants using a two-step strategy, whereby a single-crossover strain was first isolated, followed by sacB counterselection to isolate the double-crossover strain. Of 192 sucrose-resistant colonies tested, none were mutants, although the wild-type double crossover could be easily isolated. When a second copy of the wild-type glnE was integrated into the chromosome, we could isolate both wild-type and mutant double-crossover strains. Thus, the chromosomal gene could only be replaced with a disrupted copy when another functional copy of the gene was provided, demonstrating that this gene is essential under the conditions tested.
The creation of defined mutants of Mycobacterium tuberculosis is key to an understanding of individual gene function. We have developed a reliable and efficient method for generating such mutant strains by allelic replacement (homologous recombination) based on a two-step strategy (8, 12, 13).
During our initial work, we focused on genes involved in amino acid biosynthesis, one of which was glnE. Initially glnE was selected as a target for inactivation since we expected it to be a nonessential gene; there were not likely to be any polar effects resulting from inactivation, and there were suitable restriction enzyme sites for cloning. GlnE is a regulatory protein which is involved in the regulation of glutamine synthetase activity (see reviews in references 10 and 16). Glutamine synthetase, a key enzyme in nitrogen metabolism, is responsible for the incorporation of ammonia into glutamate to make glutamine at low ammonia concentrations.
Enteric bacteria only possess a type I glutamine synthetase (GS-I), which is subject to nitrogen regulation by GlnE and feedback inhibition by several products, including glutamine (reviewed in reference 10). GS-I activity is controlled through adenylylation and deadenylylation. When ammonia levels rise, this energy-intensive reaction is no longer needed to conserve nitrogen levels, and GlnE adenylylates GS-I at a conserved tyrosine residue near the C-terminal end. Streptomyces coelicolor A3(2) possesses two glutamine synthetases, GS-I and a heat-labile type II (GS-II). The GlnE adenylyl transferase of S. coelicolor has been shown to adenylyate GS-I (3). GS-II does not possess the conserved tyrosine and is not regulated by GlnE.
M. tuberculosis possesses four glutamine synthetase homologues: glnA1 and glnA2 are found in the same region of the chromosome as glnE (Fig. 1A). glnA1 encodes a GS-I and has the three motifs associated with glutamine synthetase, including the conserved tyrosine residue (15), whereas glnA2 encodes a probable GS-II. GlnA3 and GlnA4 are probable GS-I and GS-II enzymes, respectively, but the motifs are less conserved. Only one copy of glnE is found within the H37Rv genome (2).
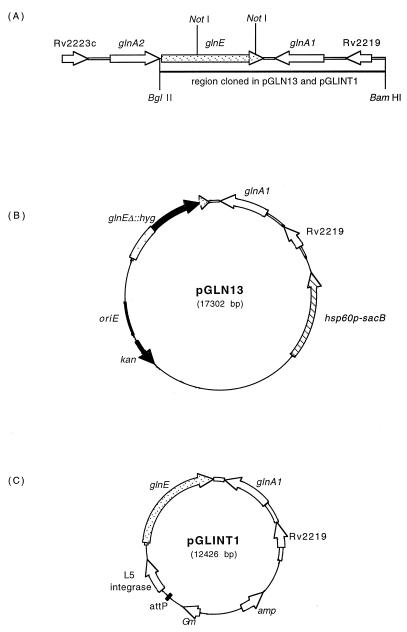
FIG. 1.
Plasmid constructs. (A) Arrangement of genes in the M. tuberculosis chromosome, showing the 6.5-kb restriction fragment used in the construction of pGLN13 and pGLINT1. (B) pGLN13 delivery vector, showing the M. tuberculosis fragment used to construct the single-crossover strain. The internal 1.7-kb NotI fragment of glnE was replaced by the hyg gene. (C) pGLINT1 integrating vector, showing the M. tuberculosis fragment cloned into the integrating vector.
Our previous work provided preliminary evidence that glnE is essential, as we were unable to replace the wild-type (wt) gene with a disrupted copy (12). The identification of essential genes can lead to the discovery of new drug targets, since attacking these gene products may kill the organism. We therefore extended our study of glnE to determine whether the gene was essential. Essential genes can only be disrupted if a second functional copy of the gene is provided elsewhere within the cell. In this paper we describe experiments to disrupt the glnE chromosomal allele in both the presence and absence of a second copy integrated into the chromosome.
MATERIALS AND METHODS
Strains and plasmids.
The M. tuberculosis strains used in this study are shown in Table 1. M. tuberculosis was grown in Middlebrook 7H9 liquid containing 10% OADC (Becton Dickinson) and 0.05% (wt/vol) Tween 80 or on solid Middlebrook 7H10 agar containing 10% (vol/vol) OADC. Kanamycin was used at 20 μg/ml, hygromycin at 100 μg/ml, and gentamicin at 10 μg/ml where appropriate.
TABLE 1.
M. tuberculosis strains
| Strain | Description | Relevant genotype |
|---|---|---|
| H37Rv | Laboratory strain (ATCC 25618) | glnE |
| GLUE1 | Single-crossover strain | glnE kan hsp60p-sacB glnEΔ::hyg |
| GLUE2 | Double-crossover strain (wt) | glnEa |
| GLUE3 | Single-crossover strain plus integrating vector | glnE kan hsp60p-sac glnEΔ::hyg glnEintb aacC1 |
| GLUE4 | Double-crossover strain (wt) plus integrating vector | glnE glnEint aacC1 |
| GLUE5 | Double-crossover strain (mutant) plus integrating vector | glnEΔ::hyg glnEint aacC1 |
Same as H37Rv but derived from GLUE1 single-crossover strain.
glnEintis the wt glnE allele integrated into the chromosome using the integrating vector.
Construction of vectors.
The glnEΔ::hyg disrupted allele was excised from plasmid pGLN2 (12) and cloned into p2NIL (13) to make pGLN11. This allele has a 1.7-kb internal fragment of the gene replaced with the hyg gene. The marker gene hsp60p-sacB from pGOAL13 (13) was then cloned into the unique PacI site of pGLN11 to make the final suicide delivery vector pGLN13 (Fig. 1B). The wild-type (wt) glnE allele was cloned into pGEM3Zf(+) as a 6.5-kb BamHI-BglII fragment to give pGLN1. The HindIII fragment from pUC-Gm-INT (9) carrying the L5 integrase and attachment site (attP) was then cloned into pGLN1 to make the integrating vector pGLINT1 (Fig. 1C).
Isolation of single-crossover strains.
The nonreplicating vector pGLN13 (Fig. 1B) was treated with UV light in order to stimulate homologous recombination and electroporated into M. tuberculosis (12). Single-crossover strains were selected using hygromycin and kanamycin.
Isolation of double-crossover strains from a single-crossover strain.
One single-crossover strain (GLUE1) was streaked out onto medium without antibiotics and incubated for 2 weeks at 37°C. A loopful of cells was resuspended in liquid medium by vortexing with 1-mm glass beads. Serial dilutions were plated onto 2% sucrose plates plus antibiotics (Table 2). Plates were incubated at 37°C for 4 to 6 weeks. Sucrose-resistant colonies were then patch tested for kanamycin resistance.
TABLE 2.
Frequency of allelic replacementa
| Strain | Antibiotic | No. of strains
|
|||
|---|---|---|---|---|---|
| Tested | Single crossoverb | Wt double crossover | Mutant double crossover | ||
| GLUE1 | Hygromycin | 112 | 112 | NAc | 0 |
| None | 80 | 20 | 60 | 0 | |
| GLUE3 | Gentamicin | 40 | 3 | 33 | 4 |
| None | 40 | 7 | 29 | 4 | |
Strains were plated onto 2% sucrose and the indicated antibiotics. Sucrose-resistant colonies were then tested for hygromycin and kanamycin resistance.
Spontaneous sucrose-resistant mutants.
NA, not applicable, since the wt double-crossover strain would not be isolated in the presence of hygromycin.
Construction of a merodiploid strain and isolation of double-crossover strains.
GLUE1 (single-crossover strain) was electroporated with the integrating vector pGLINT1. Transformants carrying the vector were selected on gentamicin, kanamycin, and hygromycin.
An individual colony was picked for further manipulation (GLUE3). Isolation of double-crossover strains was carried out essentially as for the single-crossover strain.
RESULTS
Construction of a single-crossover strain (GLUE1).
Initial attempts to create glnE double-crossover strains were unsuccessful, although we generated a large number (several hundred) of single-crossover strains (12). This provided preliminary evidence that the gene is essential. Since double-crossover strains were not isolated using a one-step strategy, we changed to a two-step strategy (13) and tried to isolate double-crossover strains from a single-crossover strain (Fig. 2). The use of a two-step strategy enables us to determine whether a gene is essential, since single-crossover strains will be viable, but in the second step only double-crossover strains carrying the wt gene and not those carrying the disrupted allele will be found.
FIG. 2.
Construction of single-crossover and double-crossover strains of wt M. tuberculosis. (A) The nonreplicating delivery vector pGLN13 was transformed into M. tuberculosis. A single recombination event gives rise to one of two possible single-crossover strains (B). (C) The second crossover can result in either restoration of the wt gene or a mutant double-crossover strain.
pGLN13 (Fig. 1B) was electroporated into M. tuberculosis, and single-crossover strains were obtained. Using approximately 2 μg of DNA, 150 single-crossover strains were obtained. The delivery vector also contained kan and hsp60p-sacB, so these markers were present in the single-crossover strain (Fig. 2B). One single-crossover strain was selected using kanamycin and hygromycin and used in the second step to try to isolate double-crossover strains. Southern blotting confirmed that it had the single-crossover genotype 2 (Fig. 2B). This strain was streaked onto a fresh agar plate (no antibiotics) to allow the second crossover to occur.
Second-step selection for double-crossover strains from GLUE1.
Since the single-crossover strain contained the sacB gene, it should be sensitive to sucrose. Therefore, in the second step, we selected against single-crossover strains by plating onto sucrose (Table 2). In all cases a reduction of approximately 104 CFU was seen on sucrose plates. Hygromycin-resistant, kanamycin-resistant strains were scored as spontaneous sucrose-resistant, single-crossover strains; hygromycin-resistant, kanamycin-sensitive strains as mutant double-crossover strains; and hygromycin-sensitive, kanamycin-sensitive strains as wt double-crossover strains. Southern analysis of several representative colonies confirmed the expected genotypes.
For plates containing hygromycin, only double-crossover strains carrying the disrupted allele should be isolated (glnEΔ::hyg), as wt strains will be hygromycin sensitive. A total of 112 colonies were tested, and all were spontaneous sucrose-resistant, single-crossover strains. On plates without hygromycin, both the mutant and wt double-crossover strains should be viable. Of 80 colonies tested, 60 were wt double-crossover strains, with the other 20 being spontaneous sucrose-resistant single-crossover strains. Thus, once again, no mutant double-crossover strains were isolated. This confirmed that the second crossover was occurring, but we were not isolating mutants on the medium used.
Construction of a merodiploid strain and second-step selection for double-crossover strains.
In order to confirm essentiality, we constructed a merodiploid strain carrying a second wt copy of the gene in a different region of the chromosome (GLUE3) (Fig. 3). An L5-based integrating vector carrying an intact copy of glnE (pGLINT1; Fig. 1C) was electroporated into the single-crossover strain GLUE1 (Fig. 2B). Gentamicin-resistant colonies were isolated, and the presence of the integrating plasmid was confirmed by Southern analysis.
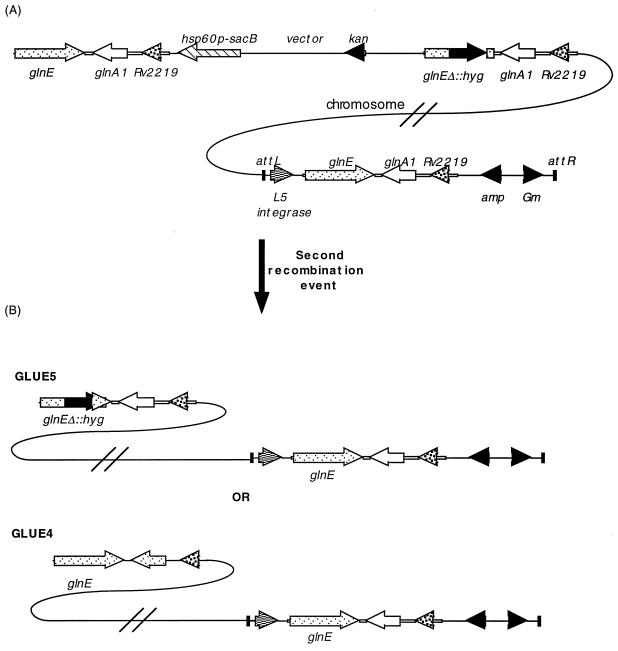
FIG. 3.
Construction of a merodiploid strain and isolation of double-crossover strains. (A) The integrating vector was transformed into the single-crossover strain (single crossover 2 in Fig. 2B), providing an extra copy of the glnE gene elsewhere on the chromosome. A second crossover event results in either of two double-crossover strains (B) being isolated. Both double-crossover strains will still retain a functional glnE gene in the integrated vector.
We then tried to isolate double crossovers from this strain as before (Table 2). A loopful of GLUE3 cells was resuspended, diluted, and plated onto sucrose plates. No hygromycin was included so that both wt and mutant strains could be isolated. Of 40 sucrose-resistant colonies tested from the gentamicin plate, 33 were wt double-crossover strains and 4 were mutant double-crossover strains. We repeated the experiment without gentamicin and obtained similar results, obtaining 29 of 40 wt double-crossover strains and 4 of 40 mutant double-crossover strains. Mutants were analyzed by Southern blotting, showing that in the mutant double-crossover strains, the wt glnE allele had indeed been replaced, while the integrated copy was retained. These results demonstrated that homologous recombination was occurring at this locus to obtain both double-crossover strains, although the frequency of recombination which gave rise to strains carrying the disrupted allele was lower. In total, 0 of 192 (<0.5%) mutant double-crossover strains were isolated from GLUE1 and 8 of 80 (10%) mutant double-crossover strains were isolated from GLUE3. Our inability to obtain the disrupted allele in the absence of a second functional copy shows that this gene is essential in the growth conditions that we used.
DISCUSSION
Previous results suggested that glnE is an essential gene since we could not obtain gene replacement using a one-step strategy, where double-crossover strains are selected immediately after electroporation. Using a two-step strategy in this work, we were still unable to obtain the glnEΔ::hyg disruption, providing more evidence that the gene is essential. Polar effects of a glnEΔ::hyg mutation are unlikely, as the downstream gene, glnA1, is transcribed in the opposite direction. The construction of a merodiploid strain containing an integrated copy of glnE allowed us to provide formal proof of essentiality under the conditions employed; i.e., homologous recombination leading to gene disruption could only occur in the presence of a functional copy of the gene. The integrated copy of glnE did not take any part in recombination in the strains we analyzed; this is not surprising, since the integrated copy is found at a large distance from the normal chromosomal location (over 200 genes away from the integration site of L5) and any recombination would result in a large, presumably lethal deletion.
It is important to begin to develop guidelines for defining essentiality in M. tuberculosis, as targeted mutagenesis is now possible. This is much more difficult to demonstrate than in fast-growing bacteria. One possibility for failing to isolate a mutant is that, although viable, the growth rate is slower. M. tuberculosis normally takes 2 to 3 weeks to form visible colonies in the presence of sucrose. We extended this incubation time to 6 weeks to allow the identification of mutants with growth rates two to three times slower than wt. Beyond this, other issues, such as dehydration of the plates, become problems. We therefore considered this our somewhat arbitrary cut-off point, at which we concluded that there were no viable colonies. It is still possible that the mutants are viable but have a very greatly reduced growth rate. However, our observations suggest that this is not the case. When plating the cells onto sucrose plates, we have sometimes seen initial cell growth to form barely visible colonies, which then disappear only to leave pock marks on the agar. This suggests that the mutant double crossover has occurred before plating and that initial growth is sustained by the remaining intracellular pool of glutamine/glutamate but is then rapidly followed by cell death and lysis.
Salmonella enterica serovar Typhimurium glnE mutants have been shown to grow better in nitrogen-limited medium, presumably since the internal glutamate pool is less depleted under these conditions (17). However the S. coelicolor glnE mutant has no such growth defect (3). 7H10 medium, used for culturing M. tuberculosis, contains a large amount of l-glutamate (0.5 g/liter) and a substantial amount of ammonia in the form of ammonium sulfate (0.5 g/liter) and ferric ammonium citrate (0.04 g/liter). The availability of suitable solid medium for growth of M. tuberculosis poses a problem in this respect (1), since the only other commonly used defined solid medium is Dubos (Difco). This contains ferric ammonium citrate (a source of ammonia) and also has Casitone, which has previously been shown to be lethal to M. tuberculosis auxotrophs (12, 14). Therefore, the provision of suitable medium and/or supplements presents a difficulty, and the possibility that glnE mutants may be viable in other media or growth conditions remains.
The frequency of the second crossover to give the wt allele was eightfold higher (a total of 62 compared to 8). The fact that it is unequal can partly be explained by the fact that the length of homologous DNA on either side of the disruption was different, 1.1 and 3.8 kb. Since one side was three times longer than the other, a higher frequency of recombination to regenerate the wt allele would be expected. As expected, the initial single-crossover strain resulted from a single crossover on the long (3.8 kb) side. In addition, there are other factors which influence the frequency of recombination at any particular DNA site. In our experience, the frequency of recombination can differ by several orders of magnitude between different loci, but the reasons for this are not apparent (12).
In enteric bacteria, glnE is not essential, and Escherichia coli mutants can grow without supplements. In contrast, glnA mutants require glutamine for growth, since glutamine synthetase is essential.
However, the situation in enteric bacteria is quite different from that in M. tuberculosis. First, while enteric bacteria only possess one glutamine synthetase (type I) whose activity is regulated by GlnE, M. tuberculosis, like Streptomyces spp., also has a type II glutamine synthetase, which is not subject to regulation by adenylylation; thus, the situation is not directly comparable.
Second, GlnA1 is expressed at a high level in M. tuberculosis, constituting a large proportion of the total secreted protein (4). When expressed in Mycobacterium smegmatis, the M. tuberculosis enzyme is exported (although the native M. smegmatis glutamine synthetase is not) (5) and also leads to increased survival in macrophages (11). Inhibitors of the extracellular GS-I or antisense oligonucleotides to glnA1 are inhibitory to M. tuberculosis cell growth (6, 7). GlnA1 appears to synthesize l-glutamine (4) and has been implicated in the biosynthesis of poly-l-glutamine/glutamate, which forms up to 10% of the cell wall (6, 7). This unusual polymer is found in the pathogenic mycobacteria but not in the nonpathogenic species and may be involved in virulence.
Third, mycobacteria have d-iso-glutamine at position 2 of the peptide side chain of the peptidoglycan, compared with d-glutamate in enteric bacteria. The peptidoglycan side chain is initially synthesized with d-glutamate and subsequently amidated to d-iso-glutamine. Presumably a glutamine synthetase other than GlnA1 is required for d-iso-glutamine synthesis.
We do not know why glnE is essential in M. tuberculosis. Bacteria use glutamine-glutamate interconversion by glutamine synthetase as an important method for maintaining their nitrogen balance. Disruption of glnE should lead to GS-I being constitutively active (although there are other mechanisms of control); therefore, this may upset the intracellular pool of glutamate/glutamine. Thus, glnE mutants may have very low levels of glutamate. Glutamate plays a central role in cell metabolism, being the precursor for many different molecules, including purines and pyrimidines. We speculate that, in M. tuberculosis, glnE is required to maintain the intracellular balance of glutamate/glutamine, and in the absence of regulation, GlnA activity would deplete the intracellular pool of glutamate. This is supported by the fact that S. coelicolor glnE mutants have a lowered ratio of glutamate to glutamine (3). This may be more pronounced in M. tuberculosis because it synthesizes large quantities of poly-l-glutamine, possibly leading to a severe shortage of l-glutamate for central metabolism. Isolation of mutants was not possible, even though the medium that we used contained high levels of l-glutamate, suggesting that this amino acid is not transported efficiently into the cell. The transport capabilities of M. tuberculosis are little known, although a search of the genome reveals the presence of glnH and glnQ homologues, which are involved in glutamine transport in other bacteria. This suggests that l-glutamine may be transported into the cell, but whether this also applies to l-glutamate is unknown.
We have shown that the use of merodiploid strains to demonstrate the essentiality of a gene in M. tuberculosis is possible. Our work demonstrates that glnE is essential for the viability of M. tuberculosis in certain culture conditions. Further work will determine the role of glnE in the regulation of glutamine synthetase activity.
ACKNOWLEDGMENTS
Tanya Parish was funded by the Glaxo Wellcome Action TB project.
We thank the referees for their interesting and useful comments.
REFERENCES
- 1.Allen B. Mycobacteria: general culture methodology and safety considerations. In: Parish T, Stoker N G, editors. Mycobacteria protocols. Vol. 101. Totowa, N.J: Humana Press; 1998. pp. 15–30. [DOI] [PubMed] [Google Scholar]
- 2.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Fink D, Falke D, Wohlleben W, Engels A. Nitrogen metabolism in Streptomyces coelicolor A3(2): modification of glutamine synthetase I by an adenylyltransferase. Microbiology. 1999;145:2313–2322. doi: 10.1099/00221287-145-9-2313. [DOI] [PubMed] [Google Scholar]
- 4.Harth G, Clemens D L, Horwitz M A. Glutamine synthetase of Mycobacterium tuberculosis: extracellular release and characterization of its enzymatic activity. Proc Natl Acad Sci USA. 1994;91:9342–9346. doi: 10.1073/pnas.91.20.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harth G, Horwitz M A. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J Biol Chem. 1997;272:22728–22735. doi: 10.1074/jbc.272.36.22728. [DOI] [PubMed] [Google Scholar]
- 6.Harth G, Horwitz M A. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins as potential novel drug targets. J Exp Med. 1999;189:1425–1435. doi: 10.1084/jem.189.9.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harth G, Zamecnik P C, Tang J Y, Tabatadze D, Horwitz M A. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-l-glutamate/glutamine cell wall structure, and bacterial replication. Proc Natl Acad Sci USA. 2000;97:418–423. doi: 10.1073/pnas.97.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinds J, Mahenthiralingam E, Kempsell K E, Duncan K, Stokes R W, Parish T, Stoker N G. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145:519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- 9.Mahenthiralingam E, Marklund B I, Brooks L A, Smith D A, Bancroft G J, Stokes R W. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellular. Infect Immun. 1998;66:3626–3634. doi: 10.1128/iai.66.8.3626-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller B H, Shinnick T M. Evaluation of Mycobacterium tuberculosis genes involved in resistance to killing by human macrophages. Infect Immun. 2000;68:387–390. doi: 10.1128/iai.68.1.387-390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parish T, Gordhan B G, McAdam R A, Duncan K, Mizrahi V, Stoker N G. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology. 1999;145:3497–3503. doi: 10.1099/00221287-145-12-3497. [DOI] [PubMed] [Google Scholar]
- 13.Parish T, Stoker N G. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 14.Pavelka M S, Jacobs W R. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis BCG, and Mycobacterium tuberculosis H37Rv by allelic exchange. J Bacteriol. 1999;181:4780–4789. doi: 10.1128/jb.181.16.4780-4789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesole G, Bozzetti M P, Lanave C, Preparata G, Saccone C. Glutamine synthetase gene evolution: a good molecular clock. Proc Natl Acad Sci USA. 1991;88:522–526. doi: 10.1073/pnas.88.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reitzer L J, Magansanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1987. pp. 302–321. [Google Scholar]
- 17.Yan D L, Ikeda T P, Shauger A E, Kustu S. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:6527–6531. doi: 10.1073/pnas.93.13.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]