Abstract
The virulence of Staphylococcus aureus is controlled by the accessory gene regulator (agr) system, including an extracellular inducer encoded by agrD. Variable agr PCR restriction fragment length polymorphism (RFLP) patterns of unique S. aureus strains (n = 192) were determined for a region comprising agrD and parts of the neighboring agrC and agrB genes. Twelve unique RFLP patterns were identified among S. aureus strains in general; these patterns were further specified by sequencing. All sequences could be catalogued in the three current agr groups. A major proportion of the S. aureus strains belong to agr group 1, whereas only 6% of the methicillin-susceptible S. aureus strains and 5% of the methicillin-resistant S. aureus strains belong to agr groups 2 and 3, respectively. The homology between groups varied from 75 to 80%, and within groups it varied from 96 to 100%. Different levels of sequence variability were observed in the different agr genes. agr-related bacterial interference among colonizing S. aureus strains in the noses of persistent and intermittent human carriers was studied. S. aureus strains belonging to different agr groups were encountered in the same individual. This may suggest that the activity of the agrD gene product does not define colonization dynamics, which is further substantiated by the rarity of agr group 2 and 3 strains.
Staphylococcus aureus is a major human pathogen, causing a wide range of diseases including septicemia, meningitis, endocarditis, osteomyelitis, septic arthritis, toxic shock syndrome, and food poisoning (20). The pathogenic diversity of the bacterium reflects its ability to successfully survive in many different host tissues during infection. The pathogenic capacity of S. aureus is clearly dependent on its production of exoproteins. The synthesis of these virulence factors is globally regulated by an S. aureus quorum-sensing system called the accessory gene regulator (agr). The expression of genes in agr varies in response to changes in cell density (15). This global regulatory system utilizes a posttranscriptionally modified signal peptide that is excised from the AgrD peptide. This thiolactone-containing peptide accumulates in the external environment during postexponential growth of the S. aureus population. It is postulated that the signal peptide interacts with the transmembrane receptor protein (AgrC) of a classical two-component regulatory system (AgrC, AgrA) that in turn activates the transcription of the agr locus (5, 9). This induces the down-regulation of genes encoding surface proteins and the up-regulation of genes encoding secreted virulence factors (2, 5).
Ji et al. (4) recently described interference among different strains of staphylococci with regard to synthesis of their virulence factors and other extracellular proteins. These authors found that the AgrD peptide produced by a given strain of S. aureus activates its own agr locus but may inhibit the expression of agr in other strains. It is suggested that this inhibitory effect is correlated with the ability of a strain to compete with other strains for sites of colonization or infection. This phenomenon of AgrD-dependent cross-inhibition suggests the presence of significant variability of the domain encoding for the AgrD signal peptide (4). Indeed, it was shown that the autoinducer (AgrD) and its modifying protein (AgrB) and receptor (AgrC) harbored sequence variation. This affects the specificity of the receptor-ligand interaction. On the basis of autoinducer-receptor specificity, S. aureus can be divided into at least three different agr groups (10).
Here, we report on the prevalence and nature of agrD polymorphism, as determined for a large collection of methicillin-resistant S. aureus (MRSA) and methicillin-sensitive S. aureus (MSSA) strains. agr restriction fragment length polymorphism (RFLP) types were determined and classified into agr groups. The dissemination of agr RFLP types between MRSA and MSSA was studied. The potential relevance of agrD polymorphism in relation to colonization of distinct strain types in the noses of intermittent versus persistent S. aureus carriers was also analyzed.
MATERIALS AND METHODS
Strain collection.
Strains of MRSA and MSSA (n = 192) were pooled from eight existing collections (Table 1). All strains were well typed by using a wide variety of phenotypic and genotypic procedures: pulsed-field gel electrophoresis (PFGE), randomly amplified polymorphic DNA (RAPD), binary typing, and more. For cultivation, bacteria from glycerol stocks stored at −80°C were inoculated on Columbia III agar supplemented with 5% sheep blood (Becton Dickinson, Etten-Leur, The Netherlands) and incubated at 37°C for 24 h. All strains were identified as S. aureus by standard microbiological methods (6). MSSA strains from collection 1, isolated during the colonization studies, were derived from nine healthy nasal carriers of S. aureus. When 5 to 8 out of the 10 serial samples were positive, a person was identified as an intermittent carrier (n = 6). Persistent carriers (n = 3) had 9 or 10 positive cultures out of the 10 samples (17).
TABLE 1.
Characterization of the 192 S. aureus strains used in this study
| Collection | Geographic origin | No. of strains | Description of the collection | Reference |
|---|---|---|---|---|
| 1 | Denmark | 33 | Nasal carriership (MSSA strains) | 17 |
| 2 | Worldwide | 59 | Geographically and temporally unrelated MRSA strains | 7 |
| 3 | USa | 1 | Type strain of a local MRSA outbreak in NYCc hospital | 18 |
| 4 | Spain and Portugal | 11 | Iberian (MRSA) clone | 18 |
| 5 | US | 27 | Community-acquired MRSA strains, isolated from outpatients from an NYC hospital | 18 |
| 6b | US | 49 | Multicenter collection of MRSA (n = 32) and MSSA (n = 17) strains | 16 |
| 7 | The Netherlands | 10 | Representative MRSA (n = 6) and MSSA (n = 4) strains from 10 outbreaks in Dutch hospitals | 19 |
| 8 | US | 2 | MSSA strains obtained from 2 hospitalized patients (NYC) | 18 |
US, United States.
Centers for Disease Control and Prevention, Atlanta, Ga.
NYC, New York City.
DNA isolation.
Five to 10 colonies were suspended in a buffer containing 150 μl of 25 mM Tris-HCl (pH 8.0), 10 mM EDTA and 50 mM glucose. To prepare spheroplasts, 75 μl of a lysostaphin solution at a concentration of 100 μg/ml (Sigma Chemical Corporation, St. Louis, Mo.) was added. The mixture was incubated for 1 h at 37°C. DNA isolation was done as described by Boom et al. (3). Briefly, guanidine hydrothiocyanate was added for cell lysis and DNA was purified by affinity chromatography with Celite (Janssen Pharmaceuticals, Beerse, Belgium). DNA was eluted from the Celite particles with 100 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA. The DNA concentration was estimated by electrophoresis in the presence of ethidium bromide (0.3 μg/ml) (13). Stock solutions of DNA were adjusted to a concentration of 50 ng/μl and stored at −20°C until further use.
PCR amplification of the agrD and partial agrC and agrB sequences.
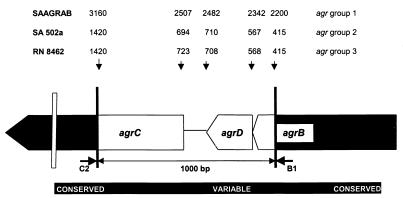
Approximately 50 ng of DNA was amplified in a 100-μl reaction mixture consisting of 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin, and 0.1% Triton X-100. Deoxyribonucleotide triphosphates (0.2 mM; Amersham Pharmacia Biotech, Roosendaal, The Netherlands) as well as 0.2 U of Taq polymerase (SuperTaq; HT Biotechnology, Cambridge, United Kingdom) were present in the reaction mixture. The locations of the primers agrB1 and agrC2 within the agr locus are indicated in Figure 1. The National Center for Biotechnology Information (NCBI) code and sequences of the primers (50 pmol of primer per reaction) were as follows: agrC2, 5′-CTT GCG CAT TTC GTT GTT TGA-3′, SAAGRAB sequence, nucleotides (nt) 3208 to 3189 (GenBank accession no. X52543); agrB1, 5′-TAT GCT CCT GCA GCA ACT AA-3′, SAAGRAB sequence, nt 2142 to 2161. The PCR mixture was overlaid with 100 μl of mineral oil to prevent evaporation. Amplification of the DNA fragments was performed in a thermocycler (model 60; Biomed, Theres, Germany) with predenaturation at 94°C for 4 min, followed by 40 cycles of 1 min at 94°C, 2 min at 50°C, and 3 min at 74°C and ending with a postelongation step at 74°C for 3 min. Amplicons were purified by an additional ethanol precipitation step. The DNA pellet was lyophilized and redissolved in distilled water. The yield of amplicons was determined by electrophoresis in the presence of ethidium bromide (0.3 μg/ml) (13).
FIG. 1.
Schematic map of the S. aureus agr locus showing the amplified region. The amplicon contained the variable sequence, defining the distinct agr groups (4). Primers C2 and B1 were selected from the conserved sequences. The GenBank nucleotide position numbers of the three agr sequences from the control strains, representing the three agr groups, are indicated above the corresponding positions on the map.
Restriction of the PCR products.
Restriction analysis of the amplicons was performed with RsaI (Roche Molecular Biochemicals, Almere, The Netherlands) and AluI (Roche Molecular Biochemicals) according to the manufacturer's instructions. The AluI and RsaI digests were visualized on 3% NuSieve agarose gels (FMC Bioproducts, SanverTech, Heerhugowaard, The Netherlands) stained with ethidium bromide.
DNA sequencing and homology analysis.
The 373 DNA-sequencing system (Perkin-Elmer, Foster City, Calif.) was used for sequencing those amplicons that generated a unique RFLP pattern. At first, amplicons were cloned with the TOPO-TA cloning kit (Invitrogen, Leek, The Netherlands). Dye terminator chemistry was applied (8, 14) using the manufacturer's cycle sequencing protocol (Amersham Pharmacia Biotech). Briefly, DNA was amplified in the presence of a thermostable polymerase and primers agrB1 and agrC2. The extended, fluorescently labeled fragments were separated by polyacrylamide gel electrophoresis. Labels were excited by a laser as they passed the detector near the bottom of the gel. The first 100 nt were deleted because of unreliable sequencing results (GenBank; SAAGRAB sequence no. 2100–2200 and 3160–3300, SA502a sequence no. 300–415 and 1420–1550, and RN8462 sequence no. 280–415 and 1420–1550). Reliability of the sequencing was assessed by including different clones with the same RFLP pattern and checking the RFLP pattern versus the primary sequence. These comparisons confirmed the lack of PCR errors (results not shown). The sequences were compared with the data from the nucleotide and protein sequence database of NCBI and analyzed for similarity with BLAST (basic local alignment search tool) program (1). Sequence alignment and analysis of sequence polymorphism among the different agr genes were performed with Megalign Lasergene software (DNASTAR Inc., Madison, Wis.).
RESULTS
agr RFLP.
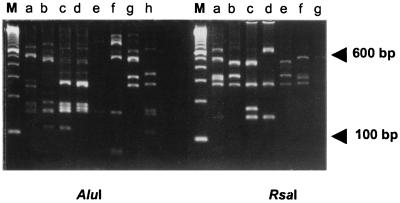
The agr restriction patterns of 192 epidemiologically and genetically unique S. aureus strains were determined. Only 12 unique-combination restriction patterns (AluI and RsaI digests) (Fig. 2) were detected, indicating a relatively high degree of sequence conservation. The frequency and distribution of the different RFLP patterns among the agr groups (see also next section) (4) are outlined in Table 2. Among the MRSA strains (71.4% of the total S. aureus collection), 10 distinct RFLP patterns (AA, CC, DD, DE, ED, FB, FE, GF, GG, and HE) could be identified. The most predominant agr RFLP types, AA and DE, represented 43.0 and 40.8% of the MRSA strains, respectively. Based on DNA sequencing (see also below) all types appeared to belong to agr group 1, except for agr type GG (prevalence of 5.1%; agr group 3). Seven different RFLP patterns (AA, BA, BB, DD, DE, ED, and FB) were found among the MSSA strains of which AA (prevalence, 23.6%) and ED (prevalence, 36.4%) appeared to be the most prevalent lineages. Apart from RFLP type BB (prevalence, 14.5%; agr group 2), all types were classified as belonging to agr group 1. One Dutch MRSA strain (Table 1, collection 7) generated an abnormally sized amplicon. Sequencing revealed that the Tn4001 transposon was inserted in the 5′ part of the agrC gene (inserted between positions 2546 and 2547 of the SAAGRAB sequence). No phenotype analysis was performed for this strain, so we cannot discuss the activity of this deviating agr locus.
FIG. 2.
RFLP analysis of the agr locus. Amplified DNA was digested with AluI, yielding patterns a to h (left) and with RsaI, producing patterns a to g (right). Twelve different combined AluI-RsaI combination patterns were detected. Lanes M, molecular size markers.
TABLE 2.
Frequency and distribution of agr RFLP patterns within agr groups of the well-typed epidemiologically unrelated MRSA (n = 137) and MSSA (n = 55) strainsa
| agr group | No. of strains (MSSA/MRSA) with RFLP pattern:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | BB | BA | CC | DD | DE | ED | FB | FE | GG | GF | HE | |
| 1 | 13/59 | 3/0 | 0/2 | 8/2 | 2/56 | 20/7 | 1/1 | 0/1 | 0/1 | 0/1 | ||
| 2 | 8/0 | |||||||||||
| 3 | 0/7 | |||||||||||
Strains were genetically typed using PFGE, RAPD analysis, binary typing, mecA/Tn554 probing, or a combination thereof. For AluI eight individual RFLP patterns (a to h; Fig. 2) were identified, whereas the use of RsaI allowed for the identification of seven RFLP patterns (a to g; Fig. 2). Overall, 12 combined patterns were defined.
Sequence polymorphism in the agr locus.
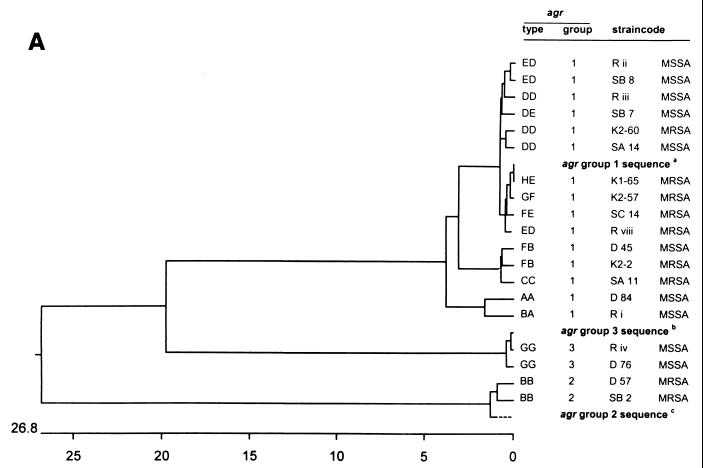
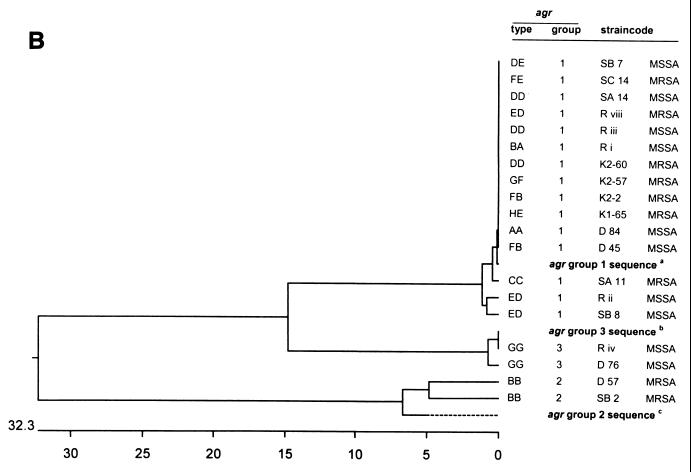
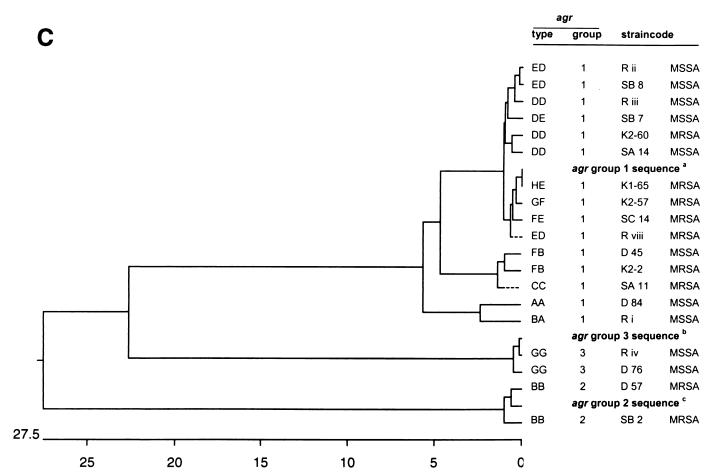
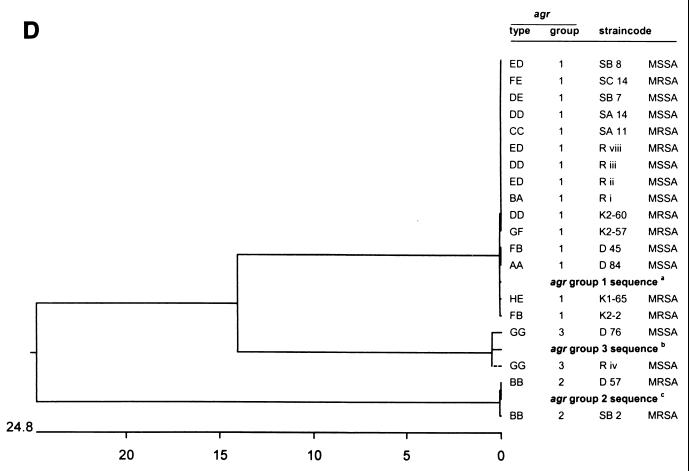
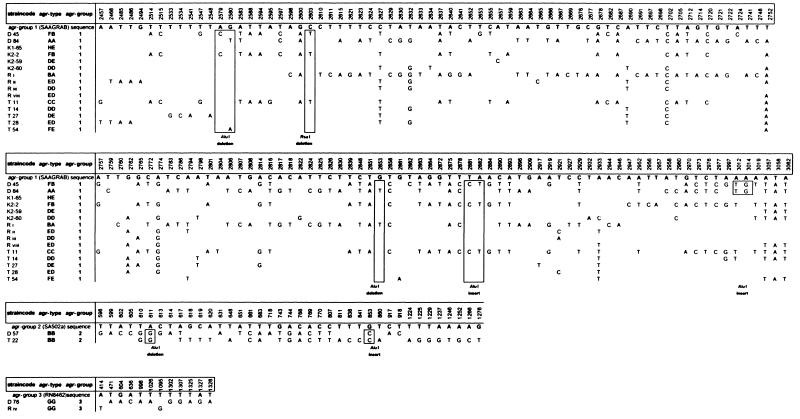
The nucleotide sequences of the 12 unique agr RFLP types were determined and compared with the GenBank data. The sequences of agr RFLP types BB, DD, DE, ED, FB, and GG were obtained for two or three different strains (Fig. 3 and 4). All agr RFLP types could be classified into the three previously known agr groups (Table 2). The vast majority (92%) of the agr RFLP types belong to group 1. Two distinct agr RFLP types (BB and GG) could be classified as groups 2 and 3, respectively. The frequency and positions of mutations in the agr locus for the unique RFLP types are displayed in Fig. 4. The positions and numbers of point mutations for strains D45 (RFLP type FB), K2-2 (RFLP type FB), and T11 (RFLP type CC) were similar. The sequences of RFLP types DD, DE, ED, and FE are closely related to the SAAGRAB sequence and rarely displayed a very small number of mutations. The nucleotide sequence of strain K1-65 (RFLP type HE) is identical to the SAAGRAB sequence (GenBank). Gain and loss of AluI and RsaI restriction sites are shown in Fig. 4. RFLP polymorphism cannot be deduced completely from the data presented in Fig. 4, since the AluI and RsaI restriction sites on both termini of the amplicon were not included in the sequence analysis. The inter- and intrasequence polymorphism of the agr groups is outlined in Fig. 3A. The sequence variability within an agr RFLP type ranged from 0 to 1% (types ED, DD, and FB). The distances between agr RFLP types within an agr group varied from 0 to 4%. The sequence diversity between agr groups 1 and 3 was 20%, while the agr group 2 cluster differed by 26.8% from the former two groups. The divergent variability among agrD, partial agrC, and agrB genes is displayed in Fig. 3B to D, respectively. The agrD sequence varied from 0 to 1% among the agr types within each agr group, except for agr group 2 (type BB), where the agrD gene varied from 5 to 7%. The first 600 nt of the agrC 5′ part varied from 0 to 6% among the different agr RFLP types. The intragroup variability of the agrC gene ranged from 22.6% for groups 1 and 3 to 28.8% for group 2. The last 140 bp from the agrB 3′ domain displayed a high level of similarity (99 to 100%) among the diverse RFLP types. The agrB sequence variability within the three distinct agr groups ranged from 14% for groups 1 and 3 to 24.8% for group 2.
FIG. 3.
Dendrograms displaying the cluster analysis of the intra- and extra-agr group sequence homologies from the 12 distinct agr RFLP patterns. The numbers on the horizontal axes display the percentages of similarity between the primary sequences of the distinct RFLP patterns. (A) Clustering of the total amplicon sequences; (B) clustering of the agrD sequences; (C) clustering of the agrC sequences; (D) clustering of the agrB sequences. agr group reference sequences were obtained from the data bank of the NCBI. aNCBI code SAAGRAB, accession no. X52543; bNCBI code SA502a, accession no. AF001782; cNCBI code RN8462, accession no. AF001783. The Tn4001 insertion within the agr sequence of strain R viii was deleted for cluster analysis.
FIG. 4.
Overview of the DNA sequences of the prevalent agr RFLP types subdivided into agr groups. The nucleotide positions of the detected sequence mutations, based on variation from the agr consensus sequences (three agr group control strains; GenBank), are indicated above. Identical residues are not indicated. Position numbers of the point mutations correspond with those indicated on the agr gene map (Fig. 1). Boxed areas, loss and gain of restriction sites for the enzymes AluI and RsaI.
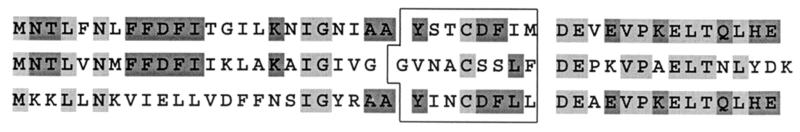
The amino acid sequences of all agr types studied displayed variability values similar to those observed for the nucleotide sequences (data not shown). The sequences of the amino acid residues of the AgrD propeptide within the unique agr RFLP types were compared and are displayed in Table 3. The group 1 AgrD propeptide sequence is completely conserved, except for types ED and CC. One amino acid, located downstream the signal peptide, was affected. A higher degree of variability was detected within the group 2 AgrD propeptide sequence, whereas only a single RFLP type was found. The AgrD signal peptide of this group consists of nine amino acids and displays homology with the AgrD nonapeptide sequence of Staphylococcus epidermidis Tü3298 (Table 3) (12). The AgrD propeptide sequences were compared and are outlined in Fig. 5. agr groups 1 and 2 displayed a high level of similarity upstream of the AgrD signal peptide. The AgrD propeptide sequences of agr groups 1 and 3 were conserved within and downstream of the signal peptide.
TABLE 3.
Comparative analysis of the amino acid sequence variability within the prevalent unique agr RFLP types and agr groupsa
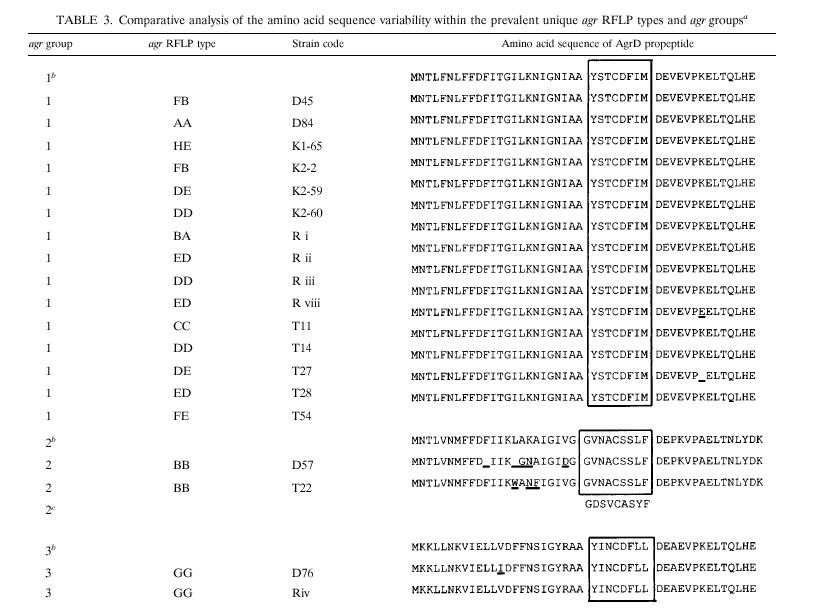 |
The control sequences obtained from GenBank were as follows: agr group 1, SAAGRAB sequence, accession no. X52543; agr group 2, SA502a, accession no. AF001782; agr group 3, RN8462, accession no. AF001783. The sequences of the AgrD signal peptides are boxed. The sequences were aligned by MegAlign software (DNAStar). The signal peptide sequence of S. epidermidis Tü 3298 was derived from the study of Otto et al. (12). Sequence differences are underlined.
Control sequence.
S. epidermidis signal peptide sequence.
FIG. 5.
Comparative analysis of the AgrD propeptide sequences. Box, signal peptides; light grey, identical residues in all three sequences; dark grey, identical residues in two sequences.
agr polymorphism among S. aureus strains of nasal carriers.
The S. aureus strains (n = 3), obtained from the volunteers were analyzed for agr polymorphism, and the results were compared with those obtained with other pheno- and genotyping methods (Table 4). Analysis of the agr polymorphism of the S. aureus strains from the persistent carriers reveals concordance with the results of the other typing techniques, except for protein A polymorphism. S. aureus strains isolated over several years from persistent carriers displayed an agr group switch in two of three carriers studied. Carrier 84 switched from agr group 1 to agr group 2, and carrier 126 switched from agr group 2 to agr group 1. Such switches are also observed among the intermittent carriers of S. aureus (see carriers 9 [switch from group 1 to 3] and 88 [switch from group 2 to 3]). The agr RFLP types within a given agr group of a carrier displayed the same level of variability as for RAPD analysis and coagulase (coag) gene typing. agr polymorphism within a single agr group of a persistent carrier was detected, as confirmed by a 3 to 4% divergence at the sequence level (data not shown). A predominant genotype could be identified. The pheno- and genotypically identical predominant strains (RAPD type BBB, coag gene type 14B; Table 4) displayed agr polymorphism (RFLP types AA and ED) but not agr group switching.
TABLE 4.
agr polymorphism among S. aureus strains isolated from both persistent and intermittent nasal carriersa
| Person no. | Strain no. | Date of isolation (day-mo-yr) | agr type | agr group | RAPD type | No. of protA repeats | Coag gene type | Phage type |
|---|---|---|---|---|---|---|---|---|
| Persistent carriers | ||||||||
| 53 | 44 | 09-03-92 | DD | 1 | DDD | 4 | 9C | 641 |
| 53 | 45 | 31-08-92 | FB | 1 | AAA | 7 | 12A | 793 |
| 53 | 47 | 01-11-93 | DD | 1 | GGG | 10 | 10F | 693 |
| 53 | 48 | 15-08-94 | DD | 1 | GGG | 9 | 10F | 542 |
| 84 | 54 | 09-03-92 | AA | 1 | JIH | 8 | 15I | 692 |
| 84 | 55 | 18-04-93 | AA | 1 | JIH | 7 | 15I | 692 |
| 84 | 56 | 01-11-93 | ED | 1 | BBB | 5 | 14B | 811 |
| 84 | 57 | 15-08-94 | BB | 2 | KJI | 9 | 9C | 292 |
| 126 | 58 | 19-10-92 | BB | 2 | LKC | 8 | 11N | 793 |
| 126 | 59 | 18-04-93 | ED | 1 | BBB | 7 | 14B | 812 |
| 126 | 60 | 15-08-94 | ED | 1 | BBB | 7 | 14B | 811 |
| Intermittent carriers | ||||||||
| 85 | 61 | 24-02-92 | AA | 1 | JIH | 7 | 15I | 311 |
| 85 | 62 | 09-03-92 | ED | 1 | BBB | 8 | 14B | 812 |
| 85 | 63 | 06-01-93 | ED | 1 | BBB | 8 | 14B | 232 |
| 85 | 64 | 15-08-94 | AA | 1 | JIH | 9 | 15I | 362 |
| 2 | 69 | 25-06-92 | ED | 1 | BBB | 9 | 14B | 813 |
| 2 | 70 | 19-10-92 | ED | 1 | BBB | 9 | 14B | 691 |
| 2 | 71 | 18-04-93 | AB | 1 | AAA | 7 | 12A | 793 |
| 2 | 72 | 14-02-94 | ED | 1 | BBB | 9 | 14B | 811 |
| 9 | 73 | 25-06-92 | ED | 1 | BBB | 5 | 14B | 811 |
| 9 | 74 | 19-10-92 | BA | 1 | CCC | 7 | 9C | 292 |
| 9 | 75 | 01-11-93 | BA | 1 | CCC | 7 | 9C | 233 |
| 9 | 76 | 15-08-94 | GG | 3 | MLJ | 5 | —b | 191 |
| 32 | 79 | 20-01-92 | ED | 1 | BBB | 3 | 14B | 811 |
| 32 | 80 | 25-06-92 | ED | 1 | BBB | 3 | 14B | 811 |
| 32 | 81 | 15-08-94 | DD | 1 | NMK | 9 | 9J | 793 |
| 20 | 84 | 09-03-92 | AA | 1 | PNC | 8 | 10G | 562 |
| 20 | 85 | 27-04-92 | AA | 1 | CCC | 10 | 9C | 293 |
| 20 | 86 | 25-06-92 | AA | 1 | BBB | 10 | 14B | 811 |
| 20 | 87 | 06-01-93 | ED | 1 | AAA | 7 | 12A | 793 |
| 113 | 88 | 09-03-92 | BB | 2 | JFH | 7 | 10I | 692 |
| 113 | 89 | 19-10-92 | AA | 1 | BBB | 10 | 14B | 811 |
| 113 | 90 | 14-02-94 | ED | 1 | OOL | 9 | 9C | 631 |
Data on the carrier status and RAPD, protein A (protein A (protA), coag gene, and phage types are adapted from van Belkum et al. (17). The RAPD three-letter code summarizes the typing results for the primer used (first letter, primer AP-1; second letter, primer AP-7; third letter, primer ERIC-2). Numbers of protein A repeats were derived from an analysis of the so-called X region by PCR. Amplicons were digested with RsaI. The number of direct repeats in this region was determined after electrophoresis. Coag gene types were derived from a PCR-RFLP analysis of the coag gene and subsequent detection of the amplicon with AluI. Phage types are given in an abbreviated form according to standards applied in the Statens Seruminstitut, Copenhagen, Denmark.
—, no restriction site for the amplicon.
DISCUSSION
The synthesis of S. aureus virulence factors is controlled by a cell density-sensing system based on the action of a signal peptide secreted in the environment by the organism itself. When a strain enters the postexponential phase, it produces signal peptides that may inhibit or induce the expression of the agr operon in other strains, depending on the agr group of the strain. The amino acid sequences of the ligand (AgrD) and receptor (AgrC) for such mutually interfering strains differ considerably. This phenomenon suggests the presence of selectively variable domains (4) and points to intricate structure-function relationships at the protein level. Structure-activity studies have demonstrated that the presence of a thiolactone group and the biochemical structure of the signal peptide (AgrD) are essential for stimulating biological activity (10). Cross-inhibition is less dependent on the structure of the signal peptide (12). Hypothetically, cross-inhibition of agr gene expression represents an example of bacterial interference that could be associated with colonization resistance or even competition of strains during infection.
agr clonality for MRSA and MSSA.
A major part (84%) of the MRSA strains harbored agr RFLP type AA or DE. The vast majority (98%) of RFLP type AA strains originated in the United States and were isolated in the 1980s. The other RFLP type (DE) is present in the majority (94%) of European strains isolated in the early 1960s. This geographical and temporal conservation of agr RFLP type among MRSA strains fits well with former observations regarding the clonal population structure of European and American MRSA (11). All MRSA strains belong to agr group 1, except for a small cluster of strains (n = 7) originating in Ontario, Canada (agr group 3). Ninety percent of the MSSA strains could be classified as agr group 1 as well. The remaining eight strains were indexed as agr group 2, and all had agr RFLP type BB. Strains that belong to the latter group were geographically unlinked but genetically closely related. Strains appeared to be similar based on phage, ribo-, and multilocus enzyme electrophoresis typing. However, this strain cluster displayed diverged PFGE and RAPD patterns as if its members had evolved over time (data not shown). The index strain of a MRSA outbreak (strain collection 7) harbored part (approximately 1,000 bp) of the Tn4001 transposon sequence, an aminoglycoside resistance determinant in S. aureus strains. Only the transposase gene was inserted in the 5′ part of agrC. The Tn4001 sequence was absent in other outbreak strains. The potential effect of the transposon insertion on the activity of the agr operon was not analyzed and remains unclear, but it seems as if the 5′ part of agrC is not essential for staphylococcal viability.
agr polymorphism at the sequence level.
The agr sequences from the agr RFLP types present in our S. aureus strains could be classified in one of the three different agr groups defined by Ji et al. (4). The main part (92.2%) of the S. aureus strains belong to agr group 1. Consequently, the evolutionary significance of the other agr groups is unclear. The amino acid sequences of AgrD from agr groups 2 and 3 are clearly different from that from agr group 1. Groups 2 and 3 both share domains with group 1 but do not share domains with each other, except for sequences which are present in all three agr groups. The amino acid homology among the three different AgrD propeptide sequences suggests recombination.
Cluster analysis of the DNA sequences determined for the agr RFLP types demonstrated that agrD and parts of agrB and agrC have different evolutionary clock speeds. The observed stability of the agrD signal peptide sequence within each agr group confirms the results from the biochemical study with coagulase-negative staphylococci of Otto et al. (12) dealing with the relevance of the AgrD structure with respect to the ligand-receptor interaction specificity for agr activation. Overall, agr groups 1 and 3 are more closely related (80% sequence homology) than agr groups 1 and 2 (74% sequence similarity). Considering the agr sequences, there is no indication of major recombination events. It is interesting to see that AgrC sequence variation exceeds that of AgrD. Thus it could be speculated that the diversity of the AgrC receptor protein has a more profound influence than that of AgrD in the recognition reaction between noncanonical AgrD and AgrC proteins. This would imply that the reactivity of the peptides is largely defined by receptor polymorphism and not autoinducer peptide variability. However, this requires additional verification by biological experimentation.
agr incompatibility and colonization.
We studied the possible role of bacterial interference in relation to nasal S. aureus carriership. We therefore analyzed the agr locus polymorphism and the subsequent assignment to agr “incompatibility” groups (1, 2, and 3) among staphylococcal strains isolated from persistent and intermittent carriers. In both persistent and intermittent nasal carriers of S. aureus, strains with different genetic profiles, includng those identified by RAPD analysis, coag gene genotyping, and phage typing, and different agr groups alternate over time. Since carriers of S. aureus rarely carry two clones at the same time (17), agr grouping may well play a role. However, contrary to previous suggestions summarized above (4, 10), we here show that the carrier status is not restricted to a single agr group. RFLP analysis of the agr locus closely mimicked the results obtained with whole-genome typing. Although the numbers of individuals included in the present analysis are limited and although we did not determine biological agr activity, our data show both alternation and persistence of agr types. This is not in favor of a prominent biological function of the agr system in nasal colonization dynamics. This observation is further supported by the fact that the vast majority of S. aureus strains belong to agr group 1.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Balaban N, Novick R P. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc Natl Acad Sci USA. 1995;92:1619–1623. doi: 10.1073/pnas.92.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 5.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloos W E, Bannerman T L. Staphylococcus and Micrococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 282–298. [Google Scholar]
- 7.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 8.Kretz K, Callen W, Hedden V. Cycle sequencing. PCR Methods Appl. 1994;3:S107–S112. doi: 10.1101/gr.3.5.s107. [DOI] [PubMed] [Google Scholar]
- 9.Lina G, Jarraud S, Ji G, Greenland T, Pedraza A, Etienne J, Novick R P, Vandenesch F. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol. 1998;28:655–662. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- 10.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick R P, Muir T W. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musser J M, Kapur V. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol. 1992;30:2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto M, Sussmuth R, Vuong C, Jung G, Gotz F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Smith L M, Sanders J Z, Kaiser R J, Hughes P, Dodd C, Connell C R, Heiner C, Kent S B, Hood L E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 15.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R, Jarvis W R, Kreiswirth B, Eisner W, Maslow J, McDougal L K, Miller J M, Mulligan M, Pfaller M A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Belkum A, Riewarts Eriksen N H, Sijmons M, van Leeuwen W, van den Bergh M, Kluytmans J, Espersen F, Verbrugh H. Coagulase and protein A polymorphisms do not contribute to persistence of nasal colonisation by Staphylococcus aureus. J Med Microbiol. 1997;46:222–232. doi: 10.1099/00222615-46-3-222. [DOI] [PubMed] [Google Scholar]
- 18.van Leeuwen W, van Belkum A, Kreiswirth B, Verbrugh H. Genetic diversification of methicillin-resistant Staphylococcus aureus as a function of prolonged geographic dissemination and as measured by binary typing and other genotyping methods. Res Microbiol. 1998;149:497–507. doi: 10.1016/s0923-2508(98)80004-1. [DOI] [PubMed] [Google Scholar]
- 19.van Leeuwen W, Verbrugh H, van der Velden J, van Leeuwen N, Heck M, van Belkum A. Validation of binary typing for Staphylococcus aureus strains. J Clin Microbiol. 1999;37:664–674. doi: 10.1128/jcm.37.3.664-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldvogel F A. Staphylococcus aureus (including toxic shock syndrome) In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1994. pp. 1754–1777. [Google Scholar]