Abstract
Telomeric G-quadruplexes have been a promising target for developing antitumor drugs with fewer side effects. The intracellular environment is usually in a state of molecular crowding. Studying the interaction mechanism among ligands and telomeric G-quadruplexes under crowded conditions is important for designing drugs that target telomeric G-quadruplexes. In the present study, the telomeric G-quadruplex Tel24 (TTAGGG)4 was found to fold into a conformational ensemble of parallel and (3 + 1) hybrid-2 conformations in solution with molecular crowding conditions created by PEG200. G-quadruplex-ligand 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl] acridinium methosulfate (RHPS4) preferentially stabilized the (3 + 1) hybrid-2 conformation and shifted the conformational ensemble equilibrium of Tel24 towards the hybrid conformation. We also found that the (3 + 1) hybrid-2 conformation of Tel24 was more likely to form as compared to the parallel conformation in the conformational ensemble of Tel24. Overall, this study provides new insights into the conformation of telomere G-quadruplexes and their interactions with ligands in a physiological environment.
Tel24 G-quadruplex can form a conformational ensemble consisting of parallel and (3 + 1) hybrid-2 conformations. RHPS4 preferentially stabilized the hybrid-2 conformation and shifted the conformational ensemble equilibrium.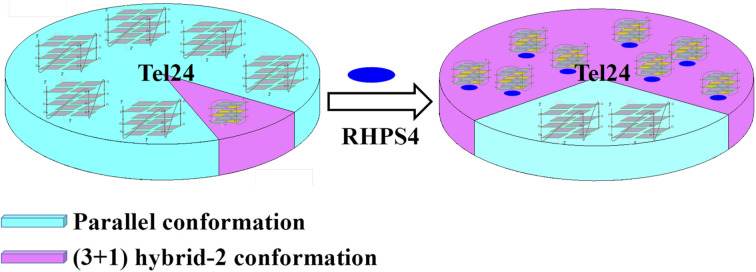
Introduction
In the presence of monovalent metal ions such as K+ or Na+, four guanine-rich repeats can fold into a G-quadruplex structure by stacking of two or three G-quartet layers, each containing four coplanar guanines stabilized by Hoogsteen hydrogen bonds.1–3
G-Quadruplexes play an important role in mediating biological processes, have significant biological activity and are often used as a target for screening antitumor and antiviral drugs.4–8 Telomere maintenance is essential for the continued division of tumor cells.9 The 3-terminal of human telomeres contains ∼200 nt of single-stranded overhanging TTAGGG repeats, which could fold into a stable G-quadruplex structure consisting of consecutive G-quadruplex units connected by TTA linkers.10–13 Telomeric G-quadruplexes have been explored as targets for antitumor drugs.14–16 Small molecules targeting telomeric G-quadruplexes can inhibit tumor cell immortality by blocking telomerase catalyzed telomere extension.17,18
Telomeric G-quadruplexes were reported to form antiparallel, (3 + 1) hybrid-1, and (3 + 1) hybrid-2 conformations in a dilute solution containing Na+ or K+.19–21 Macromolecules occupy 20% to 30% of cell volume, resulting in a highly crowded macromolecular environment within the cell.22,23 Therefore, studying the telomeric G-quadruplex structure and their interaction with small molecules under molecular crowding conditions provides more information for designing the targeting drug molecules.
Molecular crowding has been reported to affect the conformation and stability of G-quadruplexes, and inhibits G-quadruplex interactions with ligands.24–29 Molecular crowding conditions created by polyethylene glycol, with an average molecular weight of 200 (PEG200), induced a conformational change of the G-quadruplex from an antiparallel to a parallel structure.24 Molecular crowding conditions simulated by solvents with different molecular weights have different mechanisms of action affecting G-quadruplexes. The dehydration effect is a key factor in ethylene glycol-induced enhanced stability of G-quadruplex, whereas PEG8000 stabilizes the G-quadruplex mainly through its interaction with the G-quadruplex.25 While molecular crowding stabilizes G-quadruplex with certain selectivity. PEG200 has been reported to stabilize RNA G-quadruplexes with three and four G-quartets but not with two G-quartets.26 Telomeric G-quadruplexes are reported to form parallel conformation under molecular crowding conditions.30,31 However, the conformation of telomeric G-quadruplex within the cell may not be single due to the complexity of the intracellular environment. The interaction of small molecules with G-quadruplexes should be considered for combining multiple conformations.
RHPS4, a telomerase inhibitor, has significant antitumor activity by targeting the telomeric G-quadruplex.32,33 In the present study, we explored the effect of RHPS4 on conformational changes of telomeric G-quadruplexes (Tel24) under molecular crowding conditions created by PEG200. Tel24 was found to fold into a conformational ensemble consisting of parallel and (3 + 1) hybrid-2 conformations in 100 mM K+ solution containing 20 wt% PEG200. In fact, the binding of RHPS4 to the Tel24 is a recognition between RHPS4 and an ensemble consisting of multiple conformations. RHPS4 preferentially stabilized the (3 + 1) hybrid-2 conformation and shifted the conformational ensemble equilibrium of Tel24 to the hybrid conformation.
Further research reveals that in the conformational ensemble of Tel24, the (3 + 1) hybrid-2 conformation was easier to form than the parallel conformation. This study is expected to be a breakthrough in discovering ligands with a higher ability to stabilize telomeric G-quadruplexes.
Materials and methods
Materials
The oligonucleotide of Tel24 was synthesized by TSINGKE Biological Technology Co., Ltd (Wuhan, China). RHPS4 was purchased from Topscience Bio-Tech Co., Ltd (Shanghai, China).
Circular dichroism spectroscopy
CD spectra were collected from 220 to 320 nm with a 1 nm bandwidth on a circular dichroism spectrophotometer (Jasco, Japan). All the samples were prepared in 10 mM tris–HCl (pH 7.4) buffer containing 100 mM KCl and different concentrations of PEG200. The concentration of oligonucleotides was 10 μM, and the solutions of oligonucleotides were initially heated at 95 °C for 5 min and then cooled to room temperature and incubated at 4 °C overnight to eliminate any possible single-stranded structures. To prepare the final DNA-complex samples, the RHPS4 was added to the annealed DNA samples and incubated at 25 °C for 10 min.
Fluorescence spectroscopy
The oligonucleotide of Tel24 was annealed in 10 mM Tris–HCl buffer (pH 7.5), 100 mM KCl and different concentrations of PEG200 (w/v) by heating at 95 °C for 5 min, followed by gradual cooling at room temperature and later incubated at 4 °C overnight. Fluorescence spectra were scanned by Spectrofluorophotometer (Hitachi, Japan) at room temperature. The mixture of Tel24 and RHPS4 was incubated for 10 min at 25 °C before the spectra collection. Emission spectra between 520 nm and 750 nm of RHPS4 were recorded at λex = 511 nm. The excitation slit and emission slit were both set at 5 nm. The concentration of Tel24 sequences varied from 0 to 6.0 μM (0, 0.2, 0.5, 0.7, 1.0, 1.5, 2.0, 2.5, 3.0, 4.0, 5.0, 6.0 μM), while the concentration of RHPS4 was 3.0 μM. The fluorescence variation of RHPS4 with the titration of Tel24 was used to derive the dissociation constant (KD) value according to the method of Benesi–Hildebrand,34 using the following equation: 1/(ΔF) = 1/(b[DNA][H]0Ka) + 1/(b[H]0). ΔF represents the change in the fluorescence intensity (ΔF = F − F0); F denotes the fluorescence intensity of RHPS4 at 550 nm in the presence of a different concentration of Tel24; F0 is the fluorescence intensity of the free compound RHPS4 at 550 nm. [DNA] is the total added Tel24 G-quadruplex forming oligonucleotide concentration, and [H]0 is the total concentration of compound RHPS4. Moreover, Ka is the binding constant, while the dissociation constant KD is equal to 1/Ka.
Results
Effect of RHPS4 on the conformation of Tel24 under molecular crowding conditions
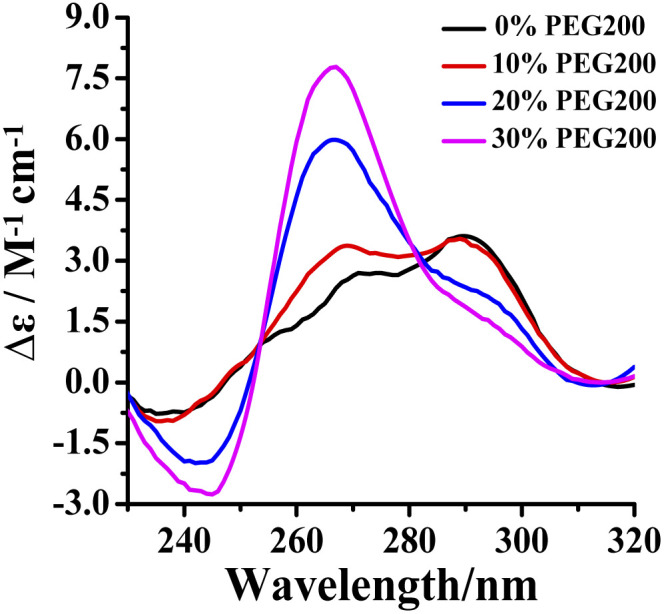
The intracellular volume is occupied by 20–30 wt% of large molecules in a physiological environment. In order to investigate the effect of RHPS4 on the conformation of Tel24 in a physiological environment, circular dichroism (CD) spectra analysis has been used in 100 mM K+ solution with molecular crowding conditions created by different concentrations of crowding agent PEG200. In dilute solution, the CD spectrum of Tel24 showed a negative peak at about 240 nm, a shoulder near 270 nm, and a positive peak around 290 nm, respectively. Such features are consistent with G-quadruplexes of (3 + 1) hybrid-2 conformation (Fig. 1). The addition of PEG200 enabled a decline in positive and negative peaks at around 290 nm and 240 nm, respectively, while a positive peak appeared at about 265 nm. At the 30 wt% (w/v) concentration of PEG200, a positive peak disappeared at 290 nm, and a shoulder peak was formed, resulting in a spectrum signature similar to the parallel quadruplexes. It suggested that a molecularly crowded environment can induce the formation of Tel24 in parallel conformations. These results are similar to the conformational change of GGG(TTAGGG)3 in PEG200.30 A higher concentration of PEG200 (40 wt%) further promoted the transformation of the (3 + 1) hybrid-2 structure of Tel24 into a parallel structure (Fig. S1†). However, compared with the CD characteristic peaks of the c-MYC Pu22 and KRAS G-quadruplexes in parallel structures, the CD signal peak of Tel24 in the 40 wt% PEG200 solution did not show a negative signal peak at 295 nm. This result indicated that Tel24 in a solution containing 40 wt% PEG200 still does not display a completely parallel structure.
Fig. 1. Conformation of Tel24 in a solution containing different concentrations of PEG200 was assessed by CD spectroscopy. The concentration of Tel24 was 10 μM.
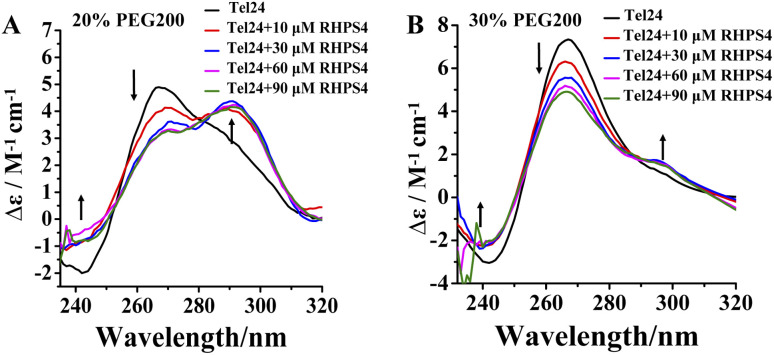
RHPS4 induced a conformational conversion of Tel24 in solutions with 20–30 wt% PEG200 as manifested by increasing the peak at near 240 nm and 295 nm and disappearing the peak at about 265 nm (Fig. 2). The conformational transition of Tel24 in 20 wt% PEG200 solution changed to a (3 + 1) hybrid-2 structure (Fig. 2A). The conformation of Tel24 in 30 wt% and 40 wt% PEG200 solutions did not change significantly. At the same time, an obvious decline was noticed for the positive peak at 265 nm, and an increase was observed for the shoulder peak at 295 nm (Fig. 2B and S2, ESI†). These results suggested that adding RHPS4 might change the conformation distribution of Tel24 under molecular crowding conditions and promote the formation of the (3 + 1) hybrid-2 G-quadruplex to some extent.
Fig. 2. Effects of the RHPS4 on the CD spectra of Tel24 in solutions containing different concentrations of PEG200. (A) 20 wt% PEG200, (B) 30 wt% PEG200.
No conformational change was observed in the parallel conformation of the c-KIT1 G-quadruplex in a molecularly crowded state (Table S1 and Fig. S3, ESI†). The addition of RHPS4 significantly reduced the positive peak at 265 nm of c-KIT1. In comparison, the positive peak remained unchanged at 295 nm. This result indicated that RHPS4 did not influence the conformations of c-KIT1 G-quadruplexes in a single parallel conformation.
Topology changes of Tel24 and Tel24 + RHPS4 under molecular crowding solutions during melting
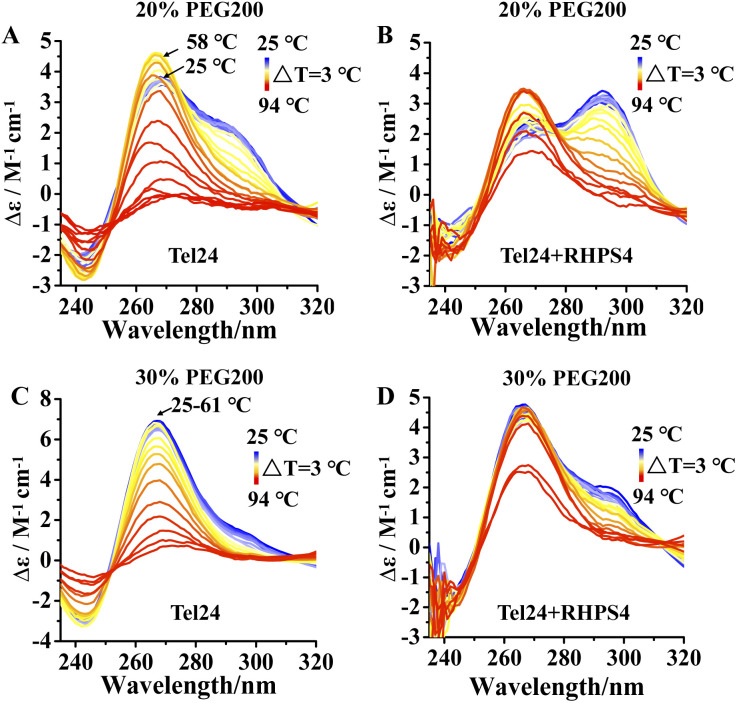
We further probed the conformation changes for Tel24 and Tel24 + RHPS4 under molecular crowding conditions on approaching the melting. Fig. 3 showed the CD spectra of Tel24 and Tel24 + RHPS4 in solution containing 20 wt% to 30 wt% PEG200, respectively, in the temperature range 25–94 °C, with a step of ΔT = 3 °C. At room temperature, the spectrum of Tel24 in 20 wt% PEG200 solutions is characterized by a positive peak at about 265 nm, a shoulder peak near 290 nm, and a negative peak at around 240 nm. In the range of 25–58 °C, a significant decrease was observed for the shoulder peak at 290 nm, while a substantial increase was found for the positive peak at 265 nm. When the temperature reached 58 °C, the CD signal peak began to decrease at 265 nm (Fig. 3A). In contrast, all characteristic peaks of Tel24 in solutions without PEG200 decreased consistently with increasing temperature (Fig. S4, ESI†). It is well known that the positive peak at 265 nm and 290 nm are characteristic of the parallel conformation and hybrid conformation, respectively.19,35 Combined with the results in Fig. 1, it suggested that Tel24 in 20 wt% PEG200 solution may be a conformational ensemble consisting of parallel and (3 + 1) hybrid-2 conformation. Moreover, the parallel conformation of Tel24 is more stable than (3 + 1) hybrid-2. The rate of decrease of the characteristic peaks at 265 nm and 290 nm were significantly slower after the addition of RHPS4 (Fig. 3B). Therefore, it indicated that RHPS4 stabilizes both parallel and (3 + 1) hybrid-2 conformations of Tel24.
Fig. 3. Temperature-dependent unfolding assessed by CD. (A) Spectrum of Tel24 in 20 wt% PEG200 solution, (B) spectrum of Tel24 + RHPS4 in 20 wt% PEG200 solution, (C) spectrum of Tel24 in 30 wt% PEG200 solution, (D) spectrum of Tel24 + RHPS4 in 30 wt% PEG200 solution. The concentration of Tel24 and RHPS4 was 10 μM and 90 μM, respectively. The arrow in panel A indicates that the CD signal peak at 265 nm increased at 25–58 °C and decreased at 58 °C. The arrow in panel C indicates that the CD signal peak at 265 nm is unchanged at 25–61 °C and started to decrease at 61 °C.
The characteristic peak at 265 nm of Tel24 in a 30 wt% PEG200 solution did not change significantly over a range of 25–61 °C. In comparison, the characteristic peak at 290 nm decreased continuously (Fig. 3C). It may be because the parallel conformations dominated the conformational ensemble of Tel24 in the 30 wt% PEG200 solution. Similarly, the rate of decline of characteristic peaks at 265 nm and 290 nm was significantly slowed by the addition of RHPS4 (Fig. 3D).
RHPS4 exhibited higher binding intensity to the (3 + 1) hybrid-2 conformation of Tel24
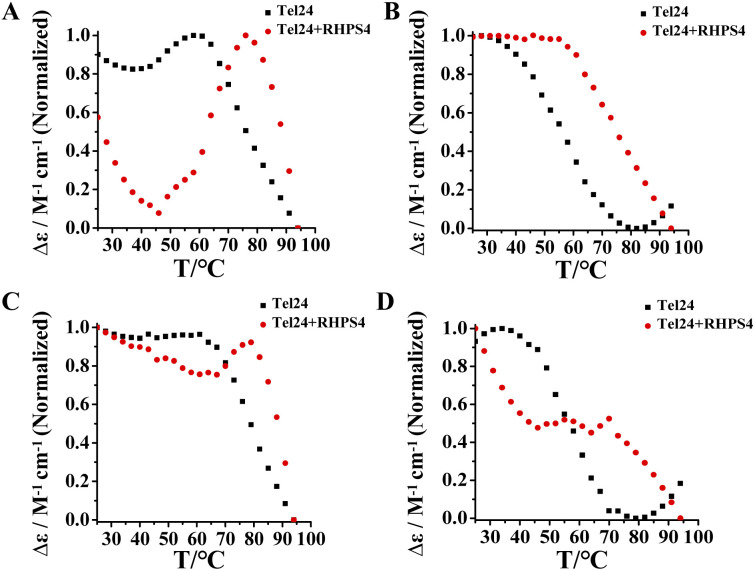
The above results suggested that Tel24 in 20–30 wt% PEG200 solution can form a conformational ensemble consisting of parallel and (3 + 1) hybrid-2 conformations. Consequently, these findings attracted our interest in exploring which conformation in the conformation ensemble of Tel24 was preferentially bonded by RHPS4. Evaluation of the stability of RHPS4 to (3 + 1) hybrid-2 conformation and parallel conformation of Tel24 by comparing the ellipticities measured at 265 nm and 295 nm (Fig. 4). The CD studies showed that although the melting curves at 265 nm and 295 nm did not reach a plateau after adding RHPS4 (Fig. 4), the melting curve at 295 nm was more affected by RHPS4. It suggested that RHPS4 exhibited higher stability to the (3 + 1) hybrid-2 conformation of Tel24 than the parallel conformation. This observation indicated that RHPS4 shifted the conformational ensemble equilibrium of Tel24 towards the (3 + 1) hybrid-2 topology by preferentially stabilizing the (3 + 1) hybrid-2 conformation at room temperature.
Fig. 4. Single wavelengths were melting profiles extracted from the experimental data set at two selected wavelengths. The CD curves display temperature dependence. (A) Spectrum of Tel24 and Tel24 + RHPS4 in 20 wt% PEG200 solution at 265 nm; (B) pectrum of Tel24 and Tel24 + RHPS4 in 20 wt% PEG200 solution at 295 nm; (C) spectrum of Tel24 and Tel24 + RHPS4 in 30 wt% PEG200 solution at 265 nm; (D) spectrum of Tel24 and Tel24 + RHPS4 in 30 wt% PEG200 solution at 295 nm. The concentration of Tel24 was 10 μM. The concentration of RHPS4 was 90 μM.
The fluorescent response with the titration was utilized to derive the association constants (KD) between RHPS4 and Tel24 in solution with or without PEG200 (Fig. S5, ESI†). Compared with Tel24 in 20 wt% PEG200 solutions (KD = 2.50 ± 0.26 μM), Tel24 in dilute solution exhibited higher binding affinity with RHPS4 (KD = 1.84 ± 0.10 μM). It suggested that RHPS4 has a higher affinity to Tel24 in the (3 + 1) hybrid-2 conformation.
Hybrid conformation was easier to form as compared to parallel conformation in the conformation ensemble of Tel24
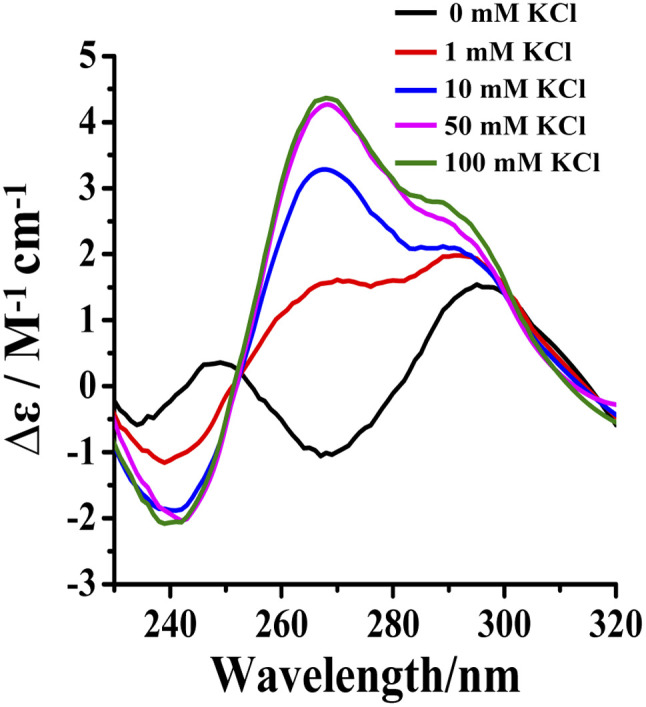
Potassium ions induce the formation of G-quadruplexes.19–21 Tel24 were found to fold into antiparallel (a negative peak at 265 nm, two positive peaks at 240 nm and 295 nm, respectively) and (3 + 1) hybrid-2 conformations in 20 wt% PEG200 solution with low concentrations of KCl (0–1 mM), while it folds into a conformational ensemble of parallel and (3 + 1) hybrid-2 conformation in solution with 10–100 mM KCl (Fig. 5). The intensity of the characteristic peak of Tel24 at 290 nm reached a threshold value in 1 mM KCl solution. However, in 1–100 mM KCl solution, the characteristic peak intensity of Tel24 showed no increase at 290 nm, while a significant increase was detected at 265 nm. Therefore, it predicted that Tel24 first formed (3 + 1) hybrid-2 conformation in 20 wt% PEG200 solution and then created a parallel conformation.
Fig. 5. Conformation of Tel24 in solutions containing 20 wt% PEG200 and different concentrations of KCl assessed by CD spectroscopy. The concentration of Tel24 was 10 μM.
RHPS4 was found to induce Tel24 to form an antiparallel conformation in low concentrations of KCl, while on the other hand, it promoted the parallel conformation conformational ensemble of Tel24 to form hybrid conformation in high concentrations of KCl (Fig. S6, ESI†). Hence, it indicated that RHPS4 interacts with Tel24 in various conformations.
Discussions and conclusions
In summary, Tel24 displayed multiple conformational distributions under different conditions. The conformational ensemble of Tel24 is inherently dynamic and should be a synthesis of parallel and (3 + 1) hybrid-2 structures under molecular crowding conditions. The binding of RHPS4 to the Tel24 is a recognition between RHPS4 and an ensemble consisting of multiple conformations. The studies of CD melting revealed that RHPS4 exhibited a higher stabilizing ability to the (3 + 1) hybrid-2 of Tel24 than the parallel conformation. RHPS4 shifted the conformational ensemble balance of Tel24 towards the (3 + 1) hybrid-2 conformation, which indicated that the (3 + 1) hybrid-2 conformation of Tel24 is the more favored conformation of RHPS4. Following the binding of RHPS4, the conformational ensemble of RHPS4 undergoes a population shift, redistributing the conformational states.
The (3 + 1) hybrid-2 conformation of Tel24 was more likely to form as compared to the parallel conformation in the Tel24 conformational ensemble. Moreover, the (3 + 1) hybrid-2 G-quadruplex was more compact than the parallel G-quadruplex.30 It suggested that the (3 + 1) hybrid-2 conformation may be lower in energy, and RHPS4 readily binds to the lower energy conformation in the ensemble. This study suggests that multiple conformations should be considered when designing G-quadruplex ligands.
Funding
This research was supported by The National Natural Science Foundation of China (Grant No. 31900011) and Open Project Funding of the State Key Laboratory of Biocatalysis and Enzyme Engineering (Grant No. SKLBEE2018010) and Research and Innovation Initiatives of WHPU (2019y05) and Technical Reserve Project from School of Modern Industry for Selenium Science and Engineering, Wuhan Polytechnic University & Enshi Se-Run Material Engineering Technology Co., Ltd. (No. Sel202104).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors thank the research group of Prof. Benmei Wei (School of Chemical and Environmental Engineering, Wuhan Polytechnic University) for providing us with the Chirascan circular dichroism spectrophotometer.
Electronic supplementary information (ESI) available: Details of experimental information, Table S1 and Fig. S1–S6. See https://doi.org/10.1039/d2ra03959a
Notes and references
- Yu H. Gu X. Nakano S. Miyoshi D. Sugimoto N. J. Am. Chem. Soc. 2012;134:20060–20069. doi: 10.1021/ja305384c. [DOI] [PubMed] [Google Scholar]
- Parkinson G. N. Lee M. P. Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Collie G. W. Haider S. M. Neidle S. Parkinson G. N. Nucleic Acids Res. 2010;38:5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsel-Hertsch R. Di Antonio M. Balasubramanian S. Nat. Rev. Mol. Cell Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Lipps H. J. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S. Hurley L. H. Neidle S. Nat. Rev. Drug Discovery. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat P. Zhong J. Lekieffre L. Cowieson N. P. Clancy J. L. Preiss T. Balasubramanian S. Khanna R. Tellam J. Nat. Chem. Biol. 2014;10:358–364. doi: 10.1038/nchembio.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara M. Chem. Commun. 2021;57:8063–8066. doi: 10.1039/d1cc02995a. [DOI] [PubMed] [Google Scholar]
- Moyzis R. K. Buckingham J. M. Cram L. S. Dani M. Deaven L. L. Jones M. D. Meyne J. Ratliff R. L. Wu J. R. Proc. Natl. Acad. Sci. U. S. A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan R. J. Karlseder J. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S. Parkinson G. N. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- Xu Y. Ishizuka T. Kurabayashi K. Komiyama M. Angew. Chem., Int. Ed. Engl. 2009;48:7833–7836. doi: 10.1002/anie.200903858. [DOI] [PubMed] [Google Scholar]
- Hu M. H. Chen S. B. Wang B. Ou T. M. Gu L. Q. Tan J. H. Huang Z. S. Nucleic Acids Res. 2017;45:1606–1618. doi: 10.1093/nar/gkw1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C. Liu Z. Hou H. Ding J. Chen X. Xie C. Song Z. Hu Z. Feng M. Mohamed H. I. Xu S. Parkinson G. N. Haider S. Wei D. Nucleic Acids Res. 2020;48:11259–11269. doi: 10.1093/nar/gkaa870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji B. Bhattacharya S. Chem. Commun. 2014;50:6422–6438. doi: 10.1039/c4cc00611a. [DOI] [PubMed] [Google Scholar]
- Chen C. Y. Wang Q. Liu J. Q. Hao Y. H. Tan Z. J. Am. Chem. Soc. 2011;133:15036–15044. doi: 10.1021/ja204326w. [DOI] [PubMed] [Google Scholar]
- Kaulage M. H. Maji B. Pasadi S. Ali A. Bhattacharya S. Muniyappa K. Eur. J. Med. Chem. 2018;148:178–194. doi: 10.1016/j.ejmech.2018.01.091. [DOI] [PubMed] [Google Scholar]
- Ambrus A. Chen D. Dai J. Bialis T. Jones R. A. Yang D. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J. Punchihewa C. Ambrus A. Chen D. Jones R. A. Yang D. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu K. N. Phan A. T. Kuryavyi V. Lacroix L. Patel D. J. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. J. Curr. Opin. Struct. Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- Huff H. C. Maroutsos D. Das A. Protein Sci. 2019;28:928–940. doi: 10.1002/pro.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddi B. Phan A. T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011 Jun 29;133(25):9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- Trajkovski M. Endoh T. Tateishi-Karimata H. Ohyama T. Tanaka S. Plavec J. Sugimoto N. Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations. Nucleic Acids Res. 2018;46(8):4301–4315. doi: 10.1093/nar/gky250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S. Tateishi-Karimata H. Takahashi S. Ohyama T. Sugimoto N. Effect of Molecular Crowding on the Stability of RNA G-Quadruplexes with Various Numbers of Quartets and Lengths of Loops. Biochemistry. 2020;59(28):2640–2649. doi: 10.1021/acs.biochem.0c00346. [DOI] [PubMed] [Google Scholar]
- Ou Z. Z. Wang Y. Q. Gao Y. Y. B Wang X. M Qian Y. Li Y. Wang X. S. Targeting human telomeric and c-myc G-quadruplexes with alkynylplatinum(II) terpyridine complexes under molecular crowding conditions. J. Inorg. Biochem. 2017;166:126–134. doi: 10.1016/j.jinorgbio.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Matsumoto S. Tateishi-Karimata H. Ohyama T. Sugimoto N. Effect of DNA modifications on the transition between canonical and non-canonical DNA structures in CpG islands during senescence. RSC Adv. 2021;11(59):37205–37217. doi: 10.1039/d1ra07201c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T. T. Sato S. Yasukawa R. Takeuchi R. Ozaki S. Fujii S. Takenaka S. The Interaction of Cyclic Naphthalene Diimide with G-Quadruplex under Molecular Crowding Condition. Molecules. 2020;25(3):668. doi: 10.3390/molecules25030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y. Kan Z. Y. Wang Q. Yao Y. Liu J. Hao Y. H. Tan Z. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- Trajkovski M. Endoh T. Tateishi-Karimata H. Ohyama T. Tanaka S. Plavec J. Sugimoto N. Nucleic Acids Res. 2018;46:4301–4315. doi: 10.1093/nar/gky250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatak P. Cookson J. C. Dai F. Smith V. Gartenhaus R. B. Stevens M. F. Burger A. M. Br. J. Cancer. 2007;96:1223–1233. doi: 10.1038/sj.bjc.6603691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner T. P. Williams H. E. Gluszyk K. I. Roe S. Oldham N. J. Stevens M. F. Moses J. E. Searle M. S. Org. Biomol. Chem. 2009;7:4194–4200. doi: 10.1039/b910505k. [DOI] [PubMed] [Google Scholar]
- Eugene N. Agnieszka S. Noël B. Analysis of Fluorometric Titration Curves. J. Phys. Chem. A. 2000;104(22):5388–5395. [Google Scholar]
- Del Villar-Guerra R. Trent J. O. Chaires J. B. Angew. Chem., Int. Ed. Engl. 2018;57:7171–7175. doi: 10.1002/anie.201709184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.