Abstract
The gram-negative pathogen Porphyromonas gingivalis requires hemin for growth. Hemoglobin bound to haptoglobin and hemin complexed to hemopexin can be used as heme sources, indicating that P. gingivalis must have a means to remove the hemin from these host iron-binding proteins. However, the specific mechanisms utilized by P. gingivalis for the extraction of heme from heme-binding proteins and for iron transport are poorly understood. In this study we have determined that a newly identified TonB-dependent hemoglobin-hemin receptor (HmuR) is involved in hemoglobin binding and utilization in P. gingivalis A7436. HmuR shares amino acid homology with TonB-dependent outer membrane receptors of gram-negative bacteria involved in the acquisition of iron from hemin and hemoglobin, including HemR of Yersinia enterocolitica, ShuA of Shigella dysenteriae, HpuB of Neisseria gonorrhoeae and N. meningitidis, HmbR of N. meningitidis, HgbA of Haemophilus ducreyi, and HgpB of H. influenzae. Southern blot analysis confirmed the presence of the hmuR gene and revealed genetic variability in the carboxy terminus of hmuR in P. gingivalis strains 33277, 381, W50, and 53977. We also identified directly upstream of the hmuR gene a gene which we designated hmuY. Upstream of the hmuY start codon, a region with homology to the Fur binding consensus sequence was identified. Reverse transcription-PCR analysis revealed that hmuR and hmuY were cotranscribed and that transcription was negatively regulated by iron. Inactivation of hmuR resulted in a decreased ability of P. gingivalis to bind hemoglobin and to grow with hemoglobin or hemin as sole iron sources. Escherichia coli cells expressing recombinant HmuR were shown to bind hemoglobin and hemin. Furthermore, purified recombinant HmuR was demonstrated to bind hemoglobin. Taken together, these results indicate that HmuR serves as the major TonB-dependent outer membrane receptor involved in the utilization of both hemin and hemoglobin in P. gingivalis.
The ability of a pathogen to scavenge essential nutrients within a particular environmental niche in the host is essential for the initiation and the establishment of an infection. Of these essential nutrients, iron plays a crucial role. Within the human host, the majority of iron is found in the form of heme proteins, including hemoglobin, or ferritin. Due to the abundance of heme proteins in the host, they are a valuable source of iron for bacterial pathogens. As a consequence, pathogenic organisms have developed diverse mechanisms for the acquisition of heme under the iron-limiting environment of the host (1, 9, 10, 12, 15, 19, 28, 40). The best-described system by which gram-negative bacteria acquire heme involves direct binding of free heme or heme proteins to specific outer membrane receptors (9). Energy for the transport of iron and/or heme ligands via these specific heme and hemoglobin receptors across the outer membrane into the periplasmic space is dependent on TonB, in association with the ExbB and ExbD proteins (5, 30). Recently, an additional system for the acquisition of heme involving an extracellular heme binding protein that functions to capture and shuttle heme to a specific outer membrane receptor has been described. In Serratia marcescens, the secreted protein HasA extracts heme from either hemopexin-heme or hemoglobin and delivers it to the outer membrane receptor HasR (17). Similar systems have been described in Haemophilus influenzae and Pseudomonas aeruginosa (10, 23).
Porphyromonas gingivalis, the etiological agent of adult periodontal disease, requires iron in the form of hemin for growth (13, 14) due to its inability to synthesize protoporphyrin IX, which it requires as the prosthetic group of cytochrome b. The latter serves as an electron sink during amino acid fermentation (8). Hemoglobin bound to haptoglobin and hemin complexed to hemopexin can be used as iron sources by P. gingivalis, indicating that this microorganism has a mechanism for removing the hemin from these host iron-binding proteins (4). In addition, P. gingivalis is capable of utilizing transferrin, found in serum, and lactoferrin, found on mucosal surfaces, for growth (13, 14). The characteristic black pigmentation produced by P. gingivalis colonies is due to the accumulation of μ-oxo dimers of hemin on the cell's surface (37). We have previously determined that P. gingivalis is capable of transporting the intact hemin molecule into the cell by an energy-dependent process (14). The energy dependence of hemin transport in P. gingivalis suggests that a TonB analog may function to transduce energy for the transport of hemin.
Hemin binding by P. gingivalis appears to occur through both high- and low-affinity receptors (13), and recent studies suggest that a common pathway may be utilized for the transport of hemin and hemoglobin (13); however, little is known regarding the specific P. gingivalis receptors for either ligand's binding. Hemin-binding proteins either induced by hemin limitation (4) or repressed by excess of this compound (38) have been described, but their role in hemin transport has not been further defined. Recently, two P. gingivalis TonB-dependent receptors, HemR and Tla, have been described (1, 19). The hemR gene from P. gingivalis 53977 exhibits homology to genes involved in iron acquisition in other bacterial species; however, conclusive evidence for the role of HemR in iron uptake from hemin or hemoglobin has not been reported (19). The Tla protein is required for growth of P. gingivalis with low levels of hemin; however, its role as a specific hemin receptor has not been defined.
Although previous studies have documented the ability of P. gingivalis to utilize hemoglobin as a sole iron source, receptors involved in the binding of this compound to the P. gingivalis cell have not been identified. Recent studies have reported that the lysine- and arginine-specific gingipains Kgp and HRgpA (31) can bind and subsequently cleave hemoglobin (11, 24; Sroka et al., submitted; C. A. Genco, A. E. Sroka, and J. Potempa, unpublished data). It is not clear which part of the Kgp complex participates in hemoglobin binding, since reports indicate that either the catalytic domain or the hemagglutinin domain is involved (11, 12, 21, 28, 29; Sroka et al., submitted; Genco et al., unpublished). Depending on the strain and cultivation conditions, a variable amount of gingipains remain attached to the outer membrane or are secreted into the growth medium (16). While Kgp can function in hemoglobin binding, it may be premature to categorize it as an outer membrane receptor. The amino acid sequence of Kgp has no similarity to the TonB-dependent outer membrane proteins, indicating that a separate TonB-dependent outer membrane protein is responsible for binding and transport of heme from hemoglobin into the cell.
Previous studies in our laboratory have demonstrated that a common mechanism exists for the transport of both hemin and hemoglobin in P. gingivalis. In this study we report the characterization of the structural gene for a novel P. gingivalis TonB-dependent outer membrane receptor (HmuR) which is required for both hemoglobin and hemin binding and utilization in P. gingivalis. Inactivation of hmuR resulted in a diminished ability of P. gingivalis to bind hemoglobin and to grow with hemoglobin or hemin as sole iron sources. Furthermore, E. coli cells expressing the membrane-bound recombinant HmuR (rHmuR) were shown to bind both hemoglobin and hemin, and purified rHmuR was demonstrated to bind hemoglobin.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The P. gingivalis and Escherichia coli strains used in this study are indicated in Table 1. P. gingivalis wild-type strains were maintained on anaerobic blood agar (ABA) plates (Remel, Lenexa, Kans.). P. gingivalis strains WS1 was maintained on ABA plates supplemented with 1 μg of erythromycin per ml. All P. gingivalis cultures were incubated at 37°C in an anaerobic chamber (Coy Laboratory Products, Ann Arbor, Mich.) with 85% N2, 5% H2, and 10% CO2 for 3 to 5 days. Following incubation at 37°C, cultures were inoculated in Anaerobe Broth MIC (Difco, Detroit, Mich.) and incubated at 37°C (under anaerobic conditions) for 24 h. E. coli was typically maintained in Luria-Bertani (LB) medium (Sigma, St. Louis, Mo.), supplemented with appropriate antibiotics and incubated aerobically with shaking.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| P. gingivalis | ||
| A7436 | Wild type | Lab collection |
| 381 | Wild type | Lab collection |
| W50 | Wild type | T. van Dyke, Boston University, Boston, Mass. |
| ATCC 33277 | Wild type | Lab collection |
| ATCC 53977 | Wild type | P. Baker, Bates College, Lewiston, Maine |
| WS1 | A7436, hmuR::erm | This study |
| E. coli | ||
| DH5α | recA1 lacZYA-argF supE44 | Promega |
| TOP10F′ | F′ [laclq Tn10(Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 (ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| BL21(DE3)pLysE | F−ompT hsdSB(rB− mB−) gal dcm (DE3)pLysE (Camr) | Invitrogen |
| Plasmids | ||
| pGEM3z(−) | Ampr | Promega |
| pWS1 | pGEM3z(−) containing 485-bp N terminus of hmuR and the B. fragilis ermF cassette within the PstI site of the hmuR gene | This study |
| pCRT7/CT-TOPO | Ampr | Invitrogen |
| pTO1 | pCRT7/CT-TOPO containing the hmuR gene without the signal peptide sequence | This study |
| pTO2 | pCRT7/CT-TOPO containing the hmuR gene with the signal peptide sequence | This study |
To examine the ability of P. gingivalis to grow with different iron sources, P. gingivalis strains A7436 and WS1 were grown on anaerobic blood agar at 37°C for 3 days and then inoculated into Schaedler broth supplemented with 150 μM dipyridyl to chelate iron and incubated at 37°C under anaerobic conditions for 24 h. This served as the inoculum into Schaedler broth supplemented with 150 μM dipyridyl plus hemin (1.5 μM), hemoglobin (1.5 μM), or ferric chloride (100 μM). Prior to the addition of hemoglobin, 0.1% human serum albumin was added to sequester free heme. For some experiments, cultures were grown in basal medium (BM; Trypticase peptone, 10 g; tryptophan, 0.2 g; NaCl, 2.5 g; sodium sulfite, 0.1 g, and cysteine 0.4 g [per liter]) (13).
Isolation of the P. gingivalis hmuR locus.
The P. gingivalis hmuR gene and upstream sequences were initially identified on a 5.3-kb DNA fragment from the A7436 cosmid library (36). The carboxy-terminal sequence was obtained by sequencing a second P. gingivalis strain A7436 clone which contained downstream DNA sequences. The hmuR DNA sequence was further confirmed by DNA sequence analysis of a PCR fragment corresponding to the entire hmuR gene. PCR amplification of P. gingivalis A7436 genomic DNA using primers F1 and R1 (Table 2) was carried out with Vent DNA polymerase (New England Biolabs, Beverly, Mass.) at 94°C for 1 min, 40°C for 2 min, and 72°C for 2 min for two cycles in a DNA Thermacycler 480 (Perkin-Elmer, Norwalk, Conn.). This was followed by 25 cycles at 94°C for 1 min, 55°C for 2 min, and 72°C for 2 min. The resulting PCR fragment was sequenced as described below. Southern blot analyses and genomic DNA isolations were performed as previously described (38).
TABLE 2.
Primers and probes used in this study
| Primer pair or probea | Sequence | Descriptionb |
|---|---|---|
| F1 | ATAAGTTAAGAGGGAAATATG | Amplifies entire 1.94-kb hmuR gene |
| R1 | CATTTCGCACCCATGCCGAAG | |
| F2 | ACTGGAATTCGTGTAGTAACAAAGCAG | Amplifies 505 bp (8 to 493 nt) of hmuR gene; PCR product is probe 2 |
| R2 | ACTGAAGCTTTGATGATATTTGATAACACC | |
| F3 | ACGTGAATTCGTGTAGTAACAAAGCAG | Amplifies 855 bp (8 to 853 nt) of hmuR gene |
| R3 | GCTGATACGCCAGTTGGCA | |
| F4 | GAAATGGATCAGGCTATCTAC | Amplifies 1.2-kb junction fragment of hmuY-hmuR |
| R4 | GCTGATACGCCAGTTGGCA | |
| F5 | GGTAAGCACCTGAAGACTTATG | Amplifies 469 bp (84 to 552 nt) of the sod gene |
| R5 | CCAGTCAACAATACTCCAAAGA | |
| F6 | GAAATGGATCAGGCTATCTAC | Amplifies 300 bp (85 to 384 nt) of the hmuY gene |
| R6 | GAGTTCTCCATCCTGATA | |
| F7 R7 | ATGGCCAACCCTCCGGCCCAACCTA GAAAGTGATCCGAACCAACCCGTAT | Amplifies the hmuR gene without the signal peptide and without stop codon |
| F8 | ATGAAAAGTCTAGTAACAAAGCAGG | Amplifies the hmuR gene with signal peptide and without stop codon |
| R7 | GAAAGTGATCCGAACCAACCCGTAT | |
| Probe 1 | ClaI-ClaI-digested internal fragment (696 bp) of the hmuR (nt 791 to 1436) gene (has a HindIII site at nt 1387) |
F, forward; R, reverse.
nt, nucleotide(s).
DNA sequencing and computer analysis.
DNA sequencing of P. gingivalis A7436 clones and the PCR fragment corresponding to the entire hmuR gene was performed using the PRISMTM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin Elmer, Foster City, Calif.) and 373A DNA sequencer. Computer analysis was performed as outlined by the Intelligenetics Suite and BLAST programs.
GenBank accession numbers.
The sequences of the hmuR and hmuY genes were deposited into GenBank under accession numbers U87395 and 300705, respectively. The partial sequence of hmuR (previously designated hemB) was previously deposited under the same accession number and subsequently modified.
RT-PCR.
P. gingivalis cultures were grown to the mid-logarithmic phase in anaerobic broth supplemented with 165 μM dipyridyl or anaerobic broth with dipyridyl plus hemin (1.5 μM). Total RNA was isolated using the RNagents Kit (Promega, Madison, Wis.). Samples were initially treated with DNase prior to reverse transcription-PCR (RT-PCR). To 1.0 μg of total RNA was added 1 μl of 10× DNase I buffer, 1 μl of DNase (Promega) and diethyl pyrocarbonate (DEPC)-treated water to achieve a final volume of 10 μl. Samples were incubated at room temperature for 15 min. DNase I was inactivated by the addition of 1 μl of 25 mM EDTA to the reaction mixture. The samples were then heated to 65°C for 10 min and placed on ice. Primers used in PCR included hmuR- and sod-specific primers, as well as a primer representing an hmuY-hmuR-specific junction fragment (Table 2). To the RNA samples was added 25 μl of 2× reaction mix, 100 ng of each primer, 1 μl of reverse transcriptase-Taq mix, and DEPC-treated water to a final volume of 50 μl. The samples were overlaid with mineral oil and placed in a DNA Thermocycler (Perkin-Elmer). cDNA synthesis was performed at 50°C for 30 min, followed by predenaturation at 94°C for 2 min. PCR amplification was carried out using the following parameters: denaturation at 94°C for 1 min, annealing at 54°C for 2 min, and elongation at 72°C for 2 min, for 30 cycles.
Construction and isolation of a P. gingivalis hmuR mutant.
Primers F2 and R2 were used to amplify the region corresponding to bp 8 to 493 of the hmuR gene, yielding a DNA fragment of 485 bp (Table 2). To the forward primer, four nonspecific bases and an EcoRI restriction site were added. To the reverse primer, four nonspecific bases and an HindIII site were added. These additions increased the final size of the PCR product to 505 bp. This PCR fragment was cloned into pGEM3z (Promega), and the hmuR fragment was then interrupted by the insertion of the ermF gene (32) into the PstI site of the hmuR DNA fragment. The resulting plasmid (pWS1) was transformed into E. coli JM109 (Promega), and the insertion of ermF (with flanking sequences) (31) into the hmuR fragment was confirmed by DNA sequencing. pWS1 was introduced into P. gingivalis A7436 by electroporation briefly as follows. P. gingivalis A7436 was inoculated into anaerobe broth to an initial optical density at 660 nm (OD660) of 0.1 and incubated anaerobically for 6 h (final OD660 = 0.4). The P. gingivalis culture was then centrifuged at 10,000 × g for 10 min and washed with electroporation buffer (1 mM MgCl2, 10% glycerol), and the pellet was mixed with 200 ng of pWS1 DNA and placed in a 2.5-cm electroporation cuvette. Electroporation was carried out at 25 μF, 200 Ω, and 2.5 V and resulted in time constants of 3.1 to 3.4 s. The P. gingivalis A7436 alone was also electroporated and used as a negative control. After electroporation, 800 μl of the anaerobic broth was added, and the cells were incubated overnight at 37°C under anaerobic conditions. Samples were centrifuged, 900 μl of supernatant was removed, the pellet was resuspended in the remaining 100 μl of supernatant, and the culture was plated onto an ABA plate containing 1 μg of erythromycin per ml. The plates were incubated under anaerobic conditions at 37°C for 7 to 10 days as described above. Individual transformants were isolated, and insertion of the ermF gene in the P. gingivalis hmuR mutants (WS1, WS2, WS4, and WS5) was confirmed by Southern blot analysis. The mutation in the hmuR gene was further confirmed in P. gingivalis WS1 by PCR analysis using primers specific for the 5′ and 3′ portions of the hmuR gene.
Construction of the HmuR expression plasmid.
The hmuR gene was PCR amplified from 100 ng of total genomic DNA obtained from P. gingivalis A7436 (94°C for 30 s, 60°C for 30 s, and 72°C for 2 min, followed by 30 min at 72°C; 25 cycles). The forward primers (F6 and F7, Table 2) were designed to produce hmuR either with or without its native signal peptide sequence. The reverse primer (R6, Table 2) was designed to remove the native stop codon and preserve the reading frame through the C-terminal tag. The amplified products were purified and cloned into the vector pCRT7/CT-TOPO (Invitrogen, Carlsbad, Calif.), which contains sequences coding for the V5 epitope and polyhistidine (His6) regions. The resulting plasmids (pTO1 and pTO2) were transformed into E. coli TOP10F′, and transformants were selected on LB plates containing 100 μg of ampicillin per ml. The hmuR insert was confirmed by restriction analysis, PCR, and DNA sequence analysis.
Expression of rHmuR in E. coli.
E. coli BL21(DE3)pLysE cells (Invitrogen) were transformed with pTO1 or pTO2, and transformants were selected on LB medium or minimal medium (M9) containing 100 μg of ampicillin and 34 μg of chloramphenicol per ml. Then, 1 ml of the overnight culture was inoculated into fresh 10 ml of LB medium or M9 supplemented with both antibiotics and grown at 37°C to an OD600 of 0.5 to 0.6. To induce the expression of the cloned P. gingivalis hmuR gene, isopropyl β-d-thiogalactopyranoside (IPTG; Sigma) was added to a final concentration of 0.5 to 1.0 mM, and growth was continued for 5 h. Samples were removed at hourly intervals, centrifuged, and frozen at −20°C. E. coli cells harboring a plasmid expressing the lacZ gene, pCRT7/CT-LacZ (Invitrogen, Carlsbad, Calif.) was utilized as a positive control, and the vector alone was used as a negative control.
SDS-PAGE and Western blotting.
Samples taken before and 1 to 5 h after IPTG induction were suspended in 2 × Laemmli sample buffer, boiled for 5 min and examined by polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) on 12% gels (22). The proteins were either stained with Coomassie brilliant blue R-250 (CBB; Bio-Rad, Hercules, Calif.) or were transferred (43) onto nitrocellulose membranes (Bio-Rad) in 30 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer (pH 11.0; Sigma) for 1 h at constant current of 0.2 A. Western blotting was carried out according to the method of Burnette (7) with slight modifications. Membranes were incubated for 2 h at room temperature in 50 mM Tris-HCl (pH 7.5) containing 0.15 M NaCl (TBS) and 3% skim milk. After washing with TBS containing 0.05% Tween 20 (TTBS), anti-fusion protein antibody conjugated with horseradish peroxidase (mouse anti-V5-HRP; Invitrogen) in TBS containing 1% skim milk was added, and this was incubated for 2 h at room temperature. Membranes were washed with TTBS and in the final step with TBS. Chemiluminescence detection was performed within 1 min at room temperature using the ECL System (Amersham Pharmacia Biotech, Piscataway, N.J.). Autoradiography films (Amersham Pharmacia Biotech) were exposed for 1 to 5 min and then developed. Electrophoresis of rHmuR purified from membrane fraction, for N-terminal sequencing, was carried out according to the method of Schagger and von Jagow (35) and transferred onto a polyvinylidene difluoride (PVDF) (Bio-Rad) membrane as indicated above.
Purification of rHmuR.
Following a 5-h IPTG induction period E. coli BL21(DE3)pLysE cells harboring pTO1 or pTO2 were harvested by centrifugation for 20 min at 8,000 × g. The pellet was resuspended in 20 mM phosphate buffer (pH 7.4) containing 0.14 M NaCl (PBS), supplemented with protease inhibitors (Complete EDTA-free; Roche Molecular Biochemicals, Indianapolis, Ind.), frozen and thawed three times, and passed through a French press. After centrifugation for 15 min at 25,000 × g, the pellet (containing inclusion bodies) was resuspended in PBS containing protease inhibitors. The remaining supernatant was centrifuged for 1 h at 70,000 × g to obtain the total membrane fraction. To purify rHmuR, frozen samples containing inclusion bodies or samples containing membrane fractions were thawed, and purification was performed according to Invitrogen's procedure using Ni-chelate chromatography under denaturing conditions. The protein was eluted from the column with urea buffer (pH 4.0), dialyzed against PBS containing decreasing concentrations of urea and 0.1% octyl-d-glucopyranoside (OG; Sigma) and finally dialyzed against PBS containing 0.5 M urea and 0.1% OG. After centrifugation samples were concentrated using Centriprep-10 (Amicon, Beverly, Mass.), and the protein concentration was determined by the bicinchoninic acid method (39).
Hemoglobin binding to rHmuR.
rHmuR purified using Ni-chelate chromatography was transferred onto a nitrocellulose membrane and probed with 100 ng of human hemoglobin (Sigma) per ml, which was biotinylated (18) according to the Pierce's protocol (Pierce, Rockford, Ill.). Hemoglobin binding to rHmuR was determined using streptavidin conjugated with horseradish peroxidase (Roche Molecular Biochemicals) and chemiluminescence detection as described above.
Binding of hemoglobin and hemin by E. coli cells expressing HmuR.
Detection of rHmuR on the surface on E. coli strain BL21(DE3)pLysE was carried out by dot blot assay using antibodies to the fusion protein as discussed above. E. coli expressing rHmuR deposited in inclusion bodies (cells transformed with pTO1) or membrane bound (cells transformed with pTO2), and cells containing plasmid alone were harvested before and after IPTG induction, washed with PBS, and adjusted to an OD600 of 1.0. Aliquots of the cell suspension (0.8 ml) were mixed with 0.2 ml of human hemoglobin dissolved in PBS (final concentration, 5 μM) or hemin dissolved in dimethyl sulfoxide (final concentration, 10 μM). Samples were incubated for 1 h at 37°C and centrifuged, and the OD400 of the resulting supernatant was determined. Adsorbed hemoglobin or hemin was evaluated by determining the decrease of the absorbance of the supernatant and was recorded as the percentage of the initial hemoglobin or hemin. Samples containing hemoglobin, hemin, or cells only were incubated under the same conditions and served as appropriate controls.
RESULTS
Characterization of the P. gingivalis hmuR gene.
To identify genes required for iron transport from hemin and hemoglobin in P. gingivalis, we initially utilized transpositional mutagenesis with the Bacteroides fragilis transposon Tn4351 and identified a mutant of P. gingivalis (MSM-3) which grew poorly with hemin or hemoglobin as sole iron sources (14). Further characterization of P. gingivalis MSM-3 revealed that introduction of Tn4351 resulted in the mobilization of the endogenous insertion sequence element IS1126 in the P. gingivalis MSM-3 genome (36). Characterization of the first additional IS1126 insertion site revealed that it had inserted into the promoter region of the gene encoding the P. gingivalis Kgp protein (kgp). The hemin-hemoglobin defect in P. gingivalis MSM-3 was thus attributed to the inactivation of kgp (36). To characterize the second additional IS1126 insertion site, an oligonucleotide specific to its flanking sequences was used to probe a P. gingivalis A7436 cosmid library. Nucleotide sequencing of a positive clone resulted in the fortuitous identification of a novel P. gingivalis gene (hmuR), which is characterized in this study. The initial 1,050 bp of the P. gingivalis hmuR gene was identified as part of a 5.3-kb DNA fragment from the P. gingivalis A7436 cosmid library. The DNA sequence corresponding to the carboxy terminus of hmuR was obtained following sequencing of a second clone containing downstream sequences. The sequence of the entire hmuR gene from strain A7436 was further confirmed following sequencing of a PCR fragment obtained from strain A7436 using primers F1 and R1 (see Table 2). The hmuR gene from strain A7436 is composed of 1,941 nucleotides and encodes for a 73-kDa predicted protein with a pI of 8.8. Analysis of the HmuR predicted protein using the SignalP program revealed a likely signal peptide cleavage site between Ala24 and Ala25. Further analysis using the Kyte and Doolittle plot program demonstrated that HmuR is hydrophobic, as is typical of outer membrane receptors (data not shown).
The P. gingivalis hmuR gene shares homology with genes whose products have been shown to be TonB-dependent outer membrane receptors involved in iron acquisition. These include the Y. enterocolitica HemR (55% identity), which is a member of a well-defined hemin uptake operon, the Shigella dysenteriae ShuA (54% identity); the E. coli CirA, FhuE, and ChuA (42, 39, and 51% identities, respectively); the Campylobacter coli CfrA (41% identity); and the V. cholerae IrgA (39% identity). Two regions of the translated open reading frame (ORF) of HmuR (residues 33 to 39 and 135 to 170) exhibited extensive sequence similarity to TonB boxes I and IV; homology between the P. gingivalis hmuR gene and the TonB-dependent receptors was most pronounced in the region which corresponds to TonB IV (Fig. 1A).
FIG. 1.
Conserved TonB Box IV, amino acid motifs, and amino acid residues in the P. gingivalis HmuR protein. (A) Homology between the P. gingivalis HmuR and the TonB box IV regions of several different heme and hemoglobin receptors and siderophore and vitamin B12 receptors. The E. coli TonB consensus sequence is also depicted. (B) Homology between the P. gingivalis HmuR protein and the carboxy-terminal region of several different heme and hemoglobin receptors and siderophore and vitamin B12 receptors. Conserved amino acids between the P. gingivalis HmuR and the consensus sequence are indicated by boldface letters. The numbers indicate the position in the unprocessed protein of the first amino acid listed. Pg, P. gingivalis; Vv, V. vulnificus; Vc, V. cholerae; Sd, S. dysenteriae; Ye, Y. enterocolitica; Hi, H. influenzae; Hd, H. ducreyi; Nm, N. meningitidis; Pf, P. fluorenscens; and Pa, P. aeruginosa.
As we were conducting these studies, a gene from P. gingivalis 53977 (hemR), which also exhibits homology to genes involved in iron acquisition from several gram-negative organisms, was identified (19). Comparison of the hmuR and hemR sequences revealed that the N-terminal region of the hmuR gene was identical to the initial 516 bases of the P. gingivalis hemR gene. However, after bp 516, no identity was observed between the P. gingivalis hmuR and hemR genes (36). HemR exhibits homology to Vibrio cholerae IrgA (41%), Y. enterocolitica HemR (25%), E. coli BtuB (36%), E. coli CirA (35%), E. coli IutA (29%), E. coli FecA (29%), E. coli FhuA (25%), and Y. enterocolitica FoxA (27%). Interestingly, we found that the carboxy-terminal region of HmuR exhibited significant sequence similarity to proteins involved in heme and hemoglobin binding and utilization (Fig. 1B). These include the major hemoglobin receptors in N. gonorrhoeae and N. meningitidis, HmbR and HpuB (35 and 41% identity, respectively), and the hemoglobin receptors in H. ducreyi HgbA (48% identity) and H. influenzae HgpB (41% identity) (Fig. 1B and references 8, 25, 26, 34, and 40). Amino acid comparisons of the conserved domains of these heme and hemoglobin receptors, as well as several siderophore and vitamin B12 receptors, revealed a highly conserved receptor domain containing invariant histidine residues and FRAP and NPNL amino acid boxes (6). These residues were also conserved in the P. gingivalis HmuR hemoglobin receptor (Fig. 1B). The conserved histidine residues were present in the P. gingivalis HmuR protein at positions 95 and 434, an Arg-Ala-Pro sequence from residues 421 to 423, and an Asp-Pro-Asp-Leu motif from residues 442 to 445. We also identified a number of conserved glutamic acid residues which were common to P. gingivalis HmuR and to several of the heme and hemoglobin receptors (Fig. 1B).
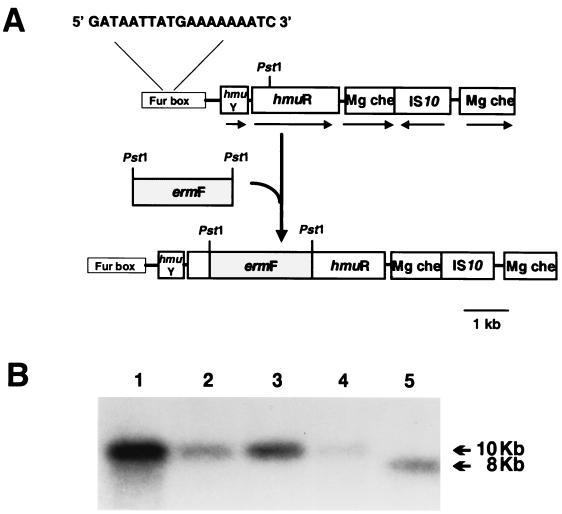
Upstream of the hmuR gene we identified an ORF of 429 bp predicted to encode a 143-amino-acid (aa) protein which we designated hmuY. Sequence analysis of HmuY revealed an ATP-GTP-binding loop (aa 21 to 28), suggesting that it may function as an ATPase. The hmuY gene exhibited 99% identity with a previously identified ORF (ORF1) located upstream of the P. gingivalis hemR gene in strain 53977 which has been proposed to function as a DNA binding protein (19). Located 228 bp upstream of the hmuY start codon, a 19-bp putative Fur box was identified (5′-GATAATTATGAAAAAAATC-3′; see Fig. 4). This Fur box is identical to that found upstream of ORF1 (18) and exhibits 68% identity (13 of 19 bases identical) to the E. coli consensus Fur box sequence. Internal regions of HmuY exhibited 76 and 89% identities with two peptides previously demonstrated to bind hemin as assessed by SDS-PAGE and TMBZ analysis (20). Located 36 bp downstream of hmuR in P. gingivalis A7436, we identified an ORF which shares homology with the gene encoding Mg chelatase (mg che). Interestingly, we found that in strain A7436 this gene was disrupted by an insertion sequence exhibiting 100% identity to the E. coli IS10 element (see Fig. 4).
FIG. 4.
Construction of the hmuR mutant WS1. (A) The P. gingivalis hmuR insertional mutant (WS1) was constructed following insertion of the ermF cassette (with flanking sequences) in the PstI site of hmuR. The direction of transcription is indicated by an arrow, and the PstI restriction site(s) of hmuR and ermF are noted. Also indicated is the map of hmuR region from P. gingivalis A7436. Upstream of the hmuR gene we identified an ORF of 438 bp (hmuY), predicted to encode a 145-aa protein. The promoter region of hmuY contains a putative Fur consensus binding sequence (13 of 19 bases identical to the E. coli Fur box; the Fur box is not drawn to scale). Located downstream of hmuR, an ORF which encodes a putative Mg chelatase (mg che), which was disrupted by a gene encoding a IS10-like element, was identified. (B) Southern blot analysis of chromosomal DNA from P. gingivalis hmuR mutants probed with an hmuR-specific probe (see Table 2). Lanes 1 to 4, genomic DNA from four separate transformants (WS1, WS2, WS4, and WS5); lane 5, genomic DNA from A7436. Introduction of the ermF cassette adds ∼2.0 kb, causing a shift in the hmuR band.
Presence of hmuR in different P. gingivalis strains.
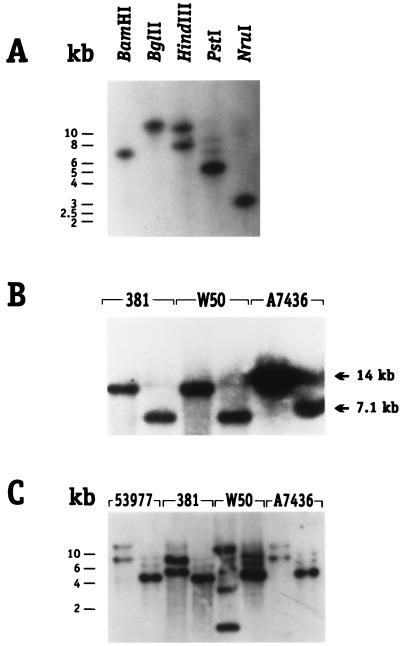
To confirm the presence of a single copy of hmuR in P. gingivalis, a probe derived from the carboxyl terminus (probe 1, Table 2) which is specific for hmuR was used in Southern blot analysis. Digestion of DNA derived from P. gingivalis A7436 with various restriction enzymes confirmed that hmuR is present in a single copy in this strain (Fig. 2A). A search of the unfinished P. gingivalis strain W83 genomic sequence database of The Institute for Genome Research (TIGR [http://www.tigr.org]), also revealed the presence of an ORF that exhibited 99% homology to the hmuR gene from strain A7436 (data not shown). To further confirm that hmuR was present in other P. gingivalis strains, Southern blot analysis with an N-terminal probe (probe 2, Table 2) was performed as shown in Fig. 2B. We observed a similar banding pattern in the P. gingivalis strains examined (A7436, W50, and 381), indicating that the N-terminal region of the hmuR gene is highly conserved. Since the probe used also recognizes a sequence present within the hemR gene, we cannot, however, rule out the possibility that observed reactivity is due to hemR sequences, which may exist in strains W50 and 381.
FIG. 2.
Southern blot analysis of DNA from P. gingivalis strains using an hmuR-specific probe. (A) P. gingivalis A7436 genomic DNA digested with various enzymes as indicated. The probe used was a carboxy-terminal probe (probe 1, Table 2). (B) Southern blot analysis of chromosomal DNA from P. gingivalis strains 381, W50, and A7436 digested with HindIII (first lane for each strain) and PstI (second lane for each strain). The probe used was an amino terminal probe (probe 2, Table 2). Fragment sizes are indicated with arrows. (C) Southern blot analysis of chromosomal DNA from P. gingivalis strains 53977, 381, W50, and A7436 digested with HindIII (first lane for each strain) and PstI (second lane for each strain). The probe used in both panels A and C was a carboxy-terminal ClaI-ClaI fragment of hmuR which contains an HindIII site at nucleotide 1387 (probe 1, Table 2).
The lack of homology between the 3′ ends of hemR gene from strain 53977 and hmuR gene from strain A7436 led us to speculate that genomic variation may exist within the carboxy termini of hmuR genes of different P. gingivalis strains. To assess the genomic variability in the hmuR gene and to determine if hmuR was present in strain 53977, probe 1 (Table 2) was utilized in Southern blot analysis with DNA from strains 53977, 381, W50, and A7436. As shown in Fig. 2C, we observed variability in bands corresponding to the carboxy terminus of hmuR in the P. gingivalis strains examined. The probe derived from this portion of the gene hybridized to 8.0- and 13.5-kb HindIII and 4.0-kb PstI fragments in strains A7436 and 53977 DNA, 7.5- and 10.0-kb HindIII and 3.6-kb PstI fragments in strain 381 DNA, and 4.5- and 7.5-kb HindIII and 4.0-kb PstI fragments in W50 DNA. These results indicate that there is a genetic variability in the carboxy terminus of hmuR among these P. gingivalis strains. Our results also suggest that additional variability may exist outside of the hmuR gene.
Transcription of hmuR and hmuY in response to iron limitation.
The promoter region of hmuY contains a putative Fur consensus binding sequence (13 of 19 bases identical to the E. coli Fur consensus box) which could serve to regulate the expression of both the hmuY and the hmuR genes. This is further supported by the absence of −10 and −35 promoter sequences upstream from the putative transcriptional start site of the P. gingivalis hmuR gene. To examine the regulation of hmuY and hmuR genes, RT-PCR analysis was performed with RNA preparations from P. gingivalis grown in iron-depleted and iron-replete conditions. Prior to conducting the RT-PCR experiment, all primers were used in standard PCR reactions to test for functionality and to determine the proper annealing and extension conditions. P. gingivalis was passaged without iron or hemin in anaerobic broth with an iron chelator (165 μM dipyridyl), and this served as the inoculum into anaerobic broth with dipyridyl and anaerobe broth containing dipyridyl and hemin. RNA was isolated from these cultures, and primers specific to the initial 845 bp of the hmuR gene (F3 and R3, see Table 2) and 469 bp of the P. gingivalis sod gene (F5 and R5, Table 2) were used in RT-PCR analysis (Fig. 3). We found that under iron depletion an hmuR transcript was synthesized and that the level of the hmuR transcript appeared to be greater than that observed in organisms grown without added iron but with added hemin. The increased transcription of hmuR does not appear to be due to growth-dependent expression, since the level of the sod transcript was similar under iron-depleted and heme-replete conditions. This finding correlates with a recent study in which Lynch and Kuramitsu (26) demonstrated that the transcription of the P. gingivalis sod gene was dependent on growth but was not affected by iron depletion. Our studies also demonstrated repression of the hmuR transcript when P. gingivalis A7436 was grown with 100 μM ferric chloride (data not shown).
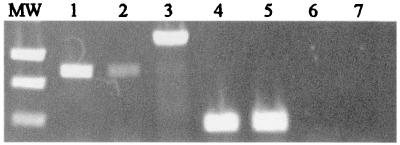
FIG. 3.
RT-PCR analysis of hmuY and hmuR transcription. Total RNA was extracted from P. gingivalis grown under iron-replete (lanes 2, 4, and 6) and iron-depleted (lanes 1, 3, 5, and 7) conditions. RT-PCR was performed using the primers indicated in Table 2. Lane M, molecular weight standards; lanes 1 and 2, hmuR; lane 3, hmuY-hmuR junction; lanes 4 and 5, sod; lanes 6 and 7, Taq negative control using primers to amplify 300 bp of hmuY (F6 and R6, Table 2).
To determine if hmuY and hmuR were cotranscribed, we used primers which would amplify an hmuY-hmuR-specific junction fragment (F4 and R4, Table 2) in RT-PCR with RNA obtained from P. gingivalis A7436 grown in anaerobic broth with dipyridyl. A fragment representing the hmuY-hmuR specific junction transcript was amplified using these primers (Fig. 3), indicating that both genes are cotranscribed. Taken together, these results indicate that the hmuY and hmuR genes are cotranscribed and suggest that transcription is increased under iron-limiting conditions.
Characterization of a P. gingivalis hmuR mutant.
Based on results obtained from the amino acid sequence analysis of HmuR, we postulated that HmuR could function as an iron-regulated TonB-dependent outer membrane receptor for the acquisition of iron from hemin and/or hemoglobin in P. gingivalis. To define the function of the hmuR gene in P. gingivalis, we constructed a P. gingivalis hmuR mutant by insertional inactivation using the ermF cassette (Fig. 4) and confirmed the insertion of the ermF cassette by Southern blot analysis. We observed an ∼2-kb shift in the DNA band corresponding to the hmuR gene in four separately isolated P. gingivalis transformants (Fig. 4B). P. gingivalis strain WS1 was chosen for further analysis, and the insertion of the ermF cassette in the hmuR gene was further confirmed by PCR analysis using 5′ and 3′ hmuR-specific primers (data not shown).
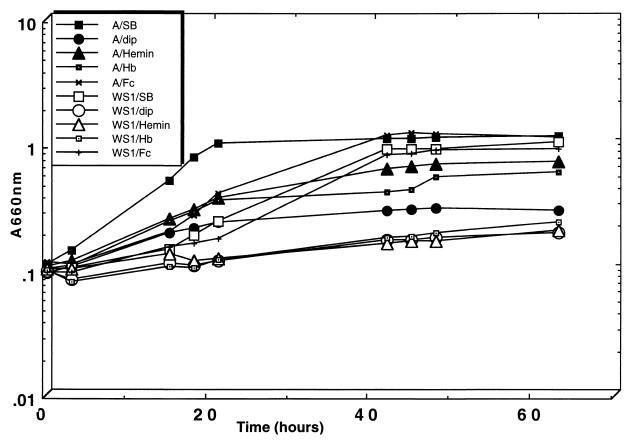
The ability of P. gingivalis WS1 to grow with hemin and hemoglobin as sole sources of iron was then examined. P. gingivalis cultures were grown for 24 h in Schaedler broth medium with 150 μM dipyridyl to chelate iron, and this served as the inoculum into Schaedler broth plus dipyridyl or Schaedler broth plus dipyridyl supplemented with hemin (1.5 μM), hemoglobin (1.5 μM), or ferric chloride (100 μM). Growth of P. gingivalis strain A7436 in Schaedler broth plus dipyridyl supplemented with hemin, hemoglobin, or ferric chloride resulted in a typical growth curve with final OD660 values of 0.71, 0.59, and 1.1, respectively, after 63 h of growth (Fig. 5). In contrast, P. gingivalis WS1 exhibited diminished growth with either hemin or hemoglobin, with final OD660 values of 0.19 and 0.23, respectively (Fig. 5). The poor growth of P. gingivalis WS1 with hemin or hemoglobin does not appear to result from a generalized growth defect since this strain grew similarly to P. gingivalis A7436 in Schaedler broth plus dipyridyl supplemented with 100 μM ferric chloride (final OD660 of 0.91). This finding indicates that HmuR is specific for the uptake of heme-containing compounds such as hemin and hemoglobin, but the uptake of inorganic iron (ferric chloride) is mediated by another mechanism. In addition, these results indicate that hemin and hemoglobin utilization in P. gingivalis occur through a common HmuR-mediated mechanism. We also found that hemoglobin was an effective competitor for the transport of radiolabeled hemin in P. gingivalis A7436 (data not shown), further supporting a common mechanism for hemin and hemoglobin utilization in P. gingivalis.
FIG. 5.
Growth of P. gingivalis A7436 (A) and WS1 with hemin, hemoglobin, and ferric chloride. Cultures were initially starved in Schaedler broth supplemented with 150 μM dipyridyl for 24 h. This was used to inoculate Schaedler broth alone (SB), Schaedler broth plus 150 μM dipyridyl (dip), Schaedler broth plus 150 μM dipyridyl plus 1.5 μM hemin (Hemin), Schaedler broth plus 150 μM dipyridyl plus 1.5 μM hemoglobin (Hb), or Schaedler broth plus 150 μM dipyridyl plus 100 μM ferric chloride (Fc). The results are representative of two experiments.
Disruption of hmuR correlates with diminished hemoglobin binding.
To determine if the inability of P. gingivalis WS1 to grow with hemoglobin was due to a decreased ability to bind hemoglobin, we examined the binding of P. gingivalis whole cells to hemoglobin by using a spectrophotometric assay (29). P. gingivalis cells were grown anaerobically overnight in BM. The percent absorbance was calculated relative to the control strain A7436, which was set at 100%. P. gingivalis WS1 exhibited a significant decrease in hemoglobin binding compared to the parent strain A7436. P. gingivalis WS1 bound 34% less hemoglobin than did the parental strain A7436 (data not shown). The observation that the hmuR mutant did not exhibit a total decrease in hemoglobin binding may be due to the presence of multiple hemoglobin binding proteins in P. gingivalis, including Kgp and HRgpA (12, 21, 24, 28, 29), as has been described for other gram-negative organisms (23, 27, 40). This idea was supported by the observation that the P. gingivalis Kgp mutant (strain MSM-3) also bound less hemoglobin than the wild-type strain A7436 (data not shown).
Expression of rHmuR and characterization of hemin and hemoglobin binding.
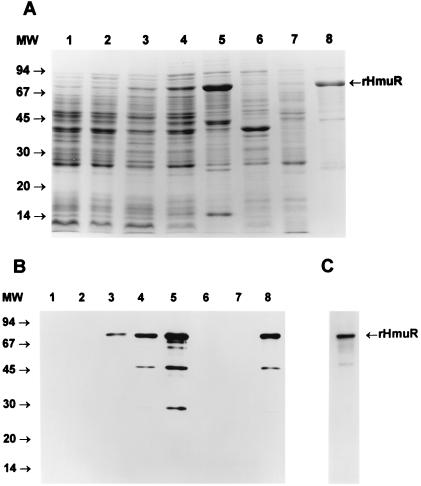
To further confirm the ability of HmuR to bind hemoglobin, we overexpressed the protein in E. coli and examined hemoglobin binding by recombinant strains. Plasmids containing hmuR either with (pTO2) or without (pTO1) its native signal peptide were subsequently transformed into E. coli. The HmuR expression level of the resulting E. coli BL21(DE3)pLysE strain harboring pTO1 was monitored by SDS-PAGE (Fig. 6A) and after transfer onto nitrocellulose membrane by detection with antibody against the fusion protein (Fig. 6B). Basal level expression of rHmuR was exhibited prior to the addition of IPTG in E. coli grown in LB medium (data not shown), as well as in M9 medium (Fig. 6B); however, an increase in the expression of the protein after IPTG induction was exhibited in E. coli grown in M9 medium. We did not detect new protein bands following IPTG induction in bacteria transformed with the vector alone (Fig. 6A), and no protein bands were visible on the immunoblot after probing with the anti-fusion protein antibody (Fig. 6B). Following IPTG induction, the expressed rHmuR together with the fusion tag attached to the C terminus of the protein possessed a molecular mass of approximately 80 kDa. We also observed several additional protein bands which may correspond to degradation products of rHmuR (Fig. 6A). This was further confirmed by Western blot analysis using antibodies to the fusion protein (Fig. 6B). The ability of the purified HmuR protein to bind hemoglobin was next examined by a solid-phase assay. As shown in Fig. 6C, rHmuR isolated from inclusion bodies bound human hemoglobin.
FIG. 6.
Expression, purification, and hemoglobin binding activity of rHmuR localized in inclusion bodies. (A) Expression of rHmuR. The gene encoding the protein lacking the signal peptide was cloned into pCRT7/CT-TOPO and expressed in E. coli BL21(DE3)pLysE. CBB-stained SDS-PAGE gel of cells harboring the vector alone (lane 1, uninduced; lane 2, induced), cells expressing rHmuR (lane 3, uninduced; lane 4, induced), inclusion body fraction (lane 5), membrane fraction (lane 6), soluble fraction (lane 7), and rHmuR purified using Ni-chelate chromatography (lane 8). The positions of molecular size markers (in kilodaltons) are on the left. (B) Identification of rHmuR. Whole-cell lysates and purified rHmuR were electrophoresed using SDS-PAGE and transferred onto a nitrocellulose membrane. E. coli harboring the vector alone (lane 1, uninduced; lane 2, induced), cells expressing rHmuR (lane 3, uninduced; lane 4, induced), inclusion bodies fraction (lane 5), membrane fraction (lane 6), soluble fraction (lane 7), rHmuR purified using Ni-chelate chromatography (lane 8). The positions of molecular size markers (in kilodaltons) are on the left. The immunoblot was probed with anti-fusion protein antibody and detected using chemiluminescence staining. (C) Hemoglobin binding by rHmuR. rHmuR purified by Ni-chelate chromatography was electrophoresed using SDS-PAGE and transferred onto a nitrocellulose membrane. The blot was probed with biotinylated human hemoglobin.
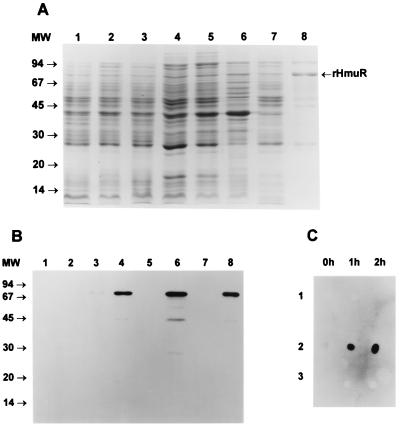
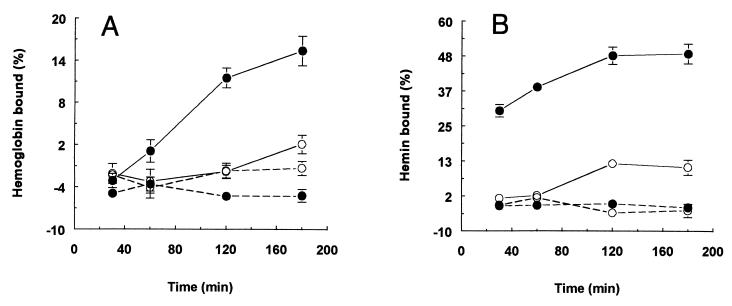
We next expressed HmuR containing its native signal peptide to export and localize this protein in outer membranes of E. coli cells. SDS-PAGE and Western blot analysis showed that rHmuR was associated with the membrane fraction (Fig. 7A and B). As shown in Fig. 7C, rHmuR was expressed on the surface of E. coli BL21(DE3)pLysE strain harboring pTO2 as detected by antibodies to the fusion protein. Low basal level expression of rHmuR was exhibited prior to the addition of IPTG in E. coli grown in LB (data not shown), as well as in M9 medium (Fig. 7B). The membrane bound rHmuR expression level of the resulting E. coli harboring pTO2 was lower compared with rHmuR deposited in inclusion bodies in E. coli transformed with pTO1 (Fig. 7A). This result was expected, as the addition of the C-terminal His tag blocked the C-terminal Phe residue, which has been shown to be highly conserved and necessary for the stable incorporation of a protein into the outer membrane. Following IPTG induction the expressed rHmuR, together with the fusion tag attached to the C terminus of the protein, possessed a molecular mass of approximately 80 kDa. We also observed several additional protein bands which may correspond to degradation products of rHmuR (Fig. 7A), and this was further confirmed by Western blot analysis using the anti-fusion protein antibody (Fig. 7B). The amino acid sequence of membrane-bound rHmuR was determined by N-terminal sequencing of the protein by Edman degradation, after the transfer onto PVDF membranes of rHmuR purified by Ni-chelate chromatography. The resulting amino acid sequence was ANPPAQPT and matches 100% to the native HmuR following signal peptide cleavage (data not shown). Binding of hemoglobin and hemin by whole E. coli cells expressing rHmuR was examined using a spectrophotometric assay. As expected, only E. coli cells expressing membrane-bound rHmuR were found to bind hemoglobin and hemin (Fig. 8). We did not observe hemoglobin or hemin binding by E. coli cells in which rHmuR was deposited in inclusion bodies (Fig. 8) or by E. coli harboring the plasmid alone (data not shown). These results indicate that in E. coli BL21(DE3) pLysE, rHmuR is exported to the membrane, where it can bind both hemoglobin and hemin.
FIG. 7.
Expression, purification, and surface exposure of membrane-bound rHmuR. (A) Expression of rHmuR. The gene encoding the protein with the signal peptide was cloned into pCRT7/CT-TOPO and expressed in E. coli BL21(DE3)pLysE. Lanes are designated in the same manner as in Fig. 6A. (B) Identification of rHmuR. Whole-cell lysates and purified rHmuR were electrophoresed using SDS-PAGE and transferred onto a nitrocellulose membrane. Lanes are designated in the same manner as in Fig. 6B. The immunoblot was probed with anti-fusion protein antibody and detected using chemiluminescence staining. (C) Identification of rHmuR on the surface of E. coli BL21(DE3)pLyE cells (panel 1, E. coli harboring vector alone; panel 2, E. coli expressing membrane-bound rHmuR; panel 3, E. coli expressing rHmuR deposited in inclusion bodies). The dot blot was probed with antibodies against the fusion protein using cells before and 1 and 2 h after IPTG induction.
FIG. 8.
Hemoglobin and hemin binding by E. coli expressing rHmuR. E. coli BL221(DE3)pLysE cells expressing rHmuR membrane bound (solid line) and rHmuR deposited in inclusion bodies (dotted line) grown in M9 media were harvested before (○) and after IPTG induction (●) and suspended in PBS. Human hemoglobin (A) and hemin (B) were added to final concentrations of 5 and 10 μM, respectively.
DISCUSSION
In this study we have determined that a newly identified TonB-dependent receptor, HmuR, is involved in the binding and utilization of hemoglobin and hemin in P. gingivalis. This is based on sequence analysis comparison, which reveals a high degree of homology of HmuR to TonB-dependent outer membrane receptors involved in the acquisition of iron from hemoglobin, characterization of the P. gingivalis hmuR mutant, and the ability of recombinant HmuR protein to bind hemoglobin and hemin. The hmuR gene containing its native signal peptide was used to express rHmuR, which was exported to the outer membrane in E. coli cells. We found that E. coli cells expressing rHmuR bound both hemoglobin and hemin. Using the hmuR gene without its native signal sequence allowed us to express and purify larger quantities of partially renatured rHmuR, and the purified protein was demonstrated to bind hemoglobin. Taken together, these results support the role of HmuR as a required P. gingivalis hemoglobin-hemin receptor.
In H. influenzae, the expression of the hemoglobin receptor HgpA is controlled by phase variation via strand slippage across “CCAA” repeats (33). Analysis of the P. gingivalis hmuR gene revealed the presence of 12 CCAA repeats at intervals of various lengths, suggesting that hemin-hemoglobin utilization via HmuR could be regulated by a similar mechanism. However, variability in the ability of P. gingivalis to utilize hemoglobin has not been examined, and it remains to be determined if hemin-hemoglobin utilization via HmuR in P. gingivalis is under phase variation.
The observation that the hmuR mutant did not exhibit a total lack in hemoglobin binding appears to be due to the presence of intact kgp and rgpA genes in this strain. We have previously demonstrated that a P. gingivalis kgp mutant grows poorly with hemin or hemoglobin as sole iron sources (14). Studies in our laboratory have also demonstrated that soluble Kgp and HRgpA bind hemoglobin and that binding is mediated through the 40- and 44-kDa polypeptides of the Kgp and HRgpA complexes (Sroka et al., submitted), respectively. Likewise, hemoglobin binding to Kgp and HRgpA has also been reported by other investigators, although conflicting studies defining the region of the protein involved in hemoglobin binding have been reported (11, 21, 28; Sroka et al., submitted). Although Kgp can be found associated with the P. gingivalis outer membrane, at this point it appears premature to classify Kgp as a receptor. The amino acid sequence of Kgp has no similarity to TonB dependent outer membrane proteins. Rather Kgp may function as a soluble hemoglobin binding protein which, similar to hemophores, captures hemoglobin and delivers it to a second outer-membrane-associated receptor, possibly the hemoglobin receptor HmuR. The best characterized of the hemophore systems is that of the S. marcescens secreted protein, HasA, which extracts heme from hemoglobin and hemopexin-heme and delivers it to the outer membrane receptor HasR (17). Unlike siderophores, HasA is not internalized with its ligand during uptake. HasA has a very high affinity for heme; however, it is unclear how the heme is released from HasA onto HasR. Both apo HasA and holo HasA interact with HasR, indicating that HasA does not interact with HasR solely via the heme molecule. A similar extracellular hemin-binding protein (HasAp) has recently been described in P. aeruginosa (23).
We found that the hmuR mutant exhibited a decreased ability to grow with hemin and that E. coli cells expressing HmuR could bind hemin. We also demonstrated that hemoglobin can compete for the binding and accumulation of hemin in P. gingivalis (data not shown), further suggesting that hemin and hemoglobin transport can occur via a common pathway. Thus, in addition to its role in hemoglobin utilization, HmuR appears to function in hemin transport in P. gingivalis. Hemin binding in P. gingivalis has been observed to occur through both high- and low-affinity binding sites, and it has been proposed that this is mediated by separate outer membrane receptors (14). In addition to the TonB-dependent hemoglobin receptor, HmuR, P. gingivalis also appears to possess two additional putative TonB-dependent hemin receptors (HemR and Tla). It is possible that HemR and Tla could function to bind hemin directly; however, conclusive evidence for the roles of HemR and TlaA in hemin binding has not been reported. A P. gingivalis tla mutant was demonstrated to grow with high levels of hemin, but growth was decreased with low levels of this iron source. These results indicate that Tla is involved in hemin transport; however, it is not known if Tla functions in heme capture or in heme binding via a receptor-like mechanism. A definitive role for the P. gingivalis HemR protein in hemin transport has not been delineated since Karunakaran et al. (19) were unable to construct a P. gingivalis hemR mutant. Despite the fact that previous studies have determined that hemR is present in strains 53977, 381, and W50, we were unable to PCR amplify the hemR gene from P. gingivalis A7436 (data not shown), suggesting that in this strain hemin transport can occur independently of HemR. The hmuR gene was also found in P. gingivalis strains 381, 53977, and W50, with variability observed in the carboxy terminus of hmuR in these strains. This variability observed within the gene encoding the carboxy terminus of HmuR may be due to genomic rearrangements facilitated by P. gingivalis insertion sequence elements. Such rearrangements have recently been proposed to result in the variability in the P. gingivalis gingipain gene family (2, 29).
Our results also indicate that hmuY and hmuR are cotranscribed and that transcription is increased following growth of P. gingivalis in iron-limiting conditions. In a number of diverse microorganisms, genes involved in iron acquisition and virulence are transcriptionally regulated by the availability of iron through the Fur protein (3). Fur forms a dimer with ferrous iron and binds to a 19-bp DNA sequence (Fur box), which overlaps the promoters of iron-regulated genes, resulting in the inhibition of transcription. Upstream of the P. gingivalis hmuY start site we identified a region with homology to the Fur consensus binding sequence. The recent isolation of a P. gingivalis fur homolog (C. A. Genco and W. Simpson, unpublished data), together with the identification of a Fur box upstream of the hmuY-hmuR operon supports the role of Fur-mediated transcriptional control of the P. gingivalis hmuR gene. Interestingly, we found that the increased transcription of hmuR under iron-limiting conditions also correlated with an increase in hemoglobin binding of P. gingivalis whole cells. We found that hemoglobin binding increased fourfold when P. gingivalis was grown in the presence of the iron chelator, dipyridyl (data not shown). Amano et al. (2) previously reported that hemoglobin binding to P. gingivalis whole cells is directly correlated with the successive passage of bacteria in media devoid of added heme. Thus, the increased hemoglobin binding of P. gingivalis whole cells obtained from cultures grown under iron limitation appears to result from the derepression of the hmuR gene as a result of Fur-mediated regulation. In contrast to our results, Karunakaran et al. (19) demonstrated that in P. gingivalis 53977, ORF1 (hmuY) was upregulated in the presence of hemin, while hemR was negatively regulated by hemin. In addition, these investigators demonstrated that ORF1 was part of a 1-kb transcript, while hemR was part of a 3-kb transcript. The differences in these findings may be due to the fact that hmuR and hemR are different genes and are regulated by different mechanisms or to strain-related differences in transcriptional regulation.
While our results indicate that HmuR is required for the binding and utilization of hemin and hemoglobin by P. gingivalis, little is known concerning the role(s) of other proteins in the transport of iron from these compounds. A search of the P. gingivalis W83 TIGR database allowed us to identify a putative hemin transport operon in P. gingivalis which exhibits a high degree of homology to the Y. enterocolitica hemin transport system. The Y. enterocolitica hemin-degrading protein HemS, hemin-binding protein HemT, hemin permease HemU, and ATP-binding hydrophilic protein HemV demonstrated homologies of 43, 48, 44, and 53%, respectively, with specific contigs in the P. gingivalis W83 database of the TIGR (41). While we recognize that the functions of these genes in P. gingivalis have not been defined, we postulate that the proteins they encode may function together with HmuR for the transport of hemin and heme from hemoglobin.
In summary, we have characterized the structural gene for a novel P. gingivalis TonB-dependent outer membrane receptor (HmuR) which functions both in hemoglobin and hemin binding and utilization in P. gingivalis. We demonstrated that the hmuY gene is found directly upstream of hmuR, that hmuY and hmuR are cotranscribed, and that transcription was negatively regulated by iron. Furthermore, recombinant HmuR was shown to bind hemoglobin, and E. coli cells expressing rHmuR were able to bind hemoglobin and hemin. We propose, based on these results, that HmuR serves as the major TonB-dependent outer membrane hemoglobin-hemin receptor in P. gingivalis. Future studies are aimed at defining the interaction between the HmuR and hemoglobin, hemin, and other substrates.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grant DE09161 from the National Institute of Dental and Craniofacial Research (to C.A.G.). Sequencing of the P. gingivalis W83 genome was accomplished with support from the National Institute of Dental and Craniofacial Research grant DE-12082.
We thank Pragnya Desai and Frank Gibson for scientific discussions and critical review of the manuscript. We also acknowledge Thonthi Karunakarun for the isolation of P. gingivalis genomic DNA and for PCR amplification of the hmuR gene from strain A7436.
REFERENCES
- 1.Aduse-Opoku J, Slaney J, Rangarajan M, Muir J, Young K, Curtis M A. The Tla protein of Porphyromonas gingivalis W50: a homolog of the R1 protease precursor (PrpR1) is an outer membrane receptor required for growth on low levels of hemin. J Bacteriol. 1997;179:4778–4788. doi: 10.1128/jb.179.15.4778-4788.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano A, Kuboniwa M, Kataoka K, Tazaki K, Inoshita E, Nagata H, Tamagawa H, Shizukuishi S. Binding of hemoglobin by Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;134:63–67. doi: 10.1111/j.1574-6968.1995.tb07915.x. [DOI] [PubMed] [Google Scholar]
- 3.Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 4.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp 26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 6.Bracken C S, Baer M T, Abdur-Rashid A, Helms W, Stojiljkovic I. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnette W N. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radio iodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 8.Carmen R J, Ramakrishnan M D, Harper F H. Hemin levels in culture medium of Porphyromonas (Bacteroides) gingivalis regulate both hemin binding and trypsinlike protease production. Infect Immun. 1990;58:4016–4019. doi: 10.1128/iai.58.12.4016-4019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C J, Sparling P F, Lewis L A, Dyer D W, Elkins C. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect Immun. 1996;64:5008–5014. doi: 10.1128/iai.64.12.5008-5014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeCarlo A A, Paramaesvaran M, Yun P L W, Collyer C, Hunter N. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J Bacteriol. 1999;181:3784–3791. doi: 10.1128/jb.181.12.3784-3791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimura S, Shibata Y, Hirai K, Nakamura T. Binding of hemoglobin to the envelope of Porphyromonas gingivalis and isolation of the hemoglobin-binding protein. Infect Immun. 1996;64:2339–2342. doi: 10.1128/iai.64.6.2339-2342.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genco C A, Simpson W, Forng R-Y, Egal M, Odusanya B M B. Characterization of a Tn4351-generated hemin uptake mutant of Porphyromonas gingivalis: evidence for the coordinate regulation of virulence factors by hemin. Infect Immun. 1995;63:2459–2466. doi: 10.1128/iai.63.7.2459-2466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genco C A. Regulation of hemin and iron transport in Porphyromonas gingivalis. Adv Dent Res. 1995;9:41–47. doi: 10.1177/08959374950090010801. [DOI] [PubMed] [Google Scholar]
- 16.Genco C A, Potempa J, Mikolajczyk-Pawlinska J, Travis J. Role of gingipains R in Porphyromonas gingivalis pathogenesis. Clin Infect Dis. 1999;28:456–465. doi: 10.1086/515156. [DOI] [PubMed] [Google Scholar]
- 17.Ghigo M J, Letoffe S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hnatovich D J, Virzi F, Rusckowski M. Investigation of avidin and biotin for imaging applications. J Nucl Med. 1987;28:1294–1302. [PubMed] [Google Scholar]
- 19.Karunakaran T, Madden T, Kuramitsu K. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S J, Chu L, Holt S C. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;21:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- 21.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Letoffe S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA hemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 24.Lewis J P, Dawson J A, Hannis J C, Muddiman D, Macrina F L. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis L A, Gray E, Wang Y-P, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 26.Lynch M C, Kuramitsu H K. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect Immun. 1999;67:3367–3375. doi: 10.1128/iai.67.7.3367-3375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morton D J, Whitby P W, Jin H, Ren Z, Stull T L. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect Immun. 1999;67:2729–2739. doi: 10.1128/iai.67.6.2729-2739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama K, Ratnayake D B, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Hemoglobin receptor protein is intragenically encoded by the cysteine proteinase encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake D B, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 30.Postle K. Ton B and the gram-negative dilemma. Mol Microbiol. 1990;4:2019–2025. doi: 10.1111/j.1365-2958.1990.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 31.Potempa J, Pavloff N, Travis J. Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol Rev. 1995;3:430–434. doi: 10.1016/s0966-842x(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen J L, Odelson D A, Macrina F L. Complete nucleotide sequence and transcription of ermF, a macrolide-lincosamide-streptogramin B resistance determinant from Bacteroides fragilis. J Bacteriol. 1986;168:523–533. doi: 10.1128/jb.168.2.523-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren Z, Jin H, Whitby P W, Morton D J, Stull T L. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J Bacteriol. 1999;181:5865–5870. doi: 10.1128/jb.181.18.5865-5870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson A R, Stojiljkovic I. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis undergoes phase variation. J Bacteriol. 1999;181:2067–2074. doi: 10.1128/jb.181.7.2067-2074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–397. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 36.Simpson W, Wang C Y, Bond V C, Potempa J, Mikolajczyk-Pawlinska J, Travis J, Genco C A. Transposition of the endogenous insertion sequence element IS1126 modulates gingipain expression in Porphyromonas gingivalis. Infect Immun. 1999;67:5012–5020. doi: 10.1128/iai.67.10.5012-5020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smalley J W, Silver J, Marsh P J, Birss A J. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the μ-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem J. 1998;331:681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smalley J W, Birss A, McKee A S, Marsch P D. Haemin-binding proteins of Porphyromonas gingivalis W50 grown in a chemostat under haemin-limitation. J Gen Microbiol. 1993;139:2145–2150. doi: 10.1099/00221287-139-9-2145. [DOI] [PubMed] [Google Scholar]
- 39.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 40.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. HmbR outer membrane receptors of pathogenic Neisseria: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J Bacteriol. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J M, Jones H A, Perry R D. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoproteins utilization. Infect Immun. 1999;67:3879–3835. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tompkins G R, Wood D P, Birchmeher K R. Detection and comparison of specific hemin binding by Porphyromonas gingivalis and Prevotella intermedia. J Bacteriol. 1997;179:620–626. doi: 10.1128/jb.179.3.620-626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]