Abstract
The rpoS gene encodes the alternative sigma factor ςS (RpoS) and is required for survival of bacteria under starvation and stress conditions. It is also essential for Salmonella virulence in mice. Most work on the RpoS regulon has been in the closely related enterobacterial species Escherichia coli. To characterize the RpoS regulon in Salmonella, we isolated 38 unique RpoS-activated lacZ gene fusions from a bank of Salmonella enterica serovar Typhimurium mutants harboring random Tn5B21 mutations. Dependence on RpoS varied from 3-fold to over 95-fold, and all gene fusions isolated were regulated by growth phase. The identities of 21 RpoS-dependent fusions were determined by DNA sequence analysis. Seven of the fusions mapped to DNA regions in Salmonella serovar Typhimurium that do not match any known E. coli sequence, suggesting that the composition of the RpoS regulon differs markedly in the two species. The other 14 fusions mapped to 13 DNA regions very similar to E. coli sequences. None of the insertion mutations in DNA regions common to both species appeared to affect Salmonella virulence in BALB/c mice. Of these, only three (otsA, katE, and poxB) are located in known members of the RpoS regulon. Ten insertions mapped in nine open reading frames of unknown function (yciF, yehY, yhjY, yncC, yjgB, yahO, ygaU, ycgB, and yeaG) appear to be novel members of the RpoS regulon. One insertion, that in mutant C52::H87, was in the noncoding region upstream from ogt, encoding a O6-methylguanine DNA methyltransferase involved in repairing alkylation damage in DNA. The ogt coding sequence is very similar to the E. coli homolog, but the ogt 5′ flanking regions were found to be markedly different in the two species, suggesting genetic rearrangements. Using primer extension assays, a specific ogt mRNA start site was detected in RNAs of the Salmonella serovar Typhimurium wild-type strains C52 and SL1344 but not in RNAs of the mutant strains C52K (rpoS), SL1344K (rpoS), and C52::H87. In mutant C52::H87, Tn5B21 is inserted at the ogt mRNA start site, with lacZ presumably transcribed from the identified RpoS-regulated promoter. These results indicate that ogt gene expression in Salmonella is regulated by RpoS in stationary phase of growth in rich medium, a finding that suggests a novel role for RpoS in DNA repair functions.
The alternative sigma factor ςS (also known as RpoS, KatF, or ς38) plays a key role in the survival of bacteria under starvation or stress conditions (for reviews, see references 14, 16, and 22). Homologs of RpoS have been found in a number of bacteria, but most work on the RpoS regulon has been in Escherichia coli. The number of genes shown to be subjected to RpoS regulation has already reached the 50 predicted by two-dimensional gel analysis of cell extracts, yet most RpoS-regulated genes and functions remain unknown.
During rapid growth in the laboratory, E. coli contains extremely little RpoS. The RpoS protein is most abundant at the onset of the stationary phase of growth, the maximum level being 30% of that of ς70 (for reviews, see references 14, 16, and 22). Indeed, onset of the stationary phase induces the RpoS regulon. Expression of RpoS is also induced when cells are exposed to certain stress conditions even during exponential phase, resulting in the activation of a number of RpoS-dependent promoters (for reviews, see references 14, 16, and 22). The cellular level of RpoS is regulated by mechanisms involving transcription, translation, and posttranslational stability. Different stress conditions differentially affect these various levels of control to create a complex regulatory profile.
Salmonellae are enteric pathogens that cause a wide range of host- and serotype-specific illnesses, including gastroenteritis and enteric fever. Salmonella enterica serovar Typhimurium (serovar Typhimurium) infection of mice results in a systemic illness similar to human enteric (typhoid) fever, with bacteria disseminating to organs rich in phagocytic cells. In serovar Typhimurium, ςS controls expression of the Salmonella virulence plasmid spv genes (10, 26). The spvRABCD gene cluster controls the growth rate of Salmonella in deep organs and is required for systemic infection and bacteremia in animals and humans (for a review, see reference 11). As expected, Salmonella rpoS mutants have a severely impaired capacity to colonize spleens of infected mice (4, 7, 20). In addition, rpoS mutations reduce the ability of serovar Typhimurium to colonize Peyer's patches of infected mice (7, 25) and decrease the persistence of virulence plasmid-cured strains in the spleen (20). These effects presumably result from the inappropriate expression of one or more unidentified rpoS-regulated chromosomal genes. The human-restricted Salmonella serovars such as Typhi, which causes typhoid fever, have no virulence plasmid, and the role of rpoS in the virulence of these serovars is unknown. However, an rpoS mutant of serovar Typhi is less cytotoxic for macrophages than the parental strain, and therefore rpoS may be involved in the virulence of serovar Typhi in humans (18). Interestingly, Salmonella rpoS mutants are efficient live vaccines (6, 7, 29), and an rpoS mutation increases the attenuation of aroA serovar Typhimurium live vaccines in BALB/c mice and athymic BALB/c mice (6, 7).
Identification of ςS-regulated genes in Salmonella may lead to characterization of novel factors contributing to the persistence of the pathogen in the environment and hosts. We therefore studied the RpoS regulon of Salmonella. This report describes a method for identifying ςS-regulated genes in serovar Typhimurium by using lacZ transcriptional fusions carried on Tn5B21 transposon insertions. In the first screening, 38 unique ςS-regulated lacZ gene fusions were isolated. We report a preliminary characterization of 21 of these mutants in Salmonella and a comparative analysis with the closely related species E. coli. Fourteen of the fusions mapped to genes present in both species, and ten of them are new members of the RpoS regulon. One of these new ςS-regulated genes has been identified as ogt, encoding a O6-methylguanine (O6MeG) DNA methyltransferase (MTase) involved in the repair of alkylation damage in DNA.
MATERIALS AND METHODS
Bacterial strains, plasmids, phages, and growth conditions.
The E. coli strains used were S17-1 (pro thi recA hsdR, chromosomal RP4-2; Tn1::ISR1 Tc::Mu Km::Tn7) (34) and MC1061 [araD139 Δ(ara-leu)7697 rpsL galU galK Δ(lacIPOZY)FX74] (3). The serovar Typhimurium mouse virulent strains used were C52 and SL1344 and their isogenic ΔrpoS::kan derivatives C52K and SL1344K, respectively (7). Bacteriophage P22HT105.1/int was used to transduce mutations between Salmonella strains (33). Green plates for screening for P22-infected cells or lysogens were prepared as described previously (37). Strains were routinely cultured at 37°C in Luria-Bertani (LB) medium (32). Antibiotics were used at the following concentrations (micrograms per milliliter): carbenicillin, 100; chloramphenicol, 30; kanamycin, 50; streptomycin 100; and tetracycline, 20.
Plasmids.
Plasmid pBDJ103 was used as a source of transposon Tn5B21 (17). Plasmid pUC19 (Cbr) (42) and the mobilizable plasmid pVK100 (Tetr Kmr) (19) were used as cloning vectors. Plasmid pVKCm (Tetr Cmr) contains the 1.1-kb HindIII-SalI fragment carrying the cat gene from pAMPCm (30) inserted into the HindIII-XhoI sites of pVK100. The 2-kb SalI fragment carrying the serovar Typhimurium rpoS gene from pSTK5 (20) was ligated into the SalI site of pVKCm to yield pVKRpoS (Cmr). DNA sequences flanking Tn5B21 insertions in serovar Typhimurium mutants were cloned by digesting genomic DNA and vector pUC19 with either HindIII or PstI (for cloning of DNA 5′ to the insertion) and with EcoRI (for cloning DNA 3′ to the insertion), ligating, and selecting for E. coli MC1061 transformants carrying the tetracycline resistance gene from Tn5B21. To construct pUCogt1 carrying the ydaL-ogt intergenic region from serovar Typhimurium, a 413-bp DNA sequence was amplified from C52 total DNA by PCR using primers YDAL1 (5′-AGGCTCGGATCCCAGCGGCTGGACATCCTCCATGGC-3′) and OGT1 (5′-TTCCGAAAGCTTCTGTTCCCACTCAATGGCCCGC-3′) such that it acquired BamHI and HindIII restriction sites at its 5′ and 3′ ends, respectively. The PCR-amplified fragment was then ligated into the BamHI-HindIII sites of pUC19 to give pUCogt1. The nucleotide sequence of the PCR-amplified fragment in pUCogt1 was checked by DNA sequencing.
Transposon mutagenesis of serovar Typhimurium.
Transposon Tn5B21 is a Tetr derivative of Tn5 which was constructed to make lacZ gene fusions (35). pBDJ103 is an Ampr ColE1 derivative which carries Tn5B21 and an Sms allele for the ribosomal protein S12 (17). The presence of the S12 gene on this plasmid can be used to select positively (in a strain carrying the rpsL allele) for loss of the plasmid. Strain C52K-Smr is a spontaneous Smr mutant of serovar Typhimurium C52K selected for growth on LB agar containing 100 μg of streptomycin per ml. pDBJ103 was introduced into serovar Typhimurium C52K-Smr by electroporation, the resulting transformants were Kmr Tetr Cbr Sms. The protocol used to generate pools of Tn5B21 insertion mutants from C52K-Smr has been described previously (17). An exponentially growing culture, derived from a single colony of serovar Typhimurium C52K-Smr(pBDJ103), was diluted, and approximately 5,000 CFU were plated onto LB agar containing 20 μg of tetracycline per ml and grown overnight at 30°C. The colonies were then replica plated onto LB agar containing 100 μg of streptomycin per ml and 20 μg of tetracycline per ml and grown overnight at 37°C to select for transposition and loss of plasmid pDBJ103. Each Tetr Smr colony obtained represents at least one unique Tn5B21 transposon insertion. All of the Tetr and Smr colonies from a single plate were pooled. Eight independent pools of serovar Typhimurium Tn5B21 mutants (from eight plates labeled 1 to 3 and E to I) were generated by this procedure. More than 99% of the CFU in each pool were Amps, confirming the loss of plasmid pDBJ103. The serovar Typhimurium pools were then screened for mutants containing rpoS-regulated lacZ fusions as depicted in Fig. 1.
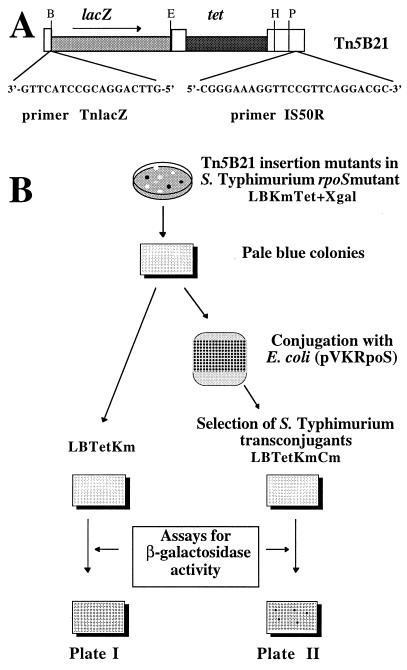
FIG. 1.
Strategy used to select Tn5B21 insertion mutants of serovar Typhimurium carrying ςS-dependent promoter-lacZ fusions. (A) Transposon Tn5B21 (35) and primers used for DNA sequencing. Restriction sites: B, BamHI; E, EcoRI; H, HindIII; P, PstI. (B) Transposon Tn5B21 insertion mutagenesis of the Smr Kmr serovar Typhimurium rpoS mutant C52K-Smr is described in Materials and Methods. Mutants containing poorly expressed lacZ gene fusions were identified as having a pale blue colony phenotype after growth in LB agar containing X-Gal. Such strains were used to inoculate 96-well enzyme-linked immunosorbent assay (ELISA) plates (0.2 ml of LB per well) and grown overnight at 37°C. Conjugation between these mutant strains and E. coli strain S17-1 carrying the mobilizable plasmid pVKRpoS (Cmr) was carried out on LB agar plates for 4 to 5 h at 37°C (donor and recipient cells were mixed in a 1:1 ratio). Serovar Typhimurium transconjugants carrying pVKRpoS were selected in LB broth supplemented with 20 μg of tetracycline per ml 25 μg of kanamycin per ml, and 30 μg of chloramphenicol per ml (LBTetKmCm). β-Galactosidase activity of serovar Typhimurium Tn5B21 mutants with and without pVKRpoS (plates II and plate I, respectively) was assayed using 96-well ELISA plates and the method of Miller (24). An automated ELISA plate reader (Labsystems Multiskan RC) was used to measure A420 after 1 h of incubation at room temperature. Mutant candidates showing increased gene fusion activity on acquisition of plasmid pVKRpoS were selected for further studies.
DNA and RNA manipulations and enzyme assays.
Standard molecular biology techniques were used (30, 32). Oligonucleotides were purchased from Genset (Paris, France). Double-stranded DNA was sequenced with a Thermo Sequenase radiolabeled terminator cycle sequencing kit and Redivue 5′ α-33P-labeled dideoxyribonucleotide triphosphates (Amersham Pharmacia). Primers TnlacZ and IS50R (Fig. 1), which anneal to the left and right ends of Tn5B21, respectively, and read into the flanking serovar Typhimurium genomic DNA insert, were used for sequencing. Homology searches were performed with both BLAST and FASTA computer programs on the website at the Institut Pasteur (http://www.pasteur.fr/). Feature annotations for E. coli sequences were found through the Colibri World Wide Web server provided by the Institut Pasteur (http://genolist.pasteur.fr/Colibri/).
Total RNA was extracted and primer extension experiments were performed as previously described (20). An oligonucleotide complementary to the 5′ end of the coding region of the ogt gene (OGT2; 5′-CCACCCATAACGGTCCTAATGGCGTGGC-3′) was used in primer extension experiments. β-Galactosidase activity was measured as described by Miller (24) and is expressed in Miller units (ΔOD420 [optical density at 420 nm] per minute per OD600).
Mouse infection.
Female BALB/c mice, which are innately susceptible to serovar Typhimurium, were obtained from the Centre d'Elevage IFFA CREDO (Domaine des Oncins, L'Arbresle, France) and were used when approximately 7 to 8 weeks old. For inoculation of mice, bacteria were freshly streaked onto LB agar plates and the antigenic formulae of serovar Typhimurium strains were confirmed by slide agglutination using rabbit antisera specific for O- and H-antigen factors (Bio-Rad). Single colonies were used to inoculate LB broth, and the cultures were incubated overnight at 37°C with gentle shaking. Each culture was then diluted in fresh medium and incubated at 37°C until reaching an OD600 of approximately 0.5. The culture was centrifuged, and cells were resuspended in phosphate-buffered saline (pH 7.2). Dilutions of this suspension in phosphate-buffered saline were used to inoculate mice. The number of CFU per milliliter in suitable dilutions was determined by plate counts. For oral inoculation, 0.2-ml aliquots were administered to mice, lightly anesthetized with ether, with 1-ml disposable syringes to which polyethylene catheters (Biotrol) were attached. Animal care and handling were in accordance with institutional guidelines.
Nucleotide sequence accession numbers.
The sequence data reported in this communication will appear in the EMBL/GenBank/DDBJ nucleotide sequence databases under accession numbers AJ291321 to AJ291334.
RESULTS AND DISCUSSION
Isolation of strains with transposon insertions in ςS-activated genes.
It may be possible to isolate RpoS-regulated lacZ gene fusions in serovar Typhimurium by using an inducible promoter to control ςS expression. However, the functional similarities of ς70 and ςS (for a review, see reference 22) might result in competition effects between the two sigma factors, leading to artifacts during the selection procedure of fusions when ςS is overexpressed (for example, selection of ς70-dependent fusion artifactually regulated by high levels of ςS). Thus, to ensure levels of ςS in the physiological range, we devised a strategy based on functional complementation with an rpoS allele on a low-copy-number vector (Fig. 1). A set of strains with lacZ gene fusions was generated in the Smr derivative of the serovar Typhimurium rpoS mutant C52K from eight independent Tn5B21 mutagenesis experiments (labeled 1 to 3 and E to I; see Materials and Methods and Fig. 1A). To isolate genes whose expression was highly dependent on RpoS, we selected strains whose basal level of lacZ expression was low in the absence of RpoS (Fig. 1B). Strains with poorly expressed lacZ gene fusions were identified as those with a pale blue colony phenotype after growth on LB agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). These strains were then screened for increased fusion gene expression following acquisition of plasmid pVKRpoS, a low-copy-number vector which contains the rpoS gene from strain C52 (Fig. 1B). A total of 39 Tn5B21 insertion strains repeatedly exhibited increased β-galactosidase activity upon introduction of plasmid-borne rpoS (Fig. 1B and data not shown).
To determine whether the 39 Tn5B21 insertions mutants were independent mutants, preliminary physical maps of the genomic regions 5′ and 3′ to the Tn5B21 insertions were determined by Southern hybridization of total DNAs from the mutants, using as probes the lacZ gene and the Tn5B21-bearing plasmid pBDJ103 (data not shown); 38 of the 39 strains appeared to be independent mutants. The two mutants that show similar physical maps were isolated from the same mutagenesis experiment and were subsequently found to contain Tn5B21 inserted in the same position in their genomes (data not shown).
Each of these putative RpoS-dependent transcriptional fusions was transduced into the rpoS mutant C52K and into the wild-type strain C52 by using phage P22 and retested for ςS dependency (Table 1). In addition, Southern hybridization experiments were conducted to check that Tn5B21 insertions mapped at the correct position in the genome of transductants (data not shown). β-Galactosidase activity of the transductant strains was assayed in stationary-phase cultures in LB medium (Table 1). Expression of the fusions was in all cases lower in the rpoS mutant C52K than in the wild-type strain C52. However, expression of the fusions in strain C52K could be restored upon introduction of plasmid pVKRpoS (Table 1). In contrast, there was no variation in the levels of β-galactosidase activity expressed by the fusions in C52K upon introduction of the control vector pVKCm (data not shown). The difference in expression of ςS-regulated gene fusions varied from 3- to 95-fold when isogenic strains with wild-type and null rpoS loci were compared. Thus, the RpoS protein can regulate transcription over a wide range. The strategy used to screen the series of fusions would be expected to prevent the selection of putative RpoS-regulated genes, for which the basal level of expression is high in the absence of RpoS and further increased upon introduction of an rpoS allele. Consistent with this, the basal levels of β-galactosidase activity in the absence of a wild-type rpoS allele were very low in the growth conditions used (Table 1).
TABLE 1.
Molecular characterization of ςS-activated gene fusions
| Tn5B21 insertion | β-Galactosidase activitya (mean ± SD)
|
E. coli homologc
|
||||
|---|---|---|---|---|---|---|
| rpoS+ | rpoS | rpoS pVKRpoS | Fold reductionb | Gene (% identity/ % similarity) | Position/length | |
| 1.32 | 196 ± 9 | 20 ± 14 | 199 ± 6 | 10 | otsA (85/89) | 236/474 |
| 1.36 | 309 ± 21 | 11 ± 0.1 | 319 ± 8 | 28 | ygaU (92/94) | 119/149 |
| 1.39 | 977 ± 133 | 65 ± 3 | 1,840 ± 121 | 15 | yciF (83/96) | 30/166 |
| 2.4 | 266 ± 18 | 8 ± 1 | 212 ± 8 | 33 | katE (82/90) | 109/753 |
| 2.9 | 126 ± 6 | 6 ± 1 | 137 ± 10 | 21 | ycgB (97/98) | 109/510 |
| 2.10 | 431 ± 24 | 12 ± 0.6 | 450 ± 77 | 36 | yeaG (100) | 550/644 |
| 2.11 | 34 ± 8 | 7 ± 0.1 | 40 ± 2 | 5 | No | |
| 3.3 | 146 ± 4 | 27 ± 0.1 | 125 ± 6 | 5 | NI | |
| 3.6 | 119 ± 5 | 8 ± 0.1 | 142 ± 3 | 15 | NI | |
| 3.7 | 473 ± 80 | 5 ± 0.1 | 527 ± 59 | 95 | NI | |
| E18 | 136 ± 9 | 4 ± 1 | 107 ± 27 | 34 | NI | |
| E26 | 61 ± 14 | 16 ± 11 | 45 ± 2 | 4 | No | |
| E44 | 85 ± 2 | 14 ± 5 | 17 ± 2 | 6 | yehY (80/89) | 287/385 |
| E45 | 253 ± 36 | 17 ± 1 | 195 ± 39 | 15 | No | |
| E46 | 95 ± 12 | 18 ± 9 | 92 ± 15 | 5 | yhjY (67/84) | 106/232 |
| F1 | 602 ± 223 | 89 ± 15 | 786 ± 86 | 7 | yciF (96/96) | 134/166 |
| F2 | 82 ± 3 | 16 ± 4 | 81 ± 2 | 5 | No | |
| F3 | 391 ± 33 | 69 ± 8 | 436 ± 80 | 6 | NI | |
| F4 | 103 ± 13 | 23 ± 18 | 95 ± 8 | 4 | poxB (94/95) | 200/572 |
| F7 | 135 ± 3 | 20 ± 6 | 124 ± 26 | 7 | NI | |
| F8 | 38 ± 3 | 15 ± 1 | 44 ± 3 | 3 | No | |
| F9 | 449 ± 157 | 32 ± 15 | 533 ± 5 | 14 | No | |
| F10 | 420 ± 155 | 29 ± 13 | 343 ± 120 | 14 | NI | |
| F11 | 159 ± 18 | 23 ± 11 | 193 ± 17 | 7 | No | |
| F12 | 318 ± 7 | 43 ± 16 | 441 ± 67 | 7 | yncC (82/87) | 34/221 |
| F13 | 65 ± 4 | 18 ± 3 | 86 ± 15 | 4 | NI | |
| F79 | 154 ± 39 | 6 ± 2 | 106 ± 6 | 26 | yjgB (93/97) | 133/339 |
| G10 | 354 ± 38 | 23 ± 7 | 276 ± 3 | 15 | yahO (66/79) | 66/91 |
| G20 | 185 ± 26 | 36 ± 4 | 175 ± 29 | 5 | NI | |
| G57 | 356 ± 113 | 21 ± 6 | 303 ± 67 | 17 | NI | |
| G87 | 58 ± 7 | 10 ± 2 | 57 ± 10 | 6 | NI | |
| H5 | 40 ± 10 | 7 ± 1 | 34 ± 1 | 6 | NI | |
| H21 | 163 ± 33 | 5 ± 1 | 126 ± 14 | 33 | NI | |
| H30 | 146 ± 28 | 24 ± 1 | 119 ± 22 | 6 | NI | |
| H87 | 165 ± 7 | 29 ± 6 | 183 ± 46 | 6 | ogt promoter region | |
| I8 | 101 ± 28 | 20 ± 5 | 76 ± 6 | 5 | NI | |
| I28 | 371 ± 88 | 44 ± 9 | 330 ± 14 | 8 | NI | |
| I36 | 326 ± 69 | 77 ± 35 | 252 ± 25 | 4 | NI | |
Comparison of β-galactosidase activities (in Miller units [24]) in strains with wild-type and null rpoS loci. Values (averages of at least three independent experiments) were calculated from stationary-growth-phase cultures in LB. rpoS+ denotes the Tn5B21 insertions in strain C52 containing a wild-type rpoS locus. rpoS denotes an isogenic strain carrying the ΔrpoS::kan locus (C52K). pVKRpoS carries the rpoS gene from strain C52 cloned into the low-copy-number vector pVK100 (19).
Decrease in β-galactosidase activity on acquisition of the null rpoS allele in strain C52. Values are rounded to the nearest whole number.
Junction fragments containing chromosomal sequences 5′ to the lacZ gene of Tn5B21 were cloned and sequenced; extents of amino acid identity and similarity were determined. No, no homology with E. coli; NI, insertion not identified. The position of each identified insertion and length of the E. coli homolog (in amino acids) are indicated. Accession numbers for the E. coli proteins were P33361 (YehY), P37663 (YhjY), P07003 (PoxB), P76114 (YncC), P27250 (YjgB), P75694 (YahO), P39169 (YgaU), P31677 (OtsA), P77391 (YeaG), P21179 (KatE), P21362 (YciF), and P29013 (YcgB).
Cellular levels of RpoS increase during entry into stationary phase in rich medium, under starvation conditions, or when strains are exposed to various stress conditions (for reviews, see references 14 and 22). However, the pattern of induction of genes regulated by RpoS may be narrower due to dependence on other global additional regulators. We monitored gene fusion activity in strains grown to stationary phase in LB rich medium (Fig. 1B), a condition allowing RpoS-dependent expression of a variety of RpoS-regulated genes in E. coli. As expected, expression of all the RpoS-regulated fusions in the rpoS+ strain C52 was induced in stationary phase of growth in rich medium (data not shown). In some cases, however, the level of expression of the fusion in C52 was low (<50 Miller units) even in the stationary phase of growth (Table 1). Growth conditions other than those used may be required for maximal expression of the RpoS-regulated gene which has been mutated in these strains.
Molecular characterization of ςS-regulated genes.
We determined the ςS-regulated loci defined by 21 of the Tn5B21 insertion mutants. We cloned the DNA fragments harboring the junction between the left end of Tn5B21 and the Salmonella chromosome and determined the nucleotide sequence of the region adjacent to Tn5B21 (upstream of the promoterless lacZ gene). The sequences were then used to search sequence databases for related genes or proteins.
Fourteen of the twenty-one mutations mapped to DNA regions very similar to E. coli regions (Table 1). These insertions mapped to open reading frames (ORFs) with the exception of insertion H87, which is located in a noncoding region (promoter region of ogt [see below]). Two insertions (F1 and 1.39) are in the same ORF (Table 1). The degree of conservation between E. coli and serovar Typhimurium in the sequenced regions of the 12 identified ORFs was from 66 to 100% identity at the protein level (Table 1) and from 71 to 87% identity at the DNA level (data not shown). These sequences are presumably orthologs. These ORFs are scattered throughout the E. coli chromosome. This approach identified the serovar Typhimurium homologs of the E. coli katE, otsA, and poxB genes, three well-characterized ςS-regulated genes (22), thereby validating the method (Table 1). katE, otsA, and poxB encode catalase HPII, trehalose-6-phosphate synthase, and a pyruvate oxidase, respectively. Our partial sequence of otsA from serovar Typhimurium strain C52 is 100% identical to the corresponding region of the serovar Typhimurium otsA sequence AF213176, and otsA has been previously shown to be regulated by ςS in serovar Typhimurium (8, 13).
Ten fusions mapped to serovar Typhimurium homologs of E. coli ORFs not previously known to require RpoS for expression (Table 1). Our partial sequence of yahO is 100% identical to nucleotides 13 to 299 of the serovar Typhimurium DNA sequence U51879 carrying the propionate catabolism operon prpBCDE and the divergently transcribed regulatory gene prpR (15). This suggests that yahO is located downstream from prpR in serovar Typhimurium as in E. coli. The functions of yehY, yhjY, yncC, yjgB, yahO, ygaU, yeaG, yciF, and ycgB are unknown. The YciF and YhjY proteins have been classified by Blattner et al. (2) as a putative structural protein and a putative protein involved in fatty acid and phospholipid metabolism, respectively. Features in the predicted amino acid sequences of yehY, yjgB, and yncC suggest that they encode an ABC transporter permease, a zinc-type alcohol dehydrogenase-like protein, and a putative transcriptional regulator of the GntR family, respectively.
Interestingly, nucleotide sequence data from the junction fragment of the other seven insertions (F2, F8, F9, F11, E26, E45 and 2.11 [Table 1]) do not match any known E. coli sequence. Preliminary examination of the remaining 17 RpoS-regulated fusions indicates that more than one-third of the RpoS-regulated fusions mapped to DNA regions not present in E. coli (data not shown). This suggests that the composition of the RpoS regulon differs markedly in the two species. This in turn is consistent with preliminary genome sequence analysis of Salmonella indicating that the genomes of E. coli and Salmonella are more different than might be suggested by the considerable concordance of their genetic maps (23, 44).
More than 50 genes have been found to be positively regulated by RpoS in E. coli (for a review, see reference 22). Previous works have identified RpoS-regulated genes in serovar Typhimurium (8, 9, 13, 31, 36), including the E. coli homologs narZ (nitrate reductase), cfa (cyclopropane fatty acid synthase), otsA (trehalose synthetase), sodC (superoxide dismutase), csg (curli biosynthesis), ORFO186 (U18997; unknown) and yohF (oxidoreductase). Of the 20 RpoS-regulated sequences that this work has identified in Salmonella, 13 are present in E. coli, and only 3 of these are known members of the RpoS regulon. The RpoS regulon may thus be larger than initially predicted. With one exception (the ogt gene, [see below]), the new members of the RpoS regulon identified in this study are homologs of ORFs in E. coli which have not been studied and not been assigned any function. This suggests that a large proportion of the bacterium's genome may be involved in adaptation to particular growth phase- or stress-related stimuli (or suboptimal conditions) and not involved in growth under optimal conditions such as those generally used in laboratories (e.g., rich or glucose-based minimal medium under aerobic growth conditions). Work is in progress in our lab to analyze phenotypes of these mutants under a variety of nonstandard growth conditions and in various genetic backgrounds.
Genetic rearrangements in the yda-ogt region of the enterobacterial chromosome.
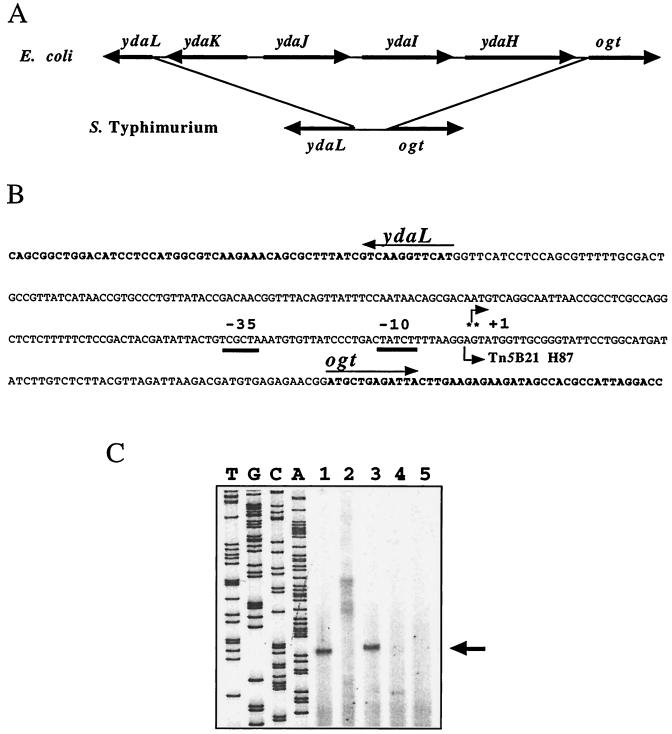
The nucleotide sequence of the 96-bp DNA region upstream from the lacZ gene of Tn5B21 in mutant C52::H87 is identical to the 5′ end of the serovar Typhimurium DNA sequence U23465. This 96-bp sequence in U23465 is located 71 nucleotides upstream from the start codon of the serovar Typhimurium ogt gene encoding O6MeG DNA MTase (46). A DNA sequence that matches the partial sequence of the ydaL gene of E. coli (89% nucleotide identity) was found 186 bp upstream from the Tn5B21 insertion H87, with the lacZ and ydaL genes being divergently transcribed. This was surprising because in E. coli, ydaL and ogt are separated by four ORFs, namely, ydaKJIH (Fig. 2A). To check whether the ogt gene was present downstream from Tn5B21 in strain C52::H87, we cloned and sequenced the DNA fragment harboring the junction between the right end of Tn5B21 and the Salmonella chromosome fragment in mutant strain C52::H87. Insertion H87 is indeed located 71 bp upstream from the translational start codon of the ogt gene of Salmonella strain C52 (Fig. 2B). The sequence of the 257-bp DNA region between the ydaL and ogt ORFs in serovar Typhimurium does not match the DNA sequence upstream from ogt in E. coli. In contrast, the nucleotide sequences of the coding regions of ogt were very similar in the two species (77% nucleotide identity and 88% amino acid identity). This shows that the ydaL-ogt DNA region of the enterobacterial chromosome has undergone genetic rearrangements during species divergence.
FIG. 2.
Genomic organization of the ogt region and mapping of the ςS-regulated promoter of ogt. (A) Comparison of the gene order near ogt in serovar Typhimurium and E. coli. (B) DNA sequence in the ydaL-ogt region in serovar Typhimurium. The 5′ ends of the ydaL and ogt genes are shown in bold. The nucleotide sequence was that from serovar Typhimurium C52 (this study). The position of the putative RpoS-dependent transcriptional start site of ogt (+1) is indicated by asterisks. The −10 and −35 regions of the rpoS-regulated promoter of ogt (ogtp1) are underlined. The position of insertion of Tn5B21 in mutant C52::H87 is indicated by a broken arrow. (C) Mapping of the 5′ end of ogt mRNA in serovar Typhimurium. A 5′ 32P-labeled primer complementary to the ogt coding strand was annealed to total RNAs isolated from LB-grown stationary-phase cultures of serovar Typhimurium wild-type (C52 and SL1344) and rpoS mutant (C52K and SL1344K) strains and of the Tn5B21 insertion mutant C52::H87. The primer was extended with reverse transcriptase, and the products were resolved by electrophoresis on a sequencing gel. The DNA sequencing ladder (lanes A, C, G, and T) was prepared using the primer as used to sequence pUCogt1. The major extended product is indicated by an arrow. Lane 1, C52; lane 2, C52K; lane 3, SL1344; lane 4, SL1344K; lane 5; C52::H87.
The ogt gene belongs to the RpoS regulon in Salmonella.
A major mutagenic adduct induced in DNA by methylating agents is O6MeG. This altered base mispairs with thymine during DNA replication, resulting in GC-to-AT transition mutations. To counteract such mutagenic effects, E. coli and serovar Typhimurium possess two DNA MTases, Ada and Ogt, that repair O6MeG lesions by directly transferring the methyl group from the methylated base to specific cysteine residues in the MTase (references 12, 27, and 46) and references therein). Exposure of E. coli cells to sublethal concentrations of DNA-methylating agents triggers the expression of a set of genes which confer increased resistance to the effects of these agents. This process, called the adaptive response, requires the Ada protein, which plays a dual role, being both a DNA repair enzyme and a transcription activator of the adaptive response. The response consists of induction of at least the ada-alkB operon, the alkA gene, and the aidB gene (for a review, see reference 21). The Ogt protein is not inducible by DNA alkylation damage and is the major MTase in unadapted cells (27, 28).
The adaptive response of serovar Typhimurium to alkylating agents seems to be less efficient than that in E. coli (12, 41). The Ada protein appears to play a major role in serovar Typhimurium tolerance to organic acid stress (1), but unlike an E. coli ada mutant, an ada mutant of serovar Typhimurium is not sensitive to the mutagenic action of DNA-methylating agents (45, 46). In contrast, an ogt mutant of serovar Typhimurium is much more sensitive than the corresponding wild-type strain to the mutagenic action of alkylating agents and to spontaneous mutagenesis, suggesting that the Ogt protein plays a major role in protecting serovar Typhimurium from the mutagenic action of both endogenous and exogenous alkylating agents (46).
It seemed likely that the lacZ fusion in strain C52::H87 was under the control of the ogt promoter. To verify this and identify the ogt promoter in Salmonella, we conducted primer extension experiments with RNAs isolated from stationary-phase cultures of wild-type strains (C52 and SL1344), rpoS mutants (C52K and SL1344K), and mutant C52::H87. A major extended product was detected with RNAs from the wild-type strains C52 and SL1344 but not with RNAs from the rpoS mutants (Fig. 2C). Therefore, the identified promoter (ogtp1) was under the control of RpoS. No extended product was detected with RNA from mutant C52::H87 (Fig. 2C). This result is consistent with the location of Tn5B21 in mutant C52::H87, just downstream from the putative RpoS-dependent promoter ogtp1 (Fig. 2B), and suggests that lacZ expression in mutant C52::H87 is under the control of ogtp1. Additional faint bands were detected in the rpoS strain C52K (Fig. 2C).
The ogtp1 −10 region (CTATCTT [Fig. 2B]) closely resembles the ςS consensus sequence. Indeed, 33 ςS-dependent promoters have a possible consensus sequence in the −10 region of CTATACT, which is very similar to the corresponding ς70 sequence of TATAAT (for a review see reference 22). No common −35 sequence element can be discerned in the ςS-dependent promoter group, and the −35 sequence of ogtp1 does not closely resemble the corresponding ς70 consensus sequence TTGACA (Fig. 2B).
These results demonstrate that RpoS regulates expression of the serovar Typhimurium ogt gene during the stationary phase in rich medium. This is consistent with previous findings suggesting that alkylating agents can accumulate in stationary phase or starved cells (references 27 and 39 and references therein). Moreover, expression of the Ada protein in E. coli has been shown to be dependent on RpoS in stationary phase (39). In Salmonella, it is not known whether RpoS controls Ada expression, and the Ada MTase does not seem to contribute to protection against mutagenesis by alkylating agents (46). Thus, RpoS may play a role in the ability of Salmonella to repair DNA damage caused by alkylating agents during stationary phase via the control of the Ogt MTase.
In preliminary experiments, transposon insertion H87 increased only two- to fivefold the number of rifampin- and nalidixic acid-resistant mutants recovered after N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) mutagenesis of serovar Typhimurium C52 (data not shown). The ogt gene is not disrupted in mutant C52::H87, and it cannot be excluded that in this mutant, basal levels of ogt mRNAs are produced from a promoter-like sequence located within the transposon. Previous work, showing that ogt plays a major role in protecting serovar Typhimurium from the mutagenic action of MNNG, used the serovar Typhimurium strain TA1535 (46). This strain is a derivative of the nonpathogenic strain LT2 with increased sensitivity to mutagens, and many LT2 isolates show altered rpoS expression (38, 43). It would be of interest to evaluate the impact of an ogt deletion on the resistance of a wild-type virulent strain of serovar Typhimurium to the effects of alkylating agents.
In conclusion, the transcription of ogt appears to be regulated by rpoS in serovar Typhimurium during the stationary phase of growth in rich medium, but further investigation is required to determine the physiological meaning of this finding.
Virulence assay.
None of the 14 strains studied, harboring Tn5B21 insertions in the conserved ςS-regulated genes (Table 1), were attenuated for virulence in Itys BALB/c mice. All mice (four per group) given either a mutant strain or wild-type strain C52 (108 bacteria orally) were dead within 12 days. Consistent with previous reports (7, 20), no animal died after receiving equivalent inocula of rpoS mutant strain C52K. rpoS mutants of Salmonella are thus highly attenuated in mice, with a 50% lethal dose at least 4 logs higher that of wild-type strains (7, 10). This presumably results largely from the reduced expression in the rpoS strain of the virulence plasmid genes spv, required for efficient growth of Salmonella in spleens and livers of infected mice (10, 20, 26). However, analysis of intestinal and splenic colonization in mice by wild-type and rpoS Salmonella strains cured for the virulence plasmid (7, 20, 25) suggested that rpoS regulates some unidentified chromosomal gene(s) involved in colonization and persistence of Salmonella in spleens and Peyer's patches.
In addition to the spv genes, at least three ςS-regulated loci may contribute to Salmonella virulence: the agf genes (analogous to the E. coli csg genes) for curli biosynthesis, narZ (analogous to E. coli narZ, the first gene of an operon encoding a nitrate reductase), and sodCII (analogous to E. coli sodC), encoding a periplasmic Cu,Zn-superoxide dismutase (5, 9, 31, 36, 40). However, mutations in these genes result in only a weak attenuating effect on Salmonella lethality for mice. Mutations in the agfB and narZ genes cause 3- and 10-fold, respectively, increases in the oral 50% lethal dose of S. typhimurium for Salmonella-susceptible Itys mice (36, 40). A sodCII mutation appears to decrease S. typhimurium lethality in Salmonella-resistant (Ityr) but not Salmonella-sensitive (Itys) mice (9). Interestingly, a second gene (sodCI) encoding a periplasmic Cu,Zn-superoxide dismutase is present in Salmonella, and mutants lacking both sodC genes are less lethal for mice than mutants possessing either sodC locus alone (9). This is an example of the potential difficulties in the analysis of gene products with redundant functions.
Therefore, although none of the conserved ςS-regulated genes identified in this study are essential for Salmonella lethality in mice, some may contribute to the mouse infection process. In addition, characterization of the Salmonella mutants carrying Tn5B21 insertions in DNA regions not present in E. coli will help elucidate the physiological function of the RpoS regulon in this pathogen.
ACKNOWLEDGMENTS
We thank M. Y. Popoff, in whose unit this work was conducted, for careful reading of the manuscript. We are grateful to B. D. Jones for the generous gift of plasmid pBDJ103.
M.I.-R. is a recipient of a postdoctoral fellowship from the Universidad Complutense. This work was supported by research funds from the Institut Pasteur and by a grant from the French Ministère de l'Education Nationale, de la Recherche et de la Technologie (Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires).
REFERENCES
- 1.Bearson B L, Wilson L, Foster J W. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol. 1998;180:2409–2417. doi: 10.1128/jb.180.9.2409-2417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban M, Cohen S N. Analysis of a gene control signal by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Chen C Y, Buchmeier N A, Libby S, Fang F C, Krause M, Guiney D G. Central regulatory role for the RpoS sigma factor in expression of Salmonella dublin plasmid virulence genes. J Bacteriol. 1995;177:5303–5309. doi: 10.1128/jb.177.18.5303-5309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collinson S K, Clouthier S C, Doran J L, Banser P A, William W K. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coynault C, Norel F. Comparison of the abilities of Salmonella typhimurium rpoS, aroA and rpoS aroA strains to elicit humoral immune responses in BALB/c mice and to cause lethal infection in athymic BALB/c mice. Microb Pathog. 1999;26:299–305. doi: 10.1006/mpat.1999.0273. [DOI] [PubMed] [Google Scholar]
- 7.Coynault C, Robbe-Saule V, Norel F. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (ςS) regulon. Mol Microbiol. 1996;22:149–160. doi: 10.1111/j.1365-2958.1996.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 8.Fang F C, Chen C-Y, Guiney D G, Xu Y. Identification of ςS-regulated genes in Salmonella typhimurium: complementary regulatory interactions between ςS and cyclic AMP receptor protein. J Bacteriol. 1996;178:5112–5120. doi: 10.1128/jb.178.17.5112-5120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang F C, DeGroote M-A, Foster J W, Bäumler A J, Ochsner U, Testerman T, Bearson S, Giard J-C, Xu Y, Campbell G, Laessig T. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci USA. 1999;96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulig P A, Danbara H, Guiney D G, Lax A J, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 12.Hakura A, Morimoto K, Sofuni T, Nohmi T. Cloning and characterization of the Salmonella typhimurium ada gene, which encodes O6-methylguanine-DNA methyltransferase. J Bacteriol. 1991;173:3663–3672. doi: 10.1128/jb.173.12.3663-3672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 15.Horswill A R, Escalande-Semerena J C. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihama A. Adaptation of gene expression in stationary phase bacteria. Curr Opin Genet Dev. 1997;7:582–588. doi: 10.1016/s0959-437x(97)80003-2. [DOI] [PubMed] [Google Scholar]
- 17.Jones B D, Falkow S. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan A Q, Zhao L, Hirose K, Miyake M, Li T, Hashimoto Y, Kawamura Y, Ezaki T. Salmonella typhi rpoS mutant is less cytotoxic than the parent strain but survives inside resting THP-1 macrophages. FEMS Microbiol Lett. 1998;61:201–208. doi: 10.1111/j.1574-6968.1998.tb12949.x. [DOI] [PubMed] [Google Scholar]
- 19.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 20.Kowarz L, Coynault C, Robbe-Saule V, Norel F. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J Bacteriol. 1994;176:6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 22.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 23.McClelland M, Wilson R K. Comparison of sample sequences of the Salmonella typhi genome to the sequence of the complete Escherichia coli K-12 genome. Infect Immun. 1998;66:4305–4312. doi: 10.1128/iai.66.9.4305-4312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Nickerson C A, Curtiss R. Role of factor RpoS in initial stage of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norel F, Robbe-Saule V, Popoff M Y, Coynault C. The putative sigma factor KatF (RpoS) is required for the transcription of the Salmonella typhimurium virulence gene spvB in Escherichia coli. FEMS Microbiol Lett. 1992;99:271–276. doi: 10.1016/0378-1097(92)90039-q. [DOI] [PubMed] [Google Scholar]
- 27.Rebeck G W, Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991;173:2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebeck G W, Smith C M, Goad D L, Samson L. Characterization of the major DNA repair methyltransferase activity in unadapted Escherichia coli and identification of a similar activity in Salmonella typhimurium. J Bacteriol. 1989;171:4563–4568. doi: 10.1128/jb.171.9.4563-4568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbe-Saule V, Coynault C, Norel F. The live oral vaccine Ty21a is a rpoS mutant and is susceptible to various environmental stresses. FEMS Microbiol Lett. 1995;126:171–176. doi: 10.1111/j.1574-6968.1995.tb07412.x. [DOI] [PubMed] [Google Scholar]
- 30.Robbe-Saule V, Schaeffer F, Kowarz L, Norel F. Relationships between H-NS, ςS, SpvR and growth phase in the control of spvR, the regulatory gene of the Salmonella plasmid virulence operon. Mol Gen Genet. 1997;256:333–347. doi: 10.1007/s004380050577. [DOI] [PubMed] [Google Scholar]
- 31.Römling U, Bian Z, Hammar M, Sierralta W D, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–78. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 35.Simon R J, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 36.Spectror M P, Garcia Del Portillo F, Bearson S M D, Mahmud A, Magut M, Finlay B B, Dougan G, Foster J W, Pallen M J. The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology. 1999;145:3035–3045. doi: 10.1099/00221287-145-11-3035. [DOI] [PubMed] [Google Scholar]
- 37.Sternberg N L, Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 38.Swords W E, Cannon B M, Benjamin W H. Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect Immun. 1997;65:2451–2453. doi: 10.1128/iai.65.6.2451-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taverna P, Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Velden A W M, Bäumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaughan P, Sedgwick B. A weak adaptive response to alkylation damage in Salmonella typhimurium. J Bacteriol. 1991;173:3656–3662. doi: 10.1128/jb.173.12.3656-3662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 43.Wilmes-Riesenberg M R, Foster J W, Curtiss R., III An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun. 1997;65:203–210. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong R M-Y, Wong K K, Benson N R, McClelland M. Sample sequencing of a Salmonella typhimurium LT2 lambda library: comparison to the Escherichia coli K12 genome. FEMS Microbiol Lett. 1999;173:411–423. doi: 10.1111/j.1574-6968.1999.tb13533.x. [DOI] [PubMed] [Google Scholar]
- 45.Yamada M, Hakura A, Sofuni T, Nohmi T. New method for gene disruption in Salmonella typhimurium: construction and characterization of an ada-deletion derivative of Salmonella typhimurium TA1535. J Bacteriol. 1993;175:5539–5547. doi: 10.1128/jb.175.17.5539-5547.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada M, Sedgwick B, Sofuni T, Nohmi T. Construction and characterization of mutants of Salmonella typhimurium deficient in DNA repair of O6-methylguanine. J Bacteriol. 1995;177:1511–1519. doi: 10.1128/jb.177.6.1511-1519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]