Abstract
Transcription factor (TF)-induced reprogramming of somatic cells across lineages and to induced pluripotent stem cells (iPSCs) has revealed a remarkable plasticity of differentiated cells and presents great opportunities for generating clinically-relevant cell types for disease modeling and regenerative medicine. The understanding of iPSC reprogramming provides insights into the mechanisms that safeguard somatic cell identity, drive epigenetic reprogramming, and underlie cell fate specification in vivo. The combinatorial action of TFs has emerged as the key mechanism for the direct and indirect effects of reprogramming factors that induce the remodelling of the enhancer landscape. The interplay of TFs in iPSC reprogramming also yields trophectoderm- and extraembryonic endoderm-like cell populations, uncovering an intriguing plasticity of cell states and opening new avenues for exploring cell fate decisions during early embryogenesis.
Keywords: reprogramming, iPSCs, enhancers, transcription factors
Introduction: overview of iPSC reprogramming
TFs are master regulators of development that determine gene expression programs and understanding how they define gene–expression programs is one of the central goals of developmental biology [1,2]. In 2006, Shinya Yamanaka’s laboratory stunned the developmental biology community by showing that ectopic expression of the TFs OCT4, SOX2, KLF4, and cMYC (OSKM) could reactivate the pluripotency gene network in terminally differentiated cells and establish iPSCs that carry the same features as embryonic stem cells [3-5]. Since then, iPSC reprogramming has offered a unique experimental system to explore the basic principles by which TFs drive cell fate specification.
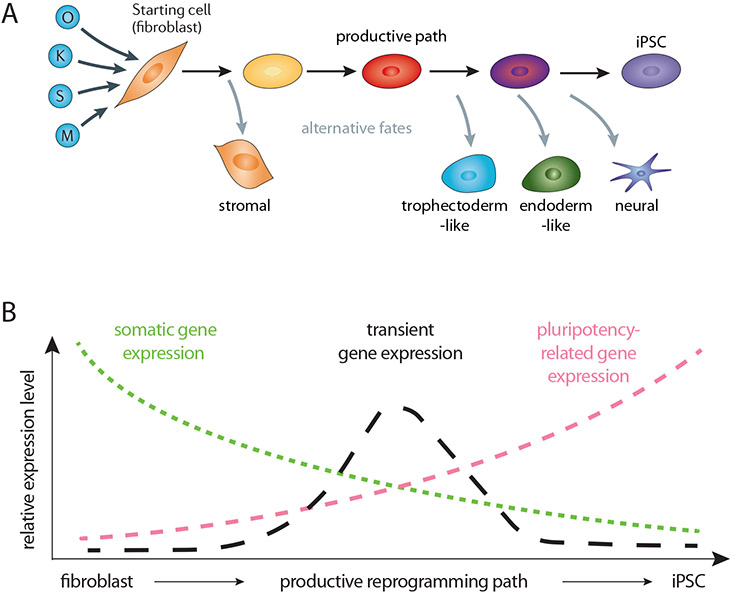
Landmark studies to uncover the features of the transition of somatic cells into iPSCs have integrated genomics approaches such as RNA-sequencing, mapping of chromatin accessibility, chromatin marks and TF binding, the isolation of reprogramming intermediates, and applied single cell transcriptomics [6-19]. These studies revealed that fibroblasts gradually progress through a continuum of states toward a mesenchymal-to-epithelial transition state from which a small proportion of cells continues to successfully reprogram to iPSCs. Many cells stall along this path or diverge from it to alternative cell states. Early in reprogramming, these alternative trajectories produce cells with a strong stromal identity characterized by increased expression of extracellular matrix genes, and later in the process trophectoderm-, extraembryonic endoderm-, and neural-like cells can arise in parallel to iPSCs [10,11,19,20] (Figure 1A). These findings indicate that cell fate specification is highly plastic during OSKM-induced somatic cell reprogramming, and that one reprogramming factor cocktail can result in numerous distinct gene expression programs. Along the iPSC path, cells lose the somatic gene expression program and activate the pluripotency expression program, which culminates in the hierarchical activation of pluripotency-related TFs (Figure 1B). These changes are accompanied by transient expression of genes from unrelated lineages [5,17,19] (Figure 1B). This pattern applies to iPSC reprogramming regardless of starting cell type and species [5,21]. In this review, we discuss the emerging general principles that allow the reprogramming factors to disassemble diverse somatic cell states and to activate the pluripotency program as well as alternative cell fates.
Figure 1: Cell state transitions and global gene expression changes in iPSC reprogramming.
A) Roadmap of iPSC induction from somatic cells. Upon expression of OSKM, a diminishing pool of cells transitions through sequential stages towards the iPSC state. In addition to cells stalling along the productive reprogramming path, the formation of alternative cell states explains the low efficiency of iPSC generation. The proportion of cell states at each stage is strongly influenced by the experimental reprogramming system and culture medium [10-12,15].
B) Key gene expression dynamics during OSKM-induced reprogramming. Regardless of the starting somatic cell type, three broad gene expression changes occur on the productive path to iPSCs: somatic program silencing, transient program expression, and pluripotency program activation. Pluripotency program activation occurs gradually with the upregulation of cell cycle, biosynthesis, chromatin remodeling genes, and culminates in the activation of endogenous pluripotency-related TFs. Somatic gene repression and pluripotency gene activation, previously thought to be separated temporally, can overlap in individual cells [12]. It is still largely unclear how the expression of the transient program relates to the silencing of the somatic program and the activation of the pluripotency program.
OSK-mediated rewiring of the enhancer landscape
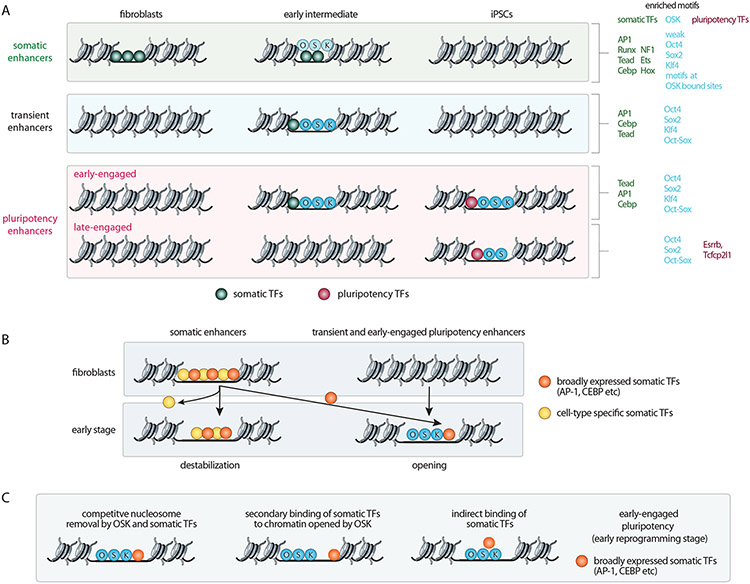
Enhancers play a central role in cell type-specific gene expression as binding platforms that integrate the function of multiple TFs [2,22]. Therefore, understanding how the reprogramming factors act on enhancers is essential for deriving the logic of their action. The genome-wide reorganization of enhancer usage during reprogramming is predominantly driven by OSK, without cMYC, which predominantly acts at promoters [18,23,24]. OSK action at enhancers leads to the inactivation of somatic enhancers, temporary activation of transient enhancers, and activation of pluripotency enhancers [6,7,16] (Figure 2A). During the earliest step of reprogramming, OSK predominantly bind somatic and transient enhancers, and engage only a small fraction of pluripotency enhancers [6] (Figure 2A). Over time, somatic and transient enhancers become silenced and no longer bound by OSK, concomitant with OSK binding to additional pluripotency enhancers [6,7,16] (Figure 2A). Thus, OSK bind to somatic, transient, and pluripotency enhancers, and produce different outcomes at these elements. As we discuss in detail below, OSK open chromatin by direct DNA binding and close chromatin active in somatic cells through indirect mechanisms, and both processes are linked through interactions with a small set of somatic TFs.
Figure 2: Enhancer reorganization during reprogramming is linked to distinct TF binding and motif patterns.
A) Key enhancer and associated TF binding changes during reprogramming. Very early in reprogramming, OSK bind a fraction of somatic enhancers as well as transient enhancers and a subset of pluripotency enhancers. At transient and early-engaged pluripotency enhancers, OSK co-bind with somatic TFs. Over time, early engaged pluripotency enhancers gain the binding of additional TFs throughout reprogramming (such as NANOG), which replaces the binding of somatic TFs. The majority of pluripotency enhancers is engaged later in the process (late-engaged) by O and S (without K) and requires additional TFs (for instance ESRRB) that are activated during the reprogramming process. In starting fibroblasts, both early and late-engaged pluripotency enhancers lack hypersensitivity (based on ATAC-seq) and reprogramming factor binding coincides with substantial nucleosome removal. Based on the presence and absence of DNA sequence motifs (as shown on the right), it is thought that OSK engage transient and pluripotency enhancers through direct DNA binding and interact with somatic enhancers largely indirectly.
B) Somatic TF redistribution model. Early in reprogramming, OSK recruit broadly expressed somatic TFs such as AP-1, CEBP, TEAD (orange symbols) to new sites in transient and pluripotency enhancers, depleting them from somatic enhancers. Since somatic cell-specific TF occupancy at somatic enhancers depends on the presence of broadly expressed somatic TFs, the binding of somatic cell-specific TFs (yellow symbols) is also decreased in this process. The redistribution of broad somatic TFs and the loss of somatic cell-specific TFs lead to the destabilization of fibroblast enhancers and the repression of the somatic gene program. In this model, OSK inactivate somatic enhancers indirectly, without the need for direct binding to somatic enhancers.
C) Putative roles for somatic TF binding at early-engaged pluripotency enhancers. From left to right: broadly expressed somatic TFs may collaborate with OSK to remove nucleosomes if their binding sites are within one nucleosome length, and therefore be required for enhancer opening early in reprogramming; somatic TFs passively bind to DNA in regions opened by OSK; and somatic TFs indirectly bind through protein-protein interaction with OSK or co-factors. In the latter two cases, somatic TFs may not have a specific function or, alternatively, may block the activation of these enhancers.
Somatic enhancer inactivation
How OSK repress somatic enhancers is not as well understood as pluripotency enhancer activation, yet extensive genomics approaches combined with loss- and gain-of-function experiments are beginning to shed light on this process [6-8,10,16,25]. A critical observation is that very early in reprogramming, somatic enhancers are perturbed genome-wide and display decreased levels of active enhancer marks (p300, H3K27ac), decreased chromatin accessibility, and decreased binding of somatic TFs [6-8]. These initial changes at somatic enhancers arise without a dramatic change in somatic TF expression levels and occur across most enhancers regardless of whether they are bound by OSK or not [6] (Figure 2A). Thus, global destabilization of somatic enhancers is not predominantly driven through their direct interaction with OSK.
Chronis et al found that the rapid loss of somatic TFs from somatic enhancers is accompanied by their redistribution to transient and early-engaged pluripotency enhancers [6] (Figures 2A/B). These new sites are bound by OSK and carry canonical motifs for OSK and somatic TFs [6-8,10]. The most parsimonious model explaining somatic enhancer inactivation therefore is that OSK redirect somatic TF binding by recruiting them to their target sites in newly opening enhancers and simultaneously removing them from somatic enhancers (Figures 2A/B), leading to widespread somatic gene silencing. Supporting the idea that the loss of somatic TFs is causal for somatic enhancer inactivation, the overexpression of somatic TFs that redistribute early in reprogramming (AP-1, CEBP, ETS, TEAD, RUNX family TFs, see below) blocks reprogramming whereas depletion enhances reprogramming [6,7,19,25]. A similar mechanism was later uncovered in T-cell development to explain a genome-wide gene expression switch of TFs guided by the master TF PU.1 [26], suggesting that the TF redistribution mechanism is broadly employed to induce cell fate transitions. Yet, many features of the redistribution process remain to be clarified. For instance, it is unknown whether redistribution of somatic TFs requires protein-protein interactions between somatic TFs and reprogramming factors, is critical for the opening of pluripotency enhancers together with OSK, or results from passive exploration of sites newly opened by OSK (Figure 2C). The subset of somatic enhancers bound by OSK may readily lose OSK during reprogramming because the reprogramming factors lack strong DNA binding motifs in those enhancers and may solely bind via interactions with somatic TFs or co-factors [6-8,10] (Figure 2A), enabling their disengagement upon loss of the somatic factors.
OSKM can induce reprogramming across different somatic cell types and species [5,10]. The identity of somatic TF co-binding with OSK at transient and pluripotency enhancers is beginning to shed light on how OSK can universally disassemble different somatic networks. Although cells express hundreds of TFs, a relatively small set of somatic TF motifs is associated with sites closing and opening early in reprogramming, including AP-1, ETS, RUNX, TEAD, CEBP TF motifs, regardless of species, starting cell type, and reprogramming method [6-8,10] (Figure 2A). Intriguingly, these TFs are expressed in many cell types. For example, AP-1 family TFs are ubiquitous transcriptional effectors with a broad role in differentiation and proliferation [27]. They are critical for enhancer selection in many cell types and collaborate with cell-type specific TFs to activate respective cell-type-specific enhancers [21,28-31]. We hypothesize that broadly expressed TFs that are critical for enhancer selection in vastly diverse cell types are exploited by OSK to shut off the starting cell program. Since somatic enhancer activation requires the collaborative action of AP-1 as well as somatic cell-specific TFs [1,21,26,28], OSK-mediated redeployment of a broadly acting somatic TF such as AP-1 may induce the loss of additional, somatic cell-specific TFs that depend on AP-1 co-occupancy (Figure 2B). Taken together, OSK may be highly effective reprogramming TFs because they can redistribute broadly acting somatic TFs. Although somatic TF redistribution appears to be the predominant mode of somatic enhancer destabilization, additional mechanisms are at play (Box1).
BOX1. Additional mechanisms of somatic program inactivation.
In addition to somatic TF redeployment, various other mechanisms are involved in controlling the activity of somatic enhancers. For instance, the recruitment of the histone deacetylase HDAC1 occurs specifically at OSK-bound somatic enhancers, which might shift the balance to co-factors towards repression [6]. Additionally, the co-repressor complex Sin3A is upregulated during reprogramming, required for iPSC induction, predominantly binds to promoters and contributes to the repression of critical somatic TFs [8]. In opposition, other mechanisms contribute to the maintenance of somatic enhancers and are barriers of reprogramming. A large number of somatic TFs, including AP-1, RUNX and CEBP, and the reprogramming factors, are modified by SUMO (small ubiquitin-like modifier). Since many TFs also contain SUMO-interacting domains, protein interaction networks are formed that stabilizes TF binding at somatic enhancers and secures somatic cell identity [55]. Accordingly, SUMO perturbation dramatically increases reprogramming efficiency [56,57]. Although the precise mechanism of how SUMO depletion enhances reprogramming is still unknown, its depletion may promote the redistribution of somatic TFs. Somatic enhancers also require the continuous recruitment of chromatin modifiers to stay active. One example is that the inhibition of MLL1, a histone H3K4 methyltransferase, results in efficient reprogramming via loss of the active histone H3K4me1 enhancer mark [58]. The loss of active chromatin modifiers and chromatin remodelers due to somatic TF redistribution may further destabilize somatic enhancers [28,46]. Regardless, these findings overall show that the inactivation of the somatic program is critical for iPSC induction and have highlighted mechanisms that maintain somatic cell identity.
Pluripotency enhancer opening
In contrast to somatic enhancers, OSK sites in pluripotency enhancers are strongly enriched for their cognate DNA motifs (Figure 2A), indicating that sequence-specific binding is critical for their selection. Some pluripotency enhancers become bound by the reprogramming factors early during reprogramming while others become bound only later (Figure 2A). Differences in chromatin accessibility are not responsible for this difference since both early and late-occupied pluripotency enhancers are embedded in nucleosomes in starting cells [6-8,10]. The recognition of nucleosomal binding sites and eviction of histone octamers are therefore required to establish the nucleosome-free region at pluripotency enhancers that is permissive for extensive TF binding and nucleation of transcriptional machinery observed in the pluripotent end state. Accessing pluripotency enhancers in closed chromatin appears to be a critical barrier, since most TFs cannot bind nucleosomal DNA [22]. Consistent with nucleosomes representing an obstacle, suppression of the histone chaperone CAF-1 enhances reprogramming by reducing the density of nucleosomes on chromatin [32].
A small number of TFs, called pioneer factors, are able to access DNA motifs wrapped in nucleosomes and to induce histone octamer displacement and to expose binding sites for additional TFs [33]. Early reports indicated that OSK are pioneer factors that can bind to nucleosomal DNA both in vivo and in vitro [24,34]. However, collaborative binding is required despite the pioneer factor activity of each reprogramming factor. Specifically, it was observed that early-engaged pluripotency enhancers are typically co-bound by OSK, and O, S, or K cannot access these sites when expressed alone in fibroblasts [6], indicating that OSK can compete with nucleosomes only when acting together. A distinguishing feature of early- and late-engaged pluripotency enhancers is that early sites are co-bound by O, S, and K, whereas late sites tend to be bound by only O and S, without K, which correlates with motif presence (Figure 2A). Thus, early in reprogramming, O and S are not sufficient to compete with nucleosomes at late pluripotency enhancers, implying that additional TFs are required for binding site selection. One such stage-specific TF is Esrrb, which only becomes expressed late in reprogramming and co-binds late pluripotency enhancers with O and S [6,8,35] (Figure 2A). Similarly, since the opening of pluripotency (and transient) enhancers by OSK in early reprogramming coincides with somatic TF recruitment, somatic TFs may be required for the selection of these enhancers (Figure 2). During developmental cell fate decisions, selection of new enhancer elements also requires combinatorial TF action [1,21,26,28,36], confirming iPSC reprogramming as a useful model for understanding the general logic of TF-guided cell fate decisions.
TFs can collaboratively bind nucleosomal DNA in multiple ways [37]. For instance, cooperativity can arise from protein-protein interactions between them, which can be enhanced by close spacing of binding sites or allosteric interactions on DNA. Consistent with this mechanism, OCT4 and SOX2 can dimerize on DNA, and this protein-protein interaction is required for reprogramming; furthermore, an Oct-Sox composite motif with juxtaposed binding sites is highly enriched in pluripotency enhancers [6-8,10,38,39] (Figure 2A). In an alternative mode, several TFs can compete with the histone octamer without the need of direct protein interactions, when their motifs are contained within the DNA sequence that is covered by the nucleosome [40].
Recent studies uncovered a range of binding modes for O and S at target sites within nucleosome-covered DNA, increasing the complexity of how these TFs open chromatin. Imaging studies with O and S engaging with nucleosomal DNA showed that binding of one factor often precedes the other [41,42]. The order is debated, and the presence of one TF can have synergistic and antagonistic effects on the binding of the other, depending on motif arrangement and position along the nucleosome [41,42]. Exciting structural studies revealed that O and S induce local DNA distortions and the detachment of DNA from the histone octamer to increase the accessibility on DNA [43,44]. Interestingly, O harbors two DNA binding domains, POU-S and POU-HD, of which the POU-S domain is sufficient to engage nucleosomal targets together with S [44]. Upon displacement of the histone octamer, it is thought that the POU-HD domain can engage the other half of the Oct4 motif [44]. Partial motifs recognized by the POU-S domain of OCT4 are enriched within the sequences curated for reprogramming factor binding sites that maintain nucleosomes in reprogramming cells [34], confirming this mode of action.
The maintenance of the open chromatin state by TFs is surprising given that TF occupancy at a binding site is intermittent with TFs cycling constantly on and off. Therefore, once the histone octamer is evicted, re-formation of nucleosomes may be inherently slow [36,41] or require other, active mechanisms, such as the action of ATP-dependent chromatin remodelling complexes. Indeed, reprogramming requires the OSK-mediated recruitment of the BAF chromatin remodeling complex [6,45]. BAF is critical for maintaining a nucleosome-free enhancer site, reinforces the binding of OSK and promotes the removal of flanking nucleosomes to enable the binding of additional TFs nearby [46,47]. Intriguingly, O, S, and K can also bind methylated DNA and induce demethylation through passive mechanisms or the recruitment of Tet enzymes [48-50], highlighting that diverse mechanisms are exploited by the reprogramming factors to open closed chromatin.
Alternative fates in reprogramming cultures
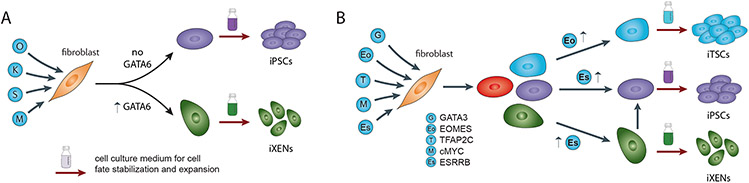
The emergence of various cell fates is an intriguing feature of iPSC reprogramming (Figure 1). For example, endodermal genes, including those encoding the TFs Gata4 or Gata6, become upregulated in OSKM-induced mouse fibroblast reprogramming cultures, and extraembryonic endoderm stem cells (iXENs) can be obtained when supporting culture medium is added [17,20]. Depletion of these endodermal TFs reduced the number of iXEN colonies while increasing iPSC colony number, indicating that iXEN formation occurs in parallel and competes with the iPSC reprogramming branch [20] (Figure 3A). It is likely that transiently induced endodermal TFs collaborate with the reprogramming factors to alter enhancer site selection and modulate the reprogramming outcome. In other studies, a small population of cells exhibiting a trophectoderm gene expression signature was detected during iPSC reprogramming of human fibroblasts [10,51]. Again, with a timely switch to appropriate culture conditions, this population can give rise to stable induced trophoblast stem cell (iTSC)-like cell lines [10,51]. However, continued reprogramming in fibroblast or iPSC medium extinguished the TSC-like identity [10,51]. Together, these results demonstrate that the signaling cues provided by the culture medium, and their downstream TFs, ultimately permit the stabilization and propagation of alternative cell fates such as iTSCs and iXENs.
Figure 3: Strategies for producing iPSCs, iTSCs and iXENs.
A) The existence of cells expressing endodermal TFs such as GATA4 and GATA6 in OSKM-induced iPSC reprogramming cultures can be exploited to, in addition to iPSCs, derive iXENs by exposing the reprogramming culture to a culture medium that supports iXENs [20]. Gata6 expression is required for iXEN formation. Similarly, iTSCs and iPSCs can be derived from human OSKM reprogramming cultures [10] (not shown).
B) iTSCs, iXENs and iPSCs can also be derived from a reprogramming culture upon expression of an alternative TF cocktail (GETMS), when appropriate media are supplied [53]. Whether XEN-like cells arise in parallel to or on the path to iPSCs remains unclear. The balance of EOMES and ESRRB influences which cell states are formed during reprogramming. High EOMES levels favor iTSC induction, whilst high ESRRB favors iXEN and iPSC induction.
Although the rules underlying this cellular plasticity of reprogramming cells are still unknown, the levels and stoichiometry of the reprogramming factors and other TFs appear critical, which is consistent with the observation that SOX2 levels define the developmental potential of early embryonic cells [14,52]. Reprogramming experiments with a non-OSKM TF cocktail also support this idea. The TFs GATA3, EOMES, TFAP2C, cMYC, and ESRRB can reprogram three stable stem cell types from mouse fibroblasts: iPSCs, iTSCs, and iXENs [53]. The balance of these TFs is the predominant factor determining the cellular outcome, with high levels of Eomes inducing iTSC identity, and Esrrb favoring iPSC and iXEN reprogramming (Figure 3B).
Conclusions
iPSC reprogramming is a rich model for understanding the TF code underlying cell fate changes in general. The future development and application of single cell multi-omics technologies combined with new lineage recording methods will provide many opportunities to address open questions. It remains to be shown how OSK interact with somatic TFs to induce their redistribution away from somatic enhancers to pluripotency enhancers; what the role of transient enhancers and the transient gene expression program is; how transient enhancers, bound directly by both somatic TFs and OSK, are silenced to give rise to iPSCs; whether somatic TFs are critical for pluripotency enhancer selection or, alternatively, may interfere with the full transcriptional activation of these sequences [31]. Similarly, whether comparable mechanisms for the decommissioning of the starting cell program also apply to direct reprogramming processes from one somatic cell into another, is an interesting question for the future. Finally, the derivation of human iPSCs, iXENs and iTSCs from one reprogramming culture has paved the way for the development of new cell models for the study of human embryogenesis [54], highlighting that insights gained from reprogramming studies will also be relevant for our understanding of early embryonic development.
Acknowledgements
W.D. is a Whitcome Graduate Student Fellow of the MBIDP at UCLA. K.P. is supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, the David Geffen School of Medicine, the NIH (GM099134), and a Faculty Scholar grant from the Howard Hughes Medical Institute.
Footnotes
Declaration of Interest
The authors declare no conflict of interest.
References
- 1.Heinz S, Romanoski CE, Benner C, Glass CK: The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 2015, 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitz F, Furlong EE: Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 2012, 13:613–626. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S: Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126:663–676. [DOI] [PubMed] [Google Scholar]
- 4.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. : Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007, 1:55–70. [DOI] [PubMed] [Google Scholar]
- 5.Nefzger CM, Rossello FJ, Chen J, Liu X, Knaupp AS, Firas J, Paynter JM, Pflueger J, Buckberry S, Lim SM, et al. : Cell Type of Origin Dictates the Route to Pluripotency. Cell Rep 2017, 21:2649–2660. [DOI] [PubMed] [Google Scholar]
- 6.Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K: Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 2017, 168:442–459 e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knaupp AS, Buckberry S, Pflueger J, Lim SM, Ford E, Larcombe MR, Rossello FJ, de Mendoza A, Alaei S, Firas J, et al. : Transient and Permanent Reconfiguration of Chromatin and Transcription Factor Occupancy Drive Reprogramming. Cell Stem Cell 2017, 21:834–845 e836. [DOI] [PubMed] [Google Scholar]
- 8.Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, Qin Y, Guo L, He J, Yu S, et al. : Chromatin Accessibility Dynamics during iPSC Reprogramming. Cell Stem Cell 2017, 21:819–833 e816. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz BA, Cetinbas M, Clement K, Walsh RM, Cheloufi S, Gu H, Langkabel J, Kamiya A, Schorle H, Meissner A, et al. : Prospective Isolation of Poised iPSC Intermediates Reveals Principles of Cellular Reprogramming. Cell Stem Cell 2018, 23:289–305 e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu X, Ouyang JF, Rossello FJ, Tan JP, Davidson KC, Valdes DS, Schroder J, Sun YBY, Chen J, Knaupp AS, et al. : Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 2020, 586:101–107. * In this study, the authors define the molecular path of iPSC induction from human fibroblasts and demonstrate that alternative cell states can be exploited to derive stable iTSCs in addition to naïve and primed iPSCs, by exposing the reprogramming culture to medium that supports a given cell type.
- 11.Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, et al. : Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming. Cell 2019, 176:928–943 e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran KA, Pietrzak SJ, Zaidan NZ, Siahpirani AF, McCalla SG, Zhou AS, Iyer G, Roy S, Sridharan R: Defining Reprogramming Checkpoints from Single-Cell Analyses of Induced Pluripotency. Cell Rep 2019, 27:1726–1741 e1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L, Lin L, Wang X, Gao M, Cao S, Mai Y, Wu F, Kuang J, Liu H, Yang J, et al. : Resolving Cell Fate Decisions during Somatic Cell Reprogramming by Single-Cell RNA-Seq. Mol Cell 2019, 73:815–829 e817. [DOI] [PubMed] [Google Scholar]
- 14.Francesconi M, Di Stefano B, Berenguer C, de Andres-Aguayo L, Plana-Carmona M, Mendez-Lago M, Guillaumet-Adkins A, Rodriguez-Esteban G, Gut M, Gut IG, et al. : Single cell RNA-seq identifies the origins of heterogeneity in efficient cell transdifferentiation and reprogramming. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP: A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell 2015, 16:323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Chen X, Li M, Liu X, Gao Y, Kou X, Zhao Y, Zheng W, Zhang X, Huo Y, et al. : Hierarchical Oct4 Binding in Concert with Primed Epigenetic Rearrangements during Somatic Cell Reprogramming. Cell Rep 2016, 14:1540–1554. [DOI] [PubMed] [Google Scholar]
- 17.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A: Dissecting direct reprogramming through integrative genomic analysis. Nature 2008, 454:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K: Role of the murine reprogramming factors in the induction of pluripotency. Cell 2009, 136:364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing QR, El Farran CA, Gautam P, Chuah YS, Warrier T, Toh CXD, Kang NY, Sugii S, Chang YT, Xu J, et al. : Diversification of reprogramming trajectories revealed by parallel single-cell transcriptome and chromatin accessibility sequencing. Sci Adv 2020, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parenti A, Halbisen MA, Wang K, Latham K, Ralston A: OSKM Induce Extraembryonic Endoderm Stem Cells in Parallel to Induced Pluripotent Stem Cells. Stem Cell Reports 2016, 6:447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK: Effect of natural genetic variation on enhancer selection and function. Nature 2013, 503:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long HK, Prescott SL, Wysocka J: Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 2016, 167:1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zviran A, Mor N, Rais Y, Gingold H, Peles S, Chomsky E, Viukov S, Buenrostro JD, Scognamiglio R, Weinberger L, et al. : Deterministic Somatic Cell Reprogramming Involves Continuous Transcriptional Changes Governed by Myc and Epigenetic-Driven Modules. Cell Stem Cell 2019, 24:328–341 e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soufi A, Donahue G, Zaret KS: Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 2012, 151:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Han Q, Peng T, Peng M, Wei B, Li D, Wang X, Yu S, Yang J, Cao S, et al. : The oncogene c-Jun impedes somatic cell reprogramming. Nat Cell Biol 2015, 17:856–867. [DOI] [PubMed] [Google Scholar]
- 26.Hosokawa H, Ungerback J, Wang X, Matsumoto M, Nakayama KI, Cohen SM, Tanaka T, Rothenberg EV: Transcription Factor PU.1 Represses and Activates Gene Expression in Early T Cells by Redirecting Partner Transcription Factor Binding. Immunity 2018, 48:1119–1134 e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eferl R, Wagner EF: AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003, 3:859–868. [DOI] [PubMed] [Google Scholar]
- 28.Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, Roberts CWM, Greenberg ME: AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol Cell 2017, 68:1067–1082 e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogan NT, Whalen MB, Stolze LK, Hadeli NK, Lam MT, Springstead JR, Glass CK, Romanoski CE: Transcriptional networks specifying homeostatic and inflammatory programs of gene expression in human aortic endothelial cells. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li QV, Dixon G, Verma N, Rosen BP, Gordillo M, Luo R, Xu C, Wang Q, Soh CL, Yang D, et al. : Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat Genet 2019, 51:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Markov GJ, Mai T, Nair S, Shcherbina A, Wang YX, Burns DM, Kundaje A, Blau HM: AP-1 is a temporally regulated dual gatekeeper of reprogramming to pluripotency. Proc Natl Acad Sci U S A 2021, 118. ** Exploring the heterokaryon system to define reprogramming mechanisms, the study shows the requirement of AP-1 for the control of somatic enhancers, and reveals that AP-1 also functions as repressor at the distal enhancer Oct4 in a phosphorylation-dependent manner.
- 32.Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, et al. : The histone chaperone CAF-1 safeguards somatic cell identity. Nature 2015, 528:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaret KS, Carroll JS: Pioneer transcription factors: establishing competence for gene expression. Genes Dev 2011, 25:2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS: Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161:555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adachi K, Kopp W, Wu G, Heising S, Greber B, Stehling M, Arauzo-Bravo MJ, Boerno ST, Timmermann B, Vingron M, et al. : Esrrb Unlocks Silenced Enhancers for Reprogramming to Naive Pluripotency. Cell Stem Cell 2018, 23:900–904. [DOI] [PubMed] [Google Scholar]
- 36.Sonmezer C, Kleinendorst R, Imanci D, Barzaghi G, Villacorta L, Schubeler D, Benes V, Molina N, Krebs AR: Molecular Co-occupancy Identifies Transcription Factor Binding Cooperativity In Vivo. Mol Cell 2021, 81:255–267 e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgunova E, Taipale J: Structural perspective of cooperative transcription factor binding. Curr Opin Struct Biol 2017, 47:1–8. [DOI] [PubMed] [Google Scholar]
- 38.Malik V, Glaser LV, Zimmer D, Velychko S, Weng M, Holzner M, Arend M, Chen Y, Srivastava Y, Veerapandian V, et al. : Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat Commun 2019, 10:3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tapia N, MacCarthy C, Esch D, Gabriele Marthaler A, Tiemann U, Arauzo-Bravo MJ, Jauch R, Cojocaru V, Scholer HR: Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency. Sci Rep 2015, 5:13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JA, Widom J: Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol 2003, 23:1623–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. : Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 2014, 156:1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Zheng EB, Zhao L, Liu S: Nonreciprocal and Conditional Cooperativity Directs the Pioneer Activity of Pluripotency Transcription Factors. Cell Rep 2019, 28:2689–2703 e2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodonova SO, Zhu F, Dienemann C, Taipale J, Cramer P: Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 2020, 580:669–672. [DOI] [PubMed] [Google Scholar]
- 44. Michael AK, Grand RS, Isbel L, Cavadini S, Kozicka Z, Kempf G, Bunker RD, Schenk AD, Graff-Meyer A, Pathare GR, et al. : Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 2020, 368:1460–1465. * In this study, the authors use a genomics readout to map all OS binding combinations on nucleosomal DNA in combination with stuctural studies to show how OS access sites in closed chromatin.
- 45.Chen K, Long Q, Xing G, Wang T, Wu Y, Li L, Qi J, Zhou Y, Ma B, Scholer HR, et al. : Heterochromatin loosening by the Oct4 linker region facilitates Klf4 binding and iPSC reprogramming. EMBO J 2020, 39:e99165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iurlaro M, Stadler MB, Masoni F, Jagani Z, Galli GG, Schubeler D: Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat Genet 2021, 53:279–287. * Studying OCT4 binding, the authors demonstrate that chromatin remodelling complexes are continously needed to preserve and restore chromatin accessibility and enable rapid TF cycling.
- 47.King HW, Klose RJ: The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanzan L, Soldati H, Ythier V, Anand S, Braun SMG, Francis N, Murr R: High throughput screening identifies SOX2 as a super pioneer factor that inhibits DNA methylation maintenance at its binding sites. Nat Commun 2021, 12:3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sardina JL, Collombet S, Tian TV, Gomez A, Di Stefano B, Berenguer C, Brumbaugh J, Stadhouders R, Segura-Morales C, Gut M, et al. : Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell 2018, 23:727–741 e729. [DOI] [PubMed] [Google Scholar]
- 50.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. : Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castel G, Meistermann D, Bretin B, Firmin J, Blin J, Loubersac S, Bruneau A, Chevolleau S, Kilens S, Chariau C, et al. : Induction of Human Trophoblast Stem Cells from Somatic Cells and Pluripotent Stem Cells. Cell Rep 2020, 33:108419. [DOI] [PubMed] [Google Scholar]
- 52.Tremble KC, Stirparo GG, Bates LE, Maskalenka K, Stuart HT, Jones K, Andersson-Rolf A, Radzisheuskaya A, Koo BK, Bertone P, et al. : Sox2 modulation increases naive pluripotency plasticity. iScience 2021, 24:102153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benchetrit H, Jaber M, Zayat V, Sebban S, Pushett A, Makedonski K, Zakheim Z, Radwan A, Maoz N, Lasry R, et al. : Direct Induction of the Three Pre-implantation Blastocyst Cell Types from Fibroblasts. Cell Stem Cell 2019, 24:983–994 e987. * This study shows that a non-OSKM reprogramming factor cocktail can induce iPSCs, iTSCs and iXENs from mouse fibroblasts and that the levels of the TFs EOMES and ESRRB determines which cell state is formed.
- 54. Liu X, Tan JP, Schroder J, Aberkane A, Ouyang JF, Mohenska M, Lim SM, Sun YBY, Chen J, Sun G, et al. : Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021, 591:627–632. ** This study exploits iPSCs and iTSCs derived from human fibroblasts upon OSKM expression to form blastoids that resemble the human blastocyst and will enable to study of human embryogenesis at scale.
- 55.Theurillat I, Hendriks IA, Cossec JC, Andrieux A, Nielsen ML, Dejean A: Extensive SUMO Modification of Repressive Chromatin Factors Distinguishes Pluripotent from Somatic Cells. Cell Rep 2020, 32:108146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cossec JC, Theurillat I, Chica C, Bua Aguin S, Gaume X, Andrieux A, Iturbide A, Jouvion G, Li H, Bossis G, et al. : SUMO Safeguards Somatic and Pluripotent Cell Identities by Enforcing Distinct Chromatin States. Cell Stem Cell 2018, 23:742–757 e748. [DOI] [PubMed] [Google Scholar]
- 57.Borkent M, Bennett BD, Lackford B, Bar-Nur O, Brumbaugh J, Wang L, Du Y, Fargo DC, Apostolou E, Cheloufi S, et al. : A Serial shRNA Screen for Roadblocks to Reprogramming Identifies the Protein Modifier SUMO2. Stem Cell Reports 2016, 6:704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Gayen S, Xiong J, Zhou B, Shanmugam AK, Sun Y, Karatas H, Liu L, Rao RC, Wang S, et al. : MLL1 Inhibition Reprograms Epiblast Stem Cells to Naive Pluripotency. Cell Stem Cell 2016, 18:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]