Abstract
The combination of the human induced pluripotent stem cell (hiPSC) and organoid technology enables the generation of human 3D culture systems, providing the opportunity to model human tissue-like structures in vitro. This protocol offers the details to generate and characterize self-assembling 3D cardiac organoids in a controlled and efficient manner based on hiPSC-derived cardiomyocytes. Cardiac organoids can be used as 3D-based assay systems and offer a wide range of applications in pharmacological and toxicological research as well as an alternative to animal experiments.
Keywords: Stem cells, Heart, Cardiac organoids, iPSCs, Disease modelling, Toxicity screening, Drug screening, Cell culture, Pharmacology
Graphical abstract
Stem cells; Heart; Cardiac organoids; iPSCs; Disease modelling; Toxicity screening; Drug screening; Cell culture; Pharmacology
1. Introduction
1.1. Before you begin
Human induced pluripotent stem cells (hiPSCs), which were reprogrammed using STEMCCA lentivirus system and overexpression of the four Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC), were used in this protocol. The hiPSC lines were characterized as previously described (Streckfuss-Bömeke et al., 2013). Experiments using hiPSCs have to adhere to ethical and legal guidelines. This work was approved by the Ethics Committee of the University Medical Center Göttingen (approval number: 21/1/11) and the Ethics Committee of the TU Dresden (EK 422092019). All cell culture experiments have to be performed in a sterile cell culture hood. HiPSCs and differentiated cultures were maintained at 37 °C, 20% O2 and 5% CO2. A regular screening for mycoplasma contamination was preformed.
1.2. Preparation of reagents
Timing: overnight, at least 12 h
-
1.Thaw Geltrex
-
a.Thaw a bottle of Geltrex in a Styrofoam box filled with wet ice overnight or alternatively at least for 12 h at 4 °C.
-
b.Freeze 15 mL conical centrifuge tubes and 1000 μL pipette tips at −20 °C overnight or at least for 12 h.
-
a.
Timing: 1 h
-
2.Aliquoting Geltrex
-
a.Place the thawed Geltrex bottle (a small frozen core might still be present) and 15 mL conical centrifuge tubes in a Styrofoam box filled with wet ice.
-
b.Carefully shake Geltrex a few times until a completely thawed homogeneous suspension has formed.
-
c.Aliquot 2 mg Geltrex with ice-cold 1000 μL pipette tips into 15 mL conical centrifuge tubes on ice rapidly and freeze immediately.
-
d.Aliquots can be stored at −80 °C for up to one year.
-
a.
CRITICAL: The concentration of Geltrex depends on the batch. The aliquot volume equivalent to 2 mg Geltrex should be adjusted based on the concentration of the individual batch. This information can be found on the supplier’s webpage.
CRITICAL: It is important to ensure that each step of the aliquoting process is performed on ice and Geltrex is kept on ice throughout the process.
1.3. Preparation of culture plates
HiPSCs and 2D-differentiation cultures are cultivated on Geltrex-coated cell culture plates to ensure optimal cell attachment and growth.
Timing: 30 min
-
3.Dilute Geltrex and Coat Culture Plates
-
a.Prepare 4 °C cold DMEM/F-12. Just in time for use, put the Geltrex aliquot from −80 °C onto wet ice.
-
b.Resuspend the frozen Geltrex aliquot directly by adding 12 mL of cold DMEM/F-12 to the 15 mL conical centrifuge tube and mix thoroughly by pipetting.
-
c.To coat the wells of cell culture plates, pipette 0.5 mL (12-well plate) or 1 mL (6-well plate) of the Geltrex-Solution (0.166 mg/mL) per well and shake gently to cover the surface area completely. Coverslips are coated in 12-well plates.
-
a.
Note: If coating coverslips, they need to be incubated with 0.1% HCl overnight followed by 70% EtOH incubation for additional 12 h. Sterilize the coverslips at 200 °C before Geltrex coating.
-
d.
Incubate the Geltrex-coated cell culture plates for 2 h at 37 °C or overnight at 4 °C.
-
e.
Gently aspirate remaining Geltrex and wash one time with DPBS.
-
f.
Geltrex-coated cell culture plates can be stored at 4 °C for up to one week.
Note: Geltrex-coated cell culture plates should be warmed to room temperature before aspirating the remaining Geltrex.
CRITICAL: Geltrex must not be thawed before adding cold DMEM/F-12.
Note: Geltrex-coated wells displaying a granular surface structure or dried out wells must not be used.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ACTN2 monoclonal mouse IgG1 | Sigma-Aldrich | Cat#A7811, RRID:AB_476766 |
| PE anti-human CD31 mouse IgG1 | BioLegend | Cat#303105, RRID: AB_314331 |
| TNNT2 monoclonal mouse IgG | ThermoFisher Scientific | Cat#MS-295-PABX, RRID:AB_61810 |
| VIMENTIN, VIM-13.2, monoclonal | Sigma-Aldrich | Cat#V525, RRID: AB_477625 |
| Alexa Fluor 488 goat anti-mouse IgG | ThermoFisher Scientific | Cat#A-11001, RRID:AB_2534069 |
| Chemicals, peptides, and recombinant proteins | ||
| Agarose | Biozym | Cat#840000 |
| Albumin, human recombinant | ThermoFisher Scientific | Cat#25200056 |
| B-27 serum free, supplement (50x) | ThermoFisher Scientific | Cat#17504044 |
| Boric acid | Roth | Cat#1024292 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | Cat#F7524 |
| CHIR99021 | Merck Millipore | Cat#361559 |
| Collagenase B | Worthington Biochemical | Cat#CLS-AFB |
| Cell Staining Buffer | BioLegend | Cat#420201 |
| DAPI (4′,6-diamidino-2-phenylindole) | Sigma-Aldrich | Cat#D9542 |
| dNTP mix (100 mM) | Bioline | Cat#BIO-39029 |
| DMEM/F-12, GlutaMax | ThermoFisher Scientific | Cat#31331028 |
| DMEM/F-12, no phenol red | ThermoFisher Scientific | Cat#21041025 |
| DMSO (dimethylsulfoxid) | Sigma-Aldrich | Cat#D2650 |
| Doxorubicin | Sigma-Aldrich | Cat#D1515-10MG |
| DPBS | ThermoFisher Scientific | Cat#14190169 |
| EDTA (ethylene diamine tetra acetic acid) | Sigma-Aldrich | Cat#E6758 |
| EtOH (ethanol) absolute | VWR | Cat#20821.310 |
| E8 basal medium, supplement | ThermoFisher Scientific | Cat#A1517001 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#F7524 |
| Fluoromount-G | eBioscience | Cat#00-4958-02 |
| Geltrex | ThermoFisher Scientific | Cat#A1413301 |
| GeneRuler 100 bp Plus DNA Ladder | ThermoFisher Scientific | Cat#0321 |
| Green GoTaq reaction buffer (5x) | Promega | Cat#M7911 |
| GoTaq G2 DNA Polymerase (5 U/μL) | Promega | Cat#M7845 |
| HCl 37% | Klinik Apotheke, Dresden | N/A |
| HDGreen | Intas | Cat#ISII-HDGreen |
| Human basic Fibroblast Growth Factor (hbFGF) | PeproTech | Cat#100-18B |
| Isopropanol | Merck Millipore | Cat#109634 |
| IWP2 | Merck Millipore | Cat#681671 |
| L-Ascorbic Acid 2-Phosphate | Sigma-Aldrich | Cat#A8960 |
| MgCl2 (25 mM) | Applichem | Cat#A103 |
| MuLV Reverse Transcriptase | ThermoFisher Scientific | Cat#N808-0018 |
| Nuclease-free water | ThermoFisher Scientific | Cat#1408217 |
| LIVE/DEAD™ Viability/Cytotoxicity Kit | ThermoFisher Scientific | Cat#L3224 |
| Olligo d(T)16 (50 μM) | ThermoFisher Scientific | Cat#N808-0128 |
| 10x PCR Buffer II | ThermoFisher Scientific | Cat#N8080006 |
| Penicillin-streptomycin solution (P/S) 100x | ThermoFisher Scientific | Cat#15140122 |
| Paraformaldehyde (PFA) | Sigma-Aldrich | Cat#158127 |
| RPMI 1640 with HEPES and GlutaMax | ThermoFisher Scientific | Cat#72400021 |
| RNAse Inhibitor (20 U/μL) | ThermoFisher Scientific | Cat#N808-0119 |
| SV total RNA isolation system Kit | Promega | Cat#Z3105 |
| Thiazovivin (TZV) | Millipore | Cat#420220 |
| TRIS ultrapure (tris-(hydroxymethyl)-aminomethane) | Applichem | Cat#A1086 |
| Triton X-100 | Sigma-Aldrich | Cat#X100 |
| 0.25% Trypsin-EDTA | ThermoFisher Scientific | Cat#25200056 |
| Versene solution (0.48 mM EDTA) | ThermoFisher Scientific | Cat#15040066 |
| 7-AAD | Abcam | Cat#ab228563 |
| Oligonucleotides | ||
| Troponin T2, cardiac type (TNNT2) For: GACAGAGCGGAAAAGTGGGA Rev: TGAAGGAGGCCAGGCTCTAT |
Eurofins | N/A |
| Gap junction protein, alpha 1 (GJA1) For: GGGTTAAGGGAAAGAGCGACC Rev: CCCCATTCGATTTTGTTCTGC |
Eurofins | N/A |
| Myosin light chain 2a, atrial isoform (MYL7) For: GAAGGTGAGTGTCCCAGAGG Rev: CTTGTAGTCGATGTTCCCCG |
Eurofins | N/A |
| Myosin light chain 2v, ventricular isoform (MYL2) For: GGCGAGTGAACGTGAAAAAT Rev: CAGCATTTCCCGAACGTAAT |
Eurofins | N/A |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) For: AGAGGCAGGGATGATGTTCT Rev: TCTGCTGATGCCCCCATGTT |
Eurofins | N/A |
| Other | ||
| 6-well (TC treated) plate | Starlab | Cat# #CC7682-7506 |
| 12-well (TC treated) plate | Starlab | Cat#CC7682-7512 |
| 96-U-shape-well plate | Brand | Cat#781660 |
| Counting chamber | LO-Laboroptik | Cat#1100000 |
| Coverslips, 22 mm | Omnilab | Cat#5161066 |
| Cryo-tubes, 2 mL | Greiner | Cat#126263 |
| Glass bottom dishes, 35 mm | Mattek | Cat#P35G-1.5-14-C |
| Isopropanol freezing container | ThermoFisher Scientific | Cat#10110051 |
| Milli-Q Water purification systems | Merck Millipore | Cat#ZIQ7000T0 |
| Petri dish, 35 mm | Sarstedt | Cat#82.1135.500 |
| Pipette tips | Sigma-Aldrich | Cat#CLS4135 Cat#CLS4136 Cat#CLS4138 Cat#CLS4140 |
| Polystyrene round-bottom tubes, 5 mL | BD Falcon | Cat#352058 |
| Polystyrene serological pipettes | Corning | Cat#CLS4487 Cat#CLS4488 Cat#CLS4489 |
| Steriflip (0.22 μm pore size filter) | Merck | Cat#SCGP00525 |
| 15 mL conical centrifuge tube | Greiner | Cat#188271 |
| 50 mL conical centrifuge tube | Greiner | Cat#227261 |
| 0.5 mL Safe-Lock tube, ambra | Eppendorf | Cat#0030121155 |
| 0.5 mL Safe-Lock tube | Eppendorf | Cat#0030123301 |
2. Materials and equipment (optional)
2.1. Thiazovivin 2 mM
Dissolve 1 mg thiazovivin (TZV) in 16.06 mL DMSO and mix well to generate a 1000x stock solution (2 mM). Aliquot the stock solution and store at −20 °C for up to one year.
CRITICAL: TZV is UV-sensitive. Avoid direct light exposure and use light-protected Safe-Lock tubes, ambra.
INFORMATION: TZV is a cell-permeable ROCK inhibitor that prevents apoptosis induced by trypsinization (Xu et al., 2010; Burridge et al., 2014).
Note: Choose an aliquot volume suitable to avoid repeated freeze-thaw cycles.
2.2. CHIR99021 12 mM
Dissolve 5 mg CHIR99021 in 894 μL of DMSO and mix well to generate a 12 mM stock solution. Aliquot the stock solution and store at −20 °C for up to 6 months.
CRITICAL: CHIR99021 is UV-sensitive. Avoid direct light exposure and use light-protected Safe-Lock tubes, ambra.
Note: Choose an aliquot volume suitable to avoid repeated freeze-thaw cycles.
2.3. IWP2 5 mM
Dissolve 10 mg IWP2 in 4.28 mL of DMSO and mix well to generate a 5 mM stock solution. Aliquot the stock solution and store at −20 °C for up to 6 months.
Note: If IWP2 does not dissolve completely, incubate for 10–15 min at 37 °C in a water bath. Choose an aliquot volume suitable to avoid repeated freeze-thaw cycles.
2.4. Collagenase B 400 U/mL
Dissolve 18,000 U of Collagenase B in 45 mL RPMI 1640 and sterile filter with 0.22 μm pore size filter (Steriflip) to generate a 400 U/mL working solution, which can be stored at 4 °C for up to two weeks.
2.5. hbFGF (5 ng/μL)
Dissolve 100 μg of hbFGF in 1 mL TRIS (5 mM, pH6.8) to generate a stock solution, which can be stored at −20 °C for up to one year. 5 ng/μL working solution is generated by 1:20 dilution with 0.1% BSA solution. Working solutions can be stored at 4 °C for up to two weeks.
Note: Choose an aliquot volume suitable to avoid repeated freeze-thaw cycles.
2.6. hiPSC culture medium
Thaw E8 supplement at RT or at 4 °C overnight. Mix one bottle of E8 medium with E8 supplement. Complete hiPSC culture medium can be stored at 4 °C for up to two weeks.
Note: Do not thaw E8 supplement in a water bath at 37 °C. TZV is optionally added freshly.
2.7. hiPSC Freezing Medium 2x
Add 20% DMSO to hiPSC Culture Medium and use the 2 mM TZV stock solution for a final concentration of 4 μM TZV (1:500 dilution). The hiPSC Freezing Medium 2x can be stored at 4 °C for up to one week.
Note: Add TZV freshly.
2.8. Cardio Differentiation Medium
Add 0.5 mg/mL human recombinant albumin and 0.2 mg/mL L-Ascorbic acid 2-phosphate to RPMI 1640 (with GlutaMax and HEPES), mix well and filter sterilely with 0.22 μm pore size filter (Steriflip).
Note: Make sure to dissolve human recombinant albumin and L-Ascorbic acid 2-phosphate completely before sterile filtration. The components dissolve better when the medium has reached room temperature.
Note: CHIR99021 or IWP2 should always be added freshly.
2.9. Cardio Culture Medium
Thaw B27 supplement at RT or at 4 °C overnight. Mix RPMI 1640 (with GlutaMax and HEPES) with B27-supplement (50x) and store at 4 °C for up to one month.
2.10. Cardio Digestion Medium
Mix Cardio Culture Medium with 20% FBS (inactivated) and 2 mM TZV stock solution (1:1000 dilution) for a final concentration of 2 μM TZV. The Cardio Digestion Medium can be stored at 4 °C for up to two weeks.
Note: FBS has to be inactivated at 56 °C for 30 min before use. TZV should always be added freshly.
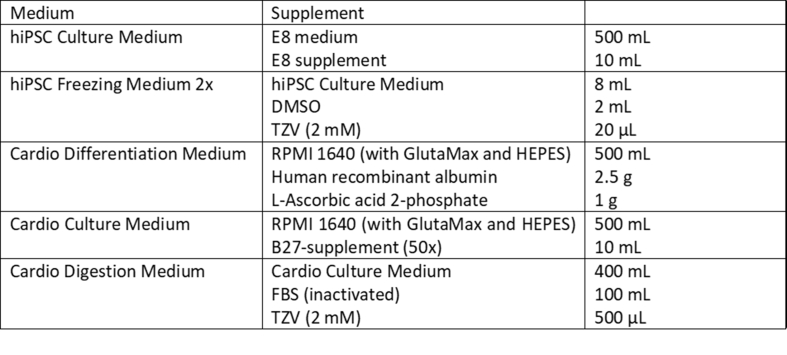
INFORMATION: An overview of media used for hiPSC cultivation, freezing, and differentiation as well as for hiPSC-CM cultivation and digestion is given in Supplementary Table 1.
2.11. Flow Cytometry Medium
Mix DMEM/F-12 (no phenol red) with 1.5% FBS, 1x penicillin-streptomycin solution (P/S) (1:100 dilution from 100x P/S), 10 ng/mL hbFGF and use 2 mM TZV stock solution (1:1000 dilution) for a final concentration of 2 μM TZV.
Note: FBS has to be inactivated at 56 °C for 30 min before use. TZV should always be added freshly.
2.12. 0.1% BSA
Dissolve 1 mg BSA in 1 mL DPBS and mix well. Aliquots can be stored at −20 °C for up to 6 months and at 4 °C for up to two weeks.
2.13. 1% BSA
Dissolve 10 mg BSA in 1 mL DPBS and mix well. Aliquots can be stored at −20 °C for up to 6 months and at 4 °C for up to two weeks.
2.14. DAPI (2 mg/mL)
Dissolve 1 mg of DAPI in 0.5 mL of Milli-Q water and mix well. Prepare a working solution by a 1:5000 dilution of the stock solution (2 mg/mL) with Milli-Q water and store at 4 °C for several months. Avoid direct light exposure.
2.15. Doxorubicin (20 mM)
Dissolve 10 mg of Doxorubicin in 862 μL of DMSO and mix well. Aliquots can be stored at −20 °C for up to 6 months. Avoid direct light exposure.
2.16. EDTA (0.5 M)
Dissolve 146.2 mg EDTA in 1 mL Milli-Q water and mix well. EDTA (0.5 M) can be stored at room temperature for several months.
2.17. 70% EtOH
Mix 700 mL EtOH absolute with 300 mL Milli-Q water. 70% EtOH can be stored at room temperature for several months.
2.18. 0.1% HCl
Add 2.7 mL HCl 37% to 997.3 mL Milli-Q water. 0.1% HCl can be stored at room temperature for several months.
2.19. Tris (0.5 M)
Dissolve 15.14 g Tris in 250 mL Milli-Q water and mix well. Adjust pH to 6.8 with 37%HCl. Tris (0.5 M) can be stored at room temperature for several months.
2.20. Tris-borate EDTA (TBE) buffer 5x
Dissolve 27.5 g boric acid, 20 mL ETDA 0.5 M and 54 g TRIS in 1 L of Milli-Q water and mix well. Tris-borate EDTA (TBE) buffer 5x can be stored at room temperature for several months.
2.21. 0.1% Triton X-100
Add 10 μL of Triton X-100 in 10 mL of 1% BSA and mix well. 0.1% Triton X-100 can be stored at 4 °C for several months.
2.22. 4% PFA
Dissolve 4 g PFA in 100 mL DPBS and mix well. 4% PFA can be stored at 4 °C for several months.
3. Step-by-step method details
3.1. hiPSC thawing
Timing: 15–20 min
Frozen hiPSCs can be thawed and cultured as described below. Frozen hiPSCs from one 6-well are thawed, resuspended and transferred onto two Geltrex-coated 6-wells (1:2).
Note: It is recommended to prepare the cell culture plates and media before thawing the cells in order to ensure a rapid workflow. Note that some of the following media must be used at indicated temperatures.
-
1.
Pre-warm the hiPSC Culture Medium to room temperature (19–23 °C) and add TZV at a final concentration of 2 μM (1 μL/mL of 1000x stock solution).
Note: Add TZV freshly to avoid UV exposure.
-
2.
Incubate Geltrex-coated 6-well plates at 37 °C for 30 min.
-
3.
Aspirate the remaining Geltrex gently and wash the wells one time with DPBS. Add 1 mL of hiPSC Culture Medium + TZV per well.
-
4.
Prepare one 15 mL conical centrifuge tube containing 10 mL cold DMEM/F-12 for each cryo-tube you are going to thaw.
Note: DMEM/F-12 is used at 4 °C.
-
5.
Thaw each cryo-tube directly at 37 °C in a water bath. Dry the tube and clean with 70% EtOH.
CRITICAL: The cryo-tubes must not be submerged in the water bath. Avoid water contact with the lid to prevent bacterial contamination.
-
6.
Carefully resuspend the cell suspension with a 1000 μL pipette and transfer gently into the prepared 15 mL conical centrifuge tubes containing 10 mL cold DMEM/F-12. Centrifuge at 200 × g for 5 min.
-
7.
Aspirate the supernatant and carefully resuspend the cell pellet in 2 mL of hiPSC Culture Medium + TZV using a 1000 μL pipette. Transfer 1 mL of the cell suspension into each of the previously prepared 6-wells containing hiPSC Culture Medium + TZV (thawing 1:2). Gently shake the plates in an 8-shaped movement pattern to distribute the cells evenly. Transfer the plates into an incubator at 37 °C and 5% CO2.
-
8.
HiPSCs should have attached to the well after 24 h. Change the medium with 2 mL/well hiPSC Culture Medium (see ‘hiPSC Culture’, step 11). For further cultivation continue with ‘hiPSC Culture’, step 9.
Note: After 24 h and successful attachment, hiPSCs are cultured without TZV.
Note: Insufficiently attached cells can be cultured for additional 24 h. Do not change the medium and add 1 mL of hiPSC Culture Medium + TZV per well. Continue with step 8 after 24 h.
3.2. hiPSC culture
Timing: 5–10 min
HiPSCs are cultured serum-free on 6-well plates. The medium has to be changed daily until the desired degree of optical confluence is reached.
-
9.
Warm hiPSC Culture Medium to room temperature (19–23 °C).
-
10.
Assess cell morphology and density daily using a brightfield microscope.
-
11.Change the medium with 2 mL of hiPSC Culture Medium per well.
-
a.Gently aspirate the old medium.
-
b.Add carefully 2 mL of fresh hiPSC Culture Medium per well.
-
a.
CRITICAL: To avoid cell detachment, carefully add fresh medium over the edge of the respective well.
3.3. hiPSC passaging
Timing: 10–20 min
At optical densities between 75 and 95% (regularly reached after 2–4 d) hiPSCs can be passaged.
Note: For hiPSC culture use 6-well plates. For cardiac differentiation use 12-well plates.
-
12.
Warm hiPSC Culture Medium to room temperature (19–23 °C) and add TZV for a final concentration of 2 μM (1 μL of the 1000x stock per mL medium).
Note: Add TZV freshly to avoid UV-exposure.
-
13.
Heat versene solution (0.48 mM EDTA) in a water bath to 37 °C.
-
14.
Incubate Geltrex-coated 6-well plates at 37 °C for 30 min.
-
15.
Aspirate the remaining Geltrex gently and wash the wells one time with DPBS. Add 1 mL of hiPSC Culture Medium + TZV per well.
-
16.
Gently aspirate the medium of your hiPSC cultures and wash quickly with 1 mL versene solution. Discard versene solution that was used for this washing step.
-
17.
Add 1 mL of fresh versene solution and incubate for 3–5 min at room temperature.
Note: Incubation times with versene solution (0.48 mM EDTA) may differ between cell lines. Adjust individually. Too short incubation can lead to poor cell detachment whereas too long incubation can lead to premature cell detachment and subsequent loss during aspiration of the versene solution.
Note: If the hiPSCs have detached prematurely, they can be transferred into 1 mL of hiPSC Culture Medium + TZV. Centrifuge at 200 × g for 5 min and remove supernatant. Resuspend the cell pellet with 1 mL hiPSC Culture Medium + TZV and proceed with step 19.
-
18.
Gently aspirate the versene solution. Detach the hiPSCs with 1 mL hiPSC Culture Medium + TZV by pipetting 2–3 times up and down using a 1000 μL pipette. Continue with step 19.
CRITICAL: Do not pipette more than 2–3 times to avoid cell death.
Note: Proceed quickly to avoid re-attachment of the cells.
-
19.
Plate the desired amount of hiPSCs onto Geltrex-coated 6-well plates (e.g. at densities 1:4–1:8 or count cells to plate a defined number) for further culture. For cardiac differentiation, plate hiPSCs at dilutions of 1:8–1:10 (or count cells to plate a defined number of cells (10,000–25,000 cells/cm2) per 12-well (e.g. 40,000–100,000 cells/12-well)) onto a Geltrex-coated 12-well plate.
-
20.
Gently shake the plates in an 8-shaped movement pattern to distribute the cells evenly. Incubate at 37 °C and 5% CO2 for further culture.
-
21.
HiPSCs should have attached to the well after 24 h. Change the medium with 2 mL/well hiPSC Culture Medium (see ‘hiPSC Culture’, step 11). For further cultivation continue with ‘hiPSC Culture’, step 9.
Pause Point: For cardiac organoid differentiation, continue with step ‘31’ (see ‘Generation of Self-assembling Cardiac Organoids Using hiPSC-derived Cardiomyocytes’). If the hiPSC are not needed for cardiac organoid differentiation at this point, the hiPSC can be frozen (see ‘hiPSC Freezing’).
3.4. hiPSC freezing
Timing: 10–20 min
HiPSCs can be frozen at optical densities of 75–95% to be thawed and used at a later time point.
-
22.
Label the amount of required cryo-tubes depending on the number of wells you are going to freeze. Confluent 6-wells are frozen 1:2 (all cells of one confluent well are split into two cryo-tubes)
-
23.
Warm hiPSC Freezing Medium 2x and hiPSC Culture Medium to room temperature (19–23 °C).
Note: Add TZV freshly to avoid UV-exposure.
-
24.
Heat versene solution (0.48 mM EDTA) in a water bath to 37 °C.
-
25.
Gently aspirate the medium of your hiPSC cultures and wash with 1 mL versene solution.
-
26.
Add 1 mL of fresh versene solution and incubate for 3–5 min at room temperature.
Note: Incubation times with versene solution (0.48 mM EDTA) may differ between cell lines. Adjust individually. Too short incubation can lead to poor cell detachment whereas too long incubation can lead to premature cell detachment and subsequent loss during aspiration of the versene solution.
Note: If the hiPSCs have detached prematurely, they can be transferred into 0.75 mL of hiPSC Culture Medium + TZV. Centrifuge at 200 × g for 5 min and remove supernatant. Resuspend the cell pellet with 0.75 mL hiPSC Culture Medium + TZV and proceed with step 28.
-
27.
Gently aspirate the versene solution. Detach the hiPSCs with 0.75 mL hiPSC Culture Medium + TZV by pipetting 2–3 times up and down using a 1000 μL pipette.
CRITICAL: Do not pipette more than 2–3 times to avoid cell death.
Note: Proceed quickly to avoid re-attachment of the cells.
-
28.
Add 0.75 mL of hiPSC Freezing Medium 2x drop-by-drop to the cell suspension of one entire 6-well plate (the final volume of each well is 1.5 mL). Mix gently by shaking the plate.
-
29.
Transfer 0.75 mL of the cell suspension into one cryo-tube (one confluent 6-well will generate 2 cryo-tubes). Transfer the cryo-tubes quickly to an isopropanol freezing container and store at −80 °C for at least 2 h. Then, place the tubes in a liquid nitrogen cryo-tank.
Note: Do not keep the hiPSCs at −80 °C for more than 12 h.
Note: The hiPSCs can be kept at −140 °C to −180 °C in the vapor phase of the liquid nitrogen cryo-tank for at least 5 years. Direct contact between the cryo-tubes and the liquid nitrogen should be avoided at all costs.
3.5. Generation of Self-assembling Cardiac Organoids Using hiPSC-derived cardiomyocytes
Timing: 14–16 days
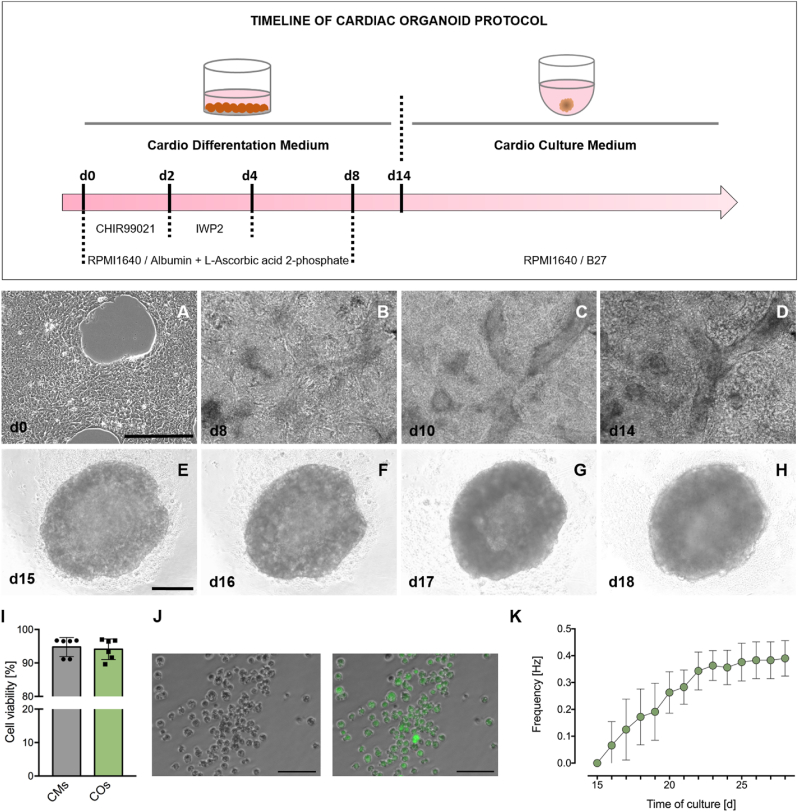
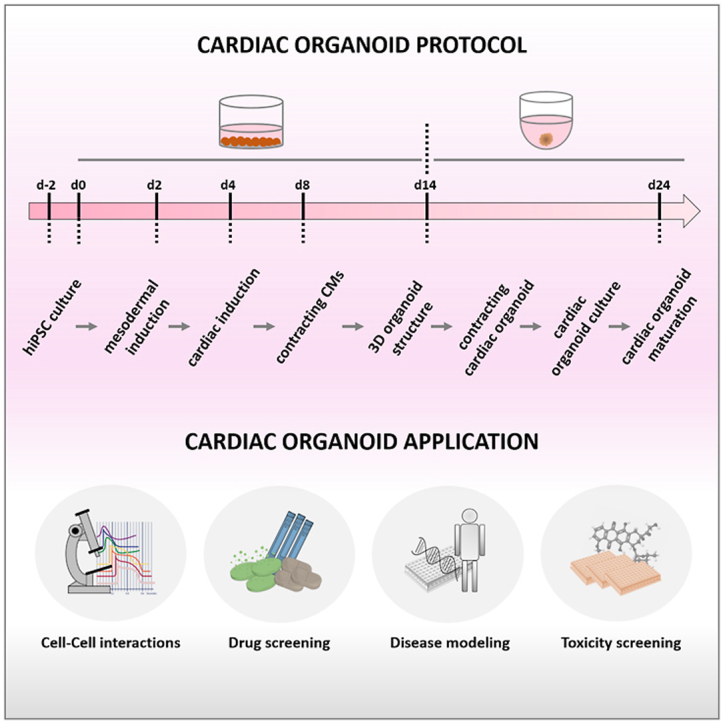
Based on the robust hiPSC-based cardiac differentiation under controlled culture conditions (Lian et al., 2012; Burridge et al., 2014; Cyganek et al., 2018), this protocol opens the possibility to generate self-assembling three-dimensional cardiac organoid cultures without a supporting scaffold at a high-throughput scale. The protocol involves cultivation in different cell culture plates, use of different media, and transient incubations with defined small molecules. Cardiac organoid differentiation and generation is shown in Figure 1. Contracting cardiomyocytes in 2D culture as well as contracting cardiac organoids are shown in videos S1 and S2, respectively.
Figure 1.
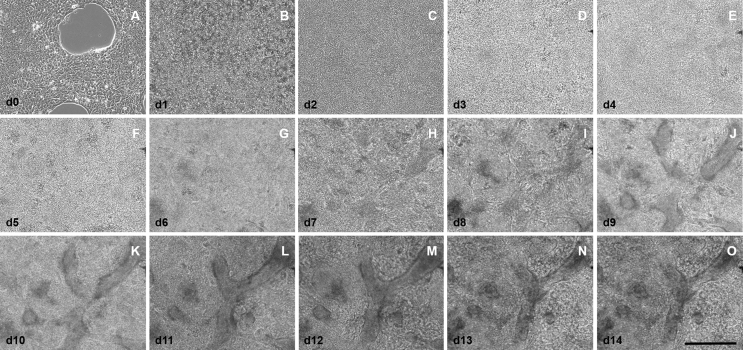
hiPSC differentiation to cardiac organoids. (Upper panel) Timeline of cardiac organoid differentiation. Cardio Differentiation Medium: RPMI1640, Albumin, L-Ascorbic acid-2-phosphate, inhibitors: CHIR99021, IWP2. Cardio Culture Medium: RPMI1640, B27. (Lower panel) Shown are morphological changes of hiPSCs during cardiac differentiation to cardiac organoids. (A) HiPSCs had a confluence of approximately 80–90% before the differentiation was initiated. (B) Preliminary contractions can be observed starting at day 8. (C) Stable contractions can be observed at day 10–14. (D) Stable contracting culture suitable for cardiac organoid generation. A-D show selected images of Supplementary Fige 1. (E–H) Compact cardiac organoids with smooth borders and contracting areas after 24 h of cell aggregation (E) and a whole cardiac organoid at day 16 (F), 17 (G) and 18 (H) of differentiation (2, 3 and 4 days into organoid culture). (I) Representative measurements of 7-AAD-negative cells in control (2D culture) and organoid-derived cardiomyocytes (after organoid dissociation). (J) Cardiac organoids were digested and isolated cardiomyocytes were stained for cell viability. (K) Beating analysis of completely contracting cardiac organoids during maturation. Scale bars: 100 μm (J) and 300 μm (A–H).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.heliyon.2022.e10365
The following are the supplementary data related to this article:
1
2
Note: For cardiac organoid differentiation and generation, hiPSCs should be in culture for at least one week. Suitable hiPSCs should be between passages 15 and 45. The proof of pluripotency at the gene and protein expression level has to be provided before use (Streckfuss-Bömeke et al., 2013).
-
30.Day-2: hiPSC passaging for cardiac organoid differentiation (=d-2 (day minus two: two days prior to induction of differentiation))
-
a.HiPSCs are passaged according to steps 12–21 (see ‘hiPSC Passaging’).
-
a.
Note: Split hiPSCs onto Geltrex-coated 12-well plates at a ratio of 1:8–1:10 (see ‘hiPSC Passaging’, step 19). The ratio may be different for each cell line. Adjust individually to reach 80–90% of optical confluence after 48 h.
-
31.Day-1: hiPSC culture for cardiac organoid differentiation
-
a.Change the medium according to steps 9–11 (see ‘hiPSC Culture’).
-
a.
Note: A medium change has to be performed to ensure 24 h of TZV-free culture before starting the cardiac organoid differentiation.
-
32.Day 0: Start of cardiac organoid differentiation (=d0)
-
a.Start cardiac organoid differentiation at an optical confluence of 80–90% (see Figure 1A and Supplementary Figure 1A).
-
a.
Note: Check cell quality (e.g. morphology and cell division) before starting the cardiac organoid differentiation (see Troubleshooting 1).
CRITICAL: Cell density at day 0 is essential for successful cardiac differentiation. Too low or too high cell densities might lead to insufficient cardiac differentiation (see Troubleshooting 2).
-
b.
Warm Cardio Differentiation Medium to room temperature (19–23 °C) and add 0.33 μL of the 12 mM stock solution of CHIR99021 per mL medium for a final CHIR99021 concentration of 4 μM.Gently remove medium by aspirating and add 2 mL of Cardio Differentiation Medium containing 4 μM CHIR99021 per well and incubate at 37 °C and 5% CO2.
Note: Add CHIR99021 freshly and avoid direct UV-exposure.
-
33.Day 2: Cardiac organoid differentiation (=d2)
-
a.Check cell viability and density.
-
a.
Note: The addition of CHIR99021 can induce cell death (up to 20%). In case of massive cell death of more than 20–40%, the differentiation should be stopped (see Troubleshooting 2).
-
b.
Warm Cardio Differentiation Medium to room temperature (19–23 °C) and add 1 μL of the 5 mM IWP2 stock per mL medium for a final IWP2 concentration of 5 μM.
-
c.
Gently remove medium by aspirating and add 2 mL of Cardio Differentiation Medium + IWP2 per well and incubate at 37 °C and 5% CO2.
Note: While changing the medium, make sure to remove dead cells and debris as well as possible. Washing the cells is not recommended at this point as it may lead to cell detachment.
-
34.Day 4: Cardiac organoid differentiation (=d4)
-
a.Check cell viability and density.
-
a.
Note: In case of massive cell death of more than 20–40%, the differentiation should be stopped (see Troubleshooting 2).
-
b.
Warm Cardio Differentiation Medium to room temperature (19–23 °C).
-
c.
Gently remove medium by aspirating and add 2 mL of Cardio Differentiation Medium per well and incubate at 37 °C and 5% CO2.
Optional: An additional medium change is possible at day 5 and especially recommended in case of high cell density (90–95%).
-
35.Day 6: Cardiac organoid differentiation (=d6)
-
a.Warm Cardio Differentiation Medium to room temperature (19–23 °C).
-
b.Gently remove medium by aspirating and add 2 mL of Cardio Differentiation Medium per well and incubate at 37 °C and 5% CO2.
-
a.
Optional: An additional medium change is possible at day 7 and especially recommended in case of high cell density (90–95%).
-
36.Day 8: Cardiac organoid differentiation (=d8)
-
a.Warm Cardio Culture Medium to room temperature (19–23 °C).
-
b.Gently remove medium by aspirating and add 2 mL of Cardio Culture Medium per well and incubate at 37 °C and 5% CO2.
-
c.Change Cardio Culture Medium daily (2 mL per well).
-
a.
Note: Preliminary contractions can be observed starting at day 8. Stable contractions can be observed at day 12–14. If a high amount of cell death or a lower number of contracting cells is observed, the cell density for the initiation of differentiation and the concentration of CHIR99021 used should be adjusted (see Troubleshooting 2).
-
37.
Day 14: Cardiac organoid generation (=d14)
CRITICAL: For cardiac organoid generation, it is mandatory to use a qualitatively stable conxtracting culture.
Note: Video S1 shows a qualitatively stable contracting culture suitable for cardiac organoid generation. If there are no or only a few contracting areas at day 14, the cultures cannot be used for cardiac organoid generation (see Troubleshooting 3).
-
a.
Warm Cardio Digestion Medium to room temperature (19–23 °C).
Note: Add TZV freshly and avoid direct UV-exposure.
-
b.
Heat collagenase B 400 U/mL und 0.25% trypsin-EDTA for 10–15 min at 37 °C in a water bath.
-
c.
Gently remove medium by aspirating. Add 1 mL of collagenase B 400 U/mL per well and incubate at 37 °C and 5% CO2 for 30 min to 2 h (time until total detachment may vary).
Note: The cells detach from the cell culture plate as a cellular monolayer. If the cells adhere to the cell culture plate after incubation, they can be detached by very careful rinsing with the applied collagenase B 400 U/mL solution. The cells should detach in large aggregates. Resuspension or frequent pipetting should be avoided.
-
d.
Use a 1000 μL pipette each to transfer the monolayer of one well into a 15 mL conical centrifuge tube and centrifuge at 200 × g for 5 min.
Note: Cells still adhering to the cell culture plate do not need to be detached additionally.
-
e.
Remove the supernatant and dissociate the cell pellet each in 1 mL of 0.25% trypsin-EDTA solution by gentle shaking. Incubate for 5 min at 37 °C.
Note: The cell pellet must be dissociated completely at the bottom of the tube. In this step, resuspension should only be performed once or twice with a 1000 μL pipette if the cell pellet is not dissociated, yet.
-
f.
Stop the reaction by adding 2 mL of Cardio Digestion Medium and centrifuge at 200 × g for 5 min.
-
g.
Remove the supernatant and resuspend the cell pellet with 2 mL of Cardio Digestion Medium. Count the cells using a cell counting chamber.
CRITICAL: The cell pellet is usually very loose at the bottom of the tube. Accidental aspiration using the vacuum suction can be prevented by using a 1000 μL pipette to remove the supernatant. Ideally, no residual supernatant should remain in the tube.
-
h.
Seed 25,000–35,000 cells per well of a 96-U-shape-well plate (maximum volume 200 μL per well).
Note: 96-U-shape-well cell culture plates do not require coating!
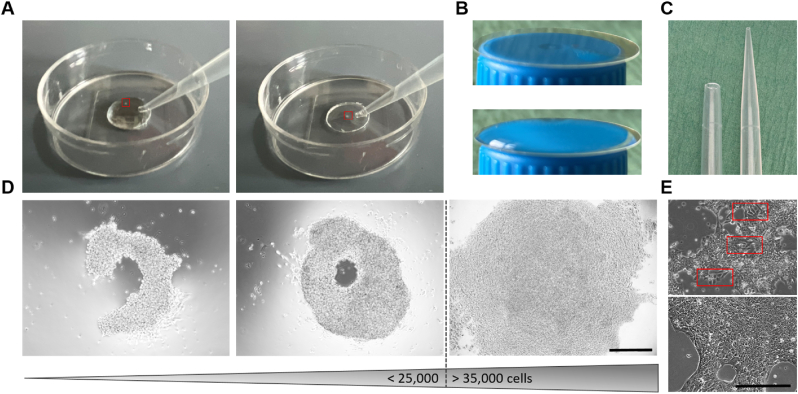
CRITICAL: Cell density is crucial for successful cardiac organoid generation. Too low or too high cell densities might lead to insufficient cardiac organoid generation (see Troubleshooting 4).
-
i.
After seeding, adjust the volume of Cardio Digestion Medium to exactly 200 μL per well and centrifuge at 200 × g for 5 min.
CRITICAL: Precise balancing of the centrifuge is crucial to ensure sufficient formation of cell aggregates.
-
j.
Incubate at 37 °C and 5% CO2.
-
38.Day 15: Cardiac organoid generation (=d15)
-
a.After 24 h, compact cardiac organoids with smooth borders and contracting areas should be present (see Troubleshooting 4). Change Cardio Culture Medium daily (200 μL per well). For further cultivation continue with ‘Cardiac Organoid Culture’, step 39.
-
a.
Note: The cardiac organoids are cultured in suspension. Do not use the vacuum suction to remove the medium.
3.6. Cardiac Organoid Culture
Timing: 10–20 min
Cardiac organoids are cultured under serum-free conditions in 96-U-shape-well cell culture plates. Change the medium every 2–3 days. Cardiac organoids can be kept in culture at least until d50.
-
39.
Warm Cardio Culture Medium to room temperature (19–23 °C).
-
40.
Gently remove the medium (ideally with a 1000 μL pipette). Pipette in a 45-degree angle near the edge of the well. Avoid direct contact of the pipette tip with the organoid. Remove medium slowly. Add 200 μL of fresh Cardio Culture Medium per well. Incubate at 37 °C and 5% CO2.
-
41.
Change Cardio Culture Medium every 2–3 days.
Note: The quality of cardiac organoids in long-term culture should be assessed on a regular basis (morphology and contraction; see Troubleshooting 4). An optimally contracting cardiac organoid at d20 is displayed in Video S2.
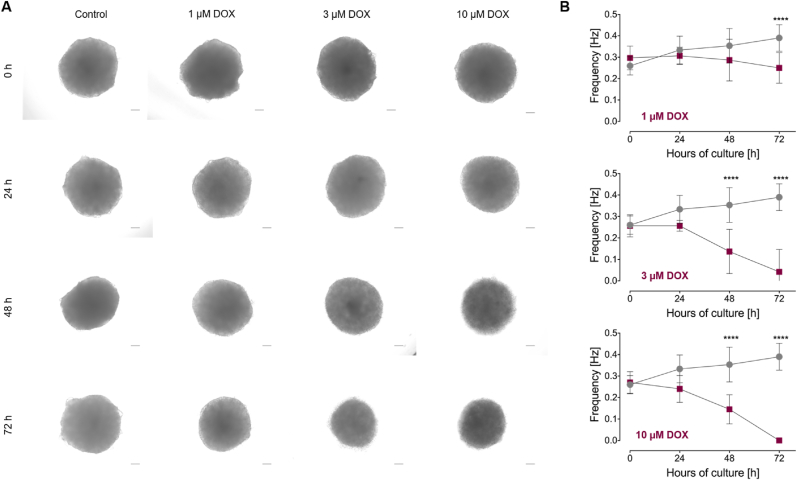
Note: For toxicity tests and drug screening, a medium change with an addition of e.g. doxorubicin (see Figure 2) can be performed here. Therefore, the medium is mixed with the appropriate amount of test substance (e.g. for a final concentration of 1 μM, 3 μM, 10 μM doxorubicin) followed by incubation for the desired time period until analysis (e.g. morphological examination, frequency analysis, staining, etc.).
Figure 2.
Morphological representation and beating frequency variation of cardiac organoids during doxorubicin exposure. Human iPSCs were differentiated into cardiac organoids according to the established protocol and incubated on day 24 of cultivation for 24 h, 48 h and 72 h with 1 μM, 3 μM or 10 μM doxorubicin (DOX). (A) Representative Phase contrast images during DOX treatment. Scale bar: 100 μm (B) Beating frequency of completely contracting cardiac organoids during DOX treatment. Results were replicated in 3 independent differentiation experiments. Mean ± SD. One Way ANOVA with Tukey's post test with ∗∗∗∗p < 0.0001. DOX: Doxorubicin.
3.7. Cardiac Organoid Digestion
Timing: 30–45 min
To determine cell viability and to perform immunofluorescence staining, cardiac organoids can be digested into single cells starting from d20.
-
42.
Warm Digestion Medium to room temperature (19–23 °C).
Note: Add TZV freshly and avoid direct UV-exposure.
Optional: If the isolated cells are going to be used for cell viability experiments, Flow Cytometry Medium must be prepared.
-
43.
Heat 0.25% trypsin-EDTA at 37 °C in a water bath for 10–15 min.
-
44.
Cardiac organoids are transferred to a 15 mL conical centrifuge tube using a cut 1000 μL pipette tip (cut off approximately ¼ of the pipette tip, see Figure 4C) and centrifuged at 200 × g for 5 min.
Figure 4.
Troubleshooting and cardiac organoid handling. (A) How to wash cardiac organoids in the 35-mm glass bottom dishes. See video S3. (B) To avoid solution running off the coverslip, the solution has to be pipetted carefully in the center of the coverslip. Adhesive forces will prevent the droplet from running off (see lower image). (C) A cut 1000 μL pipette tip (cut off approximately ¼ of the pipette tip) to transfer COs. (D) Influence of cell number on organoid generation/cell aggregation. From left to right increasing cell number was used for organoid generation. Too few cells lead to insufficient aggregation (first and second image). Too many cells lead to disaggregation and a flat organoid morphology. Cardiac organoids do not form a round, self-compacted structure after cell aggregation (third image). (E) Upper image: spontaneous hiPSC differentiation during culture. Lower image: Representative image of a good hiPSC quality. Scale bars: 300 μm.
Note: 3–6 cardiac organoids can be pooled in one 15 mL conical centrifuge tube.
CRITICAL: The cardiac organoids must be transferred with special care using cut pipette tips. For this purpose, the cardiac organoids are transferred to the tube very slowly with some medium and released very slowly from the pipette tip. Strong pipetting should be avoided.
-
45.
Remove the supernatant and resuspend the cell pellet with 1 mL 0.25% trypsin-EDTA by gentle shaking. Incubate at 37 °C for 5 min.
-
46.
Stop the reaction by adding 2 mL of Cardio Digestion Medium per tube and centrifuge at 200 × g for 5 min.
-
47.
Remove the supernatant. For immunofluorescence staining (replating and fixation of cells are mandatory) add 2 mL Cardio Digestion Medium. For cell viability experiments add 1 mL Flow Cytometry Medium.
Optional: Cell count can be determined using a counting chamber.
Pause Point: If the isolated cells are going to be used for cell viability analysis, continue with step 103 ‘Cell viability analysis of isolated cardiomyocytes from cardiac organoids’. If the cells are going to be used in immunofluorescence staining, continue with step 48 ‘Replating of isolated cardiomyocytes from cardiac organoids’.
3.8. Replating of isolated cardiomyocytes from cardiac organoids
Timing: 7–10 days
Cardiac organoids need to be dissociated and then replated onto Geltrex-coated coverslips for characterization with immunofluorescence stainings. Isolated cardiomyocytes should settle onto the Geltrex-coated coverslips after approximately 24 h, but should be kept in culture for 7–10 d before fixation.
-
48.
As described in ‘Before you begin’, coat 22-mm coverslips with Geltrex in 12-well plates.
-
49.
Warm Cardio Digestion Medium to room temperature (19–23 °C).
Note: Add TZV freshly and avoid direct UV-exposure.
-
50.
Incubate the plates containing the Geltrex-coated coverslips at 37 °C for 30 min.
-
51.
Gently aspirate the remaining Geltrex, wash with DPBS once and add 0.5 mL Cardio Digestion Medium per well.
-
52.
Digest the cardiac organoids as described in ‘Cardiac Organoid Digestion’ (steps 42–47).
-
53.
Count the cells using a counting chamber. Seed the isolated cells at desired densities in a volume of 0.5 mL Cardiac Digestion Medium into the wells.
Note: Seeding of 100,000–150,000 cells per well is optimal for a 22-mm coverslip.
-
54.
Gently shake the plates in an 8-shaped movement pattern to distribute the cells evenly. Incubate at 37 °C and 5% CO2 for further culture.
-
55.
A medium change with 2 mL of Cardio Culture Medium (without TZV) per well can be performed after 24 h.
-
56.
Culture the cardiomyocytes for 7–10 days at 37 °C and 5% CO2. Change the medium every 2–3 days. For fixation of the cells, continue with step 57 ‘Fixation of Isolated Cardiomyocytes from Cardiac Organoids’.
Note: Cardiomyocytes should display clear contractions after 7–10 d.
3.9. Fixation of Isolated Cardiomyocytes from cardiac organoids
Timing: 12–16 h
Isolated and replated cardiomyocytes need to be fixed for characterization with immunofluorescence stainings. Replated cardiomyocytes should be kept in culture on coverslips for 7–10 d before fixation. Fixed cardiomyocytes can be used directly for immunofluorescence staining or stored at 4 °C for up to one month.
Note: For fixation, seed isolated cardiomyocytes onto the Geltrex-coated coverslips. Culture for additional 7–10 d. Cardiomyocytes should display clearly visible contractions.
-
57.
Warm 4% PFA, 1% BSA and DPBS to room temperature (19–23 °C).
-
58.
Remove the medium of cardiomyocyte cultures by aspiration and wash three times with DPBS.
-
59.
Incubate cardiomyocytes with 1 mL 4% PFA per well for 20 min at room temperature (19–23 °C) and wash three times with DPBS.
-
60.
Incubate fixed cardiomyocytes in 2 mL 1% BSA per well at 4 °C overnight.
-
61.
After incubation, the fixed cardiomyocytes can be used for immunofluorescence staining. Continue with step 68 ‘Immunofluorescence of Cardiomyocytes Isolated from Cardiac Organoids’.
Note: Fixed cardiomyocytes can be stored in 1% BSA at 4 °C for up to one month.
3.10. Fixation of whole cardiac organoids
Timing: 12–16 h
To characterize and evaluate structures in cardiac organoids, whole-mount immunofluorescence can be performed starting at d20. Fixed cardiac organoids can be stored at 4 °C for up to one month.
Note: For fixation, only use cardiac organoids at d20 displaying visible contractions.
-
62.
Warm 4% PFA, 1% BSA and DPBS to room temperature (19–23 °C).
-
63.
Put 1 mL DPBS into a 35 mm petri dish.
-
64.
Transfer all cardiac organoids into the petri dish using a cut 1000 μL pipette tip (cut off approximately ¼ of the pipette tip) and wash three times with DPBS.
CRITICAL: The cardiac organoids must be transferred with special care using cut pipette tips. For this purpose, the cardiac organoids are transferred to a 35-mm Petri dish very slowly. Strong pipetting should be avoided.
-
65.
Incubate cardiac organoids in 2 mL of 4% PFA for 20 min at room temperature (19–23 °C) and wash three times with DPBS.
-
66.
Incubate fixed cardiac organoids in a 35-mm petri dish with 2 mL of 1% BSA overnight at 4 °C.
-
67.
After incubation, the fixed cardiac organoids can be used for the whole-mount immunofluorescence staining. Continue with step 81 ‘Whole-Mount Immunofluorescence of Cardiac Organoids’.
Note: Fixed cardiac organoids can be stored in 1% BSA at 4 °C for up to one month.
3.11. Immunofluorescence of Cardiomyocytes Isolated from cardiac organoids
Timing: 1–2 days
To validate the cardiac phenotype of the cardiac organoids, immunofluorescence staining can be performed. Digest cardiac organoids and replate isolated cardiomyocytes on coverslips. Stain the fixed cardiomyocytes immunocytochemically for cardiac marker proteins.
Note: Use replated and fixed cardiomyocytes on coverslips.
-
68.
Warm 1% BSA and 1% Triton X-100 to room temperature (19 °C–23 °C).
-
69.
Dilute primary antibody in 1% BSA (use 0.1% Triton X-100 for staining of nuclear antigens) (see Table 1).
Table 1.
Antibodies for immunocytochemical staining.
| Antibody | Dilution | Supplier |
|---|---|---|
| Primary antibody | ||
| ACTN2 monoclonal mouse IgG1 | 1:1,000 | Sigma-Aldrich |
| PE anti-human CD31 mouse IgG1 | 1:50 | BioLegend |
| TNNT2 monoclonal mouse IgG | 1:500 | ThermoFisher Scientific |
| VIMENTIN, VIM-13.2, monoclonal | 1:200 | Sigma-Aldrich |
| Secondary antibody | ||
| Alexa Fluor 488 goat anti-mouse | 1:400 | ThermoFisher Scientific |
Note: Per coverslip, use 200 μL of primary antibody solution.
-
70.
Wash fixed cardiomyocytes three times with DPBS.
-
71.
Incubate with 200 μL primary antibody solution overnight at 4 °C or at least for 1 h at 37 °C.
Note: The coverslip should be completely covered with the primary antibody solution (see Figure 4B). Dry spots as well as air bubbles should be avoided. Should problems with antibody solution running off the coverslip be noted, the use of a hydrophobic marker pen is encouraged.
-
72.
Dilute secondary antibody in 1% BSA (see Table 1).
Note: Avoid direct UV-exposure.
73. Wash the cells three times with DPBS
-
74.
Incubate with 200 μL of secondary antibody solution for 1–2 h at room temperature (19 °C–23 °C) in the dark.
Note: The coverslip should be completely covered with the secondary antibody solution. Dry spots as well as air bubbles should be avoided.
-
75.
Wash the cells three times with DPBS
-
76.
Incubate with mit 200 μL of DAPI working solution (1:5000) for 10 min at room temperature (19 °C–23 °C) in the dark.
-
77.
Wash the cells three times with DPBS
-
78.
Add one drop of Fluoromount-G to clean microscopic glass slide.
-
79.
Mount the coverslip gently on the microscopic glass slide (cardiomyocytes facing the mounting medium).
CRITICAL: Avoid air bubbles while mounting. Be careful not to move the coverslip on the microscopic glass slide after mounting to avoid cell destruction.
-
80.
Let the slides dry for at least 1–2 h in the dark at room temperature (19 °C–23 °C). Seal the edges with clear nail polish. The slides can be stored at 4 °C in the dark.
3.12. Whole-Mount Immunofluorescence of Cardiac Organoids
Timing: 1 day
To characterize cardiac organoids and evaluate cardiac structure at the protein level, whole-mount immunofluorescence stainings can be performed starting at d20. Cardiac organoids are fixed and stained immunocytochemically for cardiac marker expression.
Note: Use fixed cardiac organoids for whole-mount immunofluorescence stainings.
CRITICAL: Fixed cardiac organoids require particularly careful handling, as they are very sensitive to shear forces and can easily break apart.
CRITICAL: Cardiac organoids are very sensitive. Direct pipetting of solutions onto the cardiac organoid should be avoided.
-
81.
Warm 1% BSA, 1% Triton X-100 and DPBS to room temperature (19 °C–23 °C).
-
82.
Dilute primary antibody in 1% BSA and 0.1% Triton X-100 (see Table 1).
Note: Per cardiac organoid, use 200 μL of primary antibody solution.
-
83.
Transfer cardiac organoids into 35-mm glass bottom dishes using a cut 1000 μL pipette tip (cut off approximately ¼ of the pipette tip) and wash three times with DPBS.
Note: Use one cardiac organoid per 35 mm glass bottom dishes and wash carefully (see Figure 4A, Video S3).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.heliyon.2022.e10365
The following are the supplementary data related to this article:
3
CRITICAL: The cardiac organoids must be transferred with special care using cut pipette tips. Strong pipetting should be avoided.
-
84.
Wash cardiac organoids in the 35-mm glass bottom dishes three times with DPBS.
-
85.
Aspirate DPBS gently with a 200 μL pipette and incubate cardiac organoids with 200 μL of primary antibody solution for 6–8 h at room temperature (19 °C–23 °C). Make sure that the cardiac organoids remain placed in the center of the dish.
-
86.
Dilute secondary antibody in 1% BSA and 0.1% Triton X-100 (see Table 1).
Note: Avoid direct UV-exposure.
-
87.
Remove the primary antibody solution carefully with a 200 μL pipette.
-
88.
Wash the cardiac organoids three times with 200 μL DPBS using a 200 μL pipette. Make sure that the cardiac organoids remain placed in the center of the well.
Note: Cardiac organoids should not be swirled around during the washing step.
-
89.
Remove DPBS completely and incubate with 200 μL of secondary antibody solution for 6–8 h at room temperature (19 °C–23 °C) in the dark.
Note: Avoid direct UV-exposure.
-
90.
Remove the secondary antibody solution carefully.
-
91.
Wash three times with DPBS using a 200 μL pipette.
Note: Cardiac organoids should not be swirled around during the washing step.
-
92.
Remove DPBS and incubate with 200 μL DAPI working solution (1:5000) for 10 min at room temperature (19 °C–23 °C) in the dark.
Note: Avoid direct UV-exposure.
-
93.
Remove DAPI working solution using a 200 μL pipette.
-
94.
Wash the cardiac organoids three times with 200 μl DPBS using a 200 μL pipette.
Note: Cardiac organoids should not be swirled around during the washing step.
-
95.
Remove DPBS completely using a 200 μL pipette. Add a drop of Fluoromount-G and cover with a 22 mm coverslip. Make sure that the cardiac organoids remain placed in the center of the well.
CRITICAL: The coverslip should not be moved after it has been placed on the drop of Fluoromount-G. Otherwise, there is a risk of destroying the stained cardiac organoids.
Note: For representative stained organoid, refer to Figure 3A (left hand images).
-
96.
Dry for at least 1–2 h at room temperature (19 °C–23 °C) in the dark. Seal the edges with clear nail polish. The glass bottom dishes can be stored at 4 °C in the dark.
Figure 3.
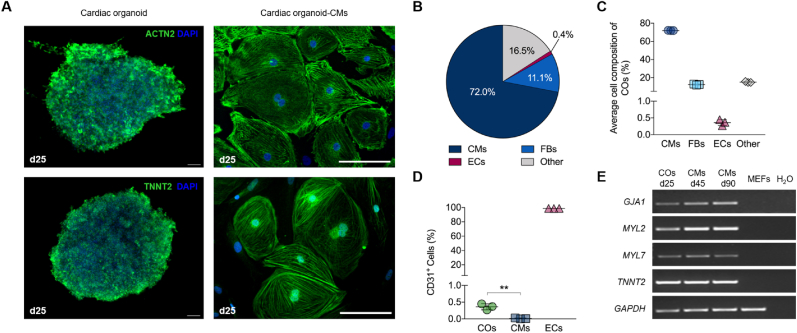
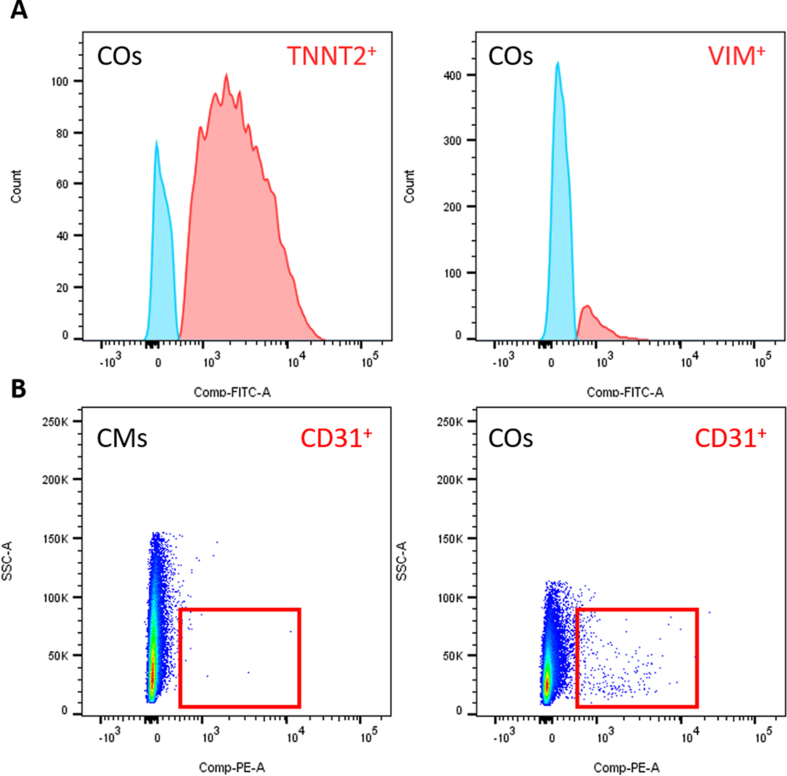
Characterization of cardiac organoids. (A) Representative images of structures within the cardiac organoids. Immunofluorescence staining in whole cardiac organoids and isolated organoid-derived cardiomyocytes (cardiac organoid-CMs after dissociation) of the following proteins was performed: Z-disc associated marker (ACTN2) and cardiac troponin T (TNNT2). Cell nuclei were stained blue with DAPI. (B) Pie chart of average cell composition in cardiac organoids (COs) using flow cytometry analyses – CMs (TNNT2), fibroblasts (FBs; Vimentin) and endothelial cells (ECs; CD31). (C) Quantification of flow cytometry data showing a robust ratio of CMs with 72%, FBs 11% and ECs <1% in COs. (D) Representative flow cytometry analysis for the endothelial cell marker CD31 in COs, 2D-cultured CMs and human umbilical vein ECs. Results were replicated in 3 experiments. Mean ± SD. One Way ANOVA with Tukey's post test with ∗∗p < 0.01. (E) RT-PCR products separated by gel electrophoresis. The following cardiac-specific marker genes were analyzed for their expression at mRNA level: GJA1 (gap junction protein, 248 bp), MYL2 (ventricular marker, 200 bp), MYL7 (atrial marker, 289 bp) and TNNT2 (cardiac troponin T). GAPDH (258 bp) was used as housekeeping gene and MEFs (mouse embryonic fibroblasts) as negative control. Scale bars: 100 μm.
3.13. Whole-mount cell viability staining of cardiac organoids
Timing: 45–60 min
To determine cell viability within the cardiac organoids, whole-mount cell viability staining can be performed starting at d20. Cell viability stainings of cardiac organoids can be performed directly without prior fixation.
CRITICAL: Cardiac organoids are very sensitive. Direct pipetting onto the cardiac organoid should be avoided.
-
97.
Warm DPBS to room temperature (19 °C–23 °C) and prepare LIVE/DEAD™ Viability/Cytotoxicity Kit components according to the manufacturer’s instructions.
-
98.
Transfer cardiac organoids into 35-mm glass bottom dishes using a cut 1000 μL pipette tip (cut off approximately ¼ of the pipette tip) and wash three times with DPBS.
Note: Use one cardiac organoid per 35 mm glass bottom dishes.
CRITICAL: The cardiac organoids must be transferred with special care using cut pipette tips. Strong pipetting should be avoided.
-
99.
Aspirate DPBS gently with a 200 μL pipette and incubate cardiac organoids with combined LIVE/DEAD® assay reagents for 30–45 min at room temperature (19 °C–23 °C) in the dark.
Note: Avoid direct UV-exposure.
-
100.
Remove LIVE/DEAD® assay reagents carefully using a 200 μL pipette. Wash cardiac organoids three times with 200 μL DPBS using 200 μL pipette.
-
101.
Remove DPBS completely using a 200 μL pipette and add 200 μL of fresh DPBS.
-
102.
Analyze labeled cardiac organoids with a fluorescence microscope.
3.14. Cell viability analysis of isolated cardiomyocytes from cardiac organoids
Timing: 45–60 min
To determine cell viability in cardiac organoids, isolated cardiomyocytes from digested cardiac organoids can be stained with 7-AAD and analyzed qualitatively by flow cytometry. Viability analysis can be performed directly starting at d20 without prior fixation.
-
103.
Digest cardiac organoids as described in steps 42–47 ‘Cardiac Organoid Digestion’.
-
104.
Warm Flow Cytometry Medium to room temperature (19 °C–23 °C).
-
105.
Resuspend the cell pellet in 1 mL Flow Cytometry Medium and determine cell count using a cell counting chamber.
-
106.
Prepare 1 mL Flow Cytometry Medium with 0.2–0.5 × 106 cells/mL and centrifuge at 200 × g for 5 min.
-
107.
Remove the supernatant and resuspend the cell pellet gently in 100 μL Flow Cytometry Medium.
-
108.
Add 1–2.5 μL 7-AAD staining solution to the cell suspension and incubate for 5 min at room temperature (19 °C–23 °C).
Note: Use 5 μL of 7-AAD staining solution for 106 cells.
-
109.
Add 400 μL of Flow Cytometry Medium and analyze by flow cytometry.
3.15. Harvesting cardiac organoids for biochemical analysis
Timing: 20–30 min
For molecular characterization by RT-PCR, pool cardiac organoids to pellets.
Note: Harvest clearly contracting cardiac organoids starting at d20.
-
110.
Prepare cold DPBS (4 °C) and liquid nitrogen and cool a table centrifuge to 4 °C.
-
111.
Label the amount of required 0.5 mL safe-lock tubes and place them on wet ice.
Note: 3–6 cardiac organoids can be pooled in one 0.5 mL safe-lock tube.
-
112.
Add 1 mL DPBS to a 35-mm petri dish and place the dish on wet ice subsequently.
-
113.
Transfer the cardiac organoids to the 35 mm petri dish using a cut 1000 μL pipette tip (cut off approximately ¼ of the pipette tip) and wash three times with DPBS.
CRITICAL: The cardiac organoids must be transferred with special care using cut pipette tips. Strong pipetting should be avoided.
-
114.
Transfer the cardiac organoids to 0.5 mL safe-lock tubes using a cut 1000 μL pipette tip (cut off approximately ¼ of the pipette tip).
-
115.
Centrifuge at 13,000 rpm (4 °C) for 2 min.
-
116.
Remove the supernatant completely using a 200 μL pipette and snap freeze the organoid pellet in liquid nitrogen. The pellet can be stored at −80 °C for at least one year.
Pause Point: To perform RNA isolation, continue with step 117 ‘RT-PCR Analysis’.
3.16. RT-PCR analysis
Timing: 1–2 days
Gene expression analysis can be performed to characterize the cardiac organoids. First, isolate RNA from organoid pellets and then perform RT-PCR. The following cardiac-specific marker genes can be tested at the mRNA level, for example, gap junction protein, alpha 1 (GJA1), myosin light chain 2a, atrial isoform (MYL7), myosin light chain 2v, ventricular isoform (MYL2) and troponin T2, cardiac type (TNNT2).
-
117.RNA isolation.
-
a.Perform RNA-Isolation e.g. with the SV Total RNA Isolation System Kit according to the manufacturer’s instructions.
-
a.
Note: Use 300–800 μL of RNA lysis buffer depending on the number of pelleted cardiac organoids (3–6).
-
b.
Elute RNA in 50–100 μL of nuclease-free water and determine the RNA concentration by photometry.
Pause Point: RNA can be stored at -80 °C for later use.
-
118.
Perform RT-PCR.
Note: Following RNA isolation, the entire or part of the mRNA is transcribed into the cDNA (complementary DNA) using the reverse transcriptase (RT) reaction.
-
a.
Prepare all components (Table 2) for RT reaction according to the manufacturer’s instructions.
-
b.
All components for one reverse transcription reaction are listed in Table 2.
-
c.
Perform RT reaction in a thermocycler using the following program (Table 3).
Table 2.
Components for reverse transcription reaction.
| Components | 20 μL final volume |
|---|---|
| 10x PCR buffer II | 2 μL |
| MgCl2 (25 mM) | 4 μL |
| dNTP mix (100 mM) | 0.8 μL |
| RNAse Inhibitor (20 U/μl) | 1 μL |
| Oligo d(T)16 (50 μM) | 1 μL |
| MuLV Reverse Transcriptase | 1 μL |
| RNA (100 ng) | 10.2 μL |
Table 3.
Thermocycler program for RT reaction.
| Time (min) | Temperature (°C) |
|---|---|
| 10 | 22 |
| 50 | 42 |
| 10 | 95 |
| ∞ | 4 |
Pause Point: cDNA can be stored at −20 °C for later use.
-
d.
Amplify resulting cDNA using specific primers. The gene-specific primer sequences and cycler conditions can be found in Key Resources Table and Table 4.
-
e.
All components for one PCR to amplify certain cDNA fragments are listed in Table 5.
-
f.
Perform reaction in a thermocycler using the following standardized program (Table 6).
-
g.
The amplified PCR products are separated by agarose gel electrophoresis and detected using UV light. Therefore, prepare 1.5–2% agarose gels and 1x TBE buffer.
Table 4.
Primer-specific properties and cycler conditions.
| Genes analyzed | Product (bp) | TA (°C) | Cycles |
|---|---|---|---|
| Troponin T2, cardiac type (TNNT2) | 305 | 56 | 35 |
| Gap junction protein, alpha 1 (GJA1) | 248 | 60 | 30 |
| Myosin light chain 2a, atrial isoform (MYL7) | 289 | 58 | 30 |
| Myosin light chain 2v, ventricular isoform (MYL2) | 200 | 56 | 30 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 258 | 60 | 30 |
Table 5.
Components for PCR reaction.
| Components | 25 μL final volume |
|---|---|
| Green GoTaq reaction buffer (5x) | 5 μL |
| dNTP mix (10 mM) | 1 μL |
| Primer forward (10 μM) | 1 μL |
| Primer reverse (10 μM) | 1 μL |
| Go Taq Polymerase (5 U/μl) | 0.125 μL |
| Nuclease-free water | 15.875 μL |
| cDNA | 1 μL |
Table 6.
Thermocycler PCR program.
| PCR Cycling Conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial denaturation | 95 °C | 3 min | 1 |
| Denaturation | 94 °C | 15 s | 30–35 cycles |
| Annealing | 56–60 °C | 30 s | |
| Extension | 72 °C | 30 s | |
| Final extension | 72 °C | 10 min | 1 |
| Hold | 4 °C | ∞ | |
Pause Point: PCR products can be stored at 4 °C.
-
h.
Use HDGreen (0.06 μL/mL) to visualize the DNA fragments. Perform electrophoresis at 120 V for 30–45 min and determine the size of the DNA fragments using the GeneRuler 100 bp Plus DNA Ladder. Detect and document DNA fragments using UV light.
3.17. Fixation of Isolated Cell from cardiac organoids
Timing: 1 h
Isolated cells need to be fixed for characterization with immunofluorescence stainings. Fixed cells can be used directly for immunofluorescence staining or stored at 4 °C for up to one month.
Pause Point: RNA can be stored at −80 °C for later use.
-
119.
Digest cardiac organoids as describe in steps 42–47 ‘Cardiac Organoid Digestion’
-
120.
Warm 4% PFA, Cell Staining buffer and DPBS to room temperature (19–23 °C).
-
121.
Resuspend the cell pellet in 1 mL DPBS and determine cell number using a cell counting chamber.
-
122.
Prepare 500 μL DPBS with 0.5–1 × 106 cells/mL and centrifuge at 200 × g for 5 min
-
123.
Wash step: Remove the supernatant, resuspend the cell pellet gently in 500 μL DPBS and centrifuge at 200 × g for 5 min. Thereafter, repeat this step twice (3x washing in total).
-
124.
Dilute primary antibody in 1% BSA (see Table 1).
-
125.
Resuspend the cell pellet in 500 μL 4% PFA and incubate for 20 min at room temperature (19 °C–23 °C) and wash three times with DPBS (see wash step)
-
126.
Resuspend the cell pellet in 500 μL Cell Staining buffer.
-
127.
Fixed cells can be used for flow cytometry analysis. Continue with step 128 ‘Flow Cytometry Analysis of Isolated Cells from Cardiac Organoids’
Note: Fixed cells can be stored in Cell Staining buffer at 4 °C for up to one month.
3.18. Flow Cytometry Analysis of Isolated Cells from cardiac organoids
Timing: 1–2 days
To determine the average cell composition of cardiac organoids, isolated and fixed cells from digested cardiac organoids can be stained immunocytochemically for cell type-specific marker proteins.
Note: Use digested cardiac organoids and fixed isolated cells.
-
128.
Use fixed isolated cells described in steps 119–127 ‘Fixation of Isolated Cell from Cardiac Organoids’.
-
129.
Warm Flow Cytometry Medium, Cell Staining buffer and DPBS to room temperature (19–23 °C).
-
130.
Resuspend the cell pellet in 1 mL DPBS and determine cell number using a cell counting chamber.
-
131.
Prepare 500 μL Cell Staining buffer with 0.5–1 × 106 cells/mL into 1.5 mL Safe-Lock tube and centrifuge at 200 × g for 5 min
Note: Prepare an additional tube as negative control.
-
132.
Wash step: Remove the supernatant, resuspend the cell pellet gently in 500 μL DPBS and centrifuge at 200 × g for 5 min. Thereafter, repeat this step twice (3x washing in total).
-
133.
Dilute primary antibody in Cell Staining buffer (see Table 1).
Note: Avoid direct UV-exposure.
-
134.
Resuspend the cell pellet in 200 μL primary antibody solution overnight at 4 °C or at least for 1 h at 37 °C.
Note: For directly labled antibodies continue with step 137.
-
135.
Dilute secondary antibody in Cell Staining buffer (see Table 1).
Note: Avoid direct UV-exposure.
-
136.
Wash three times with DPBS (see wash step)
-
137.
Resuspend the cell pellet in 100 μL secondary antibody solution or in 100 μL directly labled antibody for 1–2 h at room temperature (19 °C–23 °C) in the dark.
-
138.
Wash three times with DPBS (see wash step)
-
139.
Resuspend the cell pellet in 100 μL Flow Cytometry Medium, transfer into 5 mL Polystyrene round-bottom tubes and analyze by flow cytometry.
4. Expected outcomes
The cardiac organoid generation protocol is based on hiPSC-derived cardiomyocytes which were generated according to published cardiac differentiation protocols under defined culture conditions (Lian et al., 2012; Burridge et al., 2014; Cyganek et al., 2018). The organoids generated in this study exhibit a distinct cardiac phenotype and most importantly retain patient-specific properties. The present protocol integrates a robust 2D differentiation and ensures cardiac organoid generation without a supporting scaffold allowing high-throughput experiments. This approach involves overall less protocol steps than recent protocols at the expense of less sophisticated cardiac architecture, chamber-like structure development and germ layer interaction (Drakhlis et al., 2021; Hofbauer et al., 2021; Rossi et al., 2021). After 14 d of cardiac 2D differentiation, cardiac organoids can be generated by self-assembly in U-shape 96-well cell culture plates. In contrast to a recently published protocol by Lewis-Israeli et al. (2021), cardiac organoid generation described here does not require permanent 3D culture starting from embryonic bodies, but can be readily integrated in existing 2D culture techniques. A differentiated culture in one full well of a 12-well plate can yield approximately 0.5–1 × 106 cardiomyocytes and 16–33 corresponding cardiac organoids (approximately 30,000 cells per organoid). The generation of 96 cardiac organoids (one full 96-well U-shape cell culture plate) requires an average of differentiated cultures from three wells of a 12-well plate.
Characterization at the gene and protein level of visibly contracting cardiac organoids can be performed after 6–10 days of culture. Flow cytometry analysis of dissociated cardiac organoids demonstrated excellent cell viability (see Figure 1I) and the presence of different cardiac cell populations, with a composition of 72.0 % cardiomyocytes, 11.1 % fibroblasts and <1% endothelial cells (see Figure 3B-D; see Supplementary Figure 2), which is comparable with the data demonstrated recently (Lewis-Israeli et al., 2021). Expression analysis of cardiac markers like GJA1, MYL7, MYL2, and TNNT2 demonstrated comparable gene expression between 2D cardiomyocyte and 3D organoid cultures (Figure 3E). Immunofluorescence staining of cardiac organoids and organoid-derived cardiomyocytes revealed comparable cardiac marker protein expression. In cardiac organoids and replated cardiomyocytes isolated from the cardiac organoids, ACTN2 and TNNT2 were shown to be Z-disc-associated markers (Figure 3E). The 3D cell culture technology presented in this protocol can be employed to study cell-cell interactions, to perform drug and toxicity screenings and for disease modeling. Currently a patient individual cardiotoxicity of certain drugs is not predictable. Toxicity screenings were performed in patient-specific 2D hiPSC-CMs with promising results but with the limitation of lacking the co-localization with other cardiac cell types (Burridge et al., 2016). Investigation of doxorubicin in cardiac organoids demonstrated that our system is capable of measuring acute toxic effects as demonstrated for organoid morphology and decreased beating frequency (see Figure 2A and B).
Currently, the greatest challenge is the replication of cardiac architecture, especially the formation of ventricles, septa and heart valves. Significant progress in in vitro modeling of self-assembly of chamber-like structures has been achieved (Drakhlis et al., 2021; Hofbauer et al., 2021; Lewis-Israeli et al., 2021). However, these advances have been accompanied by a significant departure from established 2D culture protocols. Therefore, careful consideration should be given in advance to which research questions should be answered. If the experimental focus is on the generation of atrial and ventricular cardiac organoids with a focus on studying cellular composition and cell-cell interactions in the context of developmental biological processes of early cardiac embryogenesis and disease development, the protocol established here represents an efficient approach that can be easily integrated into existing protocols.
The maturity of the cardiac organoids improves with increasing culture time. The optimal culture duration needs to be determined individually based on the needs of the experimenter. In parallel to the three-dimensional organoid culture, functional studies can be performed in isolated organoid-derived replated cardiomyocytes. On a single-cell basis, various methods and protocols exist to study action potentials via patch-clamp technique or calcium dynamics via optical imaging (Li et al., 2020; Luo et al., 2020). The hiPSC-based 2D models do not form complex, organ-like structures and thus reflect human physiology only to a limited extent. The combination of hiPSC and organoid technology allows the generation of human 3D cell cultures that are closer to physiology and offer a wide range of applications in pharmacological and toxicological research as well as a viable alternative to animal experiments.
5. Limitations
The present cardiac organoid differentiation and culture protocol was established and tested with different hiPSC lines. A limiting factor for organoid generation was the varying differentiation efficiency during the first 14 days. From day 14 onward, no significant differences were observed in 3D culture. Although cardiac organoids present a model closer to human physiology than 2D cultures, there are obvious differences to the human heart, which include the time course of development, chamber-specific differentiation and the degree of maturity. In contrast to organoids, the adult human heart consists of various cell types like cardiomyocytes, fibroblasts, endothelial and smooth muscle cells as well as cells of the immune system (Segers and Lee, 2008; Kamo et al., 2015; Segers et al., 2018). Flow cytometry analysis of dissociated cardiac organoids demonstrated the presence of different cardiac cell types. However, in the future, there is a need for optimization for more efficient differentiation of endothelial cells and the introduction of other cardiac lineage specific cell types, such as endocardial and epicardial cells, smooth muscle cells and cells of the immune system. Furthermore, the size of an in vitro organoid model is limited, as exceeding the maximum possible size may lead to nutrient or oxygen starvation and subsequent cell death. There is evidence from studies on engineered human myocardial tissue that the addition of human foreskin fibroblasts induced further cardiomyocyte maturation on the protein level and that functional variability with respect to contractility decreased (Tiburcy et al., 2017). Thus the addition of chamber-specific human cardiac fibroblasts could be an even more promising approach towards an adult-like phenotype and in vitro models close to human physiology (Künzel et al., 2020). The organoids generated in this study display a distinct cardiac phenotype prompting their use to study early embryonic cardiac development, the mechanisms of monogenetic cardiac pathologies as well as drug effects in a functional setting.
6. Troubleshooting
6.1. Problem 1
Slowly proliferating hiPSCs and/or spontaneous hiPSC differentiation during culture.
6.2. Potential Solution 1
Freshly thawed hiPSCs require at least one week in culture before they can be differentiated. If the hiPSC grow very slowly during the first week, they should be kept in culture for another week. If one passage or one culture of a cell line is affected, it is recommended to thaw frozen hiPSC of these cell lines and to culture them again. If several passages and newly thawed hiPSCs of a cell line are also affected, it is recommended to check the cell line for strain cell specific characteristics (pluripotency on gene and protein expression level).
6.3. Problem 2
Insufficient cardiac differentiation reflected by partial contraction.
6.4. Potential Solution 2
The optimal cell density can vary depending on the cell line and should be determined individually for each cell line. For this purpose, set up a dilution series while passaging and start differentiation at the same time (d0).
A certain rate of cell death is to be expected at this point. However, in case of mayor cell death, try to optimize cell density at the start of the differentiation by trying cell densities between 1:12–1:3 or defined cell numbers between 40,000 and 100,000 cells/12-well. Additionally, optimal CHIR99021 concentrations can differ between individual cell lines. Optimal CHIR99021 concentrations at day 0 range between 1 and 4 μM CHIR99021 (Lewis-Israeli et al., 2021). However, we do not recommend varying IWP2 concentrations as an optimum of 5 μM has been determined previously (Lian et al., 2012).
6.5. Problem 3
No or insufficient contraction after 14 d.
6.6. Potential Solution 3
Contraction of the culture should be assessed 1–2 h after a medium change. Do not let the cells cool down under the light microscope. Ideally, contraction is assessed at 35–37 °C. If contractions fail to occur anyway and cell morphology is significantly different from the example shown in Video S1 the culture cannot be used for cardiac organoid generation. If the cells fail to contract but display a morphology similar to the example shown in Video S1, the culture should be continued for additional 2–4 d. If there is still no stable contraction, the culture cannot be used for the generation of cardiac organoids.
6.7. Problem 4
Cardiac organoids do not form a round, self-compacted structure after cell aggregation.
6.8. Potential Solution 4
To ensure an intact, round and closed structure, the cell density is crucial for cell aggregation. Too few cells will result in crescent or hoop-shaped structures (see Figure 4D left and middle image). Too many cells will hinder 3D development and lead to flat structures pressed against the bottom of the well (see Figure 4D right image). To determine the optimal cell density see “Potential Solution 2”.
6.9. Resource availability
6.9.1. Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kaomei Guan, kaomei.guan@tu-dresden.de.
6.9.2. Materials availability
This study did not generate new unique reagents.
6.9.3. Data and code availability
The published article includes all datasets generated or analyzed during this study.
Declarations
Author contribution statement
Karolina Künzel: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Stephan Künzel: Analyzed and interpreted the data; Wrote the paper.
Kaomei Guan: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
The work was supported by the Free State of Saxony and the European Union EFRE (SAB projects “PhenoCor” as well as “HERMES” to K. Guan), and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project Number 288034826 – IRTG 2251: “Immunological and Cellular Strategies in Metabolic Disease” to K. Guan.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Figure Supplement 1_HR1.
Figure Supplement 2_HR1.
Supplementary Table 1_V2.
References
- Burridge P.W., et al. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 2016;22(5):547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyganek L., et al. Deep phenotyping of human induced pluripotent stem cell–derived atrial and ventricular cardiomyocytes. JCI Insight. 2018;3(12) doi: 10.1172/jci.insight.99941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakhlis L., et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021;39(6):737–746. doi: 10.1038/s41587-021-00815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer P., et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184(12):3299–3317. doi: 10.1016/j.cell.2021.04.034. e22. [DOI] [PubMed] [Google Scholar]
- Kamo T., Akazawa H., Komuro I. Cardiac nonmyocytes in the hub of cardiac hypertrophy. Circ. Res. 2015;117(1):89–98. doi: 10.1161/CIRCRESAHA.117.305349. [DOI] [PubMed] [Google Scholar]
- Künzel S.R., et al. Modeling atrial fibrosis in vitro —generation and characterization of a novel human atrial fibroblast cell line. FEBS Open Bio. 2020:2211–5463. doi: 10.1002/2211-5463.12896. 12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Israeli Y.R., et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021;12(1):5142. doi: 10.1038/s41467-021-25329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., et al. Disease phenotypes and mechanisms of iPSC-derived cardiomyocytes from brugada syndrome patients with a loss-of-function SCN5A mutation. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.592893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., et al. Cozzarelli Prize Winner: robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 2012;109(27):E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., et al. IP3R-Mediated compensatory mechanism for calcium handling in human induced pluripotent stem cell-derived cardiomyocytes with cardiac ryanodine receptor deficiency. Front. Cell Dev. Biol. 2020;8:772. doi: 10.3389/fcell.2020.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G., et al. Capturing cardiogenesis in gastruloids. Cell Stem Cell. 2021;28(2):230–240. doi: 10.1016/j.stem.2020.10.013. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers V.F.M., Brutsaert D.L., De Keulenaer G.W. Cardiac remodeling: endothelial cells have more to say than just NO. Front. Physiol. 2018;9:382. doi: 10.3389/fphys.2018.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers V.F.M., Lee R.T. Stem-cell therapy for cardiac disease. Nature. 2008;451(7181):937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- Streckfuss-Bömeke K., et al. Comparative study of human-induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes, and skin fibroblasts. Eur. Heart J. 2013;34(33):2618–2629. doi: 10.1093/eurheartj/ehs203. [DOI] [PubMed] [Google Scholar]
- Tiburcy M., et al. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation. 2017;135(19):1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., et al. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. U. S. A. 2010;107(18):8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.
Data included in article/supp. material/referenced in article.