ABSTRACT
Mycoplasmas are minimal bacteria that infect humans, wildlife, and most economically relevant livestock species. Mycoplasma infections cause a large range of chronic inflammatory diseases, eventually leading to death in some animals. Due to the lack of efficient recombination and genome engineering tools for most species, the production of mutant strains for the identification of virulence factors and the development of improved vaccine strains is limited. Here, we demonstrate the adaptation of an efficient Cas9-Base Editor system to introduce targeted mutations into three major pathogenic species that span the phylogenetic diversity of these bacteria: the avian pathogen Mycoplasma gallisepticum and the two most important bovine mycoplasmas, Mycoplasma bovis and Mycoplasma mycoides subsp. mycoides. As a proof of concept, we successfully used an inducible SpdCas9-pmcDA1 cytosine deaminase system to disrupt several major virulence factors in these pathogens. Various induction times and inducer concentrations were evaluated to optimize editing efficiency. The optimized system was powerful enough to disrupt 54 of 55 insertion sequence transposases in a single experiment. Whole-genome sequencing of the edited strains showed that off-target mutations were limited, suggesting that most variations detected in the edited genomes are Cas9-independent. This effective, rapid, and easy-to-use genetic tool opens a new avenue for the study of these important animal pathogens and likely the entire class Mollicutes.
IMPORTANCE Mycoplasmas are minimal pathogenic bacteria that infect a wide range of hosts, including humans, livestock, and wild animals. Major pathogenic species cause acute to chronic infections involving still poorly characterized virulence factors. The lack of precise genome editing tools has hampered functional studies of many species, leaving multiple questions about the molecular basis of their pathogenicity unanswered. Here, we demonstrate the adaptation of a CRISPR-derived base editor for three major pathogenic species: Mycoplasma gallisepticum, Mycoplasma bovis, and Mycoplasma mycoides subsp. mycoides. Several virulence factors were successfully targeted, and we were able to edit up to 54 target sites in a single step. The availability of this efficient and easy-to-use genetic tool will greatly facilitate functional studies of these economically important bacteria.
KEYWORDS: CRISPR-Cas9, mycoplasma, animal pathogens, minimal cell, genome editing
INTRODUCTION
Mycoplasmas are minimal pathogens that belong to the class Mollicutes (1). They are characterized by a streamlining evolution from a Gram-positive ancestor, marked by drastic genome reduction (2–4). This evolution has led to the loss of diverse cellular functions, including cell wall production, various metabolic pathways, and efficient recombination machinery (5). These minimal bacteria are found in a wide range of host species, including humans and livestock. Many mycoplasmas are etiological agents of diseases that considerably reduce animal production yields and inflate veterinary health care costs (3, 6, 7). Currently, three of the most prevalent and economically relevant species worldwide are Mycoplasma gallisepticum, Mycoplasma bovis, and Mycoplasma mycoides subsp. mycoides (Mmm). M. gallisepticum is an animal pathogen that is listed by the World Organization for Animal Health (OIE) and is responsible for respiratory disease in poultry farms worldwide (8–10). M. bovis is responsible for bovine respiratory disease (BRD) as well as mastitis and reproductive disease (11, 12). Mmm is a member of the “M. mycoides cluster”, which is a group of five significant ruminant pathogens. It is the causative agent of contagious bovine pleuropneumonia (CBPP), an OIE-listed disease that can take chronic, acute, or hyperacute forms. In the hyperacute form, CBPP clinical signs include pericardial effusion, fever, and death (13). The control strategies for mycoplasmoses vary geographically and include animal movement regulation and the use of antibiotics, vaccines, and ultimately culling. In the context of increasing antibiotic resistance (14) and the relative lack of effective vaccines, better knowledge of the molecular basis of virulence and of host-pathogen interactions is required to improve curative protocols and design more efficient prevention methods (13, 15). However, the limited number of efficient genome engineering tools for many mycoplasmas, including the three species listed above, restricts functional genomics approaches and hinders efforts toward the production of rationally designed vaccines. During the past decade, cutting-edge genome engineering methods have been developed, and these methods rely on the cloning of the mycoplasma genome in yeast before editing with various genetic tools and back transplantation into a recipient cell (16, 17). Although offering unmatched possibilities to investigate and redesign complete genomes, these in-yeast approaches can be difficult to adapt to other species. They are still restricted to a small number of mycoplasmas and have not been adapted for any of the three pathogens considered here. Indeed, efforts to adapt these synthetic biology approaches to Mmm have been unsuccessful thus far, though they are now available for all other members of the M. mycoides cluster (18, 19). Only replicative (oriC) plasmids and transposon-based mutagenesis are available for M. bovis, and Mmm (20, 21). For M. gallisepticum, in addition to oriC plasmids (22) and transposon mutagenesis, a first tool for the targeted homologous recombination (HR) of short genomic regions has recently been reported (23).
Since 2012, CRISPR-based genetic tools have revolutionized the field of genome engineering in eukaryotes and prokaryotes. The typical CRISPR-Cas9 tool is based on the Cas9 nuclease and is guided to specific loci by single guide RNAs (sgRNAs) (24). Following DNA cleavage, repair occurs through distinct cellular mechanisms, including nonhomologous end joining (NHEJ) and homology-directed recombination (HDR). However, many bacteria, including mycoplasmas, lack efficient NHEJ and HDR repair machineries. Therefore, the CRISPR-Cas9 tools can only be used as a counterselection method (25). Base-editor systems (BEs) have offered a means by which to overcome this problem. Indeed, BEs combine a catalytically inactivated form of Cas9 (dCas9) fused with a cytosine deaminase (CBE) or an adenosine deaminase (ABE) and a uracil glycosylase inhibitor (UGI). At the molecular level, the mechanism of action of a CBE occurs as follows (Fig. S1): (i) an R-loop is opened in the target site by the dCas9-sgRNA complex; (ii) the single-strand DNA is accessible to the fused deaminase within a specific editing window; and (iii) the CBE catalyzes the deamination of cytosine into uracil, which is recognized as thymine after replication. Meanwhile, ABE converts adenine into inosil, which is recognized as guanine after replication (26–28). Base excision repair on edited nucleotides is prevented by UGI, and this improves the global editing efficiency (29). Therefore, CBE and ABE induce C:G to T:A and A:T to G:C transitions, respectively, and could allow for the insertion of a stop codon into genes of interest (26, 30).
Here, we first demonstrate the functionality of a base editor system in M. gallisepticum using a transposon to introduce the CBE and sgRNA into the bacterial cells. After optimization, three virulence genes located at various positions on the M. gallisepticum chromosome were successfully disrupted, and individual mutants were isolated. Next, a replicative plasmid and a transposon were used as vectors to evaluate this CBE in M. bovis and Mmm, and we demonstrate the high efficiency of this system for single-gene targeting in ruminant pathogens. Finally, multitarget mutagenesis using a single CRISPR guide was proven by the successful disruption of 54 insertion sequences in Mmm.
(A previous version of this manuscript was deposited on bioRxiv https://doi.org/10.1101/2022.03.09.483585. The manuscript is made available under a CC-BY-NC-ND 4.0 International license.)
RESULTS
Design and construction of a base-editor genetic tool for mycoplasma.
Two different cytosine deaminases commonly used for base editing, rAPOBEC1 from Rattus norvegicus (26) and pmcDA1 from Petromyzan marinus (26, 27) (Fig. 1A, SI-1, SI-2), were evaluated as editing tools in mycoplasmas. The coding sequences of these two proteins were codon optimized, chemically synthesized, and fused to an inactive version of Cas9 (dCas9) from Streptococcus pyogenes (SpdCas9). All genetic elements were assembled within a Tn4001-derived transposon (31), resulting in plasmids pTI4.0_SpdCas9_pmcDA1 (Fig. S2B) and pTI4.0_ rAPOBEC1_SpdCas9 (Fig. S3A).
FIG 1.
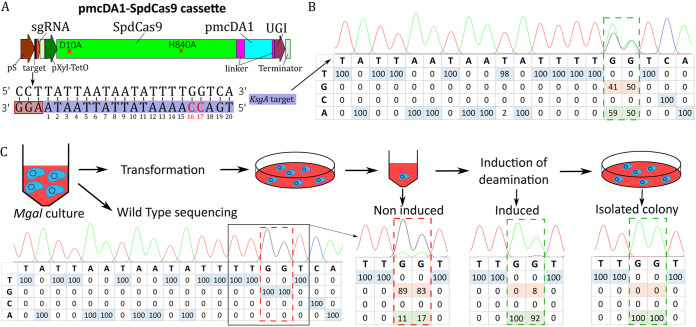
Targeting of the ksgA gene in M. gallisepticum using the pmcDA1 deaminase-encoding plasmid. (A) Scheme of the CBE expression cassette. The synthetic cassette is composed of (i) a sgRNA (red), including the 20-nucleotide target spacer (dark blue) under the control of the spiralin promoter (brown), and (ii) a codon-optimized hybrid protein composed of S. pyogenes inactivated Cas9 (SpdCas9, green), linkers (purple), the pmcDA1 deaminase protein (light blue), and uracil glycosylase inhibitor (UGI) (dark purple) under the control of the Pxyl/tetO2 inducible promoter (dark green). Fibril terminators from S. citri (light green) were added downstream of the sgRNA and hybrid protein expression cassettes. The sequence of the ksgA target is represented here. Cytosine residues that are susceptible to deamination are colored in red. The positions of bases in the target are indicated below each base, with numbering starting at the PAM sequence. (B). The percentage of bases found in the population of transformants was determined by Sanger sequencing of the complementary strand. The chromatograms were analyzed using EditR software and are represented in a table for each nucleotide position in the target sequence. Positions 16 and 17 are framed in a dotted rectangle. (C) Schematic diagram of the base-editing experiment in M. gallisepticum (Mgal) and an indication of various checkpoints (Sanger results and EditR analysis) for screening until isolated mutants are obtained.
We selected the ksgA gene of M. gallisepticum as the first target to evaluate the ability of these recombinant CBEs to produce mutations in mycoplasma. The ksgA gene is nonessential and encodes an RNA methyltransferase that renders the bacteria sensitive to kasugamycin (32). A 20-nucleotide sequence (5‘-TGAC17C16AAAATATTATTAATA/AGG-3′), upstream of the SpdCas9 compatible protospacer-adjacent motif (PAM) sequence AGG (Fig. 1A and B), was chosen as the target for ksgA inactivation. On the basis of previous reports, the two cytosines at positions 16 and 17 upstream of the PAM sequence should be in the theoretical editing window of the two CBE systems (33). Deamination of C17 is expected to change a glutamine CAA codon into a TAA stop codon. After the transformation of M. gallisepticum with either pTI4.0_SpdCas9_pmcDA1 or pTI4.0_rAPOBEC1_SpdCas9 and an overnight induction of the inducible system with anhydrotetracycline (aTC) at 5 μg·mL−1 in growth medium (25), we analyzed the impact of the CBEs on cytosine deamination within the target site by PCR and Sanger sequencing on the global population of transformants (Fig. 1A and B; Fig. S3A). We observed only limited base editing for the two cytosines at positions 16 and 17 (2%) for the pTI4.0_rAPOBEC1_SpdCas9 transformants (Fig. S3), whereas base editing was more prevalent (59% of C to T conversion at C16, 50% at C17) for the pTI4.0_SpdCas9_pmcDA1 transformants (Fig. 1). There was no conversion in the control without the sgRNA. Given these results, the deaminase pmcDA1 appears functional in M. gallisepticum.
Optimization of the CBE system and isolation of ksgA mutants in M. gallisepticum.
Given our initial results, the CBE system based on the pmcDA1 cytosine deaminase (pTI4.0_spdCas9_pmcDA1) was selected for further optimization. First, we determined the optimal inducer concentration by assessing the deamination level of targeted cytosines using the same experimental strategy and ksgA as the target after overnight induction with aTC at 0.1, 0.25, 0.5, 1, 2.5, or 5 μg·mL−1 (Fig. S4A). There was a notable leakage of expression in the absence of the inducer, with up to 25% conversion at position C16. In the presence of the inducer, the optimal concentration for base-editing was in the range of 0.1 to 0.5 μg·mL−1 aTC (>50% efficiency) (Fig. S4A). There was no detectable growth defect of M. gallisepticum with up to 1 μg·mL−1 aTC. However, the addition of aTC at 5 μg·mL−1 resulted in a marked reduction in growth, as shown by the almost complete absence of a pH shift in the broth medium after aTC induction (pH = 6.6 at induction versus 6.54, 5.41, and 5.3 after overnight induction with 5 μg.mL−1, 0.5 μg.mL−1, and no inducer, respectively). This result suggests that aTC is toxic for M. gallisepticum at high concentrations. The use of freshly prepared (<24 h) inducer was also necessary for maximally efficient base editing (data not shown).
Then, we studied the time of induction required for maximum efficiency in the CBE system. Inducer concentrations used for this experiment were 0.25 and 0.5 μg·mL−1, and base conversion was determined at two time points: after a 2 h induction or after an overnight induction (Fig. S4B). After 2 h, significant base conversion was already evident, relative to the noninduced condition, with up to 30% of base conversion at the two cytosine positions. However, overnight induction resulted in the highest efficiency, yielding a 2-fold higher conversion (approximately 60%) than that observed after 2 h of induction. Thus, the best induction conditions for the maximum efficiency of the mycoplasma CBE in M. gallisepticum were aTC at 0.1 to 0.5 μg·mL−1 and overnight incubation.
Finally, we performed a third experiment using the newly defined conditions to obtain isolated M. gallisepticum mutants using the pTI4.0_SpdCas9_pmcDA1 construct (Fig. 1C). Targeted ksgA sites were analyzed at four time points: before transformation (wild type), after transformation, after induction, and in isolated colonies selected on puromycin selective plates. In the noninduced condition, 11% of C16 and 17% of C17 were edited (Fig. 1C), indicating a leakage of the inducible promoter, as previously observed. After induction, deamination in the edited population was nearly complete, with 100% conversion of C16 and 92% conversion of C17 to thymine. Further screening of five isolated colonies confirmed that four carried mutations at the two targeted cytosines, whereas the last one showed an incomplete deamination profile, suggesting that deamination occurred during clonal expansion. Resistance to kasugamycin was confirmed by plating an isolated mutant on a Hayflick plate supplemented or not with the antibiotic (Fig. S5). Taken together, these results show that the pmcDA1-based CBE tool promotes base conversion and gene inactivation in M. gallisepticum with high efficiency.
Determination of the editing window for maximum CBE efficiency in M. gallisepticum.
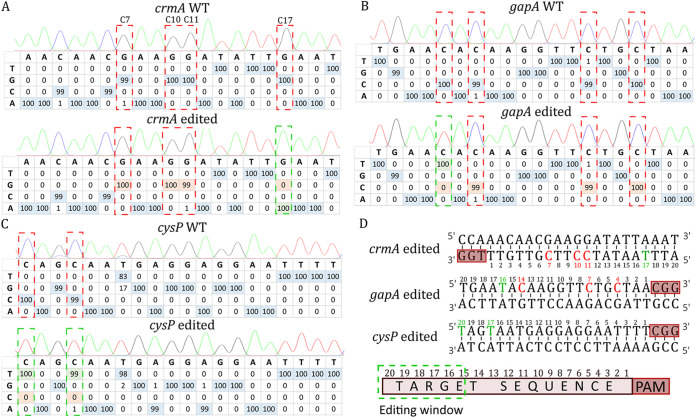
We investigated the base-editing window of our CBE system by targeting three other M. gallisepticum genes that encoded virulence factors: crmA (GCW_RS01080), gapA (GCW_RS01075), and cysP (GCW_RS01695) (Fig. 2). The crmA and gapA genes encode two primary cytoadherence proteins (34, 35). Meanwhile, the cysteine protease encoded by cysP has been shown to cleave chicken immunoglobulin G (36). In the crmA target (Fig. 2A) (5′-ATTC17AATATC11C10TTC7GTTGTT/TGG-3′), cytosine residues C7, C10, C11, and C17 were potential deamination sites. After induction, C17 was the only base to be deaminated, with 57% conversion at the population level before the second plating (Fig. 1C). The screening of isolated colonies resulted in 5 of 10 clones showing a C17 to T17 transition (Fig. 2A, crmA edited). In the gapA target (Fig. 2B) (5′-TGAAC16AC14AAGGTTC7TGC4TAA/CGG-3′), C4, C7, C14, and C16 were potential deamination sites. After induction, C16 was the sole deaminated position, with 54% conversion estimated in the transformant population. In this population, 4 of 10 isolated clones showed a C16 to T16 mutation (Fig. 2B, gapA edited). Finally, in the cysP target (Fig. 2C) (5′-C20AGC17AATGAGGAGGAATTTT/CGG-3′), C17 and C20 were potential deamination sites. After induction and population analysis, the conversion levels were 76% and 77% for these two cytosines, respectively. All 10 of the 10 isolated clones showed mutations at the two positions (Fig. 2C, cysP edited). Thus, the editing window of the CBE system in M. gallisepticum ranged from positions 16 to 20, upstream of the PAM sequence (Fig. 2D).
FIG 2.
Targeting of three virulence factors of M. gallisepticum to explore the editing window of the CBE system in M. gallisepticum. The 20-nucleotide target sites were sequenced and are represented here for the crmA (A), gapA (B), and cysP (C) genes before and after the base-editing experiments. Cytosines susceptible to deamination (or guanine in the reverse strand) are framed in red, and those that were deaminated are framed in green. The percentage of each base is shown in the tables, as determined using the Sanger sequencing.ab file and EditR software. Edited bases are highlighted in green. For the crmA target, the complementary strand was sequenced. (D) The 20-nucleotide target sites are shown for the three targets. The position of each base in the target is indicated below each nucleotide and corresponds to the nucleotide position in the target upstream of the PAM sequence. Red letters represent undeaminated cytosines, and green letters represent converted thymines. A scheme based on the three experiments and highlighting the editing window is shown at the bottom. As shown in the scheme, cytosines located at positions 16 to 20 can be CBE-targeted.
Application of the CBE system in M. bovis.
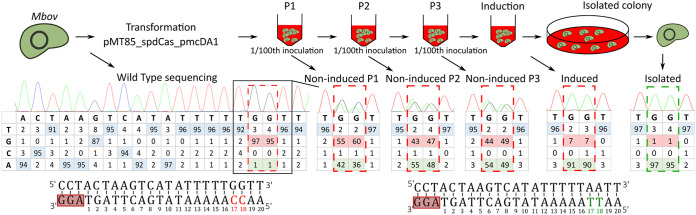
We initially evaluated the pTI4.0_spdCas9_pmcDA1, successfully used in M. gallisepticum, in M. bovis. However, we did not obtain M. bovis transformants using this CBE-carrying plasmid. This may be linked to the use of the pac resistance marker (puromycin), which has not been described in the literature for M. bovis. We therefore inserted the inducible CBE system into another transposon-based plasmid that carried a gentamicin resistance marker in order to generate pMT85_SpdCas9_pmcDA1 (Fig. S2). The resulting construct was evaluated in M. bovis by targeting the mnuA gene (MBOVPG45_0215) (Fig. 3). MnuA is a major membrane nuclease that has been suggested to play a key role in M. bovis virulence by degrading the neutrophil extracellular traps (NETs) produced by the host in response to the pathogen (37). In the target spacer (5′-AAC18C17AAAAATATGACTTAGT/AGG-3′), C17 and C18 stand as two potential deamination sites. The conversion of C17 would change a glutamine CAA codon into a TAA stop codon and disrupt the mnuA gene. M. bovis cells were transformed with pMT85_SpdCas9_pmcDA1 targeting the mnuA gene and the transformants grown in liquid gentamicin selective medium for three passages. Expression of the CBE system was induced overnight with aTC at 0.5 μg·mL−1, and M. bovis transformants were plated on selective medium to isolate colonies. Target sites were analyzed in the global population before transformation, after each passage following transformation (P1, P2, and P3) and after induction (for the cell suspension and isolated colonies) (Fig. 3). As previously found in M. gallisepticum, there was considerable leakage of the inducible promoter in the M. bovis population immediately after transformation, with ~50% of the cytosines being converted to thymines at positions C17 and C18 at P1, P2, and P3. Overnight induction with aTC increased the conversion level to 90%. The screening of three independent colonies showed that both cytosines (C17 and C18) in the Mbov_mnuA mutants had been converted to thymine. We then carried out a nuclease phenotypic assay to demonstrate that the mutations introduced into the mnuA gene led to its inactivation (38). Indeed, the tested Mbov_mnuA mutant was unable to hydrolyze DNA, whereas the wild type strain degraded it all (Fig. S6). These results demonstrate the portability of this CBE-based method to produce targeted mutations in M. bovis.
FIG 3.
Disruption of the MnuA nuclease-encoding gene by base editing in M. bovis. A schematic diagram based on a base-editing experiment in M. bovis. After transformation of M. bovis (Mbov) with the plasmid pMT85_SpdCas9_pmcDA1, cells were propagated in liquid media supplemented with gentamicin. After three passages in liquid medium (P1 to P3), an inducer was added to the cell suspension. Aliquots were collected at each passage and after induction to monitor the target site sequence by Sanger sequencing and EditR analysis. Sequencing chromatograms of the complementary strand are presented along with the percentage of the base for each position of the target sequence. As shown in the diagram, cytosines susceptible to deamination are framed in red, and the edited bases are framed in green.
Application of the CBE system in the Mycoplasma mycoides subsp. mycoides genome.
Next, we introduced the sgRNA and CBE expression cassettes into the replicative oriC plasmid pMYCO1 to demonstrate the flexibility of the mycoplasma CBE system. This plasmid is routinely used in several species of the M. mycoides cluster (21), including Mmm. The resulting pMYCO1_SpdCas9_pmcDA1 plasmid (Fig. S2) was used to target the nonessential gene glpO (5′-C20AAC17AAC14AATAC9GATAAC3AT/TGG-3′), which encodes the metabolic enzyme L-α-glycerophosphate oxidase, which is putatively involved in mycoplasma virulence (39). Indeed, GlpO catalyzes the oxidation of glycerol-3-phosphate, leading to the release of hydrogen peroxide (H2O2), a product known to contribute to cytopathic effects in host tissues. C3, C9, C14, C17, and C20 were potential deamination sites, with the conversion of the last three positions leading to three TAA stop codons. After the transformation of Mmm, we tested various inducer concentrations and induction times and monitored the conversion of the cytosines in the cell population (Fig. S7). Before the induction of CBE expression, the conversion of cytosines was observed as in M. gallisepticum and M. bovis, reaching up to 30% at position C17. The best results were obtained after overnight induction with aTC at 0.5 μg·mL−1. We observed the conversion for each cytosine within the target region and those located 14 nt and 22 nt upstream of the PAM sequence. The observed deamination at position C14 in Mmm_glpO had not been observed in Mgal_gapA (Fig. 2), suggesting an extended editing window. However, low conversion frequencies (20 to 30%) were observed at the editing window extremities (i.e., C14 and C22), whereas higher efficiencies were observed between C17 and C20 (60 to 70%). After subcloning on agar plates, the mutants showed diverse editing profiles at the four cytosine positions (mixed population and different fully deaminated cytosine combinations). Nevertheless, 2 of 10 isolated clones showed the expected four mutations (C14, C17, C20, and C22). A phenotypic assay to evaluate H2O2 production confirmed the inactivation of the glpO gene in these clones (Fig. S7E). Finally, after three passages in liquid medium without selective pressure and one passage in solid medium for isolation, we recovered plasmid-free Mmm mutants edited at glpO. These are the first reported site-specific mutants generated in Mmm.
Inducing multiple mutations in Mycoplasma mycoides subsp. mycoides with a single CRISPR guide.
We tested the limits of the mycoplasma CBE system by targeting transposase-encoding genes that are associated with two families of insertion sequences, IS1634 and IS3, present 58 and 26 times in the Mmm T1/44 genome, respectively (Fig. 4 and Fig. S8). As transposase gene sequences are highly conserved within each family, we attempted to target a maximum number of copies by using a single sgRNA per family. For the IS1634 transposase, a 20-nucleotide spacer was designed to perform the inactivation of 55 of 58 copies (5′-AGAC17C16AGATTGTTATAGGTA/TGG-3′). Deamination of C16 on the reverse strand would change a glutamine CAG codon into a TAG stop codon at position 204 of the protein (Fig. 4A). For the IS3 transposase, two 20-nucleotide spacers were designed for the sgRNAs, the first targeting the start codon of all 26 copies of the transposase (sgRNA1, 5′-C20ATATAAAAACCCCATTTCC/TGG-3′) and the second allowing the introduction of a stop codon in 22 of the 26 copies (sgRNA2, 5′-AC19AAGTGGAATACTATAAGT/TGG-3′) (Fig. S8). After transformation and induction, all target sites were assessed by PCR and Sanger sequencing (Fig. 4B and Fig. S8). We observed low base-editing efficiency for the IS1634 target after the first induction, with only 5% conversion for C16 and 4% for C17. Two additional induction steps were carried out, resulting in an increase in base conversion of C16 and C17, yielding 37% and 40% after the second induction and 72% and 75% after the third induction, respectively. After the plating and PCR/sequencing screening of 20 isolated clones, two showed sequencing profiles that suggested that all IS1634 sites had been mutated (Fig. 4C). Finally, full genome sequencing showed that 52 and 54 transposases out of 55 were inactivated in these two clones. For IS3 inactivation, 66% and 77% conversion were observed in population analyses after three inductions using sgRNAs with spacers one and two, respectively (Fig. S8B, S8D). After the plating and subsequent PCR screening of 10 clones, one fully deaminated clone was obtained for each of the two sgRNAs (Fig. S8C, S8E). Full genome sequencing confirmed mutations at 22 out of 26 with sgRNA1 and 22 out of 22 target sites with sgRNA2 (Table S3). These results demonstrate the ability of the mycoplasma CBS system to edit numerous target sites in a single step.
FIG 4.
Multitargeting of IS1634 copies in the Mmm genome. (A) Schematic diagram of the Mmm T1/44 genome with 58 complete or truncated copies of the IS1634 transposases represented as solid lines. In the right panel, the targeted site within the IS1634 transposase gene is represented as a red bar. (B) Schematic diagram of the three induction steps of the mycoplasma CBE system for targeting 55 IS1634 sites using a single gRNA. The nucleotide sequence of the target is represented with the PAM sequence and framed by a red rectangle. Cytosines susceptible to deamination are shown in red. Fresh cultures of Mmm cells harboring the pMYCO1_SpdCas_pmcDA1 plasmid are represented as red-colored tubes. Cultures in exponential and stationary growth phases are indicated by orange-and yellow-colored tubes, respectively. Three consecutive culture steps were performed (including the starting culture) by a 1/100 dilution in fresh medium. At each step, induction was performed by adding aTC and incubating the cells until stationary-phase was reached. Deamination of target cytosines was evaluated by PCR and Sanger sequencing. The percentage of each base at each position was estimated from chromatograms using EditR software. The two positions of interest are highlighted in a dashed red rectangle. (C) Sanger sequencing results and base distribution in a selected isolated clone. Fully mutated positions are framed in green in the table and shown in green in the target sequence.
Evaluation of off-target mutations by whole-genome sequencing.
We investigated potential undesired mutations and evaluated the off-target activity of the CBE system in mycoplasmas by carrying out whole-genome sequencing on seven edited (mutant) clones, using both short-read (Illumina) and long-read (Oxford Nanopore Technologies) sequencing platforms: Mgal_ksgA, Mbov_0215, Mmm_glpO, Mmm_IS1634_cl3.1.6, Mmm_IS1634_cl3.1.11, Mmm_IS3_cl4.1.2, and Mmm_IS3_cl5.1.18 (Table S3). We detected 5 to 36 undesired mutations in the sequenced genomes. Mutations were further analyzed and classified into three categories (Fig. S9A, Table S3): (i) sgRNA-guided off-target mutations possibly generated by the CBE system, if the targeted sequence was similar to the sgRNA spacer sequence and adjacent to an NGG PAM sequence; (ii) potentially unguided spurious deamination by pmcDA1 (30, 40) for all C to T or G to A mutations in genome regions with no similarity to the sgRNA spacer sequence; and (iii) other mutations that could not have been induced by the CBE system and that potentially occurred during passaging. Based on these criteria, 0 to 5 sgRNA-guided off-target mutations were predicted, representing 0% to 17% of the total mutations, with the exception of Mmm_glpO, for which 2 of 5 mutations were classified in this category. Two examples of off-target interactions were seen: the ksgA sgRNA targeting DNA in M. gallisepticum (mutation at position 1,313 in the genome) and the glpO sgRNA targeting DNA in Mmm (mutation at position 220,532 in the genome) are presented (Fig. S9B, S9C; Table S3). Comparing the undesired mutations distribution between the two independent Mmm_IS1634 clones that were obtained with the same sgRNA and fully sequenced, we noticed that only 1 in 33 mutations was in common (and classified as an “other mutation”). This indicated that, at least in Mmm and with this sgRNA, the background mutations were not reproducible and changed from one clone to another. Such a distribution suggests that the vast majority of mutations were not caused by an sgRNA-driven off-target activity of the base editing tool. Spurious deamination represented 20% to 84% of mutations, whereas other mechanisms accounted for 11% to 50% of mutations. Interestingly, the number and percentage of spurious deamination that occurred when using the mycoplasma CBE system in Mmm were much higher when a three-induction protocol was used to maximize the efficiency of IS1634 and IS3 targeting (11 to 24 mutations) than when the single-induction protocol was used to target glpO (1 mutation). Thus, sgRNA-guided off-target mutations were relatively rare, whereas spurious deaminations accumulated with extended CBE activity.
DISCUSSION
The functional genomics of Mollicutes has long been hampered by the absence of efficient genetic tools by which to generate targeted mutations. Here, we adapted a new genetic tool for Mollicutes that is based on CRISPR for direct mutagenesis. We evaluated its efficiency in three economically relevant animal pathogens.
CBE systems have proven to be highly efficient in diverse eukaryotic cells, including those of plants and humans (41, 42), as well as in prokaryotic organisms, including Klebsiella (43), Pseudomonas (44), Clostridium (45), Streptomyces (46), and Agrobacterium (47) species. Here, we designed and constructed two mycoplasma CBE systems, but our preliminary results suggested that only the pmcDA1-based CBE was active in M. gallisepticum (Fig. 1; Fig. S3). Both cytosine deaminases have been used successfully in bacteria (46, 48), but optimizing the expression levels of BE components, including sgRNAs, has been shown to be key in avoiding cell toxicity and in reaching a high rate of base conversion (49, 50). In this study, transformation experiments of M. gallisepticum with plasmids carrying the pmcDA1-based and rAPOBEC1-based CBEs did not suggest cell toxicity effects. However, the rAPOBEC1-based CBE was not efficient in M. gallisepticum, and further investigations are required to clarify why. In contrast, the pmcDA1-based CBE was highly efficient in generating point mutations in M. gallisepticum, M. bovis, and Mmm (Fig. 1 and 3; Fig. S7). Such contrasted efficiencies of rAPOBEC1 and pmcDA1-based CBEs have also been reported in other organisms (51).
Taking advantage of such a tool, we succeeded in targeting up to 54 IS1634 targets in the Mmm chromosome with a single CRISPR guide. Multiple targeting of repeated sequences has been reported previously in eukaryotic and prokaryotic systems (48, 52, 53), but to our knowledge, this is the first time that so many target sites have been modified in bacteria, with the last record in E. coli targeting 41 loci (48). Noticeably, achieving such multiple targeting required tuning the protocol, including multiple rounds of induction of the base-editing system expression. In the best conditions, the global mutation efficacy in the bacterial population for a single target ranged from 50% to 100% (in gapA and ksgA, respectively) and led to the easy isolation of mutants. When targeting 54 sites, we observed a decrease of the global efficacy, with only 5% conversion in the bacterial population after the first induction. However, this efficacy reached 38% and 73% after a second and a third round of induction, respectively, and the screening of 10 to 20 clones was sufficient to successfully isolate clones with all or nearly all targets modified. The efficacy and specificity of CRISPR-Cas-based genetic tools rely on the ability of sgRNA to anneal with the target sequence. In our study, all of the sgRNAs we designed and evaluated in M. gallisepticum, M. bovis and Mmm gave comparable results, each time resulting in the expected mutation. More data may reveal some gRNA effect in terms of efficacy, but we have not observed such so far.
In order to evaluate potential off-target mutations induced by the mycoplasma CBE system, we selected seven mutants for whole-genome sequencing (Fig. S9, Table S3). An analysis of the detected mutations showed that only a few of them could have resulted from sgRNA-guided off-target deamination events. Spurious deamination of pmcDA1, which preferentially deaminates TC motifs (30), appeared to increase with extended induction periods, suggesting that the control of CBE expression could be crucial to reducing the frequency of undesired mutations. Such spurious deamination induced by CBE systems has already been reported in other bacteria, including Corynobacterium glutamicum and Bacillus subtilis, in which 9 and 19 SNVs, respectively, could be attributed to deaminase activity (54, 55). In the CBE system adapted here, the expression of the SpdCas9-pmcDA1-UGI hybrid protein is driven by the promoter Pxyl/tetO2, which can be induced by aTC. Although we observed a clear increase in the frequency of deaminated bases in the three studied mycoplasmas after induction, the conversion process was already observed before induction (Fig. 1 and 3; Fig. S7). This indicates a certain level of promoter leakage in the three species, in accordance with another report on M. pneumoniae (56). Several strategies can be proposed to limit the background of undesired mutations, including reducing the induction time and using an improved inducible promoter, such as that recently developed for M. pneumoniae (56). In addition, the use of high-fidelity Cas9 variants or CBE variants (33) may also improve the specificity of the genetic tool. Finally, the elimination of the CBE system after mutagenesis is also crucial for avoiding the accumulation of undesired mutations over time. This can be achieved for the CBE systems based on the pMT85_2Res and pMYCO1 backbones. Indeed, all genetic elements flanked by the res sequences in the pMT85_2Res can be removed from the chromosome using dedicated resolvase activity (57), and the oriC plasmids can be lost after a few passages in nonselective medium (58). Alternatively, CBE constructs can be enhanced with a CRE-Lox system, which is functional in some mycoplasma species (23, 56, 59).
In the era of synthetic biology, with the fast expansion of the toolbox for modifying the genomes of living organisms, base-editing tools have attracted interest because of specific features: independence of endogenous cellular DNA repair pathways, lower cytotoxic effects than with other Cas9-based tools, high efficacy, which means that there is no need for selection markers, possibility of multitargeting, and the lack of a scar at the edited locus. These properties make BE promising tools for clinical usage in human therapeutics (60) and for multiple applications in plants (51, 61) and microorganisms of medical or biotechnological relevance (62–64). Because of their high efficacy, BE are particularly attractive for bacteria that are difficult to transform, as, ultimately, one single transformant can be enough to obtain a mutant. This is the case for mycoplasmas and other bacteria belonging to the Mollicutes class. These minimal bacteria have a limited set of enzymes involved in their DNA repair pathways, and endogenous HR is, for most species, not efficient enough to be used as the basis for “everyday mutagenesis”. In practice, reproducible HR-based mutagenesis with suicide plasmids is only available for Mycoplasma genitalium (65–67), although some mutants were obtained by HR in between chromosome and replicative plasmids for some other species, including Spiroplasma citri (68–70), Mycoplasma pulmonis (71), and M. gallisepticum (22, 72). The low efficiency of HR on the target locus and background integration at the oriC (68, 69, 71, 73) makes these tools poorly efficient and therefore not of practical interest. As a consequence, transposon mutagenesis remains the only practical way to generate mutants in many species, including M. gallisepticum (74–77), M. bovis (78, 79), Mycoplasma hyopneumoniae (80), Mycoplasma hyorhinis (81), and many others. While large transposon-based mutant libraries have been obtained to identify essential genes in M. genitalium (82, 83), M. pneumoniae (84), Mycoplasma mycoides subsp. capri (85), and Mesoplasma florum (86), the relatively low transformation efficiency of current protocols for some other species, such as Mycoplasma mobile (87) or S. citri (88), is a strong limit for the targeting of a specific gene or group of genes. Because natural recombination events and transformation efficiency are limited for many mycoplasma species, the use of exogenous recombination systems was attempted. Importing the RecA protein from E. coli improved the obtention of bacterial recombinants in Mycoplasma mycoides subsp. capricolum (89) and in M. hyorhinis (90), but not in M. hyopneumoniae (91). Recently, a RecET-like system from B. subtilis was used for targeted gene inactivation or gene replacement in Mycoplasma pneumoniae (25) and M. gallisepticum (23). However, for now, all of these systems remain limited in efficacy, and their use in a large spectrum of mycoplasmas still needs to be evaluated. In contrast with the situation in eukaryotic cells, CRISPR-Cas9-mediated chromosomal cleavage is often lethal in bacteria, presumably due to the lack of double-strand break repair systems in most of the genera. For this reason, the original S. pyogenes CRISPR-Cas9 system was first used in M. pneumoniae as a counterselection tool, in combination with the exogenous recombination system mentioned above (25). Transcription interference assays using dCas9 (CRISPRi) have been achieved successfully in M. pneumoniae, the synthetic M. mycoides subsp. capri JCVI-syn1.0 (32), M. gallisepticum and Mycoplasma hominis (92). The use of the mycoplasma endogenous CRISPR-Cas9 system has been described in M. gallisepticum for targeting two genes (93, 94), but unpredictable results have been reported, with mutations localized mostly outside the gRNA targeted sequences for one gene (ksgA) and without getting genomic mutants for the second one (MnuA). The role of the endogenous Cas9 in these studies remains to be elucidated, and this is due in part to the lack of knowledge about the M. gallisepticum CRISPR-Cas9 system. Further studies are necessary to shed light on the specificity of this system and to explore its possible use as a tool for targeted mutagenesis. In the mycoplasma field, an impactful achievement is the possibility to clone a complete genome in yeast, to modify it in the yeast, and to transplant it back into a recipient cell to generate mutants. Because of the highly efficient HR in yeast, and together with advances in gene synthesis, genetic engineering is now possible at the genome scale using CRISPR-Cas and other genetic tools available in yeast. Currently, this method is available for several species related to the M. mycoides cluster (17–19), but it has not yet been extended to members of other phylogenetic groups. In order to circumvent this limit, a recombinase-assisted genomic engineering (RAGE) technology was recently developed and proved efficient to introduce a 15 kbp fragment at a specific locus of the M. pneumoniae genome, and it was able to replace 38 kbp from the genome by means of engineered versions modified either in yeast or in E. coli (59). While they do offer new possibilities of genome engineering in these bacteria, these sophisticated methods are only available for a limited number of species. Therefore, more straightforward and wide-spectrum tools are needed.
In this context, the development of a highly efficient base-editing tool is a significant step forward in the mycoplasma field. Especially, this application of CBE opens new possibilities for targeting multigene families with a limited number of sgRNA molecules. In mycoplasmas, such families are often predicted to encode surface proteins suspected to be involved in host-pathogen interactions, but current mutagenesis methods, including in-yeast genome engineering, are unable to disrupt all of the genes of a family in a reasonable amount of time. In this work, the same promoters were used to drive the expression of the sgRNA (PS) and CBE system (Pxyl/tetO2) in three species belonging to different phylogenetic groups, suggesting that both expression cassettes could be used without modification in various species of mycoplasmas and possibly other Mollicutes. Three different plasmid backbones were used here, including two Tn4001-derived transposons and one replicative oriC plasmid (Fig. S2). Transposons and oriC plasmids are currently the most widely used genetic tools available for Mollicutes. Transposon mutagenesis with Tn4001 derivatives has been used in 15 species, and oriC plasmids are available for 14 species. Thus, the mycoplasma CBE system could be easily evaluated in many species, either directly or after cloning the sgRNA and CBE expression cassettes into a compatible vector. During this study, we also found that induction could be performed immediately following plasmid transformation (i.e., immediately after the 2 h cell recovery in Hayflick medium) instead of after colony recovery and regrowth. This improvement allowed us to reduce the duration of the experiments by 14 days and resulted in the expected mutants in less than 3 weeks.
Currently, the base-editing tool described here is limited to C to T mutations. However, the efficacy of base editors is regularly enhanced by combining various enzymes. Adenine deaminase-mediated and dual deaminase-mediated base editing systems have been developed, improved, and validated successfully in various organisms (46, 50, 63, 95, 96). A second limitation comes from the use of SpdCas9 that recognizes canonical NGG-type PAM sites, which limits its target range in genomes. Many variants have now been evolved from SpCas9 to broaden PAM recognition features, including SpCas9-NRRH, SpCas9-NRTH, SpCas9-NRCH (97), SpCas9-NG (98), SpG, and SpRY (99). Cas9 from other bacterial species, with different PAM recognition specificities, including ScCas9 (Streptococcus canis) (100), SaCas9 (Staphylococcus aureus) (101), Nm1Cas9, and Nm2Cas9 (Neisseria meningitidis) (51), have also been used in ABE and CBE systems. Finally, the relatively “wide” editing window of the current CBE might be narrowed by C-terminal truncations of pmCDA1 (102). Therefore, the adaptation of base-editing tools for Mollicutes, starting with this work, is open for many further expansions. In particular, the further characterization of endogenous CRISPR-Cas9 systems of mycoplasmas and their PAM requirement may open new opportunities for the design of new base editors and for the expansion of the genetic toolbox for these minimal bacteria.
MATERIALS AND METHODS
Oligonucleotides and plasmids.
All oligonucleotides used in this study were supplied by Eurogentec and are described in Table S1. All plasmids constructed and used in this study are listed in Table S2. Detailed protocols for plasmid construction and other methods are provided as SI Materials and Methods.
Bacterial strains and culture.
M. gallisepticum strain S6 (Tax ID: 1006581) was cultivated at 37°C in modified Hayflick medium (103) in a 5% CO2 atmosphere, and puromycin at 10 μg·mL−1 and kasugamycin at 400 μg·mL−1 were used for selection. M. bovis PG45 (Tax ID: 289397) was cultivated in SP4 medium (103), and gentamicin at 100 μg·mL−1 was used for selection. Mmm T1/44 (Tax ID: 2103) was cultivated in SP5 medium (18), and puromycin at 8 μg·mL−1 was used for selection (104). Phenol red was used as a pH indicator in the mycoplasma media. Escherichia coli NEB-5α (NEB, C2987H) was used for plasmid propagation and was cultivated in Luria broth (ThermoFisher: 12795027), with the addition of ampicillin at 100 μg·mL−1 or kanamycin at 50 μg·mL−1 for selection.
Transformation of Mycoplasma species.
Mycoplasma transformation was performed using a polyethylene glycol mediated protocol (105, 106). Late log-phase mycoplasma cultures were transformed with plasmid DNA (20 μg). After transformation, cells were resuspended in 1 mL of the appropriate medium, incubated for 2 h at 37°C, and plated onto selective solid medium. After incubation at 37°C for 3 to 10 days, single colonies containing the deaminase constructs were obtained. Expression of the CBE system was induced in early logarithmic growth-phase cultures overnight, and the cultures were plated on selective medium to isolate colonies.
ACKNOWLEDGMENTS
This work was partially funded by ANR as part of the project RAMbo-V (ANR-21-CE35-0008). Genome sequencing was performed by the Genome Transcriptome Facility of Bordeaux (https://pgtb.cgfb.u-bordeaux.fr, Grants from Investissements d’avenir, Convention attributive d’aide EquipEx Xyloforest ANR-10-EQPX-16-01).
The conceptualization was completed by T.I. and P.S.-P., and the formal analysis was completed by T.I. and F.R. Funding was acquired by Y.A., C.L., A.B., and P.S.-P. The investigation was conducted by T.I., F.R., G.G., and the methodology was designed by T.I. and P.S.-P. Supervision was provided by C.L. and P.S.-P. Validation was performed by Y.A., C.L., A.B., and P.S.-P., and visualization was performed by T.I., P.S.-P. The writing of the original draft was completed by T.I., A.B., and P.S.-P., and the review and editing were completed by T.I., F.R., Y.A., C.L., A.B., and P.S.-P.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Pascal Sirand-Pugnet, Email: pascal.sirand-pugnet@inrae.fr.
Charles M. Dozois, INRS
REFERENCES
- 1.May M, Balish MF, Blanchard A. 2014. The order Mycoplasmatales, p 515–550. In The Prokaryotes. Springer Berlin Heidelberg. [Google Scholar]
- 2.Sirand-Pugnet P, Citti C, Barré A, Blanchard A. 2007. Evolution of mollicutes: down a bumpy road with twists and turns. Res Microbiol 158:754–766. 10.1016/j.resmic.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev 62:1094–1156. 10.1128/MMBR.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Citti C, Dordet-Frisoni E, Nouvel LX, Kuo CH, Baranowski E. 2018. Horizontal gene transfers in Mycoplasmas (Mollicutes). Curr Issues Mol Biol 29:3–22. 10.21775/cimb.029.003. [DOI] [PubMed] [Google Scholar]
- 5.Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barré A, Yoshizawa S, Fourmy D, de Crécy-Lagard V, Blanchard A. 2014. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet 10:e1004363. 10.1371/journal.pgen.1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Citti C, Blanchard A. 2013. Mycoplasmas and their host: emerging and re-emerging minimal pathogens. Trends Microbiol 21:196–203. 10.1016/j.tim.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Rosengarten R, Citti C, Much P, Spergser J, Droesse M, Hewicker-Trautwein M. 2001. The changing image of mycoplasmas: from innocent bystanders to emerging and reemerging pathogens in human and animal diseases. Contrib Microbiol 8:166–185. 10.1159/000060409. [DOI] [PubMed] [Google Scholar]
- 8.Hennigan SL, Driskell JD, Ferguson-Noel N, Dluhy RA, Zhao Y, Tripp RA, Krause DC. 2012. Detection and differentiation of avian mycoplasmas by surface-enhanced Raman spectroscopy based on a silver nanorod array. Appl Environ Microbiol 78:1930–1935. 10.1128/AEM.07419-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peebles ED, Branton SL. 2012. Mycoplasma gallisepticum in the commercial egg-laying hen: a historical perspective considering the effects of pathogen strain, age of the bird at inoculation, and diet on performance and physiology. J Appl Poult Res 21:897–914. 10.3382/japr.2012-00555. [DOI] [Google Scholar]
- 10.Feberwee A, de Wit S, Dijkman R. 2022. Clinical expression, epidemiology, and monitoring of Mycoplasma gallisepticum and Mycoplasma synoviae: an update. Avian Pathol 51:2–18. 10.1080/03079457.2021.1944605. [DOI] [PubMed] [Google Scholar]
- 11.Dudek K, Nicholas RAJ, Szacawa E, Bednarek D. 2020. Mycoplasma bovis infections: occurrence, pathogenesis, diagnosis and control, including prevention and therapy. Pathogens 9:640–643. 10.3390/pathogens9080640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan A, Sadler RJ, Sawford K, Andel M, Ward M, Cowled B. 2021. Mycoplasma bovis outbreak in New Zealand cattle: an assessment of transmission trends using surveillance data. Transbound Emerg Dis 68:3381–3395. 10.1111/tbed.13941. [DOI] [PubMed] [Google Scholar]
- 13.Di Teodoro G, Marruchella G, Di Provvido A, D'Angelo AR, Orsini G, Di Giuseppe P, Sacchini F, Scacchia M. 2020. Contagious bovine pleuropneumonia: a comprehensive overview. Vet Pathol 57:476–489. 10.1177/0300985820921818. [DOI] [PubMed] [Google Scholar]
- 14.Gautier-Bouchardon AV. 2018. Antimicrobial resistance in Mycoplasma spp. Microbiol Spectr 6. 10.1128/microbiolspec.ARBA-0030-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishfaq M, Hu W, Khan MZ, Ahmad I, Guo W, Li J. 2020. Current status of vaccine research, development, and challenges of vaccines for Mycoplasma gallisepticum. Poult Sci 99:4195–4202. 10.1016/j.psj.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz E, Talenton V, Dubrana M-P, Guesdon G, Lluch-Senar M, Salin F, Sirand-Pugnet P, Arfi Y, Lartigue C. 2019. CReasPy-cloning: a method for simultaneous cloning and engineering of megabase-sized genomes in yeast using the CRISPR-Cas9 system. ACS Synth Biol 8:2547–2557. 10.1021/acssynbio.9b00224. [DOI] [PubMed] [Google Scholar]
- 17.Lartigue C, Vashee S, Algire MA, Chuang RY, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, Alperovich N, Thomas DW, Merryman C, Hutchison CA, Smith HO, Venter JC, Glass JI. 2009. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325:1693–1696. 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 18.Labroussaa F, Lebaudy A, Baby V, Gourgues G, Matteau D, Vashee S, Sirand-Pugnet P, Rodrigue S, Lartigue C. 2016. Impact of donor-recipient phylogenetic distance on bacterial genome transplantation. Nucleic Acids Res 44:8501–8511. 10.1093/nar/gkw688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talenton V, Baby V, Gourgues G, Mouden C, Claverol S, Vashee S, Blanchard A, Labroussaa F, Jores J, Arfi Y, Sirand-Pugnet P, Lartigue C. 2022. Genome engineering of the fast-growing Mycoplasma feriruminatoris toward a live vaccine chassis. ACS Synth Biol 11:1919–1930. 10.1021/acssynbio.2c00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chopra-Dewasthaly R, Zimmermann M, Rosengarten R, Citti C. 2005. First steps towards the genetic manipulation of Mycoplasma agalactiae and Mycoplasma bovis using the transposon Tn4001mod. Int J Med Microbiol 294:447–453. 10.1016/j.ijmm.2004.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janis C, Lartigue C, Frey J, Wróblewski H, Thiaucourt F, Blanchard A, Sirand-Pugnet P. 2005. Versatile use of oriC plasmids for functional genomics of Mycoplasma capricolum subsp. capricolum. Appl Environ Microbiol 71:2888–2893. 10.1128/AEM.71.6.2888-2893.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S-W, Browning GF, Markham PF. 2008. Development of a replicable oriC plasmid for Mycoplasma gallisepticum and Mycoplasma imitans, and gene disruption through homologous recombination in M. gallisepticum. Microbiology 154:2571–2580. 10.1099/mic.0.2008/019208-0. [DOI] [PubMed] [Google Scholar]
- 23.Ipoutcha T, Gourgues G, Lartigue C, Blanchard A, Sirand-Pugnet P. 2022. Genome engineering in Mycoplasma gallisepticum using exogenous recombination systems. ACS Synth Biol acssynbio.1c00541. [DOI] [PubMed] [Google Scholar]
- 24.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piñero-Lambea C, Garcia-Ramallo E, Martinez S, Delgado J, Serrano L, Lluch-Senar M. 2020. Mycoplasma pneumoniae genome editing based on oligo recombineering and Cas9-mediated counterselection. ACS Synth Biol 9:1693–1704. 10.1021/acssynbio.0c00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424. 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A. 2016. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science (80-) 353. 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 28.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. 2017. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature 551:464–471. 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, Kim YB, Badran AH, Liu DR. 2017. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anzalone AV, Koblan LW, Liu DR. 2020. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38:824–844. 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 31.Mahairas GG, Minion FC. 1989. Random insertion of the gentamicin resistance transposon Tn4001 in Mycoplasma pulmonis. Plasmid 21:43–47. 10.1016/0147-619X(89)90085-1. [DOI] [PubMed] [Google Scholar]
- 32.Mariscal AM, Kakizawa S, Hsu JY, Tanaka K, González-González L, Broto A, Querol E, Lluch-Senar M, Piñero-Lambea C, Sun L, Weyman PD, Wise KS, Merryman C, Tse G, Moore AJ, Hutchison CA, Smith HO, Tomita M, Venter JC, Glass JI, Piñol J, Suzuki Y. 2018. Tuning gene activity by inducible and targeted regulation of gene expression in minimal bacterial cells. ACS Synth Biol 7:1538–1552. 10.1021/acssynbio.8b00028. [DOI] [PubMed] [Google Scholar]
- 33.Huang TP, Newby GA, Liu DR. 2021. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc 16:1089–1128. 10.1038/s41596-020-00450-9. [DOI] [PubMed] [Google Scholar]
- 34.Papazisi L, Frasca S, Gladd M, Liao X, Yogev D, Geary SJ. 2002. GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence. Infect Immun 70:6839–6845. 10.1128/IAI.70.12.6839-6845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mugunthan SP, Harish MC. 2021. Multi-epitope-based vaccine designed by targeting cytoadherence proteins of Mycoplasma gallisepticum. ACS Omega 6:13742–13755. 10.1021/acsomega.1c01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cizelj I, Berčič RL, Dušanić D, Narat M, Kos J, Dovč P, Benčina D. 2011. Mycoplasma gallisepticum and Mycoplasma synoviae express a cysteine protease CysP, which can cleave chicken IgG into Fab and Fc. Microbiology (Reading) 157:362–372. 10.1099/mic.0.045641-0. [DOI] [PubMed] [Google Scholar]
- 37.Mitiku F, Hartley CA, Sansom FM, Coombe JE, Mansell PD, Beggs DS, Browning GF. 2018. The major membrane nuclease MnuA degrades neutrophil extracellular traps induced by Mycoplasma bovis. Vet Microbiol 218:13–19. 10.1016/j.vetmic.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Sharma S, Tivendale KA, Markham PF, Browning GF. 2015. Disruption of the membrane nuclease gene (MBOVPG45_0215) of Mycoplasma bovis greatly reduces cellular nuclease activity. J Bacteriol 197:1549–1558. 10.1128/JB.00034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilo P, Vilei EM, Peterhans E, Bonvin-Klotz L, Stoffel MH, Dobbelaere D, Frey J. 2005. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. J Bacteriol 187:6824–6831. 10.1128/JB.187.19.6824-6831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Y, Leete TC, Born DA, Young L, Barrera LA, Lee SJ, Rees HA, Ciaramella G, Gaudelli NM. 2020. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat Commun 11:1–10. 10.1038/s41467-020-15887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Xu W, Wang F, Zhao S, Feng F, Song J, Zhang C, Yang J. 2019. Increasing cytosine base editing scope and efficiency with engineered Cas9-PMCDA1 fusions and the modified sgRNA in rice. Front Genet 10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim HS, Jeong YK, Hur JK, Kim JS, Bae S. 2019. Adenine base editors catalyze cytosine conversions in human cells. Nat Biotechnol 37:1145–1148. 10.1038/s41587-019-0254-4. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Wang aS, Chen W, Song L, Zhang Y, Shen Z, Yu F, Li M, Jia Q. 2018. CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl Environ Microbiol 84:1–15. 10.1128/AEM.01834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J, Lu LB, Liang TX, Yang LR, Wu JP. 2020. CRISPR-assisted multiplex base editing system in Pseudomonas putida KT2440. Front Bioeng Biotechnol 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Seys FM, Minton NP, Yang J, Jiang Y, Jiang W, Yang S. 2019. CRISPR–Cas9 D10A nickase-assisted base editing in the solvent producer Clostridium beijerinckii. Biotechnol Bioeng 116:1475–1483. 10.1002/bit.26949. [DOI] [PubMed] [Google Scholar]
- 46.Tong Y, Whitford CM, Robertsen HL, Blin K, Jørgensen TS, Klitgaard AK, Gren T, Jiang X, Weber T, Lee SY. 2019. Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proc Natl Acad Sci USA 116:20366–20375. 10.1073/pnas.1913493116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigues SD, Karimi M, Impens L, van Lerberge E, Coussens G, Aesaert S, Rombaut D, Holtappels D, Ibrahim HMM, van Montagu M, Wagemans J, Jacobs TB, de Coninck B, Pauwels L. 2021. Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc Natl Acad Sci USA 118:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banno S, Nishida K, Arazoe T, Mitsunobu H, Kondo A. 2018. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat Microbiol 3:423–429. 10.1038/s41564-017-0102-6. [DOI] [PubMed] [Google Scholar]
- 49.Kim MS, Kim H-R, Jeong D-E, Choi S-K. 2021. Cytosine base editor-mediated multiplex genome editing to accelerate discovery of novel antibiotics in Bacillus subtilis and Paenibacillus polymyxa. Front Microbiol 12:691839. 10.3389/fmicb.2021.691839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shelake RM, Pramanik D, Kim J-Y. 2022. In vivo rapid investigation of CRISPR-based base editing components in Escherichia coli (IRI-CCE): a platform for evaluating base editing tools and their components. Int J Mol Sci 23. 10.3390/ijms23031145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu R, Qin R, Xie H, Li J, Liu X, Zhu M, Sun Y, Yu Y, Lu P, Wei P. 2022. Genome editing with type II-C CRISPR-Cas9 systems from Neisseria meningitidis in rice. Plant Biotechnol J 20:350–359. 10.1111/pbi.13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong Z, Guo J, Deng L, Chen L, Wang J, Li S, Xu W, Deng Z, Sun Y, Discovery D. 2019. Base editing in Streptomyces with Cas9-deaminase fusions. BioRxiv 630137. [Google Scholar]
- 53.Bae SJ, Park BG, Kim BG, Hahn JS. 2019. Multiplex gene disruption by targeted base editing of Yarrowia lipolytica genome using cytidine deaminase combined with the CRISPR/Cas9 system. Biotechnol J 15:1–46. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Liu Y, Liu J, Guo Y, Fan L, Ni X, Zheng X, Wang M, Zheng P, Sun J, Ma Y. 2018. MACBETH: multiplex automated Corynebacterium glutamicum base editing method. Metab Eng 47:200–210. 10.1016/j.ymben.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Yu S, Price MA, Wang Y, Liu Y, Guo Y, Ni X, Rosser SJ, Bi C, Wang M. 2020. CRISPR-dCas9 mediated cytosine deaminase base editing in Bacillus subtilis. ACS Synth Biol 9:1781–1789. 10.1021/acssynbio.0c00151. [DOI] [PubMed] [Google Scholar]
- 56.Mariscal AM, González-González L, Querol E, Piñol J. 2016. All-in-one construct for genome engineering using Cre-lox technology. DNA Res 23:263–270. 10.1093/dnares/dsw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janis C, Bischof D, Gourgues G, Frey J, Blanchard A, Sirand-Pugnet P. 2008. Unmarked insertional mutagenesis in the bovine pathogen Mycoplasma mycoides subsp. mycoides SC. Microbiology (Reading) 154:2427–2436. 10.1099/mic.0.2008/017640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lartigue C, Blanchard A, Renaudin J, Thiaucourt F, Sirand-Pugnet P. 2003. Host specificity of mollicutes oriC plasmids: functional analysis of replication origin. Nucleic Acids Res 31:6610–6618. 10.1093/nar/gkg848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia-Morales L, Ruiz E, Gourgues G, Rideau F, Piñero-Lambea C, Lluch-Senar M, Blanchard A, Lartigue C. 2020. A RAGE based strategy for the genome engineering of the human respiratory pathogen Mycoplasma pneumoniae. ACS Synth Biol 9:2737–2748. 10.1021/acssynbio.0c00263. [DOI] [PubMed] [Google Scholar]
- 60.Modell AE, Lim D, Nguyen TM, Sreekanth V, Choudhary A. 2022. CRISPR-based therapeutics: current challenges and future applications. Trends Pharmacol Sci 43:151–161. 10.1016/j.tips.2021.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang X, Wang Y, Wang N. 2022. Base editors for citrus gene editing. Front Genome Ed 4:852867. 10.3389/fgeed.2022.852867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Xiao Y, Wei X, Pan J, Duanmu D. 2021. Highly efficient CRISPR-mediated base editing in Sinorhizobium meliloti. Front Microbiol 12:686008. 10.3389/fmicb.2021.686008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang P, Han T, Liu W, Ren W, Wu Y, Xiao Y. 2022. Adenine base editing system for Pseudomonas and prediction workflow for protein dysfunction via ABE. ACS Synth Biol 11:1650–1657. 10.1021/acssynbio.2c00066. [DOI] [PubMed] [Google Scholar]
- 64.Jain S, Bhowmick A, Jeong B, Bae T, Ghosh A. 2022. Unravelling the physiological roles of mazEF toxin-antitoxin system on clinical MRSA strain by CRISPR RNA-guided cytidine deaminase. J Biomed Sci 29:28. 10.1186/s12929-022-00810-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seybert A, Gonzalez-Gonzalez L, Scheffer MP, Lluch-Senar M, Mariscal AM, Querol E, Matthaeus F, Piñol J, Frangakis AS. 2018. Cryo-electron tomography analyses of terminal organelle mutants suggest the motility mechanism of Mycoplasma genitalium. Mol Microbiol 108:319–329. 10.1111/mmi.13938. [DOI] [PubMed] [Google Scholar]
- 66.Martínez-Torró C, Torres-Puig S, Marcos-Silva M, Huguet-Ramón M, Muñoz-Navarro C, Lluch-Senar M, Serrano L, Querol E, Piñol J, Pich OQ. 2021. Functional characterization of the cell division gene cluster of the wall-less bacterium Mycoplasma genitalium. Front Microbiol 12:695572. 10.3389/fmicb.2021.695572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martínez-Torró C, Torres-Puig S, Monge M, Sánchez-Alba L, González-Martín M, Marcos-Silva M, Perálvarez-Marín A, Canals F, Querol E, Piñol J, Pich OQ. 2020. Transcriptional response to metal starvation in the emerging pathogen Mycoplasma genitalium is mediated by Fur-dependent and -independent regulatory pathways. Emerg Microbes Infect 9:5–19. 10.1080/22221751.2019.1700762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lartigue C, Duret S, Garnier M, Renaudin J. 2002. New plasmid vectors for specific gene targeting in Spiroplasma citri. Plasmid 48:149–159. 10.1016/s0147-619x(02)00121-x. [DOI] [PubMed] [Google Scholar]
- 69.Duret S, Danet JL, Garnier M, Renaudin J. 1999. Gene disruption through homologous recombination in Spiroplasma citri: an scm1-disrupted motility mutant is pathogenic. J Bacteriol 181:7449–7456. 10.1128/JB.181.24.7449-7456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duret S, André A, Renaudin J. 2005. Specific gene targeting in Spiroplasma citri: improved vectors and production of unmarked mutations using site-specific recombination. Microbiology (Reading) 151:2793–2803. 10.1099/mic.0.28123-0. [DOI] [PubMed] [Google Scholar]
- 71.Cordova CMM, Lartigue C, Sirand-Pugnet P, Renaudin J, Cunha RAF, Blanchard A. 2002. Identification of the origin of replication of the Mycoplasma pulmonis chromosome and its use in oriC replicative plasmids. J Bacteriol 184:5426–5435. 10.1128/JB.184.19.5426-5435.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Markham PF, Kanci A, Czifra G, Sundquist B, Hains P, Browning GF. 2003. Homologue of macrophage-activating lipoprotein in Mycoplasma gallisepticum is not essential for growth and pathogenicity in tracheal organ cultures. J Bacteriol 185:2538–2547. 10.1128/JB.185.8.2538-2547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nieszner I, Vronka M, Indikova I, Szostak MP. 2013. Development of a site-directed integration plasmid for heterologous gene expression in Mycoplasma gallisepticum. PLoS One 8:e81481. 10.1371/journal.pone.0081481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mudahi-Orenstein S, Levisohn S, Geary SJ, Yogev D. 2003. Cytadherence-deficient mutants of Mycoplasma gallisepticum generated by transposon mutagenesis. Infect Immun 71:3812–3820. 10.1128/IAI.71.7.3812-3820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whetzel PL, Hnatow LL, Keeler CL, Dohms JE. 2003. Transposon mutagenesis of Mycoplasma gallisepticum. Plasmid 49:34–43. 10.1016/s0147-619x(02)00114-2. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Yi L, Zhang F, Qiu X, Tan L, Yu S, Cheng X, Ding C. 2017. Identification of genes involved in Mycoplasma gallisepticum biofilm formation using mini-Tn4001-SGM transposon mutagenesis. Vet Microbiol 198:17–22. 10.1016/j.vetmic.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 77.Tseng CW, Kanci A, Citti C, Rosengarten R, Chiu CJ, Chen ZH, Geary SJ, Browning GF, Markham PF. 2013. MalF is essential for persistence of Mycoplasma gallisepticum in vivo. Microbiology (Reading) 159:1459–1470. 10.1099/mic.0.067553-0. [DOI] [PubMed] [Google Scholar]
- 78.Sharma S, Markham PF, Browning GF. 2014. Genes found essential in other mycoplasmas are dispensable in Mycoplasma bovis. PLoS One 9:e97100. 10.1371/journal.pone.0097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Josi C, Bürki S, Vidal S, Dordet-Frisoni E, Citti C, Falquet L, Pilo P. 2019. Large-scale analysis of the Mycoplasma bovis genome identified non-essential, adhesion- and virulence-related genes. Front Microbiol 10:2085. 10.3389/fmicb.2019.02085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maglennon GA, Cook BS, Deeney AS, Bossé JT, Peters SE, Langford PR, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, BRaDP1T consortium . 2013. Transposon mutagenesis in Mycoplasma hyopneumoniae using a novel mariner-based system for generating random mutations. Vet Res 44:1–11. 10.1186/1297-9716-44-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trueeb BS, Gerber S, Maes D, Gharib WH, Kuhnert P. 2019. Tn-sequencing of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis mutant libraries reveals non-essential genes of porcine mycoplasmas differing in pathogenicity. Vet Res 50:55. 10.1186/s13567-019-0674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith H, Venter JC. 1999. Global transposon mutagenesis and a minimal mycoplasma genome. Science (80-) 286:2165–2286. 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 83.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc Natl Acad Sci USA 103:425–430. 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lluch-Senar M, Delgado J, Chen W, Lloréns-Rico V, O'Reilly FJ, Wodke JA, Unal EB, Yus E, Martínez S, Nichols RJ, Ferrar T, Vivancos A, Schmeisky A, Stülke J, Noort V, Gavin A, Bork P, Serrano L. 2015. Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome-reduced bacterium. Mol Syst Biol 11:780. 10.15252/msb.20145558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hutchison CA, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. 2016. Design and synthesis of a minimal bacterial genome. Science 351:aad6253. 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 86.Baby V, Lachance J-C, Gagnon J, Lucier J-F, Matteau D, Knight T, Rodrigue S. 2018. Inferring the minimal genome of Mesoplasma florum by comparative genomics and transposon mutagenesis. mSystems 3 10.1128/mSystems.00198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tulum I, Yabe M, Uenoyama A, Miyata M. 2014. Localization of P42 and F(1)-ATPase α-subunit homolog of the gliding machinery in Mycoplasma mobile revealed by newly developed gene manipulation and fluorescent protein tagging. J Bacteriol 196:1815–1824. 10.1128/JB.01418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaurivaud P, Laigret F, Garnier M, Bove JM. 2000. Fructose utilization and pathogenicity of Spiroplasma citri: characterization of the fructose operon. Gene 252:61–69. 10.1016/s0378-1119(00)00230-4. [DOI] [PubMed] [Google Scholar]
- 89.Allam AB, Reyes L, Assad-Garcia NG, Glass JI, Brown MB. 2010. Enhancement of targeted homologous recombination in Mycoplasma mycoides subspecies capri by inclusion of heterologous recA. Appl Environ Microbiol 76:6951–6954. 10.1128/AEM.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ishag HZA, Xiong Q, Liu M, Feng Z, Shao G. 2017. E. coli recA gene improves gene targeted homologous recombination in Mycoplasma hyorhinis. J Microbiol Methods 136:49–56. 10.1016/j.mimet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 91.Clampitt JM, Madsen ML, Minion FC. 2021. Construction of Mycoplasma hyopneumoniae P97 null mutants. Front Microbiol 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Evsyutina DV, Fisunov GY, Pobeguts OV, Kovalchuk SI, Govorun VM. 2022. Gene silencing through CRISPR interference in Mycoplasmas. Microorganisms 10:1159. 10.3390/microorganisms10061159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mahdizadeh S, Sansom FM, Lee S-W, Browning GF, Marenda MS. 2020. Targeted mutagenesis of Mycoplasma gallisepticum using its endogenous CRISPR/Cas system. Vet Microbiol 250:108868. 10.1016/j.vetmic.2020.108868. [DOI] [PubMed] [Google Scholar]
- 94.Klose SM, Wawegama N, Sansom FM, Marenda MS, Browning GF. 2022. Efficient disruption of the function of the mnuA nuclease gene using the endogenous CRISPR/Cas system in Mycoplasma gallisepticum. Vet Microbiol 269:109436. 10.1016/j.vetmic.2022.109436. [DOI] [PubMed] [Google Scholar]
- 95.Lapinaite A, Knott GJ, Palumbo CM, Lin-Shiao E, Richter MF, Zhao KT, Beal PA, Liu DR, Doudna JA. 2020. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science 369:566–571. 10.1126/science.abb1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liang Y, Xie J, Zhang Q, Wang X, Gou S, Lin L, Chen T, Ge W, Zhuang Z, Lian M, Chen F, Li N, Ouyang Z, Lai C, Liu X, Li L, Ye Y, Wu H, Wang K, Lai L. 2022. AGBE: a dual deaminase-mediated base editor by fusing CGBE with ABE for creating a saturated mutant population with multiple editing patterns. Nucleic Acids Res 50:5384–5399. 10.1093/nar/gkac353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miller SM, Wang T, Randolph PB, Arbab M, Shen MW, Huang TP, Matuszek Z, Newby GA, Rees HA, Liu DR. 2020. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat Biotechnol 38:471–481. 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O. 2018. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science (80-) 361:1259–1262. 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. 2020. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368:290–296. 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu T, Zeng D, Zheng Z, Lin Z, Xue Y, Li T, Xie X, Ma G, Liu Y-G, Zhu Q. 2021. The ScCas9++ variant expands the CRISPR toolbox for genome editing in plants. J Integr Plant Biol 63:1611–1619. 10.1111/jipb.13164. [DOI] [PubMed] [Google Scholar]
- 101.Veillet F, Kermarrec M-P, Chauvin L, Chauvin J-E, Nogué F. 2020. CRISPR-induced indels and base editing using the Staphylococcus aureus Cas9 in potato. PLoS One 15:e0235942. 10.1371/journal.pone.0235942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tan J, Zhang F, Karcher D, Bock R. 2019. Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat Commun 10:439. 10.1038/s41467-018-08034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Freundt EA. 1983. Culture media for classic Mycoplasmas. Methods in Mycoplasmology. [Google Scholar]
- 104.Algire MA, Lartigue C, Thomas DW, Assad-Garcia N, Glass JI, Merryman C. 2009. New selectable marker for manipulating the simple genomes of Mycoplasma species. Antimicrob Agents Chemother 53:4429–4432. 10.1128/AAC.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.King KW, Dybvig K. 1994. Transformation of Mycoplasma capricolum and examination of DNA restriction modification in M. capricolum and Mycoplasma mycoides subsp. mycoides. Plasmid 31:308–311. 10.1006/plas.1994.1033. [DOI] [PubMed] [Google Scholar]
- 106.Zhu X, Dong Y, Baranowski E, Li X, Zhao G, Hao Z, Zhang H, Chen Y, Hu C, Chen H, Citti C, Guo A. 2020. Mbov_0503 encodes a novel cytoadhesin that facilitates Mycoplasma bovis interaction with tight junctions. Microorganisms 8:164. 10.3390/microorganisms8020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods, gene and plasmid sequences, and Fig. S1 to S9. Download aem.00996-22-s0001.pdf, PDF file, 1.3 MB (1.3MB, pdf)
Table S1. Download aem.00996-22-s0002.xlsx, XLSX file, 0.01 MB (14.4KB, xlsx)
Table S2. Download aem.00996-22-s0003.xlsx, XLSX file, 0.01 MB (10.9KB, xlsx)
Table S3. Download aem.00996-22-s0004.xlsx, XLSX file, 0.03 MB (32.3KB, xlsx)