Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the infectious agent of the COVID-19 pandemic, remains a global medical problem. Angiotensin converting enzyme 2 (ACE2) was identified as the primary viral entry receptor, and transmembrane serine protease 2 (TMPRSS2) primes the spike protein for membrane fusion. However, ACE2 expression is generally low and variable across tissues, suggesting auxiliary receptors facilitate viral entry. Identifying these factors is critical for understanding SARS-Cov-2 pathophysiology and developing new countermeasures. However, profiling host-virus interactomes involves extensive genetic screening or complex computational predictions. Here, we leverage the photocatalytic proximity labeling platform μMap to rapidly profile the spike interactome in human cells and identify 8 novel candidate receptors. We systemically validate their functionality in SARS-CoV-2 pseudoviral uptake assays with both Wuhan and Delta spike variants and show that dual expression of ACE2 with either NRP2, EPHA7, SLC6A15 or MAL2 significantly enhances viral uptake. Collectively, our data show that SARS-CoV-2 synergistically engages several host factors for cell entry and establishes μMap as a powerful tool for rapidly interrogating host-virus interactomes.
Graphical Abstract

INTRODUCTION
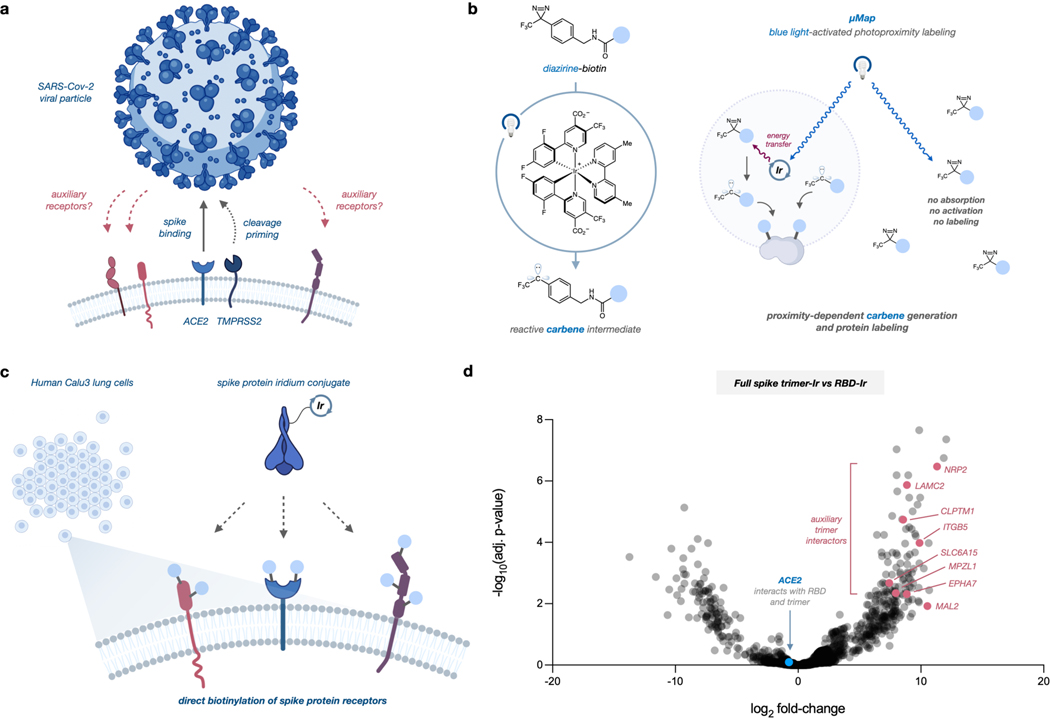
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent of the COVID-19 pandemic. Identifying viral entry and replication factors is key for understanding and resolving outbreaks, and angiotensin converting enzyme 2 (ACE2) has been identified as the obligate receptor for SARS-CoV and SARS-CoV-2.1,2 Cleavage of the viral spike protein by the transmembrane serine protease 2 (TMPRSS2) also facilitates SARS-CoV-2 membrane fusion.3–5 Although ACE2 expression is higher in nasal epithelial cells, these levels decrease throughout the lower respiratory tract.1,2 While this implies nasal initiation of infection, transition mechanisms to deep lung pathogenesis in severe COVID are unknown.6,7 Similarly, COVID infections can rapidly progress throughout the body and cause multiple organ failure,8 yet ACE2 expression is low or variable beyond gastrointestinal and respiratory tracts.9 Accordingly, it is likely that SARS-CoV-2 interacts with additional receptors, although viral entry outside of canonical pathways remains poorly characterized (Figure 1a).
Figure 1.
a) SARS-Cov-2 utilizes ACE2 and TMPRSS2 for cell uptake, but additional entry factors are likely. b) μMap photoproximity labeling via iridium photocatalysts that activate nearby diazirines into reactive carbenes. c) Iridium conjugation with SARS-Cov-2 spike protein enables rapid receptor identification. d) Quantitative proteomics volcano plot of candidate cell-surface proteins after incubation of Ir-spike proteins with Calu-3 cells and photolabeling. Dataset compares full-length spike protein against receptor-binding domain (RBD). ACE2 interacts with both constructs and is thus near zero enrichment.
Computational modeling of the receptor binding domain (RBD) has identified interactions with heparin sulfate for cell entry, and recent work validated functional associations with C-type lectin receptors10 and metabotropic glutamate receptor 2.11 Additionally, results from two studies suggest that neuropilin-1 (NRP1) enhances TMPRSS2-mediated SARS-CoV-2 entry.12,13 While these studies elucidated additional viral entry routes, they also required extensive computational effort generated from crystal structures or validated hypotheses from a priori evidence of known receptors. Genetic knock-out screens can reduce bias and allow identification of proviral proteins,14 yet these campaigns involve extensive library optimization and require suitable systems for transfection and engineering. In contrast, proximity labeling has emerged as a versatile methodology for unbiased interrogation of protein interactions via catalytic tagging of spatially connected biomolecules.15 APEX16 and BioID17 have found particular widespread application and have been recently utilized for investigating associations between SARS-Cov-2 viral components and intracellular host proteins. Although these studies demonstrated relevant interactions between the viral proteins and host signal peptidase complex18 as well as myosin heavy chain,19 these platforms require genetic engineering to introduce proximity labeling components and are limited in both spatial control and labeling resolution. To circumvent these challenges and investigate the native cell-surface interactome with greater precision, we hypothesized that our recently developed photocatalytic proximity labeling method, μMap,20 could directly profile the host-virus microenvironment and identify SARS-CoV-2 auxiliary receptors (Figure 1b,c). In contrast to peroxidase or biotin ligation strategies, μMap uses iridium (Ir) photocatalysts to convert nearby diazirines into carbenes via Dexter energy transfer and biotinylate proteins within ~4 nm radius, offering high spatial and temporal control over labeling (Figure 1b).20 This platform has been applied for small molecule target identification,21 mapping chromatin reorganization events,22 and rapidly profiling immunosynapse interactions.20 Additionally, similar Ir-based photocatalytic proximity labeling strategies have also been recently developed for cataloging mitochondrial proteins in activated macrophages23 and profiling surface proteins in breast cancer.24 Rhodamine-based oxidation labeling has also provided new methodology for validating cell-surface microenvironments, underscoring the utility of light-mediated proximity labeling for interrogating cell-surface proteomes with high spatiotemporal precision.25 Given this precedent, we sought to deploy μMap to identify novel receptors of the SARS-CoV-2 spike protein.
RESULTS AND DISCUSSION
As an initial test, we first synthesized recombinant RBD and full-length spike-Ir conjugates and attempted photolabeling on HEK293T cells overexpressing ACE2 (Figure S1). Cells were incubated with spike-Ir conjugates and then washed, and photolabeling was initiated in a 250 μM biotin-diazirine solution under blue light irradiation. Compared with a free Ir control and parental 293T cells, we observed robust biotinylation of membrane proteins from trimer conjugates only in ACE2-expressing cells (Figure S2). Notably, RBD labeling produced few new bands beyond ACE2 in these tests, yet the spike trimer displayed extensive biotinylation, revealing the possibility of novel interactions.
To perform μMap analysis of the spike interactome in a native context, we identified Calu-3 human lung cells as they express both canonical entry factors ACE2 and TMPRSS2 (Figure S1) and have been previously utilized to study COVID pathophysiology.1,2 We were also particularly interested in utilizing the full-length spike protein (~180 kDa), as the N- terminal domain bears epitopes of neutralizing antibodies, yet its functional interactions are still unknown,26,27 suggesting it binds other host factors. We executed our photolabeling workflow followed by membrane lysate isolation, streptavidin enrichment, and quantitative proteomic analysis. Compared against a free Ir control, ACE2 was strongly enriched (Figure S3, Table S1), recapitulating our initial feasibility test. To delineate auxiliary interactions between the smaller RBD (~50 kDa) and full spike, we also performed μMap with both conjugates and compared datasets. As expected, ACE2 was not enriched as it binds both protein constructs, but we identified 8 enriched membrane proteins as candidate receptors for the full spike (Figure 1d, Table S2). TMPRSS2 was not observed in any of our datasets, which is unexpected given its known role as a canonical SARS-CoV-2 entry factor along with ACE2.3–5 However, it is also known that the TMPRSS2-Spike interaction is transient after cleavage, and given that μMap targeting of cell-surface proteins employs binding and several washing steps, we hypothesize that this contributed to lack of proteomic detection. Regardless, neuropilin-2 (NRP2) was highly enriched, and has high homology with NRP1 which was reported to bind to the cleaved spike and facilitate SARS-CoV-2 entry.12,13 Cleft Lip And Palate Associated Transmembrane Protein 1 (CLPTM1) and Ephrin Receptor A7 (EPHA7) were also prioritized, the latter of which was reported as key for transmission of Kaposi’s sarcoma-associated herpes virus and rhesus monkey rhadinovirus28. We also identified Myelin and Lymphocyte Protein 2 (MAL2), recently reported as a herpes simplex virus 1 co-factor in oligodendrocytes.29 Solute Carrier Family 6 Member 15 (SLC6A15), Myelin protein zero-like protein 1 (MPZL1), Laminin Subunit Gamma 2 (LAMC2), and Integrin Subunit Beta 5 (ITGB5) were also noted as candidate receptors (Figure 1d, Table S2). LAMC2 was intriguingly identified in a microarray study as a possible SARS-CoV-2 transmission factor.30 While these 8 annotated candidate proteins are not the sole transmembrane proteins enriched in our dataset (Figure 1d, Table S2), this group was selected due to their high enrichment and likelihood of interacting with the SARS-CoV-2 spike protein on the cell surface. Importantly, all candidates are known to be expressed in esophageal and respiratory tract tissues,31 and so we next sought to functionally test these receptors for viral infection.
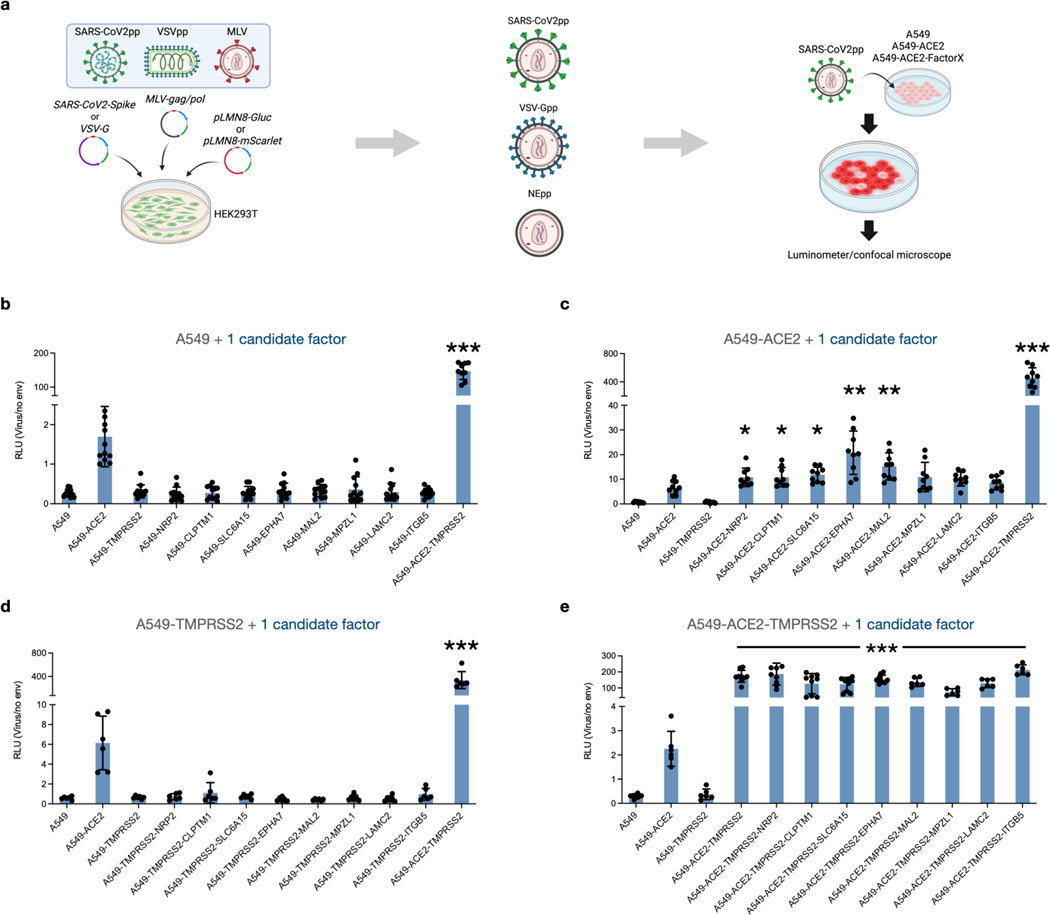
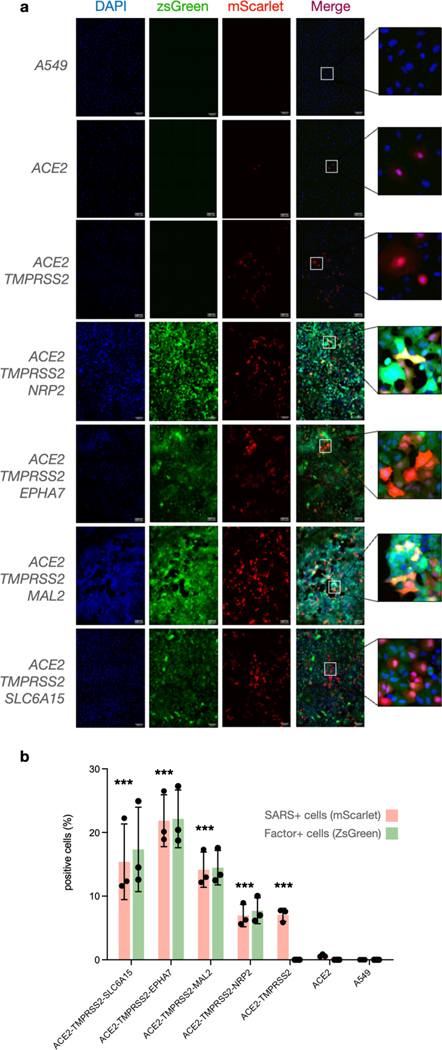
We first generated SARS-CoV-2 pseudovirus particles (SARS-CoV-2pp) encoding Gaussia luciferase as a quantifiable proxy (Figure 2a). In line with previous observations, expression of ACE2 or TMPRSS2 in HEK293T and A549 cells resulted in significant entry enhancement (Figure S4). We next constructed a panel of A549 cell lines to display various combinations of cell-surface candidate factors. Importantly, we transduced cells with bicistronic lentiviral cassettes containing a zsGreen reporter to enable fluorescence-activated cell sorting and standardized expression of each markers, and ACE2 levels were similarly gated using antibody staining (Figure S5). For additional rigor, we also measured expression of respective genes in sorted cell lines via quantitative PCR and verified similar mRNA levels (Figure S6). We next infected these lines with SARS-Cov-2pp, and while A549 cells expressing ACE2 and/or TMPRSS2 exhibited significant increases in viral uptake compared to parental cells, display of individual candidate factors provided no viral uptake enhancement (Figure 2b). However, cells stably expressing ACE2 along with individual factors exhibited significantly enhanced viral entry (Figure 2c). In particular, dual expression of ACE2 with NRP2, CLPTM1, SLC6A15, EPHA7 or MAL2 led to upwards of ~3-fold increases in viral uptake compared with cells only expressing ACE2, suggesting a synergistic cell entry mechanism. As expected, TMPRSS2 conferred large uptake increases when expressed with ACE2 (Figure 2b–e), yet no difference was observed in lines expressing TMPRSS2 and each factor (Figure 2d) or in combination with ACE2 and TMPRSS2 (Figure 2e). We hypothesized that these differences may be masked by enzymatic luciferase-based detection, so to observe uptake in greater detail, we infected ACE2-TMPRSS2 triple expression lines displaying the highest enhancement (NRP2, SLC6A15, EPHA7 and MAL2) with mScarlet reporter pseudoparticles (Figure 3). Quantifying individual populations via microscopy confirmed minimal entry in A549 cells as well as lines expressing only ACE2, but large enhancement was seen in ACE2 lines simultaneously displaying TMPRSS2 along with NRP2, MAL2, EPHA7 or SLC6A15. Non-enveloped viral controls showed no uptake (Figure S7), reflecting spike-mediated entry (Figure 3a, b). Together, these results confirm the importance of ACE2 and TMPRSS2 for SARS-Cov-2 entry, and we excitingly demonstrate that NRP2, SLC6A15, EPHA7 and MAL2 significantly enhance viral uptake when expressed with ACE2.
Figure 2. SARS-CoV-2 uptake is enhanced in cells expressing candidate receptors and ACE2.
a) Schematic for generating pseudoviral particles and infection. b-e) Virus uptake into A549 cells expressing ACE2, TMPRSS2, and/or entry factor candidates. Entry was measured as relative luminescent signal (RLU) normalized against non-enveloped particle control. Datapoints represent mean (n = 6) and error bars denote standard deviation. Independent t-tests between ACE2 and expression lines are indicated * p < 0.05, ** p < 0.01., ***, p < 0.001.
Figure 3. NRP2, MAL2, EPHA7, and SLC6A15 significantly enhance SARS-Cov-2 entry.
a) A549 cells expressing various factors were infected with SARS-CoV-2pp encoding mScarlet, stained with DAPI (blue), and visualized using fluorescence microscopy for factor expression (szGreen) and viral uptake (red). Scale bar indicates 100 μm, and insets display zoomed-in regions of each image. b) Entry quantification as percentage of red: (SARS positive)/(DAPI positive), green: percentage of (SARS positive)/(Factor positive). Datapoints represent mean from (n = 3), and error bars denote standard deviation. Independent t-tests of SARS+ values between ACE2 and ACE2–TMPRSS2 triple expression lines are indicated as significant (***, p < 0.001)
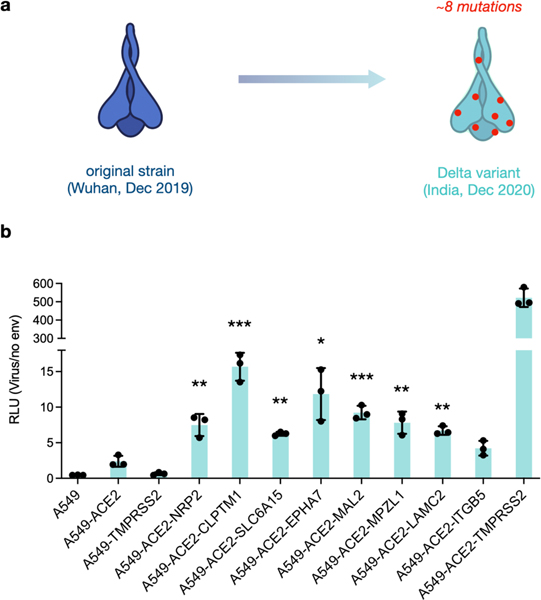
Numerous SARS-CoV-2 variants have continued to emerge that are increasingly transmissible and/or less vulnerable to antibody neutralization. In particular, the Delta (B.1.617.2) variant that arose in India in late 2020 contains 8 amino acid mutations from the original Wuhan spike (Figure 4a).32 To test uptake with our identified receptors, we generated Delta SARS-CoV2-pp and repeated our luminescent entry assay and interestingly observed significant entry enhancement across all factors except for ITGB5, with upwards of ~5–7-fold increases in ACE2-CLPTM1 and ACE2-EPHA7 lines vs cells only expressing ACE2 (Figure 4b). Conversely, the Wuhan strain only exhibited increased uptake in cell lines expressing both ACE2 in combination with NRP2, MAL2, EPHA7 or SLC6A15 (Figure 2c), suggesting the higher transmissibility of the Delta variant is due in part to broader receptor tropism.
Figure 4. Candidate receptors enhance Delta variant spike-mediated uptake.
a) Delta variant spike harbors 8 mutations compared to Wuhan strain. b) Delta pseudovirus uptake in cells expressing ACE2 and candidate factors. Entry was measured as relative luminescent signal (RLU) normalized against non-enveloped particle control. Datapoints represent mean (n = 3) and error bars denote standard deviation. Independent t-tests between ACE2 and respective lines are indicated as significant * p < 0.05, ** p < 0.01, ***, p < 0.001.
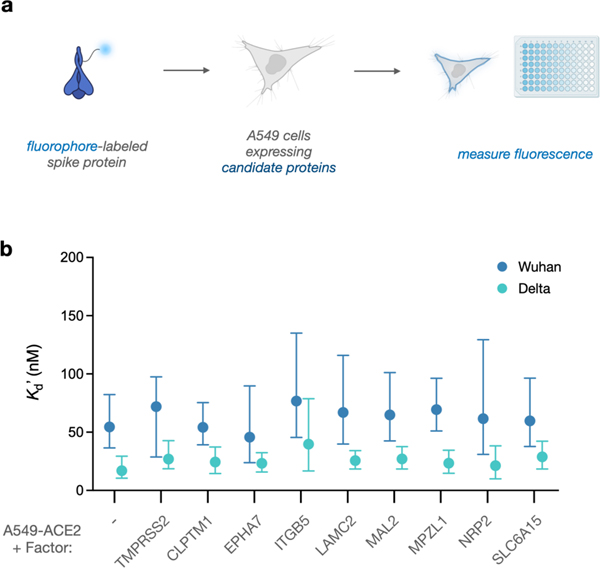
Lastly, we sought to probe whether candidate factors stably interact with SARS-Cov-2. Fluorophore-spike conjugates were generated, and each cell line was incubated with increasing amounts of the conjugate. After washing, fluorescence was measured to estimate bound protein remaining (Figure 5a). As expected, parental A549 cells displayed minimal interaction while cells expressing ACE2 exhibited robust binding with low nM affinity (Figures S8, 5b). Intriguingly, the Delta spike variant exhibited significantly enhanced binding affinity in all ACE2-expressing lines, in agreement with working hypotheses regarding its higher transmissibility (Figure S9, 5b).33 However, cell lines expressing each candidate displayed minimal binding, and dual expression with ACE2 did not increase affinity compared with single expression of ACE2 in either variant. Although we anticipated observing a binding enhancement, not all entry factors stably interact with the spike protein. In agreement with prior work, TMPRSS2-expressing cells did not stably bind (Figures S8, S9), despite the significant impact of the protease on viral uptake.1Similarly, NRP1 was found to enhance cell entry, yet binding interactions with SARS-Cov-2 were undetectable in analogous assays.12 While the candidate factors identified in this study functionally enhance uptake, our results suggest that interactions with the spike protein are transient, and synergistic associations between ACE2 and a receptor ensemble likely contribute to spike binding and subsequent particle entry.
Figure 5. Candidate receptors do not enhance SARS-Cov-2 spike binding on human cells.
a) Schematic for testing binding affinity of spike protein and candidate factors. b) Apparent dissociation constants (K’d) for SARS-Cov-2 spike from Wuhan (blue) and Delta (green) strains towards cells expressing ACE2 and various cell-surface proteins. Datapoints represent mean (n = 3), and error bars denote 95% confidence intervals.
Understanding the molecular mechanisms of SARS-Cov-2 infection across human tissues is key to resolving the current pandemic and developing countermeasures for future outbreaks. Here we show that photocatalytic proximity labeling using μMap is a rapid and powerful tool for interrogating critical receptors at the host-virus interface. We profile the Wuhan spike interactome and identify at least four proteins, NRP2, MAL2, EPHA7 and SLC6A15, as auxiliary SARS-Cov-2 entry receptors that significantly enhance uptake of both the original and Delta SARS-Cov-2 variant in human cells. The Omicron spike variant has now dominated global infections, and the molecular mechanisms of its high transmissibility and immune evasion remain poorly understood.34,35 Ongoing research in our group seeks to expand this work into emerging variants across human cell types to delineate these complex interactions. Additionally, we observed many intracellular proteins that were enriched in our datasets (Fig 1d, Tables S1, S2) that potentially interact with the SARS-CoV-2 spike protein. We hypothesize that these interactions are potentially resulting from internalized spike-Ir conjugates, and future efforts will validate and functionally investigate these potentially critical interactions. Moreover, we and the Rovis group have also recently reported red light-based strategies for photocatalyic proximity labeling,36, 37 and future efforts will deploy these platforms for studying host-virus interactions in complex in vivo environments. Together, this work demonstrates a powerful and generalizable methodology for rapidly elucidating entry mechanisms for a variety of pathogens in diverse cellular settings.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by kind gifts from Merck, BMS, Pfizer, Janssen, Genentech, and Eli Lilly. We also acknowledge the Princeton Catalysis Initiative for supporting this work. The authors thank Saw Kyin and Henry H. Shwe at the Princeton Proteomics Facility. We also thank Mohsan Saeed (Boston University) for providing A549-ACE2/TMPRSS2 cells and pLOC-ACE2-PuroR and SARS-CoV2-S_B.1.1.529_codon optimized plasmid and Celeste Nelson for providing the Calu-3, BEAS-2B and NuLi-1 cell lines. We are grateful to Christina DeCoste and Katherine Rittenbach of the Flow Cytometry Resource Facility at Princeton University for their assistance in the planning and execution of all flow cytometry experiments. We would also like to thank Gary S. Laevsky and Sha Wang of the Confocal Imaging Facility, a Nikon Center of Excellence, in the Department of Molecular Biology at Princeton University for instrument use and technical advice.
Funding Sources
This study was funded in part by grants from the National Institutes of Health (R01AI138797, R01AI107301, R01AI146917, R01AI153236 all to A.P.), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (101539 A.P.), Princeton COVID-19 research funds through the Office of the Dean for Research and a component of the National Institutes of Health (NIH) under award number UL1TR003017 (to A.P.). This work was also funded by the NIH National Institute of General Medical Sciences (R35-GM134897-02). S.D.K acknowledges the NIH for a postdoctoral fellowship (1F32GM142206-01). The Molecular Biology Flow Cytometry Resource Facility is partially supported by the Cancer Institute of New Jersey Cancer Center Support grant (P30CA072720). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Saori Suzuki‡ − Department of Molecular Biology, Princeton University, Princeton, New Jersey 08544, United States
Jacob B. Geri‡ − Merck Center for Catalysis at Princeton University, Princeton, New Jersey 08544, United States; Department of Chemistry, Princeton University, Princeton, New Jersey 08544, United States
Steve D. Knutson‡ − Merck Center for Catalysis at Princeton University, Princeton, New Jersey 08544, United States; Department of Chemistry, Princeton University, Princeton, New Jersey 08544, United States
Harris Bell-Temin – Bristol Meyers Squibb, Princeton, New Jersey 08544, United States
Tomokazu Tamura − Department of Molecular Biology, Princeton University, Princeton, New Jersey 08544, United States
David F. Fernández − Merck Center for Catalysis at Princeton University, Princeton, New Jersey 08544, United States; Department of Chemistry, Princeton University, Princeton, New Jersey 08544, United States
Gabby H. Lovett − Merck Center for Catalysis at Princeton University, Princeton, New Jersey 08544, United States; Department of Chemistry, Princeton University, Princeton, New Jersey 08544, United States
Nicholas A. Till − Merck Center for Catalysis at Princeton University, Princeton, New Jersey 08544, United States; Department of Chemistry, Princeton University, Princeton, New Jersey 08544, United States
Brigitte L. Heller − Department of Molecular Biology, Princeton University, Princeton, New Jersey 08544, United States
Jinchao Guo − Department of Molecular Biology, Princeton University, Princeton, New Jersey 08544, United States
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Proteomic datasets (XLSX)
Experimental procedures and supplementary figures (PDF)
Figure artwork was created in part using biorender.com.
REFERENCES
- 1.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181 (2), 271–280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426 (6965), 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y. & Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85 (9), 4122–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M. & Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010, 84 (24), 12658–12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S. & Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011, 85 (2), 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH 3rd, Kato T, Lee RE, Yount BL, Mascenik TM et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182 (2), 429–446.e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn JH, Kim J, Hong SP, Choi SY, Yang MJ, Ju YS, Kim YT, Kim HM, Rahman MDT, Chung MK et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Invest. 2021, 131 (13), e148517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delorey TM, Ziegler CG, Heimberg G, Normand R, Yang Y, Segerstolpe Å, Abbondanza D, Fleming SJ, Subramanian A. & Montoro DT COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595 (7865), 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M. & Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol 2020, 16 (7), e9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thépaut M, Luczkowiak J, Vivès C, Labiod N, Bally I, Lasala F, Grimoire Y, Fenel D, Sattin S. & Thielens N. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2021, 17 (5), e1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Yang G, Wang X, Wen Z, Shuai L, Luo J, Wang C, Sun Z, Liu R. & Ge J. SARS-CoV-2 uses metabotropic glutamate receptor subtype 2 as an internalization factor to infect cells. Cell Discov. 2021, 7 (1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer M, Hollandi R. et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370 (6518), 861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370 (6518), 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baggen J, Persoons L, Vanstreels E, Jansen S, Van Looveren D, Boeckx B, Geudens V, De Man J, Jochmans D. & Wauters J. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021, 53 (4), 435–444. [DOI] [PubMed] [Google Scholar]
- 15.Qin W, Cho KF, Cavanagh PE & Ting AY Deciphering molecular interactions by proximity labeling. Nat. Methods 2021, 18 (2), 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK & Ting AY Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2015, 12 (1), 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux KJ, Kim DI, Raida M. & Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196 (6), 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Wang C, Feng X, Nie L, Tang M, Zhang H, Xiong Y, Swisher SK, Srivastava M. & Chen J. Interactomes of SARSCoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. EMBO J. 2021, 40 (17), e107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Fan J, Chen Z, Zhang M, Peng H, Liu J, Ding L, Liu M, Zhao C. & Zhao P. Nonmuscle myosin heavy chain IIA facilitates SARS-CoV-2 infection in human pulmonary cells. Proc. Natl. Acad. Sci. 2021, 118 (50), e2111011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geri JB, Oakley JV, Reyes-Robles T, Wang T, McCarver SJ, White CH, Rodriguez-Rivera FP, Parker DL Jr, Hett EC & Fadeyi OO Microenvironment mapping via Dexter energy transfer on immune cells. Science 2020, 367 (6482), 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trowbridge AD, Seath CP, Rodriguez-Rivera FP, Li BX, Dul BE, Schwaid AG, Geri JB, Oakley JV, Fadeyi OO & Oslund RC Small molecule photocatalysis enables drug target identification via energy transfer. bioRxiv 2021. doi. 10.1101/2021.08.02.454797, (accessed 08-01-2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seath CP, Burton AJ, MacMillan DW & Muir TW Tracking chromatin state changes using μMap photo-proximity labeling. bioRxiv 2021. doi. 10.1101/2021.09.28.462236, (accessed 08-01-2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z, Liu Z, Xie X, Zeng R, Chen Z, Kong L, Fan X. & Chen PR Bioorthogonal photocatalytic Decaging-enabled mitochondrial proteomics. J. Am. Chem. Soc. 2021, 143 (44), 18714–18720. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Xie X, Huang Z, Lin F, Liu S, Chen Z, Qin S, Fan X. & Chen PR Spatially resolved cell tagging and surfaceome labeling via targeted photocatalytic decaging. Chem 2022, 8 (8), 2179–2191. [Google Scholar]
- 25.Müller M, Gräbnitz F, Barandun N, Shen Y, Wendt F, Steiner SN, Severin Y, Vetterli SU, Mondal M. & Prudent JR Light-mediated discovery of surfaceome nanoscale organization and intercellular receptor interaction networks. Nat. Commun. 2021, 12 (1), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JF, Sahi V, Figueroa A. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584 (7821), 450–456. [DOI] [PubMed] [Google Scholar]
- 27.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y. et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369 (6504), 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groβkopf AK, Schlagowski S, Hörnich BF, Fricke T, Desrosiers RC & Hahn AS EphA7 Functions as Receptor on BJAB Cells for Cell-to-Cell Transmission of the Kaposi’s Sarcoma-Associated Herpesvirus and for Cell-Free Infection by the Related Rhesus Monkey Rhadinovirus. J. Virol. 2019, 93 (15). e00064–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Guerrero JA, de la Nuez C, Praena B, Sánchez-León E, Krummenacher C. & Bello-Morales R. Herpes Simplex Virus 1 Spread in Oligodendrocytic Cells Is Highly Dependent on MAL Proteolipid. J Virol 2020, 94 (4). e01739–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu A, Sarkar A. & Maulik U. Study of cell to cell transmission of SARS CoV2 virus particle using gene network from microarray data. bioRxiv 2020. doi. 10.1101/2020.05.26.116780, (accessed 08-01-2022). [DOI] [Google Scholar]
- 31.Consortium G. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015, 348 (6235), 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, Widen SG, An Z, Weaver SC & Menachery VD Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022, 39 (7), 110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Zhou L, Mo M, Liu T, Wu C, Gong C, Lu K, Gong L, Zhu W. & Xu Z. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Target. Ther. 2022, 7 (1), 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hui KP, Ho JC, Cheung M. c., Ng K. c., Ching RH, Lai K. l., Kam TT, Gu H, Sit K-Y & Hsin MK SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603 (7902), 715–720. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Iketani S, Guo Y, Chan JF-W, Wang M, Liu L, Luo Y, Chu H, Huang Y. & Nair MS Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022, 602 (7898), 676–681. [DOI] [PubMed] [Google Scholar]
- 36.Buksh BF, Knutson SD, Oakley JV, Bissonnette NB, Oblinsky DG, Schwoerer MP, Seath CP, Geri JB, Rodriguez-Rivera FP & Parker DL μMap-Red: Proximity Labeling by Red Light Photocatalysis. Journal of the American Chemical Society 2022, 144 (14), 6154–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tay N; Ryu KA; Weber J; Olow A; Reichman D; Oslund R; Fadeyi O; Rovis T. Targeted Activation in Localized Protein Environments via Deep Red Photoredox Catalysis. ChemRxiv 2021. 10.26434/chemrxiv-2021-x9bjv, (accessed 08-01-2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.