Abstract
Objective:
The standard twelve-lead electrocardiogram (ECG) is a widely used tool for monitoring cardiac function and diagnosing cardiac disorders. The development of smaller, lower-cost, and easier-to-use ECG devices may improve access to cardiac care in lower-resource environments, but the diagnostic potential of these devices is unclear. This work explores these issues through a public competition: the 2021 PhysioNet Challenge. In addition, we explore the potential for performance boosting through a meta-learning approach.
Approach:
We sourced 131,149 twelve-lead ECG recordings from ten international sources. We posted 88,253 annotated recordings as public training data and withheld the remaining recordings as hidden validation and test data. We challenged teams to submit containerized, open-source algorithms for diagnosing cardiac abnormalities using various ECG lead combinations, including the code for training their algorithms. We designed and scored algorithms using an evaluation metric that captures the risks of different misdiagnoses for 30 conditions. After the Challenge, we implemented a semi-consensus voting model on all working algorithms.
Main results:
A total of 68 teams submitted 1,056 algorithms during the Challenge, providing a variety of automated approaches from both academia and industry. The performance differences across the different lead combinations were smaller than the performance differences across the different test databases, showing that generalizability posed a larger challenge to the algorithms than the choice of ECG leads. A voting model improved performance by 3.5%.
Significance:
The use of different ECG lead combinations allowed us to assess the diagnostic potential of reduced-lead ECG recordings, and the use of different data sources allowed us to assess the generalizability of algorithms to diverse institutions and populations. The submission of working, open-source code for both training and testing and the use of a novel evaluation metric improved the reproducibility, generalizability, and applicability of the research conducted during the Challenge.
1. Introduction
Cardiovascular diseases are the leading cause of death worldwide [1]. Early treatment of cardiovascular disease can prevent serious cardiac events and improve outcomes, and the electrocardiogram (ECG) is a critical screening tool for a range of cardiac abnormalities, such as atrial fibrillation and ventricular hypertrophy [2], [3]. Moreover, recent advances in ECG technologies have allowed the development of smaller, lower-cost, and easier-to-use devices with the potential to improve access to cardiac screening and diagnoses in low-resource environments. However, due to the large variety of potential diagnoses, manual interpretation of ECGs is a time-consuming task that requires highly skilled and well trained personnel [4], [5].
Automatic detection and classification of cardiac abnormalities from ECGs can assist clinicians and ECG technicians, especially in the context of the ever-increasing number of recorded ECGs. Recent progress in machine learning techniques combined with standard clinical or handcrafted features has led to the development of algorithms that may identify cardiac abnormalities [6]-[10]. However, most published methods have only been developed in or tested on a single populations and/or relatively small and relatively homogeneous populations. Moreover, few publications provide software that allows for redeveloping and evaluating these models, so the work is often not scientifically repeatable or extendable. Additionally, most algorithms focus on identifying a limited number of cardiac issues that do not represent the complexity and difficulty of pathologies present in the ECG, and the reported performance for these cardiac issues does not reflect the cost of misclassification in a multi-class classification problem, where most outcomes have very different burdens on the individual. Finally, most publications that focus on automated approaches do not consider different lead combinations. The wide diagnostic potential of more accessible devices that use subsets of the standard twelve leads is largely unknown [11]-[13].
The 2021 George B. Moody PhysioNet Challenge (formerly the PhysioNet/Computing in Cardiology Challenge) provided an opportunity to address these issues by inviting teams to develop algorithms for diagnosing 30 cardiac abnormalities from various twelve-lead, six-lead, four-lead, three-lead, and two-lead ECG recordings [14]-[16]. For more than two decades, through the PhysioNet Challenges, we have invited participants from academia and industry to address clinically important questions that have not been adequately addressed. Similarly to previous years, the Challenge ran over the course of nine months to allow for the development and refinement of the Challenge objective, data, and evaluation metric. Previous Challenges have addressed arrhythmia detection from ECGs: the 2017 Challenge considered the identification of atrial fibrillation in single-lead ECG recordings, and the 2020 Challenge considered the identification of 27 cardiac abnormalities from twelve-lead ECG recordings [14], [17]. However, this was the first Challenge that explored the diagnostic potential of reduced-lead ECGs for a variety of diagnoses [15], [16]. In particular, we have built on the data and scoring metric from the 2020 Challenge, doubled the number of ECG recordings, introduced new data sources and diagnoses, and, importantly, considered subsets of the standard twelve-lead ECG.
For the 2021 Challenge, we sourced 131,149 twelve-lead ECG recordings from ten databases from around the world; we shared two-thirds of the recordings as public training data and retained one-third of the recordings as hidden validation and test data, including recordings from two completely hidden databases. We designed an evaluation metric to capture the risks of different outcomes and misdiagnoses for 30 diagnoses, and we used it to evaluate the submitted algorithms on twelve-lead, six-lead, four-lead, three-lead, and two-lead versions of the ECG recordings. We required the teams to submit containerized, open-source code for training and testing their algorithms to ensure full scientific reproducibility. This article expands upon [15], which first described the 2021 Challenge, to provide a fuller description of the Challenge motivation, data, scoring metric, and algorithms, including a naïve voting model that leverages the strengths of the algorithms to achieve higher performance than the individual algorithms as well as follow-up algorithms to the Challenge.
2. Methods
2.1. Data
For the 2021 Challenge, we sourced data from several countries across three continents. Each database contained electrocardiogram (ECG) recordings with diagnoses and basic demographic information. We use twelve-lead ECG recordings for the public training data and twelve-lead, six-lead, four-lead, three-lead, and two-lead versions of ECG recordings for the hidden validation and test data.
2.1.1. Challenge Data Sources
We created ten databases for the 2021 Challenge [15]. Tables 1 and 2 describe the sources and splits (training, validation, and test) of these data. We publicly released the training data and the clinical ECG diagnoses for the training data, but we kept the validation and test data hidden to allow us to assess algorithmic generalizability and other common machine learning issues. For sources represented in multiple splits, the training, validation, and test data were matched as closely as possible to preserve the distributions of age, sex, and diagnoses. When there are multiple recordings from the same individual, these recordings belong to only one of the training, validation, or test sets.
Table 1:
Sources, locations, and references for each database in the Challenge.
| Database | Source | Locations(s) | Reference |
|---|---|---|---|
| Chapman-Shaoxing | Shaoxing People’s Hospital | Shaoxing, Zhejiang, China | [18] |
| CPSC | CPSC 2018 | Various Locations, China | [19] |
| CPSC-Extra | CPSC 2018 | Various Locations, China | [19] |
| G12EC | Emory University Hospital | Atlanta, Georgia, USA | [14] |
| INCART | St. Petersburg Institute of Cardiological Technics | St. Petersburg, Russia | [20] |
| Ningbo | Ningbo First Hospital | Ningbo, Zhejiang, China | [21] |
| PTB | University Clinic Benjamin Franklin | Berlin, Germany | [22] |
| PTB-XL | Physkalisch Technische Bundesanstalt | Various Countries, Europe | [23] |
| UMich | University of Michigan | Ann Arbor, Michigan, USA | [15] |
| Undisclosed | N/A | USA | [14] |
Table 2:
Numbers of patients and recordings in the training, validation, and test splits of the databases in the Challenge. The numbers of patients for the completely hidden test sets are not given.
| Database | Total Patients |
Training Set Recordings |
Validation Set Recordings |
Test Set Recordings |
Total Recordings |
|---|---|---|---|---|---|
| Chapman-Shaoxing | 10247 | 10247 | 0 | 0 | 10247 |
| CPSC | Unknown | 6877 | 1463 | 1463 | 9803 |
| CPSC-Extra | Unknown | 3453 | 0 | 0 | 3453 |
| G12EC | 15738 | 10344 | 5167 | 5161 | 20672 |
| INCART | 32 | 74 | 0 | 0 | 74 |
| Ningbo | 34905 | 34905 | 0 | 0 | 34905 |
| PTB | 262 | 516 | 0 | 0 | 516 |
| PTB-XL | 18885 | 21837 | 0 | 0 | 21837 |
| UMich | N/A | 0 | 0 | 19642 | 19642 |
| Undisclosed | N/A | 0 | 0 | 10000 | 10000 |
| Total | N/A | 88253 | 6630 | 36266 | 131149 |
Chapman-Shaoxing. The Chapman-Shaoxing database is derived from the database in [18]. We posted this database as training data.
CPSC. The CPSC database is derived from the China Physiological Signal Challenge 2018 (CPSC 2018), held during the 7th International Conference on Biomedical Engineering and Biotechnology in Nanjing, China [19]. We posted the training data from CPSC 2018 as training data, and we split the test data from CPSC 2018 into validation and test data.
CPSC-Extra. The CPSC-Extra database contains unused data from CPSC 2018 [19]. We posted this database as training data.
G12EC. The G12EC Database is a new database representing a large population from the Southeastern United States. We split this database into training, validation, and test data.
INCART. The INCART database is derived from the St. Petersburg Institute of Cardiological Technics (INCART) twelve-lead Arrythmias Database [20]. We posted this database as training data.
Ningbo. The Ningbo database is derived from the database in [21]. We posted this database as training data.
PTB. The PTB database is derived from the Physikalisch-Technische Bundesanstalt (PTB) Database [22]. We posted this database as training data.
PTB-XL. The PTB-XL database is derived from the Physikalisch-Technische Bundesanstalt XL (PTB-XL) Database [23]. We posted this database as training data.
UMich. The UMich database is a database from the University of Michigan‡. We used this database as test data.
Undisclosed. The Undisclosed database is a new database from an undisclosed American institution that is geographically distinct from the sources for the other databases. This database has never been publicly released, and it may never be publicly released. We used this database as test data.
2.1.2. Challenge Data Variables
Each ECG recording was acquired in a hospital or clinical setting and included signal data, basic demographics data, and clinical diagnoses. The specifics of the data acquisition processes depended on the source of the databases and could vary from institution to institution. We have provided a summary of the clinical variables, and we encourage the readers to check the original publications for the details of each database and to cite them directly when used in their research.
We shared the full twelve-lead ECG signal data with the public training data, and we used twelve-lead, six-lead, four-lead, three-lead, and two-lead versions of the signal data for the hidden validation and test data. Table 3 summarizes the lead combinations included in the different versions of the hidden data. The choices of lead combination were made to test the notion of over-completeness. That is, the twelve-lead ECG can be approximated by a three-dimensional (time-varying) dipole [24], to produce a wide variety of morphologies, rhythms and pathologies (such as premature ventricular contractions, ST-elevation and long-QT) [25]-[29]. The dipole model has also been used to effectively generate, filter, measure, and classify the ECG [26], [27], [29]-[32].
Table 3:
Lead combinations used for the hidden validation and test sets in the 2021 Challenge.
| Number of Leads |
Number of Independent Leads |
Lead Combination |
|---|---|---|
| 12 | 8 | I, II, III, aVR, aVL, aVF, V1, V2, V3, V4, V5, V6 |
| 4 | 3 | I, II, III, V2 |
| 3 | 3 | I, II, V2 |
| 6 | 2 | I, II, III, aVR, aVL, aVF |
| 2 | 2 | I, II |
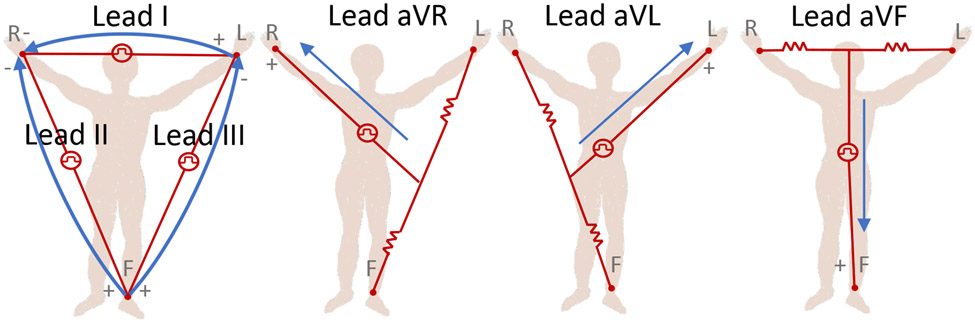
The twelve-lead ECG represents a spatial over-sampling of this dipole model, and so, one might conclude that only three orthogonal leads are required to capture all surface cardiac activity. However, the heart is not a point source dipole, and motion, physical distortion, and near-field electromagnetic effects come into play. It is well-known that precordial leads ‘image’ the ventricles far better than the limb leads, for example. For this Challenge, we asked ‘will two do?’ In other words, we wanted to know if two leads (I and II) would allow researchers to do as well as twelve leads, at least for the diagnoses being evaluated in this Challenge. However, we added three extra categories, all of which are approximately equivalent to the same two leads, in order to test whether any subtle extra information was buried in these signals, or if the addition of more leads, which would necessitate more parameters in a machine learning model, would lead to overfitting during training. We note that the addition of lead III should add no extra information, since it is part of ‘Einthoven’s Triangle’ (lead I + lead III = lead II). In addition, the augmented leads (aVR, aVL and AVF), which are unipolar (in contrast to the bipolar limb leads) while providing different ‘viewpoints’, theoretically do not add any new information since they are also formed by recording the potential difference at the right arm, left arm and left leg. There are two reasons for including these apparently redundant leads. First, they provide clinicians with a deeper intuition, which may be a reflection of the varying amplitude resolution on each lead when the vectors are represented in a low-resolution format (e.g., paper, or on-screen). Second, the vectors are referenced to different grounds. There is an assumption that there is a stable voltage reference (with negligible variation during the cardiac cycle), known as the ‘Wilson Central Terminal’ (WCT), which is obtained by averaging the three active limb electrode voltages measured with respect to the return ground electrode. In the case of the augmented leads, the reference is provided by averaging limb electrodes (‘Goldberger’s Central Terminal’); see Figure 1. However, concerns have been raised by researchers about the ambiguous value and behavior of this reference voltage, which may lead to misdiagnoses or biases in certain circumstances [33]-[37]. Notably, Bacharova et al. [33] found significant diagnostic differences based on the reference in the case of ischemia. See Jin et al. [38] for further discussion on this point.
Figure 1:
Illustrations of the three Einthoven limb leads (I, II, and III; left-most figure) and three circuits of the three Goldberger augmented leads (aVR, aVL, and aVF; three right-most figures). This figure is recreated from [39].
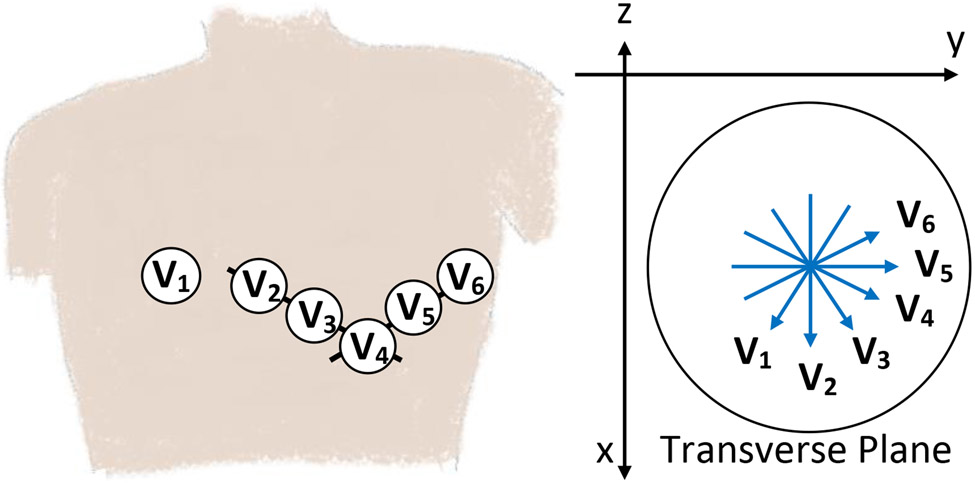
The precordial leads provide information in the transverse plane, in addition to the frontal plane information provided by the limb leads; see Figure 2. We chose to swap the augmented leads for lead V2 to provide information on the ventricular septum and anterior wall. For the two-lead case, we removed the precordial lead to stick with the most common configurations seen in ambulatory ECG monitoring. In summary, the full list of lead combinations are given in Table 3, where 12 leads provides the maximum information available in the recordings (and is the same as the 2020 Challenge [14]). The four-lead and four-lead combinations should be equivalent and provide the next largest amount of information (limb and precordial). Finally, the six-lead and two-lead combinations should be equivalent to each other and provide equivalent (and the least) information. Figure S1 illustrates the equivalence of the different lead combinations considered in the Challenge.
Figure 2:
Illustration of the electrode locations for the six precordial leads, V1 to V6, on a human torso (left figure) and projections of these lead vectors centrally located on a transverse/horizontal plane (right figure). This figure is recreated from [39].
The sampling frequency of the signals varied from 250 Hz to 1 kHz, and the duration of the signals ranged from from 5 seconds to 30 minutes. The age and sex of the subjects were provided with most recordings. Table 4 provides a summary of the age, sex, and recording information for the Challenge databases, including splits of the CPSC and G12EC databases between the training, validation, and test sets.
Table 4:
Number of recordings, mean duration of recordings, mean age of patients in recordings, sex of patients in recordings, and sample frequency of recordings for each database in the Challenge. The CPSC and G12EC databases were represented in the training, validation, and test data and include summary statistics for the entire database and splits of the database.
| Database | Number of Recordings |
Sampling Frequency (Hz) |
Mean Duration (seconds) |
Mean Age (years) |
Sex (female/male) |
|---|---|---|---|---|---|
| Chapman-Shaoxing | 10247 | 500 | 10.0 | 60.1 | 44%/56% |
| CPSC | 9803 | 500 | 16.4 | 60.0 | 47%/53% |
| - CPSC training | 6877 | 500 | 15.9 | 60.2 | 46%/54% |
| - CPSC validation | 1463 | 500 | 17.2 | 58.9 | 49%/51% |
| - CPSC test | 1463 | 500 | 17.5 | 60.0 | 47%/53% |
| CPSC-Extra | 3453 | 500 | 15.9 | 63.7 | 47%/53% |
| G12EC | 20672 | 500 | 10.0 | 60.5 | 46%/54% |
| - G12EC training | 10344 | 500 | 10.0 | 60.5 | 46%/54% |
| - G12EC validation | 5167 | 500 | 10.0 | 60.3 | 47%/53% |
| - G12EC test | 5161 | 500 | 10.0 | 60.7 | 46%/54% |
| INCART | 74 | 257 | 1800.0 | 56.0 | 46%/54% |
| Ningbo | 34905 | 500 | 10.0 | 57.7 | 43%/56% |
| PTB | 516 | 1000 | 110.8 | 56.3 | 27%/73% |
| PTB-XL | 21837 | 500 | 10.0 | 59.8 | 48%/52% |
| UMich | 19642 | 250 or 500 | 10.0 | 60.2 | 47%/53% |
| Undisclosed | 10000 | 300 | 10.0 | 63.0 | 47%/53% |
The diagnoses or labels were provided with the training data; neither the teams nor their algorithms had access to the diagnoses for the validation and test data. The quality of the labels depended on the clinical or research practices, and the datasets included labels that were machine-generated, over-read by a single cardiologist, and adjudicated by multiple cardiologists. Human experts may have used different criteria for ECG interpretation for some abnormalities; see e.g., [40]. We did not correct for differences in labeling practices except to encode the diagnoses using approximate Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT) codes for all of the datasets.
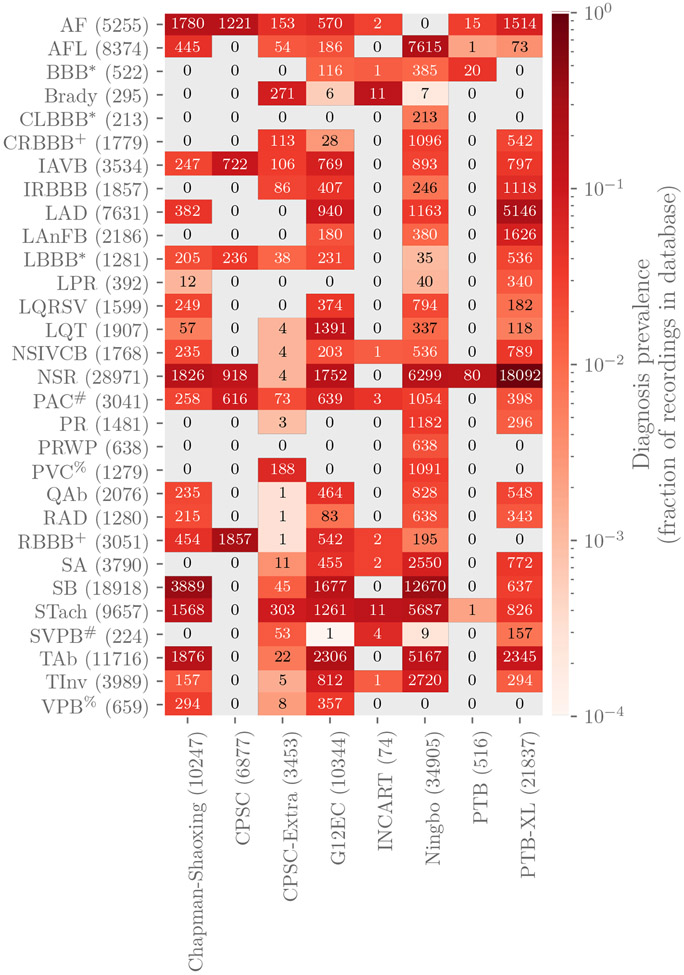
The data include 133 diagnoses or classes. All 133 diagnoses were represented in the training data, and a subset of these diagnoses were represented in the validation and test data. We evaluated the participant algorithms using 30 of the 133 diagnoses that were chosen by our cardiologists because they were relatively common, of clinical interest, and more likely to be recognizable from ECG recordings. Table 5 contains the list of the scored diagnoses for the Challenge with long-form descriptions, the corresponding SNOMED-CT codes, and abbreviations. While we only scored the algorithms using the diagnoses in Table 5 and Figure 3, we included all 133 classes in the data so that that participants could choose whether or not to use them with their algorithms.
Table 5:
Diagnoses, SNOMED CT codes, and abbreviations that were scored for the Challenge. CLBBB and LBBB, CRBBB and RBBB, PAC and SVPB, and PVC and VPB are distinct diagnoses, but we scored them as if they were the same diagnosis.
| Diagnosis | Code | Abbreviation |
|---|---|---|
| Atrial fibrillation | 164889003 | AF |
| Atrial flutter | 164890007 | AFL |
| Bundle branch block | 6374002 | BBB |
| Bradycardia | 426627000 | Brady |
| Complete left bundle branch block | 733534002 | CLBBB |
| Complete right bundle branch block | 713427006 | CRBBB |
| 1st degree AV block | 270492004 | IAVB |
| Incomplete right bundle branch block | 713426002 | IRBBB |
| Left axis deviation | 39732003 | LAD |
| Left anterior fascicular block | 445118002 | LAnFB |
| Left bundle branch block | 164909002 | LBBB |
| Prolonged PR interval | 164947007 | LPR |
| Low QRS voltages | 251146004 | LQRSV |
| Prolonged QT interval | 111975006 | LQT |
| Nonspecific intraventricular conduction disorder | 698252002 | NSIVCB |
| Sinus rhythm | 426783006 | NSR |
| Premature atrial contraction | 284470004 | PAC |
| Pacing rhythm | 10370003 | PR |
| Poor R wave progression | 365413008 | PRWP |
| Premature ventricular contractions | 427172004 | PVC |
| Q wave abnormal | 164917005 | QAb |
| Right axis deviation | 47665007 | RAD |
| Right bundle branch block | 59118001 | RBBB |
| Sinus arrhythmia | 427393009 | SA |
| Sinus bradycardia | 426177001 | SB |
| Sinus tachycardia | 427084000 | STach |
| Supraventricular premature beats | 63593006 | SVPB |
| T wave abnormal | 164934002 | TAb |
| T wave inversion | 59931005 | TInv |
| Ventricular premature beats | 17338001 | VPB |
Figure 3:
Numbers of recordings with each scored diagnosis in the training databases in the Challenge. Colors indicate the fraction of recordings with each scored diagnosis in each database, i.e., the total number of each scored diagnosis in a database normalized by the number of recordings in each database. Parentheses indicate the total numbers of records with a given label across the training data (rows) and the total numbers of recordings, including recordings without scored diagnoses, in each database (columns). The symbols *, +, #, and % indicate that distinct diagnoses were scored as if they were the same diagnosis.
All data were provided in MATLAB- and WFDB-compatible format [41]. Each ECG recording had a binary MATLAB v4 file for the ECG signal data and an associated plain text file in WFDB header format describing the recording and patient attributes, including the diagnosis or diagnoses for the recording. We did not change the original data or labels from the databases, except (1) to provide consistent and Health Insurance Portability and Accountability Act (HIPAA)-compliant identifiers for age and sex, (2) to encode the diagnoses as approximate SNOMED CT codes, and (3) to store the signal data using 16-bit signed integers for WFDB format.
2.2. Challenge Objective
We asked the Challenge participants to design working, open-source algorithms for identifying cardiac abnormalities from standard twelve-lead and several reduced-lead ECG recordings. We required that the Challenge teams submit code both for training their models and for applying their trained models, which aided the reproducibility of the research conducted during the Challenge. We ran the participants’ trained models on the hidden validation and test data and evaluated their performance using an expert-based evaluation metric that we designed for this year’s Challenge.
2.2.1. Challenge Overview, Rules, and Expectations
This year’s Challenge was the 22nd PhysioNet/Computing in Cardiology Challenge [41]. Similarly to previous Challenges, this year’s Challenge had an unofficial phase and an official phase. The unofficial phase (December 24, 2020 to April 8, 2021) provided an opportunity to socialize the Challenge and seek discussions and feedback from teams about the data, scoring, and requirements. The unofficial phase allowed 5 scored entries from each team on the hidden validation data. After a short break, the official phase (May 1, 2021 to August 15, 2021) introduced additional training data. The official phase allowed 10 scored entries from each team on the hidden validation data. After the end of the official phase, we evaluated one algorithm from each team on the hidden test data to prevent sequential training on the test data. Moreover, while teams were encouraged to ask questions, pose concerns, and discuss the Challenge in a public forum, they were prohibited from discussing or sharing their work for the Challenge to preserve the diversity and uniqueness of the approaches to the problem posed by the Challenge.
2.2.2. Classification of ECGs
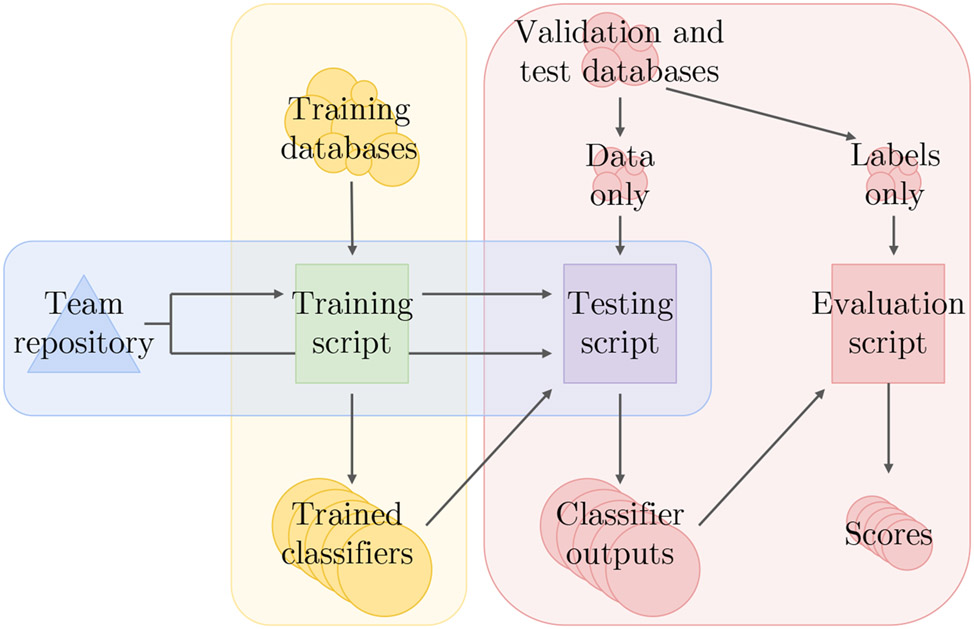
We required teams to submit their code for both training and testing their models, including any code for processing or relabeling the data. We ran each team’s training code on the training data to create a model, and we ran this model on the hidden validation and test sets. We ran the trained model on the recordings sequentially, instead of providing them all of the recordings at the same time, to apply them as realistically as possible. We then scored the outputs from the models. Figure 4 illustrates this computational pipeline.
Figure 4:
Computational pipeline for the 2021 Challenge. The vertices show the code, data, and results, and the edges show the relationships between the code, data, and results. Teams share their training and test scripts, which we run and score on the training, validation, and test sets; the test scripts running the trained models never see the labels for the validation and test sets.
We allowed teams to submit either MATLAB or Python implementations of their code. Participants containerized their code in Docker and submitted it in GitHub or Gitlab repositories. We downloaded their code and ran it in containerized environments on Google Cloud. We described the computational environment that we used to run entries more fully in [42]. We used virtual machines on Google Cloud with 10 virtual central processing units (vCPUs), 65 GB of random-access memory (RAM), and an optional NVIDIA T4 Tensor Core graphics processing unit (GPU). We imposed a 72 hour time limit for training with a GPU on the full training set and a 48 hour time limit for training without a GPU on the full training set. We used virtual machines on Google Cloud with 6 vCPUs, 39 GB of RAM, and an optional NVIDIA T4 Tensor Core GPU with a 24 hour time limit with or without a GPU for running the trained classifiers on each of the validation and test sets.
To aid teams, we shared example entries that we implemented in MATLAB and Python. The MATLAB example model was a multinomial logistic regression classifier that used age, sex, and the root-mean square of the signal for each ECG lead as features. The Python example model was a random forest classifier that also used age, sex, and the root-mean square of the signal for each ECG lead as features. We did not design these example models to be competitive but instead to provide working examples of how to read and extract features from the recordings that teams could easily run in several minutes on a personal computer.
2.2.3. Evaluation of Classifiers
We introduced a scoring metric that awarded partial credit to misdiagnoses that resulted in similar outcomes or treatments as the true diagnoses that were given by our cardiologists. This scoring metric reflected the clinical reality some misdiagnoses are more harmful than others and should be scored accordingly.
First, let be a collection of m distinct diagnoses for a database of n recordings, and let A = [aij] be a multi-class confusion matrix, where aij is a normalized number of recordings in a database that were classified as belonging to class ci but actually belonged to class cj, where ci and cj may be the same class or different classes.
Specifically, for each recording k = 1, …, n, let xk be the set of positive labels and yk be the set of positive classifier outputs for recording k. We defined A = [aij] such that
| (1) |
where
| (2) |
where the quantity ∣xk ∪ yk∣ is the number of distinct classes with a positive label and/or classifier output for recording k. We normalized the counts in the confusion matrix because each recording could have multiple true or predicted diagnoses, and we wanted incentivize teams to develop multi-class classifiers, but we did not want recordings with many diagnoses to dominate the scores that the algorithms received.
Next, let W = [wij] be a reward matrix, where wij is the reward for a positive classifier output for class ci with a positive label cj, where ci and cj may be the same class or different classes. The entries of W were defined by our cardiologists based on the similarity of treatments or differences in risks (see Figure 5). This matrix provided full credit to correct classifier outputs, partial credit to incorrect classifier outputs, and no credit for labels and classifier outputs that are not captured in the weight matrix. Also, four pairs of similar classes (i.e., CLBBB and LBBB, CRBBB and RBBB, PAC and SVPB, and PVC and VPB) were scored as if they were the same class by assigning full credit to off-diagonal entries, so a positive label or classifier output in one diagnosis in the pair was considered to be a positive label or classifier output for both diagnoses in the pair. We did not change the labels in the training, validation, or test data to make these classes identical so that we could preserve any institutional preferences and other information in these data.
Figure 5:
Reward matrix W for the 30 diagnoses scored in the Challenge. The rows and columns are the abbreviations for the ground-truth and predicted diagnoses in Table 5. Off-diagonal entries that are equal to 1 indicate similar diagnoses that are scored as if they were the same diagnosis. Each entry in the table was rounded to the first decimal place due to space constraints in this manuscript, but the shading of each entry reflects the actual value of the entry.
Finally, let
| (3) |
be a weighted sum of the entries in the confusion matrix. This score is a generalized version of the traditional accuracy metric that awards full credit to correct outputs (ones in diagonal entries of the matrix) and no credit to incorrect outputs (zeros in off-diagonal entries of the matrix). To aid interpretability, we normalized this score so that a classifier that always output the true class or classes received a score of one and an inactive classifier that always output the sinus rhythm class received a score of zero, i.e.,
| (4) |
where sI is the score for the inactive classifier and sT is the score for ground-truth classifier.
We used the same values of W for each algorithm and database, but each algorithm received different values of A and sN for each database. For any particular lead combination, the algorithm with the highest value of sN on the hidden test data for a specific lead combination won.
This scoring metric was designed to award full credit to correct diagnoses and partial credit to misdiagnoses with risks or outcomes that were similar to the true diagnosis. The resources, populations, practices, and preferences of an institution all affect how such a reward matrix W should be defined; the definition of this scoring metric from our cardiologists for the Challenge provides one such example.
3. Results
3.1. Entries
A total of 68 teams from academia and industry submitted 1,056 entries throughout the unofficial and official phases of the 2021 Challenge. During the unofficial and official phases, we trained the teams’ models on the public training data and scored the trained models on the hidden validation set. After the end of the official phase, we scored a final entry from each team on the twelve-lead, six-lead, four-lead, three-lead, and two-leads versions of the hidden test set using the Challenge evaluation metric (4). The qualifying teams with the highest score on each version of the test set won the lead combination category for the Challenge.
There were 430 successful entries, including 165 successful entries during the unofficial phase and 265 successful entries during the official phase. There were 636 unsuccessful entries, including 234 unsuccessful entries and 309 unsuccessful entries that we were unable to train during the unofficial and official phases, respectively, highlighting the importance of sharing training code for the reproducibility of the models.
A total of 39 teams met all of the conditions for the Challenge, including the submission of algorithms that we could successfully run on the training, validation, and test databases [16]. There were several reasons for disqualification of teams including the following: the training code failed to train on the training data or simply loaded a pretrained model, the trained model failed to run on the validation or test data, the team failed to submit a preprint to Computing and Cardiology by the conference preprint deadline, the team failed to attend Computing in Cardiology either remotely or in person to present and defend their work. The algorithms from teams that met all of the conditions for the Challenge are called ‘official entries’, and the other algorithms are called ‘unofficial entries’.
Deep learning (DL) approaches were common (35 algorithms, 90% of the official entries), including convolutional neural networks (CNNs) in general and ResNet-based approaches in particular [43]. Although only 4 (10%) entries used other classifiers such as random forest classifiers [44], logistic regression (LogitBoost) [45], and XGBoost [46], [47], ten of the DL algorithms (about 30%) extracted hand-crafted features for their DL models. By combining hand-crafted extracted features with the DL models, these teams tried to generate more robust multi-label classification.
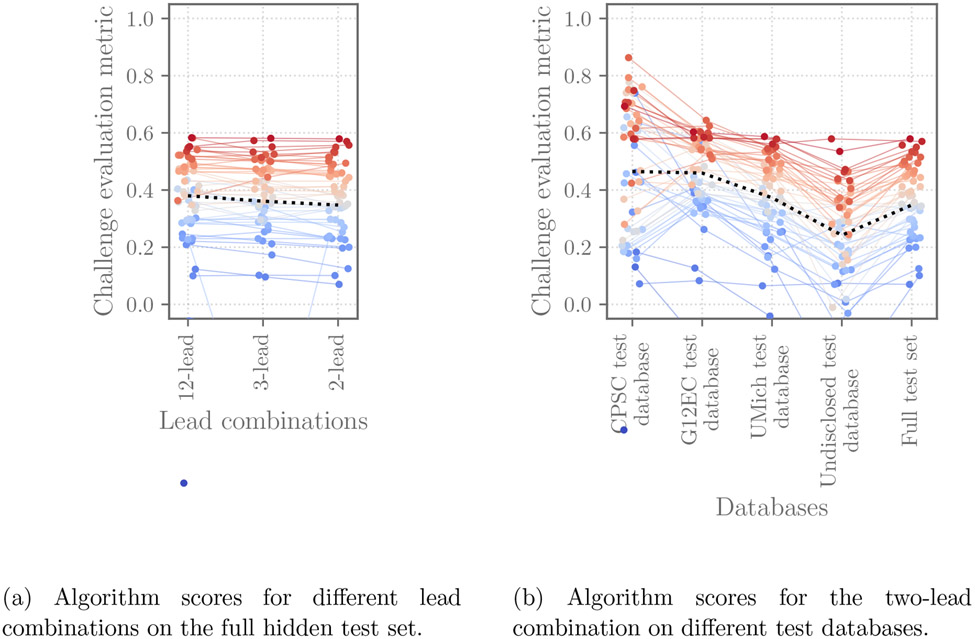
Across the different approaches, common trends appeared for the different lead combinations and data sources. Figure 6a shows the scores (the Challenge evaluation metric (4)) of the 39 official entries. It shows that the algorithms had similar performance (mean change of 2.6% for the Challenge evaluation metric) across different lead combinations on the different test databases; Figure 6 shows these scores for each of the lead combinations considered by the Challenge. Figure 6b demonstrates the scores of the 39 official entries on the individual and full test sets. It shows that the algorithms have varied performance on different test databases, including noticeably lower performance for the completely hidden test databases from sources for which no training data was provided, demonstrating the difficulty of generalizing to new databases.
Figure 6:
Scores of the 39 official entries that we were able to evaluate on the hidden validation and test databases for the Challenge and met the other conditions for the Challenge. The points indicate the score of each individual algorithm on each dataset, with the higher points showing algorithms with the highest scores on each database. The ranks on the test set are further indicated by color, with red indicating the algorithms with the best rankings on the database and blue indicating the algorithms with the worst rankings on the database. The dashed line shows the median score for each lead combination or database.
Table 6 further quantifies the changes in performance for different lead combinations and databases using the median relative change in the Challenge evaluation metric, again showing small changes across different lead combinations and larger ones across different test databases. Per-diagnosis scores and run times for the algorithms are available in [48].
Table 6:
The median relative change in the Challenge evaluation metric from the full validation set to the individual test sets and full test set for the five different lead combinations.
| Number of leads | CPSC test | G12EC test | UMich test | Undisclosed test | Full test |
|---|---|---|---|---|---|
| 12 | −10% | −3% | −14% | −39% | −17% |
| 6 | −13% | −3% | −14% | −36% | −19% |
| 4 | −13% | −3% | −12% | −37% | −17% |
| 3 | −14% | −3% | −13% | −38% | −19% |
| 2 | −13% | −3% | −13% | −41% | −18% |
Supplemental Table S1 provides a list of the 39 official entries that met all of the conditions for the Challenge, including their scores and ranks using the Challenge evaluation metric on the two-lead test set. This table also summarizes the libraries, model architectures, data processing, and optimization methods used by the algorithms, and it includes citations of the Computing and Cardiology papers for more information about the methods.
3.2. Voting algorithm
We developed and applied a naïve voting approach to combine individual algorithms into a single algorithm. This approach leveraged the different strengths of the individual algorithms while outperforming any single individual algorithm. In particular, we built a simple model that considered the classifier outputs of k different models that returned a positive vote for a diagnosis if at least αk different algorithms returned a positive vote for that diagnosis. This approach provided a majority votes-like voting model that used the data to determine a more optimal amount of consensus between methods than a simple majority.
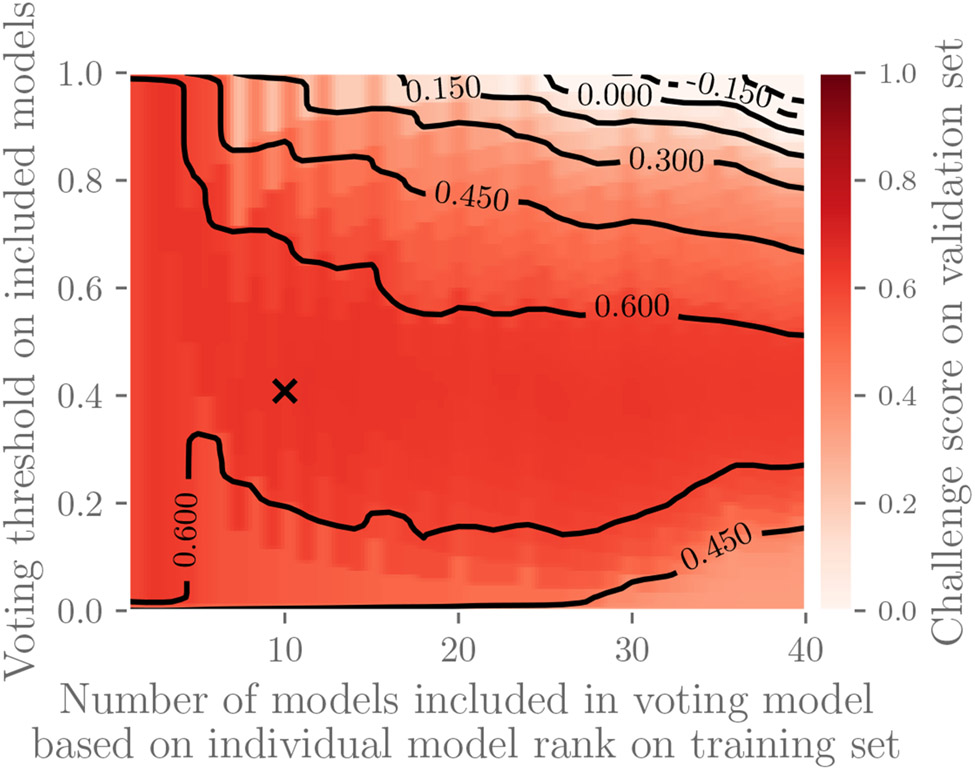
We chose the voting model parameters k and α as follows for the two-lead versions of the data and the Challenge evaluation metric; the same approach applies to other versions of the data and other scoring schemes. We first ranked the 39 official entries from highest to lowest performance according to the Challenge evaluation metric (4) on the training set, and we defined a voting model using the top k algorithms that returned a positive vote for a diagnosis if at least αk different algorithms returned a positive vote for that diagnosis. We found that k = 10 and α = 0.4 resulted in the voting model with the highest score on the validation set; Figure 7 illustrates this parameter search. We then ranked the algorithms from highest to lowest performance on the validation set, and we defined a voting model using the k = 10 highest scoring algorithms that returned a positive vote for a diagnosis if at least αk = 0.4 · 10 = 4 algorithms returned a positive vote for that diagnosis. We finally applied this model to the test data.
Figure 7:
Parameter selection for voting model. The top k models (x-axis) were included in the voting model based on performance on the training set that returns a positive vote if at least αk models (y-axis) returned positive votes. The heat map shows the performance of the resulting voting model using the Challenge evaluation metric on validation data, and the x marks the parameters that define the voting model with the highest performance on the validation data; these parameters define the voting model to be applied to the test set. This figure shows performance for two-lead versions of the data with similar results on other datasets.
This voting model received a Challenge evaluation metric of 0.60 on the two-lead version of the test data, outperforming the highest-performing individual algorithm, which received a Challenge evaluation metric of 0.58. Notably, this voting model used classifier outputs from an algorithm that was ranked 24th on the test data; some individual algorithms had lower overall performance but higher performance on some diagnoses or patient groups, which the voting model was able to utilize.
4. Follow-up Entries from the 2021 Challenge
As with most Challenges, we provided the community with another chance to evaluate their code on the test data for the 2021 Challenge. For this opportunity, we required the authors to submit updated code (or entirely new code) and a preprint describing the novelty of their updated or new approach that they planned to submit to the special issue containing this article. Tables 7, 8, and 9 provide the updated metrics for the twelve-lead, three-lead and two-lead of the full test set. The metrics include the area under the receiver operating characteristic curve (AUROC), the area under the precision recall curve (AUPRC), accuracy (defined here as the fraction of correctly classified recordings), F-measure, and the Challenge evaluation metric. We received 33 entries and successfully ran 13 entries.
Table 7:
The metrics and ranks of the teams for follow-up entries to the special issue on the twelve-lead version of the full test set, including teams’ papers and changes in score (“±”) from their original entries. “AUROC” is area under the receiver operating characteristic curve, and “AUPRC” is area under the precision recall curve. ‘Voting’ indicates the voting algorithm described in Section 3.2, which used a subset of the algorithms in the Challenge, rather than any follow-up entries. “N/A” denotes “not available”: N/Aa indicates that the voting model did not report numerical outputs, N/Ab indicates that the voting model was run only once, and N/Aa indicates failed original entries on the test set during the Challenge.
| Rank | Team [Reference] | AUROC | AUPRC | Accuracy | F-measure | Challenge metric [ ± original score] |
|---|---|---|---|---|---|---|
| - | Voting | N/Aa | N/Aa | 0.32 | 0.49 | 0.62 [N/Ab] |
| 1 | CeZIS [49] | 0.87 | 0.53 | 0.34 | 0.51 | 0.62 [+0.10] |
| 2 | ISIBrnoAIMT [50] | 0.87 | 0.45 | 0.29 | 0.41 | 0.59 [+0.01] |
| 3 | HeartBeats [51] | 0.94 | 0.52 | 0.21 | 0.44 | 0.57 [−0.01] |
| 3 | DrCubic [52] | 0.93 | 0.51 | 0.26 | 0.45 | 0.57 [+0.08] |
| 5 | AIRCAS_MEL1 [53] | 0.91 | 0.47 | 0.24 | 0.42 | 0.52 [+0.14] |
| 6 | iadi-ecg [54] | 0.87 | 0.45 | 0.27 | 0.41 | 0.48 [+0.00] |
| 6 | SMS+1 [55] | 0.87 | 0.36 | 0.21 | 0.32 | 0.48 [−0.04] |
| 8 | skylark [56] | 0.83 | 0.30 | 0.02 | 0.26 | 0.39 [+0.03] |
| 8 | itaca-UPV [57] | 0.82 | 0.32 | 0.05 | 0.29 | 0.39 [+0.05] |
| 10 | Revenger [58] | 0.83 | 0.45 | 0.37 | 0.43 | 0.38 [N/Ac] |
| 11 | Medics [59] | 0.74 | 0.26 | 0.07 | 0.26 | 0.36 [N/Ac] |
| 12 | Biomedic2ai [60] | 0.79 | 0.35 | 0.28 | 0.32 | 0.24 [−0.12] |
| 13 | WEAIT [61] | 0.52 | 0.07 | 0.01 | 0.10 | −0.12 [+0.50] |
Table 8:
The metrics and ranks of the teams for follow-up entries to the special issue on the three-lead version of the full test set, including teams’ papers and changes in score (“±”) from their original entries. “AUROC” is area under the receiver operating characteristic curve, and “AUPRC” is area under the precision recall curve. ‘Voting’ indicates the voting algorithm described in Section 3.2, which used a subset of the algorithms in the Challenge, rather than any follow-up entries. “N/A” denotes “not available”: N/Aa indicates that the voting model did not report numerical outputs, N/Ab indicates that the voting model was run only once, and N/Aa indicates failed original entries on the test set during the Challenge.
| Rank | Team [Reference] | AUROC | AUPRC | Accuracy | F-measure | Challenge metric [ ± original score] |
|---|---|---|---|---|---|---|
| 1 | CeZIS [49] | 0.87 | 0.53 | 0.34 | 0.50 | 0.61 [+0.09] |
| - | Voting | N/Aa | N/Aa | 0.32 | 0.47 | 0.60 [N/Ab] |
| 2 | ISIBrnoAIMT [50] | 0.87 | 0.44 | 0.28 | 0.40 | 0.60 [+0.02] |
| 3 | HeartBeats [51] | 0.93 | 0.50 | 0.21 | 0.46 | 0.58 [+0.05] |
| 4 | Dr_Cubic [52] | 0.93 | 0.51 | 0.26 | 0.45 | 0.56 [+0.05] |
| 5 | AIRCAS_MEL1 [53] | 0.91 | 0.45 | 0.23 | 0.41 | 0.51 [+0.08] |
| 6 | iadi-ecg [54] | 0.87 | 0.44 | 0.29 | 0.39 | 0.46 [−0.01] |
| 7 | Revenger [58] | 0.86 | 0.45 | 0.38 | 0.42 | 0.40 [+0.07] |
| 8 | skylark [56] | 0.83 | 0.29 | 0.02 | 0.26 | 0.39 [−0.06] |
| 9 | itaca-UPV [57] | 0.84 | 0.33 | 0.04 | 0.28 | 0.38 [+0.08] |
| 10 | SMS+1 [55] | 0.82 | 0.33 | 0.18 | 0.30 | 0.36 [−0.14] |
| 11 | Medics [59] | 0.79 | 0.29 | 0.08 | 0.27 | 0.32 [N/Ac] |
| 12 | Biomedic2ai [60] | 0.79 | 0.33 | 0.29 | 0.27 | 0.19 [−0.10] |
| 13 | WEAIT [61] | 0.52 | 0.07 | 0.01 | 0.10 | −0.12 [+0.50] |
Table 9:
The metrics and ranks of the teams for follow-up entries to the special issue on the two-lead version of the full test set, including the teams’ papers and changes in score (“±”) from their original entries. “AUROC” is area under the receiver operating characteristic curve, and “AUPRC” is area under the precision recall curve. ‘Voting’ indicates the voting algorithm described in Section 3.2, which used a subset of the algorithms in the Challenge, rather than any follow-up entries. “N/A” denotes “not available”: N/Aa indicates that the voting model did not report numerical outputs, N/Ab indicates that the voting model was run only once, and N/Aa indicates failed original entries on the test set during the Challenge.
| Rank | Team [Reference] | AUROC | AUPRC | Accuracy | F-measure | Challenge metric [ ± original score] |
|---|---|---|---|---|---|---|
| - | Voting | N/Aa | N/Aa | 0.30 | 0.46 | 0.60 [N/Ab] |
| 1 | CeZIS [49] | 0.87 | 0.52 | 0.33 | 0.49 | 0.59 [+0.07] |
| 1 | ISIBrnoAIMT [50] | 0.87 | 0.43 | 0.27 | 0.39 | 0.59 [+0.00] |
| 3 | HeartBeats [51] | 0.92 | 0.50 | 0.20 | 0.42 | 0.57 [+0.04] |
| 4 | Dr_Cubic [52] | 0.92 | 0.50 | 0.25 | 0.44 | 0.55 [+0.07] |
| 5 | AIRCAS_MEL1 [53] | 0.89 | 0.44 | 0.22 | 0.40 | 0.50 [+0.12] |
| 5 | SMS+1 [55] | 0.86 | 0.36 | 0.26 | 0.32 | 0.50 [+0.01] |
| 7 | iadi-ecg [54] | 0.87 | 0.42 | 0.27 | 0.38 | 0.45 [−0.01] |
| 8 | skylark [56] | 0.81 | 0.27 | 0.03 | 0.26 | 0.39 [−0.10] |
| 9 | Medics [59] | 0.78 | 0.28 | 0.08 | 0.27 | 0.38 [N/Ac] |
| 10 | itaca-UPV [57] | 0.82 | 0.31 | 0.04 | 0.26 | 0.37 [+0.03] |
| 11 | Revenger [58] | 0.84 | 0.43 | 0.35 | 0.40 | 0.35 [+0.02] |
| 12 | Biomedic2ai [60] | 0.80 | 0.33 | 0.28 | 0.29 | 0.26 [−0.08] |
| 13 | WEAIT [61] | 0.62 | 0.17 | 0.01 | 0.17 | −0.08 [+0.54] |
In general, these post-Challenge entries improved on the performance of the original entries, and they again showed small changes in performance across the different lead combinations and larger changes across the different test databases from different sources.
5. Discussion
While the 2021 Challenge sought to assess the diagnostic potential of reduced-lead ECGs, the real-world issues of using clinical data proved to be more of an obstacle to the automatic detection of cardiac abnormalities than the different choices of lead sets, highlighting the challenge of generalizing to datasets from new settings with different data collection procedures and populations. However, this common issue represents a diversity of approaches to automatically identifying cardiac abnormalities from reduced-lead ECGs.
Supplemental Table S1 summarizes the 39 algorithms submitted by teams that satisfied all of the requirements of the 2021 Challenge. It shows that deep learning (DL) approaches were common for the 2021 Challenge, which was also true of the 2020 Challenge and reflects recent trends in ECG signal processing and arrhythmia detection. Some participants adopted algorithms from other applications, but they did not necessarily perform better than custom-made machine learning algorithms. The performance of these algorithms showed that the custom model architectures, custom optimization techniques, and deliberate attempts to generalize to new databases can help to provide better diagnostic outcomes. The algorithms developed by the Challenges teams widely used convolutional neural networks (CNN) and ResNet deep neural networks with different architectures and customized models. An example of a customized model was a channel self-attention-based model developed by team cardiochallenger, which used an ensemble inception and residual architecture with a genetic algorithm to optimize thresholds for each class for maximizing the Challenge evaluation metric [62].
For handling different signal characteristics across different datasets in the training set, including different sampling rates, gains, signal quality, and signal lengths of the ECG recordings, many algorithms applied preprocessing steps to the ECG signals. The preprocessing steps included resampling, normalization, peak correction, filtering, noise reduction, and/or discarding the beginnings and ends of the signals [63]-[65]. Different normalization techniques were applied, including z-score normalization [64], [66] and minmax scaling [67]. snu_adsl reported that standardization did not necessarily improve the performance of their algorithm, but they implemented it in their preprocessing step in case the unseen dataset had unexpected characteristics.
One of the common preprocessing steps of the algorithms (implemented by 25 teams, 64% of the official algorithms) was filtering the signals using different techniques such as the Butterworth bandpass filter with a bandwidth between 1-45 Hz. [68] used a finite impulse response bandpass filter with a bandwidth between 3-45 Hz [69].
Some of the algorithms investigated the quality of the signals or used data augmentation. For instance, HaoWan_AIeC assessed the quality of each lead and created a mask for low-quality leads [70]. They also applied data augmentation by randomly cropping signals and randomly generating masks [70]. HeartlyAI applied different augmentation techniques such as cut-out, adding different types of noise, and allowing dropout of individual or groups of ECG channels [71].
Many algorithms segmented the signals into windows during preprocessing. For example, Biomedic2ai segmented the signals into 5-second windows with a stride of one second for a 4-second overlap for adjacent signals [72]. snu_adsl selected a random window with a width of 13.3 seconds and zero-padded ECG signals shorter than 13.3 seconds at the end of the signal [68]. prna set a fixed window width of 15.36 seconds, allowing the signal to be split into divisible segments sizes and zero-padding the ends of the signals as needed [69]. iadiecg extracted the middle of recordings that were longer than 10 seconds and zero-padded both sides of recordings that were shorter than 10 seconds and normalized the signals so that they had zero mean and unit variance [73]. Although these algorithms were not the highest performing algorithms, they were among the top half of the entries and obtained a Challenge evaluation metric between 0.44 and 0.46 on the two-lead full test set.
Some participants decided not to train their models on some of the training databases. The scores of the winning algorithms shows that inclusion of all of the available data may lead to better generalization on the unseen test data [62], [64], [74].
For addressing differences in data collection practices from different sources, teams applied different methods to improve generalization. For example, HaoWan_AIeC adopted MixStyle to use feature-based augmentation to generalize to different domains [70]. Team DSAIL_SNU used the WRN model architecture with 14 convolution/dense layers and a widening factor of 1 and attempted to improve generalization by using constant-weighted cross-entropy loss, additional features, MixUp augmentation, a squeeze/excitation block, and a OneCycle learning rate scheduler [75]. Another team, NIMA, whose entry was among the top three algorithms, used spatial dropouts and average pooling between each layer of two separate deep CNNs to reduce overfitting and model complexity [74].
Eleven entries (about 30% of the official teams) used a binary cross-entropy loss function for multi-label classification, but custom loss functions for this problem also helped to improve classification performance. Team ISIBrno-AIMT, the winning algorithm, optimized a ResNet architecture with a multi-head attention mechanism using a mixture of a binary cross-entropy loss function, a custom loss function that provided a differentiable approximation of Challenge evaluation metric, and an evolutionary optimization loss function that attempted to estimate the optimal probability threshold for each class [64]. BUTTeam noticed that using the Challenge evaluation metric as a loss function seemed to be unstable and lead to sub-optimal results [76]. Their approach was to first train their model with weighted cross-entropy (WCE) loss and then retrain it with the Challenge evaluation metric and a decaying learning rate [76]. Another example of a custom loss-function was a weighted, generalized softmax loss function with quadratic differences by AADAConglomerate [63].
Although 90% of the 39 official entries used DL models, about 40% of the algorithms combined hand-crafted features with their DL models [72], [77]. Team PhysioNauts was among the unofficial teams that used a ResNet model with a squeeze and excitation module with handcrafted and DL features and used a grid search and the Nelder-Mead method to optimize the Challenge evaluation metric [78]. UMCU used an adaptive pooling layer to combine the features over the temporal dimension, after which a linear layer created the final output [79].
Class imbalance was another significant issue for classification, and the larger number and varying prevalence rates of the diagnoses from different sources represented the real-world problem of clinical diagnosis. Many algorithms applied different methods to correct for class imbalance. UMCU weighted each class by dividing the maximum number of positive samples from any class by the number of positive samples from the weighted class and used a threshold of 0.5 for prediction; all values greater than 0.5 were positive, and all below 0.5 were negative [79]. Most teams performed best on the CPSC dataset, but it was the least representative dataset due to having fewer and more balanced diagnoses than the other datasets.
Almost all teams performed similarly on the different lead combination with average scores change in the Challenge evaluation metric of less than 2% from the twelve-lead to two-lead versions of the test data, which could be interpreted as responding to the Challenge question of “Will two do?” with the answer “Yes, two can do!”. Although the average scores change between different lead combinations is relatively negligible, performances varied across different diagnoses and different data sources, suggesting that better data processing and generalization techniques are required for better performance on unseen datasets.
6. Conclusions
This article explores the diagnostic potential of automated approaches for reading standard twelve-lead and various reduced-lead ECG recordings. While most algorithms had similar overall performance across the different lead combinations considered during the Challenge, including two-lead ECGs and standard twelve-lead ECGs, performance varied more widely across different diagnoses and on data from different institutions. We found no definitive evidence that over-complete lead systems provided any additional diagnostic power, although without underlying knowledge of the electronic circuitry of the individual systems used to capture the ECGs, it is impossible to be sure if inter-database differences in performances as a function of lead combinations are due to variations in: population phenotypes; clinical practice behaviors; hardware filters; manufacturer software pre-processing before data storage; or circuit configuration differences (such as derivation of the WCT). Moreover, the lack of consistent differences between lead configurations across all data is confined to the rhythms explored in this work. Other conditions, such as much more subtle ST changes perhaps, may yield different results.
Most importantly, this article describes the world’s largest open-access database of twelve-lead ECG recordings along with large hidden validation and test databases of twelve-lead and various reduced-lead recordings, providing unbiased and repeatable research on the diagnostic potential of automated approaches for reduced-lead diagnoses. The data were drawn from several countries across three continents with diverse and distinctly different populations, encompassing 133 diagnoses with 30 diagnoses of special interest for the Challenge. Additionally, we introduced a novel scoring matrix that captures the similarities between and risks of different diagnostic outcomes. Finally, we supported the development of a large corpus of open-source, repeatable, and diverse algorithmic approaches for classifying full-lead and reduced-lead ECG recordings. The algorithms and diverse data can provide benchmarks for the field and help push beyond the current theme of applying machine learning to large single-center databases, which in our experience are unlikely to generalize across populations to individuals that are underrepresented in existing datasets.
Supplementary Material
Acknowledgements
This research is supported by the National Institute of General Medical Sciences (NIGMS) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) under NIH grant numbers 2R01GM104987-09 and R01EB030362 respectively, the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR002378, as well as the Gordon and Betty Moore Foundation, MathWorks, and AliveCor, Inc. under unrestricted gifts. GC has financial interests in Alivecor, LifeBell AI and Mindchild Medical. GC also holds a board position in LifeBell AI and Mindchild Medical. AE receives financial support from the Spanish Ministerio de Ciencia, Innovacion y Universidades through grant RTI2018-101475-BI00, jointly with the Fondo Europeo de Desarrollo Regional (FEDER), and by the Basque Government through grant IT1229-19. None of the aforementioned entities influenced the design of the Challenge or provided data for the Challenge. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the above entities.
Footnotes
De-identified data collected under U-M HUM00092309: Approximately 20,000 ten-second-long twelve-lead ECGs obtained from the University of Michigan Section of Electrophysiology. The sample was randomly selected from the patients who had a routine ECG test from 1990 to 2013 to approximately match the demographics of the training databases. The dataset was de-identified and contains only basic demographics information such as age (any age over the age of 90 is denoted as 90+) and sex, the ECG waveforms and the diagnosis statements associated with the record.
References
- [1].Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Cheng S, Delling FN, Elkind MSV, Evenson KR, Ferguson JF, Gupta DK, Khan SS, Kissela BM, Knutson KL, Lee CD, Lewis TT, Liu J, Loop MS, Lutsey PL, Ma J, Mackey J, Martin SS, Matchar DB, Mussolino ME, Navaneethan SD, Perak AM, Roth GA, Samad Z, Satou GM, Schroeder EB, Shah SH, Shay CM, Stokes A, VanWagner LB, Wang N-Y, Tsao CW, and American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee, “Heart disease and stroke statistics – 2021 update: A report from the American Heart Association”, Circulation, vol. 143, no. 8, e254–e743, 2021. [DOI] [PubMed] [Google Scholar]
- [2].Kligfield P, “The centennial of the Einthoven electrocardiogram”, J. Electrocardiol, vol. 35, no. 4, pp. 123–129, 2002. [DOI] [PubMed] [Google Scholar]
- [3].Kligfield P, Gettes LS, Bailey JJ, Childers R, Deal BJ, Hancock EW, Van Herpen G, Kors JA, Macfarlane P, Mirvis DM, et al. , “Recommendations for the standardization and interpretation of the electrocardiogram: Part I: The Electrocardiogram and its technology, a scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society endorsed by the International Society for Computerized Electrocardiology”, Journal of the American College of Cardiology, vol. 49, no. 10, pp. 1109–1127, 2007. [DOI] [PubMed] [Google Scholar]
- [4].Willems JL, Abreu-Lima C, Arnaud P, Vanbemmel J, Brohet C, Degani R, Denis B, Gehring J, Graham I, vanHerpen G, Machado HC, Macfarlane PW, Michaelis J, Moulopoulos S, Rubel P, and Zywietz C, “The diagnostic performance of computer programs for the interpretation of electrocardiograms”, The New England Journal of Medicine, vol. 325, pp. 1767–1773, 1991. [DOI] [PubMed] [Google Scholar]
- [5].Shah A and Rubin S, “Errors in the computerized electrocardiogram interpretation of cardiac rhythm”, Journal of Electrocardiology, vol. 40, pp. 385–90, Sep. 2007. DOI: 10.1016/j.jelectrocard.2007.03.008. [DOI] [PubMed] [Google Scholar]
- [6].Ye C, Coimbra MT, and Kumar BV, “Arrhythmia detection and classification using morphological and dynamic features of ECG signals”, in 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, IEEE, 2010, pp. 1918–1921. [DOI] [PubMed] [Google Scholar]
- [7].Mohammadzadeh-Asl B and Setarehdan SK, “Neural network based arrhythmia classification using heart rate variability signal”, in 2006 14th European Signal Processing Conference, 2006, pp. 1–4. [Google Scholar]
- [8].Oh SL, Ng EY, Tan RS, and Acharya UR, “Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats”, Computers in Biology and Medicine, vol. 102, pp. 278–287, 2018, ISSN: 0010-4825. DOI: 10.1016/j.compbiomed.2018.06.002. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S0010482518301446. [DOI] [PubMed] [Google Scholar]
- [9].S. G, S. KP, and V. R, “Automated detection of cardiac arrhythmia using deep learning techniques”, Procedia Computer Science, vol. 132, pp. 1192–1201, 2018, International Conference on Computational Intelligence and Data Science, ISSN: 1877-0509. DOI: 10.1016/j.procs.2018.05.034. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S187705091830766X. [DOI] [Google Scholar]
- [10].Nagarajan VD, Lee S-L, Robertus J-L, Nienaber CA, Trayanova NA, and Ernst S, “Artificial intelligence in the diagnosis and management of arrhythmias”, European Heart Journal, vol. 42, no. 38, pp. 3904–3916, Aug. 2021, ISSN: 0195-668X. DOI: 10.1093/eurheartj/ehab544. eprint: https://academic.oup.com/eurheartj/article-pdf/42/38/3904/40526393/ehab544.pdf. [Online]. Available: 10.1093/eurheartj/ehab544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Aldrich HR, Hindman NB, Hinohara T, Jones MG, Boswick J, Lee KL, Bride W, Califf RM, and Wagner GS, “Identification of the optimal electrocardiographic leads for detecting Acute Epicardial Injury in Acute Myocardial Infarction”, Am. J. Cardiol, vol. 59, no. 1, pp. 20–23, 1987. [DOI] [PubMed] [Google Scholar]
- [12].Drew BJ, Pelter MM, Brodnick DE, Yadav AV, Dempel D, and Adams MG, “Comparison of a new reduced lead set ECG with the standard ECG for diagnosing cardiac arrhythmias and Myocardial Ischemia”, J. Electrocardiol, vol. 35, no. 4, Part B, pp. 13–21, 2002. [DOI] [PubMed] [Google Scholar]
- [13].Green M, Ohlsson M, Lundager Forberg J, Björk J, Edenbrandt L, and Ekelund U, “Best leads in the standard electrocardiogram for the emergency detection of acute coronary syndrome”, J. Electrocardiol, vol. 40, no. 3, pp. 251–256, 2007. [DOI] [PubMed] [Google Scholar]
- [14].Alday EAP, Gu A, Shah AJ, Robichaux C, Wong A-KI, Liu C, Liu F, Rad AB, Elola A, Seyedi S, Li Q, Sharma A, Clifford GD, and Reyna MA, “Classification of 12-lead ECGs: The PhysioNet/Computing in Cardiology Challenge 2020”, Physiological Measurement, vol. 41, no. 12, p. 124003, Jan. 2021. DOI: 10.1088/1361-6579/abc960. [Online]. Available: 10.1088/1361-6579/abc960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Reyna MA, Sadr N, Perez Alday EA, Gu A, Shah A, Robichaux C, Rad BA, Elola A, Seyedi S, Ansari S, Li Q, Sharma A, and Clifford GD, “Will two do? varying dimensions in electrocardiography: The PhysioNet/Computing in Cardiology Challenge 2021”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [16].PhysioNet/Computing in Cardiology Challenge 2021, https://physionetchallenges.org/2021/, Accessed: 2021-09-20.
- [17].Clifford GD, Liu C, Moody B, Lehman L.-w. H., Silva I, Li Q, Johnson AE, and Mark RG, “AF classification from a short single lead ECG recording: the PhysioNet/Computing in Cardiology Challenge 2017”, in 2017 Computing in Cardiology (CinC), IEEE, vol. 44, 2017, pp. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zheng J, Zhang J, Danioko S, Yao H, Guo H, and Rakovski C, “A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients”, Sci. Data, vol. 7, no. 48, pp. 1–8, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu F, Liu C, Zhao L, Zhang X, Wu X, Xu X, Liu Y, Ma C, Wei S, He Z, Li J, and Kwee ENY, “An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection”, Journal of Medical Imaging and Health Informatics, vol. 8, no. 7, pp. 1368–1373, 2018. [Google Scholar]
- [20].Tihonenko V, Khaustov A, Ivanov S, Rivin A, and Yakushenko E, “St Petersburg INCART 12-lead Arrhythmia Database”, PhysioBank, PhysioToolkit, and PhysioNet, 2008, doi: 10.13026/C2V88N. [DOI] [Google Scholar]
- [21].Zheng J, Cui H, Struppa D, Zhang J, Yacoub SM, El-Askary H, Chang A, Ehwerhemuepha L, Abudayyeh I, Barrett A, Fu G, Yao H, Li D, Guo H, and Rakovski C, “Optimal multi-stage arrhythmia classification approach”, Sci. Data, vol. 10, no. 2898, pp. 1–17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bousseljot R, Kreiseler D, and Schnabel A, “Nutzung der EKG-Signaldatenbank CARDIODAT der PTB über das Internet”, Biomedizinische Technik, vol. 40, no. S1, pp. 317–318, 1995. [Google Scholar]
- [23].Wagner P, Strodthoff N, Bousseljot R-D, Kreiseler D, Lunze FI, Samek W, and Schaeffter T, “PTB-XL, a large publicly available electrocardiography dataset”, Sci. Data, vol. 7, no. 1, pp. 1–15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Malmivuo J and Plonsey R, “Bioelectromagnetism. 17. Other ECG Lead Systems”, in. Jan. 1995, pp. 307–312, isbn: 978-0195058239. [Google Scholar]
- [25].Clifford GD and McSharry PE, “A realistic coupled nonlinear artificial ECG, BP, and respiratory signal generator for assessing noise performance of biomedical signal processing algorithms”, in Fluctuations and Noise in Biological, Biophysical, and Biomedical Systems II, SPIE, vol. 5467, 2004, pp. 290–301. [Google Scholar]
- [26].Clifford GD and Villarroel MC, “Model-based determination of QT intervals”, in 2006 Computers in Cardiology, IEEE, 2006, pp. 357–360. [Google Scholar]
- [27].Sameni R, Shamsollahi MB, Jutten C, and Clifford GD, “A nonlinear Bayesian filtering framework for ECG denoising”, IEEE Transactions on Biomedical Engineering, vol. 54, no. 12, pp. 2172–2185, 2007. [DOI] [PubMed] [Google Scholar]
- [28].Clifford GD, Nemati S, and Sameni R, “An artificial vector model for generating abnormal electrocardiographic rhythms”, Physiological Measurement, vol. 31, no. 5, p. 595, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Oster J, Behar J, Sayadi O, Nemati S, Johnson A, and Clifford GD, “Semi-supervised ECG Ventricular Beat Classification with Novelty Detection Based on Switching Kalman Filters”, IEEE Transactions on Biomedical Engineering, vol. 62, no. 9, pp. 2125–2134, 2015. DOI: 10.1109/TBME.2015.2402236. [DOI] [PubMed] [Google Scholar]
- [30].Clifford GD, Shoeb A, McSharry PE, and Janz BA, “Model-based filtering, compression and classification of the ECG”, International Journal of Bioelectromagnetism, vol. 7, no. 1, pp. 158–161, 2005. [Google Scholar]
- [31].Sameni R, Clifford GD, Jutten C, and Shamsollahi MB, “Multichannel ECG and Noise Modeling: Application to Maternal and Fetal ECG Signals”, EURASIP Journal on Advances in Signal Processing, vol. 43, no. 4, p. 14, 2007. [Google Scholar]
- [32].Sayadi O, Shamsollahi MB, and Clifford GD, “Robust detection of premature ventricular contractions using a wave-based Bayesian framework”, IEEE Transactions on Biomedical Engineering, vol. 57, no. 2, pp. 353–362, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bacharova L, Selvester RH, Engblom H, and Wagner GS, “Where is the central terminal located?: In search of understanding the use of the wilson central terminal for production of 9 of the standard 12 electrocardiogram leads”, Journal of Electrocardiology, vol. 38, no. 2, pp. 119–127, 2005, ISSN: 0022-0736. DOI: 10.1016/j.jelectrocard.2005.01.002. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S0022073605000178. [DOI] [PubMed] [Google Scholar]
- [34].Miyamoto N, Shimizu Y, Nishiyama G, Mashima S, and Okamoto Y, “The absolute voltage and the lead vector of Wilson’s central terminal”, Jpn Heart J., vol. 37, no. 2, pp. 203–214, 1996, issn: 00214868. DOI: 10.1536/ihj.37.203. [DOI] [PubMed] [Google Scholar]
- [35].Gargiulo G, Thiagalingam A, McEwan A, Cesarelli M, Bifulco P, Tapson J, and van Schaik A, “True unipolar ECG leads recording (without the use of WCT)”, Hear Lung Circ., vol. 22, S102, Jan. 2013, issn: 14439506. DOI: 10.1016/j.hlc.2013.05.243. [DOI] [PubMed] [Google Scholar]
- [36].Gargiulo GD, “True unipolar ECG machine for Wilson Central Terminal measurements”, Biomed Res Int., vol. 2015, p. 586 397, 2015, issn: 23146141. DOI: 10.1155/2015/586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Moeinzadeh H, Bifulco P, Cesarelli M, McEwan AL, O’Loughlin A, Shugman IM, Tapson JC, Thiagalingam A, and Gargiulo GD, “Minimization of the Wilson’s Central Terminal voltage potential via a genetic algorithm”, BMC Research Notes, vol. 11, no. 1, pp. 1–5, Dec. 2018, issn: 17560500. DOI: 10.1186/S13104-018-4017-Y/FIGURES/3. [Online]. Available: https://bmcresnotes.biomedcentral.com/articles/10.1186/s13104-018-4017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jin BE, Wulff H, Widdicombe JH, Zheng J, Bers DM, and Puglisi JL, “A simple device to illustrate the Einthoven triangle”, Advances in Physiology Education, vol. 36, no. 4, p. 319, 2012, issn: 15221229. DOI: 10.1152/ADVAN.00029.2012. [Online]. Available: /pmc/articles/PMC3776430/%20/pmc/articles/PMC3776430/?report=abstract%20https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3776430/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Malmivuo J and Plonsey R, “Bioelectromagnetism.”, in. 1995, pp. 277–289, isbn: 978-0195058239. [Google Scholar]
- [40].Gregg RE, Yang T, Smith SW, and Babaeizadeh S, “ECG reading differences demonstrated on two databases”, Journal of Electrocardiology, 2021. [DOI] [PubMed] [Google Scholar]
- [41].Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng C-K, and Stanley HE, “PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals”, Circulation, vol. 101, no. 23, e215–e220, 2000. [DOI] [PubMed] [Google Scholar]
- [42].Reyna MA, Josef C, Jeter R, Shashikumar SP, Westover MB, Nemati S, Clifford GD, and Sharma A, “Early prediction of sepsis from clinical data: the PhysioNet/Computing in Cardiology Challenge 2019”, Critical Care Medicine, vol. 48, pp. 210–217, February 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition”, in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, pp. 770–778. DOI: 10.1109/CVPR.2016.90. [DOI] [Google Scholar]
- [44].Ignacio PS, “Leveraging period-specific variations in ECG topology for classification tasks”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [45].Hammer A, Scherpf M, Ernst H, Weiß J, Schwensow D, and Schmid M, “Automatic classification of full- and reduced-lead electrocardiograms using morphological feature extraction”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [46].van Prehn J, Ivanov S, and Nalbantov G, “Pathologies prediction on short ECG signals with focus on feature extraction based on beat morphology and image deformation”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [47].Krivenko S, Pulavskyi A, Kryvenko L, Krylova O, and Krivenko S, “Using mel-frequency cepstrum and amplitude-time heart variability as XGBoost handcrafted features for heart disease detection”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [48].PhysioNet/Computing in Cardiology Challenge 2020, https://physionetchallenges.org/2020/, Accessed: 2021-06-08.
- [49].Antoni L, Bruoth E, Bugata P, B. P Jr., Gajdos D, Horvat S, Hudak D, Kmecova V, Stana R, Stankova M, Szabari A, and Vozarikova G, “Automatic ECG classification and label quality in training data”, Under Review, [DOI] [PubMed] [Google Scholar]
- [50].Nejedly P, Ivora A, Smisek R, Viscor I, Koscova Z, Jurak P, and Plesinger F, “Classification of ECG using ensemble of Residual CNNs with or without attention mechanism”, Under Review, [DOI] [PubMed] [Google Scholar]
- [51].Xu Z, Guo Y, Zhao T, Zhao Y, Liu Z, and Sun X, “Abnormality classification from electrocardiograms with various lead combinations”, Under Review, [DOI] [PubMed] [Google Scholar]
- [52].Li X, Li C, Xu X, Wei Y, Wei J, Sun Y, Qian B, and Xu X, “Towards generalization of cardiac abnormality classification using reduced-lead multi-source ECG signal”, Under Review, [Google Scholar]
- [53].Xia P, He Z, Zhu Y, Bai Z, Yu X, Wang Y, Geng F, Du L, Chen X, Wang P, and Fang Z, “A novel multi-scale 2-D convolutional neural network for arrhythmias detection on varying-dimensional ECGs”, Under Review, [DOI] [PubMed] [Google Scholar]
- [54].Aublin P, Behar JA, Fix J, and Oster J, “Cardiac abnormality detection based on single lead classifier voting”, Under Review, [DOI] [PubMed] [Google Scholar]
- [55].Vazquez CG, Breuss A, Gnarra O, Portmann J, and Poian GD, “Label noise and self-learning label correction in cardiac abnormalities classification”, Under Review, [DOI] [PubMed] [Google Scholar]
- [56].Nankani D and Baruah RD, “Feature fused multichannel ECG classification using channel specific dynamic CNN for detecting and interpreting cardiac abnormalities”, Under Review, [Google Scholar]
- [57].Jimenez-Serrano S, Rodrigo M, Calvo CJ, Castells F, and Millet J, “Evaluation of a machine learning approach for the detection of multiple cardiac conditions in ECG systems from 12 to 1-lead”, Under Review, [Google Scholar]
- [58].Kang J and Wen H, “A study on several critical problems on arrhythmia detection using varying-dimensional electrocardiography”, Under Review, [DOI] [PubMed] [Google Scholar]
- [59].Sawant NK and Patidar S, “Identification of cardiac abnormalities applying Fourier-Bessel expansion and LSTM on ECG signals”, Under Review, [DOI] [PubMed] [Google Scholar]
- [60].Heydarian M and Doyle TE, “Two-dimensional convolutional neural network model for classification of ECG”, Under Review, 2022. [Google Scholar]
- [61].Puszkarski B, Hryniow K, and Sarwas G, “Comparison of N-BEATS and SotA RNN architectures for heart dysfunction classification”, Under Review, 2022. [DOI] [PubMed] [Google Scholar]
- [62].Srivastava A, Hari A, Pratiher S, Alam S, Ghosh N, Banerjee N, and Patra A, “Channel self-attention deep learning framework for multi-cardiac abnormality diagnosis from varied-lead ECG signals”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [63].Linschmann O, Rohr M, Leonhardt KS, and Antink CH, “Multi-label classification of cardiac abnormalities for multi-lead ECG recordings based on auto-encoder features and a neural network classifier”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [64].Nejedly P, Ivora A, Smisek R, Viscor I, Koscova Z, Jurak P, and Plesinger F, “Classification of ECG using ensemble of Residual CNNs with attention mechanism”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [DOI] [PubMed] [Google Scholar]
- [65].Cornely AK, Carrillo A, and Mirsky GM, “Reduced-lead electrocardiogram classification using wavelet analysis and deep learning”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [66].Xia P, He Z, Zhu Y, Bai Z, Yu X, Wang Y, Geng F, Du L, Chen X, Wang P, and Fang Z, “A novel multi-scale convolutional neural network for arrhythmia classification on reduced-lead ECGs”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [67].Seki N, Nakano T, Ikeda K, Hirooka S, Kawasaki T, Yamada M, Saito S, Yamakawa T, and Ogawa S, “Reduced-lead ECG classifier model trained with DivideMix and model ensemble”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [68].Suh J, Kim J, Lee E, Kim J, Hwang D, Park J, Lee J, Park J, Moon S-Y, Kim Y, Kang M, Kwon S, Choi E-K, and Rhee W, “Learning ECG representations for multi-label classification of cardiac abnormalities”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [69].Natarajan A, Boverman G, Chang Y, Antonescu C, and Rubin J, “Convolution-free waveform transformers for multi-lead ECG classification”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [70].Yang H-C, Hsieh W-T, and Chen TP-C, “A mixed-domain self-attention network for multi-label cardiac irregularity classification using reduced-lead electrocardiogram”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [71].Sodmann PF, Vollmer M, and Kaderali L, “Segment, perceive classify multitask learning of the electrocardiogram in a single neural network”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [72].Clark R, Heydarian M, Siddiqui K, Rashidiani S, Khan A, and Doyle TE, “Detecting cardiac abnormalities with multi-lead ECG signals: A modular network approach”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [73].Aublin P, Ammar MB, Achache N, Benahmed M, Hichami AE, Barret M, Fix J, and Oster J, “Cardiac abnormality detection based on an ensemble voting of single-lead classifier predictions”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [74].Wickramasinghe NL and Athif M, “Multi-label cardiac abnormality classification from electrocardiogram using deep convolutional neural networks”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [75].Han H, Park S, Min S, Choi H-S, Kim E, Kim H, Park S, Kim J, Park J, An J, Lee K, Jeong W, Chon S, Ha K, Han M, and Yoon S, “Towards high generalization performance on electrocardiogram classification”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [76].Vicar T, Novotna P, Hejc J, Janousek O, and Ronzhina M, “Cardiac abnormalities recognition in ECG using a convolutional network with attention and input with an adaptable number of leads”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [77].Alkhodari M, Apostolidis G, Zisou C, Hadjileontiadis LJ, and Khandoker AH, “Swarm decomposition enhances the discrimination of cardiac arrhythmias in varied-lead ECG using ResNet-BiLSTM network activations”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [78].Garcia-Isla G, Muscato F, Sansonetti A, Magni S, Corino V, and Mainardi L, “Ensemble classification combining ResNet and hand-crafted features with three-steps training”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [DOI] [PubMed] [Google Scholar]
- [79].Jessen H, van de Leur R, Doevendans P, and van Es R, “Automated diagnosis of reduced-lead electrocardiograms using a shared classifier”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [80].Chollet F et al. (2015). “Keras”, [Online]. Available: https://github.com/fchollet/keras. [Google Scholar]
- [81].Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, van Kerkwijk MH, Brett M, Haldane A, del Río JF, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, and Oliphant TE, “Array programming with NumPy”, Nature, vol. 585, no. 7825, pp. 357–362, Sep. 2020. DOI: 10.1038/s41586-020-2649-2. [Online]. Available: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].T. pandas development team, Pandas-dev/pandas: Pandas, version latest, Feb. 2020. DOI: 10.5281/zenodo.3509134. [Online]. Available: 10.5281/zenodo.3509134. [DOI] [Google Scholar]
- [83].Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Kopf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J, and Chintala S, “PyTorch: An imperative style, high-performance deep learning library”, in Advances in Neural Information Processing Systems 32, Curran Associates, Inc., 2019, pp. 8024–8035. [Online]. Available: http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf. [Google Scholar]
- [84].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, and Duchesnay E, “Scikit-learn: Machine learning in Python”, Journal of Machine Learning Research, vol. 12, pp. 2825–2830, 2011. [Google Scholar]
- [85].Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, and SciPy 1.0 Contributors, “SciPy 1.0: Fundamental algorithms for scientific computing in Python”, Nature Methods, vol. 17, pp. 261–272, 2020. DOI: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Abadi Martın, Agarwal Ashish, Barham Paul, Brevdo Eugene, Chen Zhifeng, Citro Craig, Corrado Greg S., Davis Andy, Dean Jeffrey, Devin Matthieu, Ghemawat Sanjay, Goodfellow Ian, Harp Andrew, Irving Geoffrey, Isard Michael, Jia Y, Jozefowicz Rafal, Kaiser Lukasz, Kudlur Manjunath, Levenberg Josh, Mané Dandelion, Monga Rajat, Moore Sherry, Murray Derek, Olah Chris, Schuster Mike, Shlens Jonathon, Steiner Benoit, Sutskever Ilya, Talwar Kunal, Tucker Paul, Vanhoucke Vincent, Vasudevan Vijay, Viégas Fernanda, Vinyals Oriol, Warden Pete, Wattenberg Martin, Wicke Martin, Yu Yuan, and Zheng Xiaoqiang, TensorFlow: Large-scale machine learning on heterogeneous systems, Software available from tensorflow.org, 2015. [Online]. Available: https://www.tensorflow.org/. [Google Scholar]
- [87].Antoni L, Bruoth E, Bugata P, B. P Jr., Gajdos D, Horvat S, Hudak D, Kmecova V, Stana R, Stankova M, Szabari A, and Vozarikova G, “A two-phase multilabel ECG classification using one-dimensional convolutional neural network and modified labels”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [88].Ren H, Xiong M, and Hooi B, “Robust and task-aware training of deep residual networks for varying-lead ECG classification”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [89].Cai W, Liu F, Wang X, Xu B, and Wang Y, “Classifying different dimensional ECGs using deep residual convolutional neural networks”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [90].Vazquez CG, Breuss A, Gnarra O, Portmann J, and Poian GD, “Two will do: CNN with asymmetric loss, self-learning label correction, and hand-crafted features for imbalanced multi-label ECG data classification”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [91].Li X, Li C, Xu X, Wei Y, Wei J, Sun Y, Qian B, and Xu X, “Towards generalization of cardiac abnormality classification using ECG signal”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [92].Muscato FM, Corino VD, and Mainardi LT, “Ensemble learning of modified residual networks for classifying ECG with different set of leads”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [93].Bortolan G, “3-D ECG images with deep learning approach for identification of cardiac abnormalities from a variable number of leads”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [DOI] [PubMed] [Google Scholar]
- [94].Lebing Pan and WP, Li M, Guan Y, and An Y, “MTFNet: A morphological and temporal features network for multiple leads ECG classification”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [95].Demirel BU, Dogan AH, and Faruque MAA, “Two might do: A beat-by-beat classification of cardiac abnormalities using deep learning with domain-specific features”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [96].Crocker HJ and Costall AW, “An InceptionTime-Inspired convolutional neural network to detect cardiac abnormalities in reduced-lead ECG data”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [97].Jimenez-Serrano S, Rodrigo M, Calvo CJ, Castells F, and Millet J, “Multiple cardiac disease detection from minimal-lead ECG combining feed-forward neural networks with a one-vs-rest approach”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [98].Bodini M, Rivolta MW, and Sassi R, “Classification of ECG signals with different lead systems using AutoML”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [99].Uhlemann T, Prim J, Gumpfer N, Grun D, Wegener S, Krug S, Hannig J, Keller T, and Guckert M, “An ensemble learning approach to detect cardiac abnormalities in ECG data irrespective of lead availability”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [100].Warrick P, Lostanlen V, Eickenberg M, Homsi MN, Rodriguez A, and Anden J, “Arrhythmia classification of reduced-lead electrocardiograms by scattering-recurrent networks”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [101].Osnabrugge N, Keresztesi K, Rustemeyer F, Kaparakis C, Battipaglia F, Bonizzi P, and Karel J, “Multi-label classification on 12, 6, 4, 3 and 2 lead electrocardiography signals using convolutional recurrent neural networks”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [102].Baumgartner M, Kropf M, Haider L, Veeranki S, Hayn D, and Schreier G, “ECG classification combining conventional signal analysis, random forests and neural networks – a stacked learning scheme”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [103].Puszkarski B, Hryniow K, and Sarwas G, “N-beats for heart dysfunction classification”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
- [104].Singstad B-J, Muten EM, and Brekke PH, “Multi-label cardiac abnormality classification from electrocardiogram using deep convolutional neural networks”, Computing in Cardiology, vol. 48, pp. 1–4, 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.