Abstract
Gene expression of the flagellar system is tightly controlled by external stimuli or intracellular signals. A general picture of this regulation has been obtained from studies of Salmonella enterica serovar Typhimurium. However, these regulatory mechanisms do not apply to all bacterial groups. In this study, we have investigated regulation of the flagellar genetic system in Rhodobacter sphaeroides. Deletion analysis, site-directed mutagenesis, and 5′-end mapping were conducted in order to identify the fliO promoter. Our results indicate that this promoter is recognized by the factor ς54. Additionally, 5′-end mapping of the flgB and fliK transcripts suggests that these mRNAs are also transcribed from ς54 promoters. Finally, we showed evidence that suggests that fliC transcription is not entirely dependent on the presence of a complete basal body-hook structure. Our results are discussed in the context of a possible regulatory hierarchy controlling flagellar gene expression in R. sphaeroides.
The flagellum is the structure responsible for the motility of many bacteria. Some of its structural features include the basal body, the hook, and the helical filament. In Salmonella enterica serovar Typhimurium, biosynthesis of the flagellum depends on the expression of more than 40 genes. The products of these genes are required for several flagellar processes, including assembly, export, and transcriptional control (for recent reviews, see references 1 and 16).
The expression of these genes follows a hierarchical pattern that is highly regulated. At the top of the hierarchy is the flhDC operon, encoding two proteins which form the heterotetrameric positive transcriptional regulator of the class II genes. Global regulators, such as the cyclic AMP receptor protein, DnaA, and the nucleoid-associated protein H-NS, influence the expression level of this operon and consequently the formation of flagella (18, 21, 23). The expression of class II genes is dependent on the RNA polymerase-ς70 holoenzyme (Eς70) and FlhD-FlhC. Proteins involved in the formation of the hook and the basal body complex (HBB), as well as the regulatory proteins FlgM and FliA, belong to this class (11). FliA is a specific sigma factor (ς28) required for the expression of class III genes, while FlgM is an anti-sigma factor that inhibits FliA activity. The release of FliA from the inhibitory action of FlgM occurs when the HBB structure is completed, allowing FlgM export out of the cell. FliA is then free to associate with the RNA polymerase core enzyme in order to transcribe class III genes (10, 15).
The flagellar genetic system of Rhodobacter sphaeroides is poorly understood. Detailed analyses of some structural components of the flagellum have been described, but nothing is known about the factors that regulate gene expression. Recently, genetic evidence has suggested the location of functional flagellar promoters in this organism. Complementation studies have indicated the presence of promoters at the fliN-fliO intercistronic region (7), upstream of the flgBCDEF operon (T. Ballado, L. Camarena, B. González-Pedrajo, E. Silva-Herzog, and G. Dreyfus, unpublished data) and upstream of motA (6). However, no physical evidence supporting these results has been reported.
In this work we show evidence that a ς54 promoter is located at the fliN-fliO intercistronic region and is responsible for the transcription of the fliOPQR flhB operon. In addition, primer extension experiments revealed transcription start sites upstream of flgB and fliK. In these two cases, a sequence similar to that of the ς54 promoter was also identified a few base pairs upstream of the transcription start sites. These results indicate that Eς54 is responsible for the expression of genes encoding structural components of the flagellar export apparatus, the motor, the hook, and the basal body proteins. We also determined that mutations in the fliM, fliK, and fliR genes did not affect the expression of other flagellar genes dependent on ς54. In contrast, fliC mRNA was reduced in fliM or flgE strains. These results allow us to propose a regulatory hierarchy controlling the expression of the flagellar genes in R. sphaeroides.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. sphaeroides cells were grown in Sistrom's succinate-basal salt medium at 30°C (20). Heterotrophic growth conditions were achieved by growing 10-ml cultures in 250-ml Erlenmeyer flasks with strong shaking (300 rpm) in the dark. Phototrophic conditions were achieved by growing cultures in completely filled screw-cap tubes under continuous illumination. Cultures were harvested at an optical density at 600 nm of 0.5 ± 0.05 (mean ± standard deviation). When required, spectinomycin (15 μg/ml), kanamycin (25 μg/ml), or tetracycline (1 μg/ml) was added to the culture medium. Escherichia coli strains were grown aerobically at 37°C on Luria-Bertani medium. Antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 10 μg/ml; and kanamycin, 50 μg/ml.
Recombinant DNA techniques.
Routine genetic manipulations were performed as described elsewhere (2). Restriction enzymes, alkaline phosphatase, T4 ligase, and T4 polynucleotide kinase were purchased from GIBCO-BRL. Plasmid DNA was isolated from E. coli using Qiagen columns and procedures. Sequencing was carried out using a Thermosequenase kit (Amersham) on single-stranded DNA.
Conjugal mating.
Plasmid DNA was mobilized into R. sphaeroides cells by conjugation according to procedures previously reported (5).
Site-directed mutagenesis.
Site-directed mutagenesis was performed according to the method of Kunkel (14) with a uracil-containing single-stranded DNA as the template. The oligonucleotides used for mutagenesis were 5′-CTGCAACATCCGTGACGCCCGCCCGCG-3′, 5′-CTGCAACATCCGTGCTGCCCGCCCGCG-3′, 5′-GTCCCCCTCCGCTACAACATCCGTGCCG-3′, and 5′-GTCCCCCTCCGCAACAACATCCGTGCCG-3′.
RNA isolation and Northern blot analysis.
Total RNA was isolated from heterotrophic cultures as described previously (24). For Northern blotting, 20 μg of each RNA sample was separated electrophoretically on agarose-formaldehyde gels and transferred by capillary action onto nylon membranes with a pore size of 0.45 μm. Filter hybridizations were performed as described previously (2). The DNA probe used was a 1.1-kb PstI-BglII fragment from fliC, labeled with [α-32P]dCTP by nick translation.
Primer extension analysis.
Reactions were performed as described previously (2). Total RNA (50 μg for fliO and flgB reactions and 100 μg for fliK reactions) was annealed with a specific primer at 42°C in the presence of 50% formamide. Oligonucleotides used as primers for cDNA synthesis were 5′ end labeled with T4 polynucleotide kinase and 20 μCi of [γ-32P]ATP at 37°C for 30 min. Unincorporated nucleotides were removed by chromatography. The primer elongation reactions were carried out with avian myeloblastosis virus reverse transcriptase (Promega). Unlabeled primers were used to generate a nucleotide sequence ladder.
β-Glucuronidase activity assay. β-Glucuronidase assays employed 4-methylumbelliferyl-β-d-glucuronide as a substrate along with sonicated cell extracts as described previously (12). Samples of 100 μl were taken at three time points between 10 and 40 min and then mixed with 0.9 ml of stop buffer (0.2 M Na2CO3). Fluorimetric determinations were made with a Perkin-Elmer LS-5 apparatus (excitation wavelength, 360 nm; emission wavelength, 446 nm). The fluorimeter was calibrated using 4-methylumbelliferone standards. Specific enzyme activity in cell extracts was expressed as micromoles of 4-methylumbelliferone per minute per milligram of protein. Protein content was determined using the Bio-Rad protein assay kit, with bovine serum albumin as a standard.
RESULTS
Transcriptional organization of the flagellar cluster fliHIJKLMNOPQR flhB.
A large flagellar cluster was previously identified which contained 12 flagellar genes from fliH to fliR and flhB (3, 7). The transcriptional organization of this region was inferred from a complementation study on a strain carrying a polar insertion in the fliM gene (fliM::uidA-aadA). This study indicated the presence of a promoter downstream of fliN (7).
To analyze this possibility, transcriptional fusions of the fliM and fliR genes to the promoterless ′uidA gene were made. These fusions were used to replace either the fliM+ or the fliR+ genes from the chromosomes of the wild-type WS8 strain and from the PG2 strain, which carries the mutation fliK::TnphoA (8). Correct replacement in each of these four strains was confirmed by Southern blot analysis (data not shown).
The expression level of β-glucuronidase dependent on flagellar promoters was determined under heterotrophic growth conditions. Strains carrying the fliM::uidA-aadA or the fliR::uidA-aadA fusion in a wild-type background showed a high level of activity (Table 1). In contrast, when fusions were placed in the fliK::TnphoA background, only the strain carrying the fliR::uidA-aadA allele showed β-glucuronidase activity. This result can be explained in terms of a polar effect exerted by the TnphoA transposon over genes in the same transcriptional unit. Therefore, fliK, fliL, and fliM appear to belong to the same transcriptional unit, whereas fliR belongs to a different operon.
TABLE 1.
β-Glucuronidase specific activities expressed from chromosomal ′uidA fusions to fli promoters
| R. sphaeroides strain | Genotype | β-Glucuronidase activitya |
|---|---|---|
| PG2 | fliK::TnphoA | 0.02 ± 0.14 |
| NG1 | fliM::uidA-aadA | 9.8 ± 2.9 |
| SP1 | fliK::TnphoA fliM::uidA-aadA | 0.09 ± 0.2 |
| SP2 | fliR::uidA-aadA | 14.6 ± 4.5 |
| SP3 | fliK::TnphoA fliR::uidA-aadA | 12.3 ± 3.9 |
Specific activities are given in micromoles of 4-methylumbelliferone produced per minute per milligram of protein. The means and standard deviations of three independent determinations are shown.
Deletion mapping of a transcriptionally active flagellar region.
To identify the promoter controlling transcription of fliR, deletion analysis of a 4.4-kb EcoRI fragment carrying the fliR::uidA-aad allele was performed. This fragment, which incorporates sequences from the middle of fliM to the middle of flhB, was cloned into the pRK415 vector in an orientation opposite to that of the pRK415 promoters.
WS8 cells carrying this plasmid were grown to mid-log phase and the level of β-glucuronidase was determined in a cell extract. This construct produces high levels of β-glucuronidase (Fig. 1, construct 1). The ′uidA gene was cloned into pRK415 in both orientations with respect to the pRK415 promoters for use as controls (Fig. 1, constructs 6 and 7). β-Glucuronidase activity was detected only when uidA was transcribed from the known pRK415 promoters.
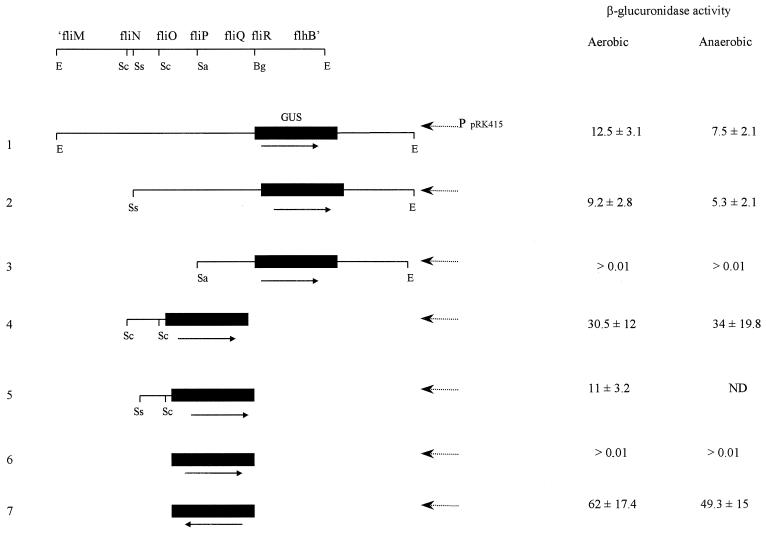
FIG. 1.
β-Glucuronidase activity from fliOp-uidA fusions. The upper part shows some of the relevant restriction sites within the 4.6-kb EcoRI fragment. Arrows indicate the direction of the uidA coding region. Broken arrows indicate the direction of the pRK415 promoters. Abbreviations for restriction sites: E, EcoRI; Sc, SacII; Ss, SstI; Sa, SalI; Bg, BglII. β-Glucuronidase activities are given as indicated in Table 1. GUS, β-glucuronidase; ND, not determined.
Nested deletions of the EcoRI fragment were performed using previously identified restriction sites. The first deletion removed the fliM coding region, leaving the SstI site in the middle of the fliN gene as a 5′ boundary (Fig. 1, construct 2). This construct shows a high level of activity. In contrast, deletion of the next 1.1 kb, up to the SalI site located in fliP, completely abolished β-glucuronidase activity (Fig. 1, construct 3). Therefore, the sequence from the SstI site to the SalI site spanning from the middle of fliN to fliP (1.1 kb) contains a functional promoter.
To further define this promoter region, a 600-bp SacII fragment, spanning from the 3′ end of fliN to the 5′ end of fliO, was cloned upstream of the ′uidA gene (Fig. 1, construct 4). This region was fully functional for promoting uidA transcription. Further deletion of 180 bp from the fliN side only modestly affected the transcriptional capability of this fragment (Fig. 1, construct 5).
In summary, these results localize a functional flagellar promoter to a 420-bp region (SstI-SacII fragment) (Fig. 1, construct 5). Previous sequence analysis of this region did not show a good match with the consensus promoter sequence for ς70; instead, a putative ς54 promoter sequence was clearly identified (7).
Site-directed mutagenesis of the ς54 consensus region.
It is known that the most conserved regions of the promoters recognized by Eς54 are the dinucleotides GG and GC located near positions −24 and −12 upstream from the transcription start site. Changes in these bases interfere with the ability of ς54 to promote transcription (4).
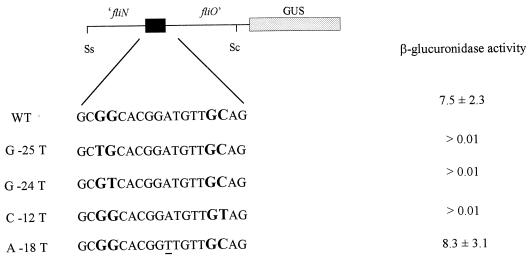
Site-directed mutagenesis was carried out on the ς54 consensus promoter region present in the SstI-SacII fragment. The putative nucleotides in the −25, −24, and −12 positions were replaced by the nucleotide T. After mutagenesis and sequencing, this region was cloned upstream of the ′uidA gene in pRK415 (Fig. 2).
FIG. 2.
β-Glucuronidase expression of wild-type and mutated fliOp promoters. The dark box represents the ς54 promoter sequence. The conserved dinucleotides GG and GC are shown in bold. Specific changes made in each construct are shown in the left column. β-Glucuronidase activities are given as indicated in Table 1. GUS; β-glucuronidase; WT, wild type.
β-Glucuronidase activity was determined for each construct using cell extracts. All mutational changes negatively affected β-glucuronidase activity (Fig. 2). The control contained a mutation at the −18 position, which is irrelevant for promoter recognition (4). In this case, β-glucuronidase activity was very similar to that of the wild type (Fig. 2). These results provide evidence that promoter activity detected in this region is dependent on the RNA polymerase associated with the ς54 factor.
It is known that integration host factor protein (IHF) binds to specific sites placed between the ς54 promoters and the activator binding sites, which are usually located upstream of the initiation site at approximately −100 to −150 bp. Therefore, it has been proposed that IHF bends DNA, favoring contact of the activator with Eς54 bound at the promoter. The 420-bp region proposed to carry a flagellar promoter was analyzed using the program Seqscan at www.bmb.psu.edu/nixon/webtools/molbiol.htm in order to identify any potential IHF binding site. No matches above the program threshold were found. However, it is still possible that a low-affinity binding site with a poor match with the consensus sequence may exist. Finally, two different palindromic sequences which may represent the activator binding site were located at −145 and −127 bp upstream of the ς54 promoter (data not shown).
5′ end mapping of the flagellar mRNA dependent on ς54 promoters.
Putative ς54 promoter sequences have been identified upstream of the flagellar genes flgB, fliK, and motA (GenBank accession number U86454) (6; Ballado et al., unpublished). The nucleotide sequence of each of these regions was aligned with the sequence of the functional ς54 promoter in the fliN-fliO intercistronic region. The nucleotide residues GGCA and TTGC are present in all sequences (data not shown). In order to obtain evidence that these sequences correspond to functional flagellar promoters, we examined if specific mRNA transcripts were produced from these putative promoters.
Primer extension assays were performed using total RNA extracted from wild-type cells. Since flagellar transcripts are scarce, we also included total RNA from WS8 cells overexpressing the region to be analyzed. This was done by cloning the specific region in pRK415 in opposite orientation to the vector promoters. In spite of the low copy number of pRK415, a slight increase (of at least two- or threefold) of the specific transcripts could be expected, facilitating its detection. Primers used in these experiments were designed to be complementary to the regions of the fliO, flgB, and fliK genes predicted to encode the N termini of the gene products.
Each reverse transcription reaction yielded a major band that defines the start site of mRNA transcription (Fig. 3). As expected, this band was stronger from RNA isolated from cells carrying the flagellar promoter sequences in pRK415. In all cases, the cDNA product was of the same size as that from wild-type mRNA.
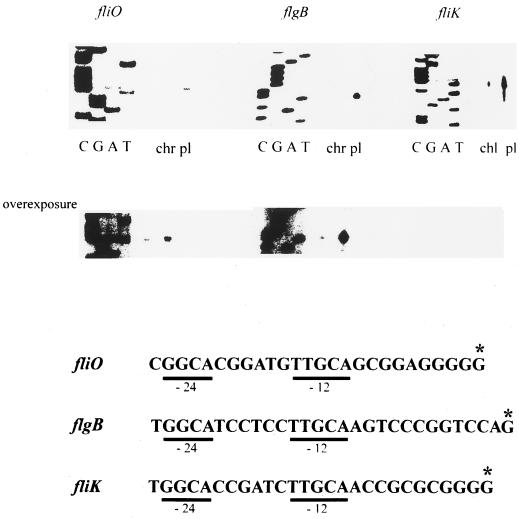
FIG. 3.
Determination of the fliO, flgB, and fliK transcriptional start sites by reverse transcriptase-mediated primer extension. Extension analysis carried out with total RNA isolated from WS8 wild-type cells is indicated as “chr.” The assay carried out with total RNA from WS8 cells transformed with the pRK415 plasmid containing the putative ς54 promoter cloned in opposite orientation to the vector promoters is indicated as “pl.” Nucleotide sequences upstream of the observed transcriptional start sites are shown. The asterisk indicates the nucleotide proposed to be the transcriptional start site. The conserved regions including the −24 and −12 positions are underlined.
The transcriptional start site for fliO was identified at 24 nucleotides (nt) upstream of the TTG start codon (data not shown) and 11 nt downstream of the dinucleotide GC of the sequence recognized by ς54. Transcription of flgB and fliK started 35 and 17 nt upstream of the ATG start codon, respectively (data not shown). In all cases, the distance between the ς54 promoter sequence and the transcription start site is consistent with the distance reported for functional ς54 promoters (4).
The 5′ end of the mRNA located upstream of fliO, together with other evidence presented here, indicates that the expression of the fliO operon depends on Eς54. Moreover, a stable 5′-end mRNA was detected upstream of other regions thought to carry a ς54-dependent promoter. Therefore, these sequences may represent the functional promoters of the flgBCDEF and the fliKLMN operons in R. sphaeroides. Nevertheless, to obtain additional evidence supporting the above conclusions it would be necessary to determine the expression level of these promoters in an rpoN background (lacking the rpoN1 and rpoN2 genes).
Expression of the flagellar fliO promoter is not dependent on growth conditions.
It has been shown for several bacterial groups that flagellar synthesis is affected by environmental conditions (9, 13). In the case of R. sphaeroides, flagellar control signals might be related to the cell cycle and possibly independent of environmental conditions. To determine if growth conditions affect the expression of fliOp, WS8 cells carrying the fusion fliOp-′uidA in pRK415 were grown heterotrophically or photoheterotrophically. No significant difference in the level of β-glucuronidase activity was observed, suggesting that the expression of some of the components involved in flagellar synthesis is not significantly affected by these growth conditions (Fig. 1). Since our results are focused on only two extreme conditions, i.e., light and oxygen, it remains to be investigated if other environmental conditions modify the expression level of the flagellar promoters.
Is flagellar gene expression in R. sphaeroides hierarchical?
In enteric bacteria, the best-characterized checkpoint in flagellar assembly is controlled by the protein FlgM (10, 15). To test if this checkpoint exists in R. sphaeroides, we investigated whether a functional HBB structure was necessary for flagellin gene expression.
Expression of fliC was evaluated by Northern blot analysis using total RNA from wild-type WS8 cells and from cells carrying a mutation in either the fliM or flgE gene. Transcripts of fliC were clearly detected in RNA preparations from wild-type and mutant strains (Fig. 4). However, the mutant strains showed a decrease in the quantity of fliC mRNA produced compared to that of WS8 cells. This result may be due to either a reduction in the amount of mRNA synthesized or a reduction in its stability. Despite this, fliC expression does not appear to be absolutely dependent on the presence of a functional HBB structure.
FIG. 4.
Northern blot analysis of fliC transcripts. Total RNA extracted from WS8 wild-type cells (lane 1), NG1 cells (fliM::uidA-aadA) (lane 2), and LC1 cells (flgE::aadA) (lane 3) was probed with a 32P-labeled fliC fragment as described in Materials and Methods. The nucleotide sequence of the 5′ end of the fliC gene, including its control region, is shown (accession number AF274346). The sequence resembling the ς28 promoter, the putative start codon of fliC, and the ribosomal binding site are shown in bold.
The regulatory region of fliC does not show any sequence similarity with the ς54 consensus promoter. However, in agreement with a previous report, a sequence similar to that of the ς28 consensus promoter (GenBank accession number Y14687) is located approximately 51 bp upstream of the putative ATG start codon (Fig. 4).
DISCUSSION
Structural components of the flagellum of R. sphaeroides have been recently characterized (3, 6, 7, 8, 19; Ballado et al., unpublished). However, still nothing is known about the factors that regulate the expression of the genes involved in motility. In this work, we identified several promoters responsible for the transcription of the flagellar genes involved in the formation of the basal body, the switch, and the hook.
This work showed that fliOp is a functional ς54 promoter. Our results support the idea that this promoter is responsible for the expression of fliO, fliP, fliQ, fliR, and probably flhB. Additional studies are in progress to define whether this operon includes flhB and beyond.
A sequence similar to that of the ς54 consensus promoter was identified upstream of the fliK gene. The functionality of this promoter sequence was supported by the identification of the fliK 5′-end mRNA, the start site of which is within the expected range for a functional ς54 promoter (Fig. 3). On the other hand, the polar effect exerted by the mutation fliK::TnphoA over fliM expression suggests that fliK and fliM belong to the same operon (Table 1). Together, these results support the idea that the ς54 promoter located upstream of fliK is responsible for transcribing the fliK, fliL, fliM, and fliN genes as a single transcriptional unit. The inclusion of the fliN gene in this operon was previously proposed from a complementation study of a strain carrying the fliM::uidA-aadA mutation (7).
A third transcriptional start site defined in this work was located upstream of flgB. A sequence similar to that of the ς54 consensus promoter is at a proper distance from the flgB mRNA initiation site. Consequently, we suggest that this sequence is the functional ς54 promoter responsible for the expression of the flgB, flgC, flgD, flgE, and flgF genes. The internal organization of this region has been recently elucidated, and genetic evidence supports the idea that these genes form an operon (Ballado et al., unpublished).
The data presented here allow us to propose that in R. sphaeroides the expression of the flagellar genes, whose products are involved in the assembly of the basal body, the switch, and the hook structures, is dependent on Eς54. This is in contrast with the situation found in both S. enterica serovar Typhimurium and E. coli, where the flagellar genes involved in the synthesis of these structures are dependent on Eς70 and the FlhD-FlhC activator complex. At a glance, R. sphaeroides flagellar gene expression would appear to be similar to that observed in Caulobacter crescentus, where the genes involved in the formation of the rod and the hook are dependent on Eς54 (22). However, several of the genes reported here as being dependent on ς54 are dependent on Eς73 in C. crescentus. Moreover, two flagellin genes of C. crescentus are dependent on Eς54 (22), whereas the regulatory region of fliC of R. sphaeroides shows a sequence resembling that of a ς28 promoter, as occurs in enteric bacteria (E. coli and Salmonella). Therefore, the flagellar genetic system of R. sphaeroides seems to combine features from these two systems.
As mentioned before, in S. enterica serovar Typhimurium the flagellar genes follow a hierarchical order of expression; therefore, the transcription of the genes placed at a low level of the hierarchy is dependent on the expression of the genes at a higher level. In this regard, in R. sphaeroides the activity of fliOp and the expression of the fliM gene, both of which were tested as the activity of the ′uidA reporter gene, have been shown to be insensitive to the presence of other mutations in flagellar genes, such as fliR, fliM, and flgE (this work and data not shown). Therefore, we suggest that all these genes belong to the same transcriptional class and may be assigned to class II. Interestingly, genetic evidence has suggested that the motA gene may be dependent on Eς54 (6). Therefore, motA could be expressed simultaneously with the genes encoding the structural components of the basal body and the hook. Additional studies are required to define whether the mot genes, together with the fli and the flg genes, can be grouped within the same transcriptional class.
According to current knowledge of the ς54 factor (17), the existence of an activator protein must be considered. This protein may represent the flagellar class I in R. sphaeroides. Although its identity remains to be elucidated, it could be suggested that this protein would belong to the family of ς54-dependent activator proteins.
Finally, the class III genes could be transcribed by Eς28, as seems to be the case for the fliC gene (Fig. 4). The reduction in the amount of the fliC mRNA observed in the flgE and fliM strains may represent the control that class II genes exert over the expression of ς28-dependent genes. This negative control does not seem to be absolute and allows a certain level of fliC expression even in the absence of a functional HBB structure. Supporting the idea that class III genes are expressed from Eς28 promoters, we have found that fliD, which is expected to belong to this class, also shows a ς28 consensus promoter a few base pairs upstream of the putative initiation codon (data not shown).
In summary, our results support the idea that in R. sphaeroides the expression of the flagellar genes is controlled according to a hierarchical pattern. Class II genes are dependent on Eς54, whereas class III genes seem to be dependent on Eς28. The control that class II genes exert over the expression of class III genes does not seem to be tight, and the molecular bases underlying this weak regulation are under investigation in our laboratory.
ACKNOWLEDGMENTS
We are indebted to Laura Velázquez for her comments and critical review of the manuscript. We thank T. Ballado for technical assistance. We also thank Luis Servín-González and E. Silva-Herzog for helpful discussions.
This work was supported in part by DGAPA grant IN221598 to G.D. and L.C.
REFERENCES
- 1.Aizawa S-I. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 3.Ballado T, Campos A, Camarena L, Dreyfus G. Flagellar genes from Rhodobacter sphaeroides are homologous to genes of the fliF operon of Salmonella typhimurium and to the type-III secretion system. Gene. 1996;170:69–72. doi: 10.1016/0378-1119(95)00855-1. [DOI] [PubMed] [Google Scholar]
- 4.Barrios H, Valderrama B, Morett E. Compilation and analysis of ς54-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deepan S H, Sockett R E. Analysis of the motA flagellar motor gene from Rhodobacter sphaeroides, a bacterium with a unidirectional, stop-start flagellum. Mol Microbiol. 1995;17:961–969. doi: 10.1111/j.1365-2958.1995.mmi_17050961.x. [DOI] [PubMed] [Google Scholar]
- 7.García N, Campos A, Osorio A, Poggio S, González-Pedrajo B, Camarena L, Dreyfus G. The flagellar switch genes fliM and fliN of Rhodobacter sphaeroides are contained in a large flagellar gene cluster. J Bacteriol. 1998;180:3978–3982. doi: 10.1128/jb.180.15.3978-3982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González-Pedrajo B, Ballado T, Campos A, Sockett R E, Camarena L, Dreyfus G. Structural and genetic analysis of a mutant of Rhodobacter sphaeroides WS8 deficient in hook length control. J Bacteriol. 1997;179:6581–6588. doi: 10.1128/jb.179.21.6581-6588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heuner K, Brand B, Hacker J. The expression of the flagellum of Legionella pneumophila is modulated by different environmental factors. FEMS Microbiol Lett. 1999;175:69–77. doi: 10.1111/j.1574-6968.1999.tb13603.x. [DOI] [PubMed] [Google Scholar]
- 10.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 11.Ikebe T, Iyoda S, Kutsukake K. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet Syst. 1999;74:179–183. doi: 10.1266/ggs.74.179. [DOI] [PubMed] [Google Scholar]
- 12.Jefferson R A, Burgess S M, Hirsh D. β-Glucuronidase from Escherichia coli as a fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapatral V, Olson J W, Pepe J C, Minnich S A. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol Microbiol. 1996;19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 16.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 17.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima T, Koyanagi R, Katayama T, Miki T, Sekimizu K. Decrease in expression of the master operon of flagellin synthesis in a dnaA46 mutant of Escherichia coli. Biol Pharm Bull. 1997;20:327–331. doi: 10.1248/bpb.20.327. [DOI] [PubMed] [Google Scholar]
- 19.Shah D S H, Armitage J P, Sockett R E. Rhodobacter sphaeroides WS8 expresses a polypeptide that is similar to MotB of Escherichia coli. J Bacteriol. 1995;177:2929–2932. doi: 10.1128/jb.177.10.2929-2932.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sistrom W R. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J Gen Microbiol. 1962;28:607–616. doi: 10.1099/00221287-28-4-607. [DOI] [PubMed] [Google Scholar]
- 21.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol. 1997;24:239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 23.Yanagihara S, Iyoda S, Ohnishi K, Iino T, Kutsukake K. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet Syst. 1999;74:105–111. doi: 10.1266/ggs.74.105. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y S, Kaplan S. Effects of light, oxygen, and substrates on steady-state levels of mRNA coding for ribulose-l,5′-bisphosphate carboxylase and light-harvesting and reaction center polypeptides in Rhodopseudomonas sphaeroides. J Bacteriol. 1985;162:925–932. doi: 10.1128/jb.162.3.925-932.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]