Abstract
Background
Adoptive transfer of genetically engineered T cells expressing antigen-specific T-cell receptors (TCRs) is an appealing therapeutic approach for Epstein-Barr virus (EBV)–associated malignancies of latency type II/III that express EBV antigens (LMP1/2). Patients who are HLA-A*01:01 positive could benefit from such products, since no T cells recognizing any EBV-derived peptide in this common HLA allele have been found thus far.
Methods
HLA-A*01:01–restricted EBV-LMP2–specific T cells were isolated using peptide major histocompatibility complex (pMHC) tetramers. Functionality was assessed by production of interferon gamma (IFN-γ) and cytotoxicity when stimulated with EBV-LMP2–expressing cell lines. Functionality of primary T cells transduced with HLA-A*01:01–restricted EBV-LMP2–specific TCRs was optimized by knocking out the endogenous TCRs of primary T cells (∆TCR) using CRISPR-Cas9 technology.
Results
EBV-LMP2–specific T cells were successfully isolated and their TCRs were characterized. TCR gene transfer in primary T cells resulted in specific pMHC tetramer binding and reactivity against EBV-LMP2–expressing cell lines. The mean fluorescence intensity of pMHC-tetramer binding was increased 1.5–2 fold when the endogenous TCRs of CD8+ T cells was knocked out. CD8+/∆TCR T cells modified to express EBV-LMP2–specific TCRs showed IFN-γ secretion and cytotoxicity toward EBV-LMP2–expressing malignant cell lines.
Conclusions
We isolated the first functional HLA-A*01:01–restricted EBV-LMP2–specific T-cell populations and TCRs, which can potentially be used in future TCR gene therapy to treat EBV-associated latency type II/III malignancies.
Keywords: Epstein-Barr virus, EBV-associated malignancies, lymphoma, nasopharyngeal carcinoma, virus-specific T cells, TCR-gene transfer
Here we identify the first HLA-A*01:01–restricted Epstein-Barr virus Latent Membrane Protein 2 (EBV-LMP2)–specific T-cell population and show that these T-cell populations and T cells modified to express the LMP2-specific T-cell receptor showed IFN-γ secretion and cytotoxicity toward EBV-LMP2–expressing malignant cell lines.
Epstein-Barr virus (EBV) is associated with the development of a broad range of malignancies, including Burkitt lymphoma, Hodgkin lymphoma (HL), B-, T-, and natural killer (NK) cell lymphomas, posttransplant lymphoproliferative disorder (PTLD), nasopharyngeal carcinoma (NPC), and gastric carcinoma (GC) [1, 2]. Although the outcome for most patients with EBV-positive (EBV+) lymphomas and NPC is favorable, patients with refractory or relapsed lymphomas have a poor prognosis. Likewise, malignancies of epithelial origin such as advanced GC are known to have a very poor prognosis [3].
In healthy individuals, EBV enters a latent phase after primary infection. Upon infecting a resting naive B cell, EBV first enters the immunogenic latency phase III where it expresses all viral proteins (eg, EBV nuclear antigen 1–3 [EBNA1–3] and latent membrane proteins [LMPs] 1 and 2) [4]. This results in the activation of the naive B cell, followed by entrance to the second latency phase (II) with a restricted gene expression of only EBNA1, LMP1, and LMP2. This induces the activated B cell to differentiate into a memory B cell, resulting in the establishment of the latency phase I, where only EBNA1 and BARF1 RNAs are expressed [5]. Malignancies associated with this virus often exhibit one of these latency phases. EBV-driven PTLD is associated with latency phase III, resulting in expression of all immunogenic antigens by EBV-infected B cells [1, 6]. Treatment of PTLD with EBV-specific T cells has been proven successful after allogeneic hematopoietic stem cell transplantation, with low rates of graft-vs-host disease [7]. Although Burkitt lymphomas only express weakly immunogenic EBV antigens (latency type I), EBV+ lymphomas and malignancies of epithelial origin, including HL, diffuse large B-cell lymphoma, GC, and NPC, additionally express latency type II proteins LMP1 and LMP2 [8, 9]. Treatment of EBV+ latency type II lymphomas using adoptively transferred EBV-LMP1/2–specific T cells was recently demonstrated [10–12].
It has been reported that patients expressing HLA-A*01:01 and/or HLA-B*37:01 have an increased risk of developing EBV+ HL and infectious mononucleosis, whereas patients expressing HLA-A*02:01 have a decreased risk of developing EBV+ HL [13, 14]. Strikingly, no EBV-specific T cells recognizing any of the EBV antigens in the context of HLA-A*01:01 have been characterized to date, whereas HLA-A*02:01–restricted EBV-specific T-cell responses have been frequently found [13, 15]. It was therefore suggested that HLA-A*01:01–restricted EBV-specific T cells are absent or only present at very low frequencies in the normal T-cell repertoire, although the reasons for this were not clear.
In this study, we aimed to isolate HLA-A*01:01–restricted EBV-specific T cells from healthy HLA-A*01:01–positive EBV+ individuals to allow development of T-cell therapy strategies for patients who harbor an EBV+ malignancy and have an HLA-A*01:01 genotype. Although such T cells were found to be present at extremely low frequencies in peripheral blood of healthy individuals, we succeeded in the characterization and isolation of several HLA-A*01:01–restricted EBV-LMP2–specific T-cell receptors (TCRs) that can be used for TCR gene therapy.
MATERIALS AND METHODS
Collection of Donor Peripheral Blood Mononuclear Cells
After informed consent according to the Declaration of Helsinki, healthy donors homozygous for HLA-A*01:01 were selected from the Sanquin database and the biobank of the Leiden University Medical Center Department of Hematology. Peripheral blood mononuclear cells (PBMCs) were isolated by standard Ficoll Isopaque separation and used either directly or thawed after cryopreservation in the vapor phase of liquid nitrogen. Donor characteristics (HLA typing and EBV serostatus) are provided in Supplementary Table 1.
Isolation and Expansion of HLA-A*01:01–Restricted EBV-Specific T Cells
PBMCs (30 × 106) from healthy donors were first incubated with in-house produced (see Supplementary Material and Methods) peptide-MHC tetramer (pMHC) complexes for 30 minutes at 4°C before adding peridinin chlorophyll protein complex–labeled CD8 (BD, Franklin Lakes, New Jersey) and fluorescein isothiocyanate–labeled CD4 and CD14 (BD) antibodies at 4°C for 30 minutes. The pMHC tetramers used and generated are shown in Table 1. Tetramer-positive CD8+, CD4–, and CD14– T cells were sorted into U-bottom microtiter plates for the generation of T-cell populations. Virus-specific T cells were first specifically expanded as described in the Supplementary Material and Methods. T cells that use a specific TCR-variable β (TCR-Vβ) family were sorted subsequently from the virus-specific T-cell populations using the monoclonal antibodies from the TCR-Vβ kit (Beckman Coulter, Fullerton, California) and were then nonspecifically expanded as described in the Supplementary Material and Methods.
Table 1.
Peptide Major Histocompatibility Complex Tetramers Used for the Isolation of Epstein-Barr Virus–Specific T Cells Restricted to HLA-A*01:01 From Peripheral Blood of Healthy Donors
| Viral Antigen | Epitope | netMHCa Affinity, nM | Isolated, No.b |
|---|---|---|---|
| BZLF-1 | FTPDPYQVPF | 36.51 | 0/6 |
| EBNA3A | YTDHQTTPT | 66.17 | 0/6 |
| LMP2 | ESEERPPTPY | 97.43 | 5/6 |
| EBNA3A | FLQRTDLSY | 123.91 | 0/6 |
| LMP2 | LTEWGSGNRTY | 271.52 | 0/6 |
Abbreviation: MHC, major histocompatibility complex.
anetMHC server 4.0.
bNumber of successful isolations out of a number of donors.
TCR Gene Transfer Into Primary T Cells and Jurkat E6 Cells
TCR variable alpha (α) and TCR variable beta (β) sequences used by EBV-LMP2ESE–specific T-cell populations were determined using ARTISAN polymerase chain reaction (PCR) adapted for TCR PCR as previously described [16, 17]. Primary CD4+ and CD8+ T cells were isolated with magnetic activated cell sorting (MACS) using CD4 and CD8 T-cell isolation kits with LS columns from Miltenyi Biotec (Bergisch Gladbach, Germany). Additional CD25 beads (Miltenyi) were added during CD4+ T-cell isolation to deplete regulatory T cells. CD4+ and CD8+ T cells were nonspecifically activated for 48 hours using an autologous feeder mixture and phytohemagglutinin as described in the Supplementary Material and Methods. After 48 hours, these cells were transduced with retroviral supernatant that contained the TCR-α and TCR-β sequence (see Supplementary Material and Methods) in rectronectin-coated 24-well plates (100 000 cells per well). In specific experiments, the endogenous TCR of primary T cells was knocked out according to a previously described protocol [18] (TRAC/TRBV knockout; ∆TCR) prior to transduction. The endogenous TCR of Jurkat E6 (Clone E6-1 ATCC TIB-152) cells, cultured in stimulator medium, was knocked out using the same approach, and these cells were used in all Jurkat E6 experiments.
MACS enrichments using APC-labeled mTCR-Cβ antibodies (BD) and anti-APC microbeads (Miltenyi) were performed to purify TCR-transduced populations. Transduction efficiencies and purities after MACS enrichments were assessed by staining transduced cells with APC-labeled mTCR-Cβ–specific antibodies (BD) for 30 minutes at 4°C. Prior to mTCR-Cβ staining, cells were stained with phycoerythrin (PE)–labeled HLA-A*01:01/pMHC-LMP2ESE pMHC tetramers to determine the specificity of the introduced TCRs. As a control, cells were stained with PE-labeled HLA-A*01:01/pMHC-CMV-pp50VTE or PE-labeled HLA-A*02:01/pMHC-CMV-pp65NLV pMHC tetramers.
Cytokine Production Assays to Determine Functionality
Interferon gamma (IFN-γ) production by virus-specific T cells or TCR-transduced CD4+/CD8+ T cells was measured and quantified using standard enzyme-linked immunosorbent assays according to the manufacturer’s instructions (Sanquin Reagents). Responder T cells were co-cultured with stimulator cells (see Supplementary Material and Methods) at a ratio of 1:5 (R:S) in T-cell medium supplemented with 25 IU interleukin (IL) 2/mL for 16 hours at 37°C. EBV-transformed lymphoblastoid cell lines (LCLs) were kept in culture for 5 days without new medium prior to experiments to upregulate the LMP2 expression in specific experiments [19].
RESULTS
Isolation of HLA-A*01:01–Restricted EBV-Specific CD8+ T Cells by pMHC Tetramer Enrichment
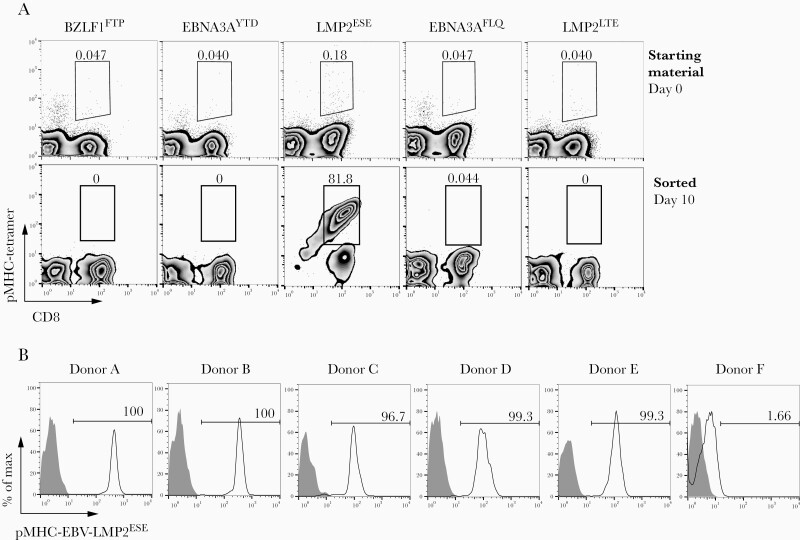
To investigate whether HLA-A*01:01–restricted EBV-specific T cells are present in peripheral blood of healthy EBV-seropositive donors, HLA-A*01:01–binding peptides derived from different immunogenic EBV antigens (EBNA3A, BZLF1, and LMP2) were identified based on an MHC class I peptide binding prediction algorithm [20, 21] (Table 1). HLA-A*01:01/pMHC tetramer complexes were subsequently synthesized (BZLF1FTP, EBNA3AYTD, LMP2ESE, EBNA3AFLQ, and LMP2LTE), and HLA-A*01:01/pMHC tetramer–positive CD8+ T cells were sorted by flow cytometry from PBMCs of 6 HLA-A*01:01–positive healthy donors. HLA-A*01:01–restricted EBV-specific T cells were detected at extremely low frequencies (not exceeding background staining in most cases) in total PBMCs from all 6 donors (representative example for 1 donor in Figure 1A, upper panel). After flow cytometric cell sorting, only EBV-LMP2ESE–specific T cells (Table 1) could be expanded from 5 of 6 donors (representative example for 1 donor in Figure 1A, lower panel). After a second round of sorting, pure EBV-LMP2ESE–specific pMHC+/CD8+ T-cell populations were obtained (Figure 1B). Sequence analysis of the TCR-β variable chain (TRBV) showed that these EBV-LMP2ESE–specific T-cell populations used different TCRs. T cells from donors A, B, and C used the same TCR-β variable and joining chain, but with small differences in the CDR3β region (Table 2). All populations were clonal except those from donor B and donor C. Two subpopulations could be purified from donor B using FACSorting based on expression of TRBV6-3 (population B1) and TRBV13 (population B2). Subpopulations from donor C could not be purified due to expression of the same TRBV6-3 (Table 2). Sequence analysis revealed that TRBV6-3 expressing TCRs from donor A and B used the same TCR-α chain with an identical CDR3α sequence, whereas the TCRs from the 2 populations in donor C used the same α variable and joining chain, but with small differences in the CDR3α region. Surprisingly, 4 of 5 donors harbored a T-cell population expressing TRBV6-3, indicating that diversity within the EBV-LMP2ESE–specific T-cell repertoire is limited.
Figure 1.
Peptide major histocompatibility complex (pMHC) tetramer staining of HLA-A*01:01–restricted Epstein-Barr virus (EBV)–specific T cells. Peripheral blood mononuclear cells (PBMCs) from 6 healthy EBV+ HLA-A*01:01–positive donors were incubated with different pMHC tetramers that were predicted to be strong binders specific for EBV and restricted to HLA-A*01:01. A, Representative examples for donor B are shown. Total PBMCs were stained with pMHC tetramers and CD8. Tetramer-positive cells were sorted and expanded for 2 weeks. Only Epstein-Barr virus Latent Membrane Protein 2 specific T cells targetting the ESEERPPTPY peptide (EBV-LMP2ESE) could be expanded. B, Histograms of pMHC tetramer EBV-LMP2ESE (black line) or unstained (gray) of all sorted EBV-LMP2ESE–specific T-cell populations after a second enrichment and 2 weeks of expansion.
Table 2.
Characteristics of Epstein-Barr virus Latent Membrane Protein 2 specific T cells targetting the ESEERPPTPY peptide Isolated From Peripheral Blood of Healthy HLA-A*01:01–Positive Donors
| Donor ID | TRBV | CDR3-β | TRBJ | TRAV | CDR3-α | TRAJ |
|---|---|---|---|---|---|---|
| A | TRBV6-3 | CASSWEGQYNEQFF | TRBJ2-1 | TRAV12 | CVVTGYSSASKIIF | TRAJ3 |
| B1 | TRBV6-3 | CASSSEGQFNEQFF | TRBJ2-1 | TRAV12 | CVVTGYSSASKIIF | TRAJ3 |
| B2 | TRBV13 | CASSFWAVTGELFF | TRBJ2-2 | TRAV4 | CLVGDM_RGSTLGRLYF | TRAJ18 |
| C1 | TRBV6-3 | CASSPEGVFNEQFF (73.5%) | TRBJ2-1 | TRAV30 | CGTGSGGGADGLTF | TRAJ45 |
| C2 | TRBV6-3 | CASSYGIYEQFF (24.4%) | TRBJ2-1 | TRAV30 | CGTEDGRGGADGLTF | TRAJ45 |
| D | TRBV6-3 | CASSYGWAEAFF | TRBJ1-1 | TRAV2 | CAGNNARLMF | TRAJ31 |
| E | TRBV12-3/4 | CASSSSWTSGSGETQYF | TRBJ2-5 | TRAV26 | CIVSGGKLIF | TRAJ23 |
Amino acids replaced by an underscore could not be determined due to an additional nucleotide insertion retrieving an even number of nucleotides. Donor B harbored 2 distinct populations that could be separated by fluorescence-activated cell sorting based on expression of TRBV6-3 (population B1) and TRBV13 (population B2). Donor C also harbored 2 distinct populations, but these could not be separated due to expression of the same TRBV6-3 gene. The frequencies within this C1 + 2 population are shown in parentheses.
Abbreviations: CDR3, complementary determining region 3; TRAJ, T-cell receptor alpha joining; TRAV, T-cell receptor alpha variable; TRBJ, T-cell receptor beta joining; TRBV, T-cell receptor beta variable.
Specific Exogenous and Endogenous EBV-LMP2ESE Recognition
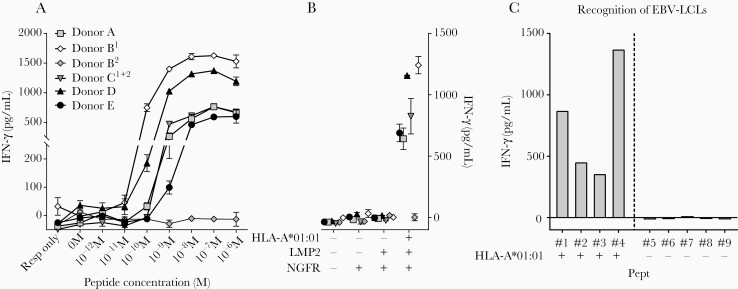
Next, we aimed to determine the functionality of these EBV-LMP2ESE–specific T-cell populations. The T-cell populations were stimulated with K562 cells transduced with HLA-A*01:01 and pulsed with various concentrations of EBV-LMP2ESE peptide. Five of 6 EBV-LMP2ESE–specific T-cell populations recognized up to 10–10 M of exogenously pulsed peptide (Figure 2A). Similarly, when stimulated with K562 cells transduced with both HLA-A*01:01 and the full coding sequence of EBV-LMP2, all T-cell populations except TRBV13-expressing T cells from donor B recognized endogenously processed and presented EBV-LMP2ESE peptide (Figure 2B). To analyze recognition of naturally processed and presented EBV-LMP2ESE peptide, EBV-LCLs were cultured for 5 days without refreshing media to increase LMP2 expression [19] (Supplementary Figure 1). Production of IFN-γ upon coculture of HLA-A*01:01–positive EBV-LCLs demonstrated that EBV-LMP2–specific T cells were capable of recognizing EBV-LMP2ESE peptide processed and presented under physiological conditions (Figure 2C).
Figure 2.
Recognition of exogenously and endogenously presented Epstein-Barr virus Latent Membrane Protein 2 ESEERPPTPY (EBV-LMP2ESE) peptide by EBV-LMP2–specific T cells. A, Six different EBV-LMP2ESE–specific T-cell populations were stimulated with HLA-A*01:01–transduced K562 cells loaded with titrated concentrations of the respective peptide for 16 hours. Interferon gamma (IFN-γ) production was measured by standard enzyme-linked immunosorbent assay. B, EBV-LMP2ESE–specific T cells were stimulated with K562 cells that were retrovirally transduced with the full coding sequence of LMP2 and/or HLA-A*01:01. K562 cells transduced with only the marker gene NGFR were used as additional control. C, Representative example of an EBV-LMP2ESE–specific T-cell population (donor A) that was stimulated with HLA-A*01:01–positive and HLA-A*01:01–negative EBV-transformed lymphoblastoid cell lines (LCLs) that expressed LMP2 under physiological conditions. Shown are mean with standard deviation of 1 experiment carried out in triplicate (A/B).
TCR Gene Transfer in Primary CD4+ and CD8+ T Cells
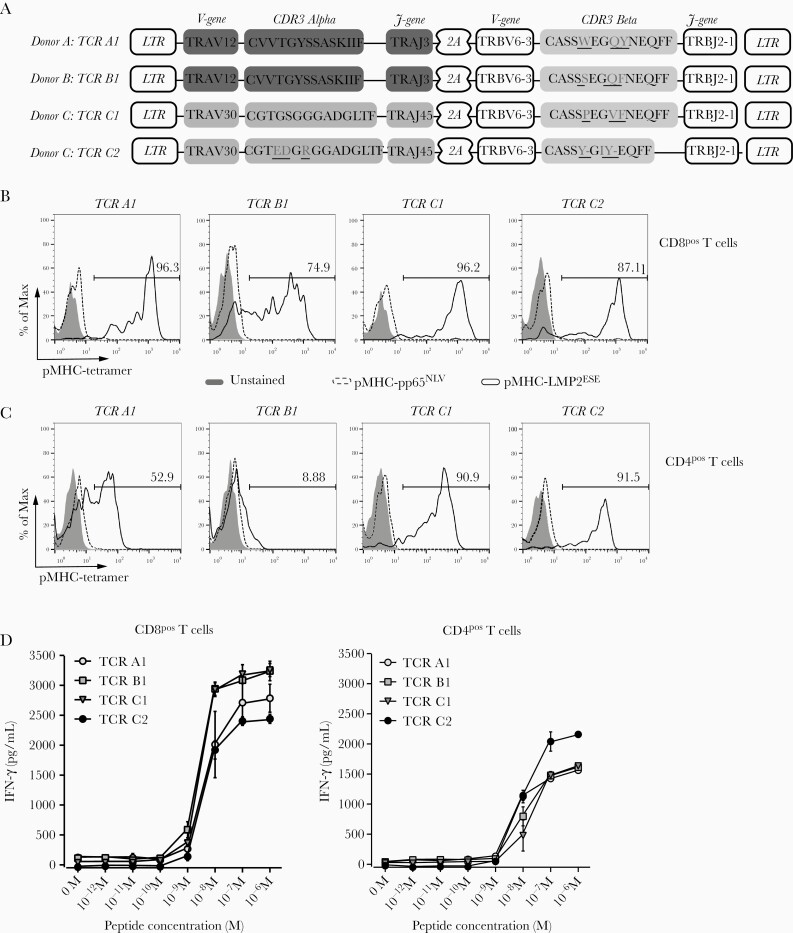
To study the introduction of EBV-LMP2ESE–specific reactivity by TCR gene transfer, different EBV-LMP2ESE–specific TCRs were cloned. Since 4 of 5 functional T-cell populations harbored EBV-LMP2ESE–specific T cells expressing TRBV6-3 and TRBJ2-1 (ie, donor A, B, and C), we compared the EBV-LMP2ESE–specific reactivity of these TCRs. TCRs were codon optimized and modified with a murine constant domain to increase preferential pairing of the introduced TCR-α and TCR-β chain and cloned into a modified MP71-flex vector (Figure 3A). First, we introduced EBV-LMP2ESE–specific TCRs into CD8+ primary T cells. CD8+ T cells were isolated from peripheral blood of 2 unrelated healthy donors using MACS separation (>95% pure; data not shown). High transduction efficiencies (range, 35%–42%) were obtained for all TCRs, resulting in 89%–98.4% transduced T cells after purification. Specific binding of HLA-A*01:01/pMHC-EBV-LMP2ESE tetramers was demonstrated for all EBV-LMP2ESE–specific TCR-transduced CD8+ T cells (Figure 3B). However, CD8+ T cells transduced with TCR-B1 exhibited heterogenous staining with the HLA-A*01:01/pMHC-EBV-LMP2ESE tetramer. TCR-transduced CD8+ T cells did not stain with a control pMHC tetramer containing an irrelevant CMV peptide.
Figure 3.
T-cell receptor (TCR) gene transfer introduced Epstein-Barr virus Latent Membrane Protein 2 ESEERPPTPY (EBV-LMP2ESE) specificity and reactivity into primary CD8+ and CD4+ T cells. Primary CD8+ and CD4+ T cells were isolated from healthy EBV-negative HLA-A*01:01–positive donor G and donor H, using magnetic activated cell sorting (MACS) isolations. T cells were transduced with retroviral supernatant containing the constructs of the TRBV6-3/TRBJ2-1 expressing EBV-LMP2ESE–specific T-cell receptors (TCRs). Transduced T cells were purified and enriched based on expression of the murine-TCR-Cβ domain using MACS isolation. Shown are results and data from donor H. A, Design of the MP71-flex retroviral expression vector. Underscores reflect differences between CDR3 regions. B and C, Shown are histograms of a specific HLA-A*01:01/peptide major histocompatability complex (pMHC)–LMP2ESE tetramer (black line) or irrelevant HLA-A*01:01/pMHC-pp65NLV tetramer (dotted line) staining of CD8+ (B) or CD4+ (C) T cells transduced and enriched based on expression of EBV-LMP2ESE–specific TCRs. Numbers in the middle right represent percentage of EBV-LMP2ESE –specific T cells binding HLA-A*01:01/pMHC-LMP2ESE tetramer. D, EBV-LMP2ESE–specific TCR-transduced CD8+ (D) and CD4+ (E) T-cell populations were stimulated with HLA-A*01:01–transduced K562 cells loaded with titrated concentrations of the respective peptide for 16 hours. Interferon gamma (IFN-γ) was measured by standard enzyme-linked immunosorbent assay. Data are representative of 3 separate experiments, performed in 2 different donors. Abbreviations: 2A, self-cleaving peptide side; LTR, long terminal repeat.
To determine their dependence on CD8 for binding to pMHC-EBV-LMP2ESE tetramers, all 4 EBV-LMP2ESE–specific TCRs were introduced in CD4+ T cells. Similar transduction efficiencies and enrichments were obtained as for CD8+ T cells. Lower intensities of pMHC-EBV-LMP2ESE–specific tetramer staining were observed for CD4+ T cells transduced with EBV-LMP2ESE–specific TCRs compared to similarly transduced CD8+ T cells. TCR-B1–transduced CD4+ T cells virtually lacked pMHC-EBV-LMP2ESE–specific tetramer staining (Figure 3C). This shows that 3 of 4 TCRs do not fully depend on the co-receptor CD8 to bind pMHC-EBV-LMP2ESE–specific tetramer, but the intensity of tetramer staining is significantly reduced in the absence of CD8.
Finally, we investigated the functionality of primary CD8+ and CD4+ T cells transduced with the EBV-LMP2ESE–specific TCRs. Both CD8+ and CD4+ T cells produced IFN-γ (and granulocyte macrophage colony-stimulating factor and IL-4; data not shown) upon stimulation with HLA-A*01:01–transduced K562 cells exogenously loaded with different concentrations of EBV-LMP2ESE peptide (Figure 3D). EBV-LMP2ESE–specific T-cell populations recognized up to 10–8 M and 10–9 M of exogenously pulsed peptide. In summary, these data show that all EBV-LMP2ESE–specific TCRs, despite differences in the level of pMHC-tetramer staining, recognize exogenously loaded EBV-LMP2ESE peptide. These findings were confirmed when EBV-LMP2ESE–specific TCRs were introduced in Jurkat E6 cells with and without CD8 (Supplementary Figure 2).
Reduction of Endogenous TCR Expression Increases EBV-LMP2ESE–Specific Tetramer Binding in CD8+ T Cells
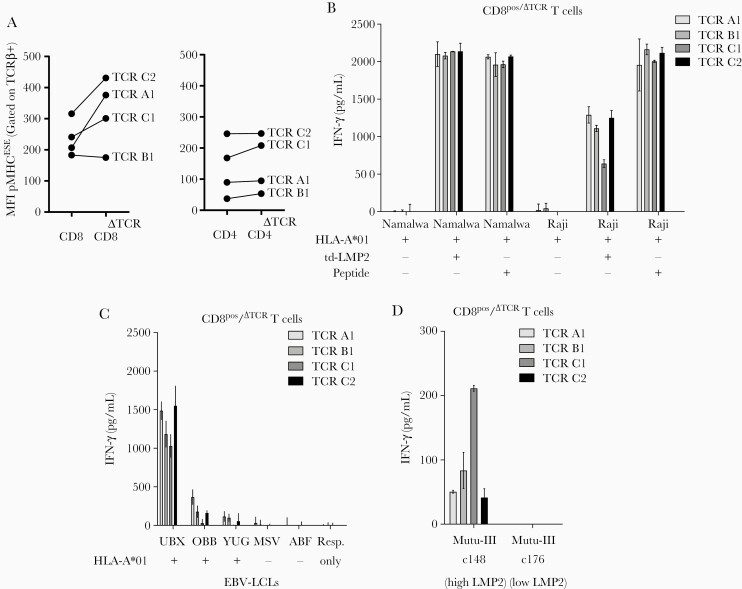
Knocking out the endogenous TCR of primary T cells can increase the functionality of the introduced TCR [18, 22]. Therefore, we knocked out the endogenous TCR (∆TCR) of CD4+ and CD8+ T cells using CRISPR-Cas9 technology targeting the TRAC/TRBC loci prior to transduction with EBV-LMP2ESE–specific TCRs (Supplementary Figure 3). Similar transduction efficiencies were observed as before, ranging from 20% to 40%, and all populations were successfully enriched for the introduced TCR (data not shown). The mean fluorescence intensity (MFI) of EBV-LMP2ESE tetramer binding was substantially increased in 3 of 4 TCR-transduced CD8+/∆TCR T cells. In contrast, in CD4+/∆TCR T cells the MFI of EBV-LMP2ESE tetramer binding was not increased (Figure 4A; Supplementary Figure 4), while the MFI of the expression of the introduced (mTCR-Cβ) was similar for CD4+ and CD8+ T cells (data not shown).
Figure 4.
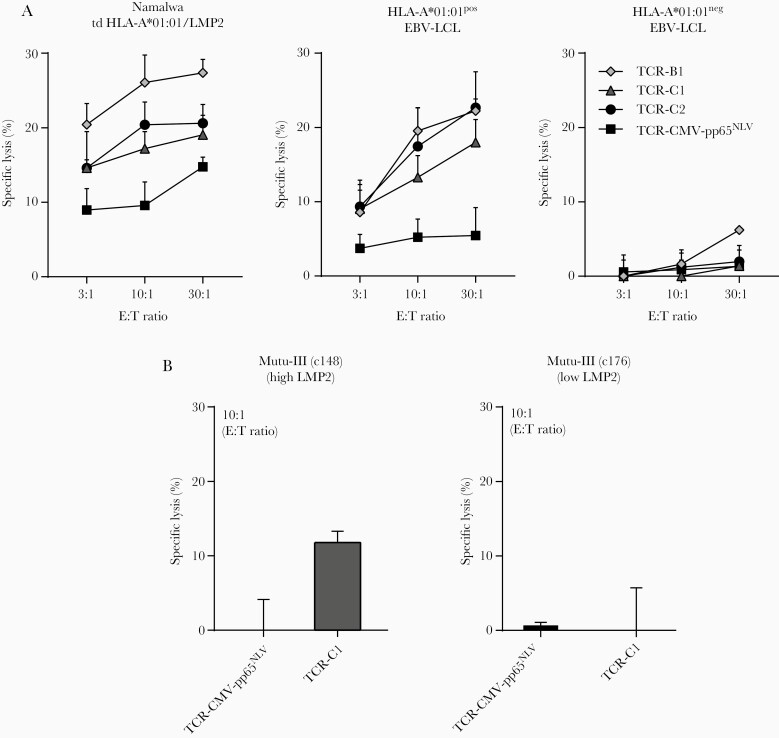
CD8+/ΔTCR T cells transduced with Epstein-Barr virus Latent Membrane Protein 2 ESEERPPTPY (EBV-LMP2ESE)–specific T-cell receptors (TCRs) effectively recognize endogenously processed and presented LMP2ESE peptide. A, Mean fluorescence intensity (MFI) of peptide major histocompatibility complex (pMHC) EBV-LMP2ESE tetramer binding was assessed for CD8+ (left) and CD4+ (right) T cells with and without (ΔTCR) endogenous expressing TCRs. B, CD8+/ΔTCR T cells transduced with EBV-LMP2ESE–specific TCRs were stimulated for 16 hours with HLA-A*01:01–transduced EBV-associated malignant cell lines Namalwa and Raji in a responder-to-stimulator ratio of 1:5. These cell lines were additionally transduced with LMP2 and the cell lines without LMP2 were exogenously pulsed with 10–6 M of EBV-LMP2ESE peptide. C, CD8+/ΔTCR T cells transduced with EBV-LMP2ESE–specific TCRs were stimulated for 16 hours with HLA-A*01:01–positive and HLA-A*01:01–negative EBV-transformed lymphoblastoid cell lines (LCLs) that express LMP2 under physiological conditions in a responder-to-stimulator ratio of 1:5. D, CD8+/ΔTCR T cells transduced with EBV-LMP2ESE–specific TCRs were stimulated for 16 hours with 2 HLA-A*01:01–transduced Burkitt lymphoma mutu-III cell lines that were expected to express LMP2 under physiological conditions. However, mutu-III subclone c148 expressed LMP2 >100-fold higher compared to subclone c176. Data shown are from separate experiments carried out in triplicate (B, C, and D) with T cells from donor H.
Functionality was assessed by stimulating CD8+/∆TCR T cells with different EBV-associated cell lines and measurement of IFN-γ production. It has been shown that some EBV-associated malignant cell lines, excluding EBV-LCLs and the Burkitt lymphoma cell line mutu-III, lose their EBV genome in vitro [23, 24]. Therefore, we used EBV-LCLs and 2 subclones of the mutu-III cell line that should express EBV-LMP2 at physiological levels and transduced the EBV-associated malignant cell lines Namalwa and Raji with HLA-A*01:01 and the full-coding EBV-LMP2 sequence. All CD8+/∆TCR T cells expressing EBV-LMP2ESE–specific TCRs recognized the malignant cell lines transduced with EBV-LMP2 and HLA-A*01:01 (Figure 4B), but not those without transduction of LMP2. In accordance, LMP2 expression was relatively higher in LMP2-transduced Namalwa cells compared to LMP2-transduced Raji cells (Supplementary Figure 5). All CD8+/∆TCR T cells expressing EBV-LMP2ESE–specific TCRs recognized HLA-A*01:01+ EBV-LCLs that expressed LMP2 at physiological levels, while no recognition of HLA-A*01:01-negative EBV-LCLs was observed (Figure 4C; Supplementary Figure 5). The EBV-related Burkitt mutu-III subclone c148 showed the highest physiological LMP2 expression, whereas the mutu-III subclone c176 had a >100 fold lower LMP2 expression (Supplementary Figure 5). In accordance with this, all CD8+/∆TCR T cells expressing EBV-LMP2ESE–specific TCRs recognized the HLA-A*01:01–transduced subclone c148, but not subclone c176 (Figure 4D).
Primary CD8+/∆TCR T Cells Transduced With EBV-LMP2ESE-Specific TCRs Lyse LMP2 Expressing Target Cells
Finally, we investigated the ability of these EBV-LMP2ESE–specific TCR-transduced T cells to lyse EBV-LMP2+ target cells. CD8+/∆TCR T cells transduced with CMV-pp65NLV–specific TCRs were used as a negative control. Efficient lysis by CD8+/∆TCR T cells transduced with EBV-LMP2ESE–specific TCRs was observed for cell lines pulsed with EBV-LMP2ESE peptide (Supplementary Figure 6). Specific lysis was also observed when tested against an HLA-A*01:01/LMP2–transduced cell line and HLA-A*01:01+ EBV-LCLs (Figure 5A). Modest lysis by CD8+/∆TCR T cells transduced with EBV-LMP2ESE–specific TCR-C1 was observed of the HLA-A*01:01–transduced mutu-III subclone c148 with the highest LMP2 expression (Figure 5B). As expected, no or only limited lysis of LMP2-expressing target cells was observed with the CMV-pp65NLV–specific TCR transduced CD8+ T cells (Figure 5), and no lysis was observed of HLA-A*01:01-negative EBV-LCLs. In summary, these findings demonstrate that HLA-A*01:01–restricted EBV–specific T cells do exist, and shows that their TCR was capable of producing IFN-γ and lysis of LMP2-expressing cells upon transduction into primary CD8+ T cells.
Figure 5.
CD8+/ΔTCR T cells transduced with Epstein-Barr virus Latent Membrane Protein 2 ESEERPPTPY (EBV-LMP2ESE)–specific T-cell receptors (TCRs) lyse target cells presenting endogenous LMP2 peptide. A, Representative examples are shown of CD8+/ΔTCR T cells transduced with EBV-LMP2ESE–specific TCRs that were tested for their lytic capacity against malignant cells transduced with HLA-A*01:01 and LMP2 or against HLA-A*01:01–positive or –negative EBV-transformed lymphoblastoid cell lines (LCLs). CD8+/ΔTCR T cells transduced with a TCR targeting HLA-A*02:01/cytomegalovirus (CMV)–pp65NLV were used as control. B, Primary CD8+/ΔTCR T cells transduced with EBV-LMP2ESE–specific TCR-C1 were tested for their lytic capacity against 2 HLA-A*01:01–transduced subclones of the malignant Burkitt lymphoma cell line mutu-III. Mutu-III subclone c148 expressed LMP2 >100-fold higher compared to subclone c176. CD8+/ΔTCR T cells transduced with a TCR targeting HLA-A*02:01/CMV-pp65NLV were used as negative control. Data are shown from 1 experiment carried out in triplicate. Abbreviations: td; transduced, E:T, Effector:Target.
DISCUSSION
In this study we successfully isolated the first HLA-A*01:01–restricted EBV-specific T cells that recognized the LMP2 epitope ESEERPPTPY. These T cells could be isolated from 5 of 6 healthy donors, and TCR sequencing analyses revealed limited TCR diversity. All EBV-LMP2ESE–specific T-cell populations, except the TRBV13-expressing subpopulation from donor B, were functional against stimulator cells that express LMP2 under physiological conditions. Furthermore, retroviral introduction of EBV-LMP2ESE–specific TCRs into CD8+ and CD4+ T cells permitted specific recognition of EBV-LMP2–expressing cell lines, and knockout of the endogenous TCR (ΔTCR) using CRISPR-Cas9 technology increased pMHC-EBV-LMP2ESE–specific tetramer binding in CD8+ T cells. These CD8+/ΔTCR T cells lyse EBV-LMP2–expressing target cells, illustrating that these novel TCRs may be exploited to enhance the immunotherapy of HLA-A*01:01+ EBV-associated malignancies.
So far, no HLA-A*01:01–restricted EBV-specific T-cell populations have been described, which lead to the assumption that they do not exist or that they are present at only very low frequencies [15]. Peptide-binding predictions revealed 5 strong binding HLA-A*01:01–restricted peptides (Table 1). The frequencies in total PBMCs of the analyzed HLA-A*01:01–restricted EBV-specific T-cell populations did not exceed background staining and T cells recognizing the LMP2ESE peptide could only be successfully isolated and expanded after a round of positive selection, again underscoring the very low frequencies of HLA-A*01:01–restricted EBV-specific T cells and confirming the reported difficulty to isolate HLA-A*01:01–restricted EBV-specific T cells [14]. The TCR repertoire of the EBV-LMP2ESE–specific T cells appeared to be very skewed, with preferential expression of TRBV12-3/4 (n = 1/5 donors), TRBV13 (n = 1/5), or TRBV6-3 (n = 4/5). TRBV13-expressing EBV-LMP2ESE–specific T cells were found to be not functional, although this population did exhibit proper pMHC-EBV-LMP2ESE–specific tetramer binding. Such dysfunctional T cells may result in an overestimation of the functional HLA-A*01:01–restricted EBV-specific T cells when pMHC tetramers are used. However, others report that pMHC tetramers can also fail to detect functional T cells, resulting in an underestimation [25].
The TCR repertoire of the TRBV6-3-expressing T-cell populations was found to be extremely skewed, with almost identical CDR3β sequences. Although these TCRs were very similar, differences in pMHC-EBV-LMP2ESE tetramer binding were observed when these TCRs were introduced in primary CD8+ and CD4+ T cells. A lower intensity of pMHC-EBV-LMP2ESE tetramer binding was observed for all TCRs transduced in CD4+ T cells, suggesting that these TCRs are not completely CD8 independent. Differences in pMHC tetramer binding of the different TCRs transduced in CD8+ T cells could be the result of competition for CD3 with the endogenous TCR [26, 27]. However, when we knocked out the endogenous TCR, a subtle increase in intensity of tetramer staining for the introduced TCR was seen, but this did not result in increased functional reactivity. Introduction of EBV-LMP2ESE–specific TCRs in CD4+ T cells also resulted in recognition of peptide-pulsed stimulator cells, but no recognition of LMP2-expressing cells was observed. In conclusion, both CD8+ and CD4+ T cells can be modified by TCR gene transfer to contribute to recognition of LMP2-expressing target cells, which could be beneficial for TCR gene transfer immunotherapies.
It is evident that virus-specific TCRs recognize antigens processed and presented in HLA by virus-infected cells. Both the TCR and the expression of the viral antigen play an important role in recognition of virus-infected cells. We demonstrated that all primary CD8+ T cells transduced with EBV-LMP2ESE–specific TCRs showed good recognition of peptide-pulsed and LMP2 expressing cell lines, suggesting sufficient avidity of the TCR. However, limited recognition was observed of EBV-LCLs expressing LMP2 at physiological levels and there was no clear correlation between EBV-LMP2–specific reactivity and LMP2 expression. In contrast to the full-coding LMP2-transduced cell lines, EBV-LCLs are known to express LMP2 as 2 splice variants (ie, LMP2a and LMP2b) [28]. Our quantitative PCR assay was not able to distinguish LMP2a from LMPb. Since the LMP2ESE epitope is located in the LMP2b splice variant, it is possible that the EBV-LCLs OBB and YUG were less well recognized because of the unpredictable ratio between these 2 variants [29]. Additionally, differential expression per cell could result in less overall recognition, only showing recognition of cells that sufficiently express LMP2. We showed that LMP2 expression can be increased by exhausting EBV-LCLs [19], resulting in recognition and lysis. EBV-LCL–specific lysis by our EBV-LMP2ESE–specific T cells did not exceed 40%, which resembles results shown by others for EBV-LMP1/2–specific T cells restricted to HLA-A*02:01 [30] and HLA-A*11:01 [23]. In line with this, others reported that EBV-EBNA1–specific T cells were not able to lyse EBV-LCLs within short-term cytotoxicity assays, but they could prevent the longer-term outgrowth of these EBV-LCLs [31].
Overall, our findings demonstrate that although present in very low frequencies, HLA-A*01:01–restricted EBV-LMP2–specific T cells do exist and are capable of killing LMP2-expressing malignant cells and LMP2 expressing EBV-LCLs. Therefore, gene transfer of these LMP2-specific TCRs may be exploited to enhance the immunotherapy of HLA-A*01:01–positive EBV-associated malignancies of latency type II/III.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The sponsor of this study is a nonprofit organization that supports science in general and had no role in gathering, analyzing, or interpreting the data
Financial support. This work was supported by Sanquin Research and the Landsteiner Laboratory for Blood Cell research (PPO 15–37/L number 2101).
Contributor Information
Wesley Huisman, Department of Hematology, Leiden University Medical Center, The Netherlands; Department of Hematopoiesis, Sanquin Research and Landsteiner Laboratory for Blood Cell Research, Amsterdam, The Netherlands.
Ilse Gille, Department of Hematology, Leiden University Medical Center, The Netherlands.
Lieve E van der Maarel, Department of Hematology, Leiden University Medical Center, The Netherlands.
Lois Hageman, Department of Hematology, Leiden University Medical Center, The Netherlands.
Laura T Morton, Department of Hematology, Leiden University Medical Center, The Netherlands.
Rob C M de Jong, Department of Hematology, Leiden University Medical Center, The Netherlands.
Mirjam H M Heemskerk, Department of Hematology, Leiden University Medical Center, The Netherlands.
Derk Amsen, Department of Hematopoiesis, Sanquin Research and Landsteiner Laboratory for Blood Cell Research, Amsterdam, The Netherlands.
J H Frederik Falkenburg, Department of Hematology, Leiden University Medical Center, The Netherlands.
Inge Jedema, Department of Hematology, Leiden University Medical Center, The Netherlands.
References
- 1. Shannon-Lowe C, Rickinson AB, Bell AI. Epstein-Barr virus-associated lymphomas. Philos Trans R Soc Lond B Biol Sci 2017; 372:20160271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neparidze N, Lacy J. Malignancies associated with Epstein-Barr virus: pathobiology, clinical features, and evolving treatments. Clin Adv Hematol Oncol 2014; 12:358–71. [PubMed] [Google Scholar]
- 3. Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol 2009; 472:467–77. [DOI] [PubMed] [Google Scholar]
- 4. Amon W, Farrell PJ. Reactivation of Epstein-Barr virus from latency. Rev Med Virol 2005; 15:149–56. [DOI] [PubMed] [Google Scholar]
- 5. Dugan JP, Coleman CB, Haverkos B. Opportunities to target the life cycle of Epstein-Barr virus (EBV) in EBV-associated lymphoproliferative disorders. Front Oncol 2019; 9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montone KT, Hodinka RL, Salhany KE, Lavi E, Rostami A, Tomaszewski JE. Identification of Epstein-Barr virus lytic activity in post-transplantation lymphoproliferative disease. Mod Pathol 1996; 9:621–30. [PubMed] [Google Scholar]
- 7. Gottschalk S, Heslop HE, Rooney CM. Adoptive immunotherapy for EBV-associated malignancies. Leuk Lymphoma 2005; 46:1–10. [DOI] [PubMed] [Google Scholar]
- 8. Greijer AE, Ramayanti O, Verkuijlen SA, Novalić Z, Juwana H, Middeldorp JM. Quantitative multi-target RNA profiling in Epstein-Barr virus infected tumor cells. J Virol Methods 2017; 241:24–33. [DOI] [PubMed] [Google Scholar]
- 9. Brooks L, Yao QY, Rickinson AB, Young LS. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol 1992; 66:2689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McLaughlin LP, Rouce R, Gottschalk S, et al. EBV/LMP-specific T cells maintain remissions of T- and B-cell EBV lymphomas after allogeneic bone marrow transplantation. Blood 2018; 132:2351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bollard CM, Gottschalk S, Torrano V, et al. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. J Clin Oncol 2014; 32:798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roskrow MA, Suzuki N, Gan Yj, et al. Epstein-Barr virus (EBV)-specific cytotoxic T lymphocytes for the treatment of patients with EBV-positive relapsed Hodgkin’s disease. Blood 1998; 91:2925–34. [PubMed] [Google Scholar]
- 13. Niens M, Jarrett RF, Hepkema B, et al. HLA-A*02 is associated with a reduced risk and HLA-A*01 with an increased risk of developing EBV+ Hodgkin lymphoma. Blood 2007; 110:3310–5. [DOI] [PubMed] [Google Scholar]
- 14. Jones K, Wockner L, Brennan RM, et al. The impact of HLA class I and EBV latency-II antigen-specific CD8(+) T cells on the pathogenesis of EBV(+) Hodgkin lymphoma. Clin Exp Immunol 2016; 183:206–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brennan RM, Burrows SR. A mechanism for the HLA-A*01-associated risk for EBV+ Hodgkin lymphoma and infectious mononucleosis. Blood 2008; 112:2589–90. [DOI] [PubMed] [Google Scholar]
- 16. van Bergen CA, van Luxemburg-Heijs SA, de Wreede LC, et al. Selective graft-versus-leukemia depends on magnitude and diversity of the alloreactive T cell response. J Clin Invest 2017; 127:517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koning MT, Nteleah V, Veelken H, Navarrete MA. Template-switching anchored polymerase chain reaction reliably amplifies functional lambda light chain transcripts of malignant lymphoma. Leuk Lymphoma 2014; 55:1212–4. [DOI] [PubMed] [Google Scholar]
- 18. Morton LT, Reijmers RM, Wouters AK, et al. Simultaneous deletion of endogenous TCRαβ for TCR gene therapy creates an improved and safe cellular therapeutic. Mol Ther 2020; 28:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wasil LR, Tomaszewski MJ, Hoji A, Rowe DT. The effect of Epstein-Barr virus latent membrane protein 2 expression on the kinetics of early B cell infection. PLoS One 2013; 8:e54010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016; 32:511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 2003; 12:1007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schober K, Müller TR, Gökmen F, et al. Orthotopic replacement of T-cell receptor α- and β-chains with preservation of near-physiological T-cell function. Nat Biomed Eng 2019; 3:974–84. [DOI] [PubMed] [Google Scholar]
- 23. Zheng Y, Parsonage G, Zhuang X, et al. Human leukocyte antigen (HLA) A*1101-restricted Epstein-Barr virus-specific T-cell receptor gene transfer to target nasopharyngeal carcinoma. Cancer Immunol Res 2015; 3:1138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zuo L, Yu H, Liu L, et al. The copy number of Epstein-Barr virus latent genome correlates with the oncogenicity by the activation level of LMP1 and NF-κB. Oncotarget 2015; 6:41033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rius C, Attaf M, Tungatt K, et al. Peptide-MHC class I tetramers can fail to detect relevant functional T cell clonotypes and underestimate antigen-reactive T cell populations. J Immunol 2018; 200:2263–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomas S, Stauss HJ, Morris EC. Molecular immunology lessons from therapeutic T-cell receptor gene transfer. Immunology 2010; 129:170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heemskerk MH, Hagedoorn RS, van der Hoorn MA, et al. Efficiency of T-cell receptor expression in dual-specific T cells is controlled by the intrinsic qualities of the TCR chains within the TCR-CD3 complex. Blood 2007; 109:235–43. [DOI] [PubMed] [Google Scholar]
- 28. Ikeda M, Ikeda A, Longan LC, Longnecker R. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 2000; 268:178–91. [DOI] [PubMed] [Google Scholar]
- 29. Lynch DT, Zimmerman JS, Rowe DT. Epstein-Barr virus latent membrane protein 2B (LMP2B) co-localizes with LMP2A in perinuclear regions in transiently transfected cells. J Gen Virol 2002; 83:1025–35. [DOI] [PubMed] [Google Scholar]
- 30. Cho HI, Kim UH, Shin AR, et al. A novel Epstein-Barr virus-latent membrane protein-1-specific T-cell receptor for TCR gene therapy. Br J Cancer 2018; 118:534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee SP, Brooks JM, Al-Jarrah H, et al. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J Exp Med 2004; 199:1409–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.