PURPOSE
Advances in biological measurement technologies are enabling large-scale studies of patient cohorts across multiple omics platforms. Holistic analysis of these data can generate actionable insights for translational research and necessitate new approaches for data integration and mining.
METHODS
We present a novel approach for integrating data across platforms on the basis of the shared nearest neighbors algorithm and use it to create a network of multiplatform data from the immunogenomic profiling of non–small-cell lung cancer project.
RESULTS
Benchmarking demonstrates that the shared nearest neighbors-based network approach outperforms a traditional gene-gene network in capturing established interactions while providing new ones on the basis of the interplay between measurements from different platforms. When used to examine patient characteristics of interest, our approach provided signatures associated with and new leads related to recurrence and TP53 oncogenotype.
CONCLUSION
The network developed offers an unprecedented, holistic view into immunogenomic profiling of non–small-cell lung cancer, which can be explored through the accompanying interactive browser that we built.
INTRODUCTION
Multiomics profiling is becoming increasingly widespread in patient cohort studies as the throughput and resolution of measurement platforms improve.1-3 Fully harnessing advances in biologic measurement technologies across platforms to realize the potential of systems medicine requires concomitant progress in integrating diverse data types to enable holistic exploration and analysis.4,5 Insights from integrated analysis across platforms can provide a more complete picture of underlying biology compared with analyzing different platforms in isolation.6 Particularly when applied to the study of cancer, holistic approaches to analyze multiplatform data sets can unlock insights for therapeutic targets and biomarkers by providing a fuller view of phenomena driving disease pathogenesis, evolution, and post-therapy recurrence.
CONTEXT
Key Objective
How can multiplatform data sets be explored in an integrated and interactive manner?
Knowledge Generated
We built the immunogenomic profiling of non–small cell-lung cancer (ICON) data network, which connects measurements on the basis of the interplay between transcriptomic, proteomic, and flow cytometry data; to facilitate hypothesis generation, we created an interactive visualization application for exploration of the ICON data network alongside patient metadata and measurements from multiple platforms. We demonstrate that querying the shared nearest neighbors-based network reveals leads related to relevant tumor characteristics by considering recurrence and TP53 oncogenotype as examples.
Relevance
Our method for multiplatform data integration and exploration can be broadly applied to forthcoming large-scale patient profiling studies to generate actionable insights for translational research. Moreover, the interactive ICON data browser made broadly available through this publication is a unique contribution to the field and can serve as a valuable resource for future cancer research.
In this study, we present a novel approach for integrating data across platforms on the basis of the shared nearest neighbors (SNN) algorithm.7 We constructed integrated networks of multiplatform biological data by connecting transcriptomic measurements highly correlated with the same measurements from different platforms. Transcripts whose top correlates from orthogonal platforms are similar suggest potential interaction between them and shared participation in overarching biological functions.
We applied the SNN-based network-building approach that we developed to the immunogenomic profiling of non–small-cell lung cancer (NSCLC; ICON) project, an ambitious undertaking to comprehensively characterize immunogenomic diversity in resected NSCLCs across diverse omics platforms. Performing community detection on the resulting ICON data network enabled the identification of modules, collections of closely related transcripts, and focused the exploration of the integrated network, which can be easily explored using the interactive data browser that we developed. The integrated ICON data network offers an unparalleled view into the immune landscape of NSCLC, and the approach for its generation along with its accompanying data browser can serve as templates for the analysis of forthcoming large-scale patient profiling studies to facilitate translational research.
METHODS
Tissue samples were collected from patients enrolled in the ICON project and were processed and analyzed per established protocols (Data Supplement). Given their depth, the platforms of RNA sequencing (RNA-seq),8 reverse phase protein array (RPPA), and flow cytometry (FC)9 were selected for network building via the SNN-based approach that we developed, which links transcriptomic measurements by their correlation with measurements from selected platforms, in this case RPPA and FC.
Network parameters were determined using log-likelihood scores (LLSs),10,11 and Infomap community detection was performed on the finalized network to identify more interpretable subnetworks,12 termed modules. Modules were annotated using established gene sets13 and tested for association with selected metadata features by linear modeling14 using tabulated single sample gene set enrichment (ssGSEA) scores.15 An interactive data browser of the network was constructed using Python Dash16 (see the Data Sharing Statement for more information).
For a detailed methodology, see the Data Supplement.
RESULTS
Overview of ICON and SNN-Based Network Approach
The ICON data set (Fig 1A) represents a uniquely deep and rich profiling of both the genomic and immune landscapes of NSCLC. It is composed of tumor and adjacent uninvolved lung tissue samples collected from nearly 150 patients at the time of surgical resection. Samples underwent RNA-seq,8 whole-exome sequencing17 (WES), T-cell receptor (TCR) sequencing,18 multiplex immunofluorescence (mIF) for immune cells,19 FC for immune cells,9 and RPPA profiling. Given the richness of the ICON cohort data (Fig 1B), we were motivated to establish connections between measurements from different platforms to create a comprehensive and integrated view of resected NSCLC.
FIG 1.
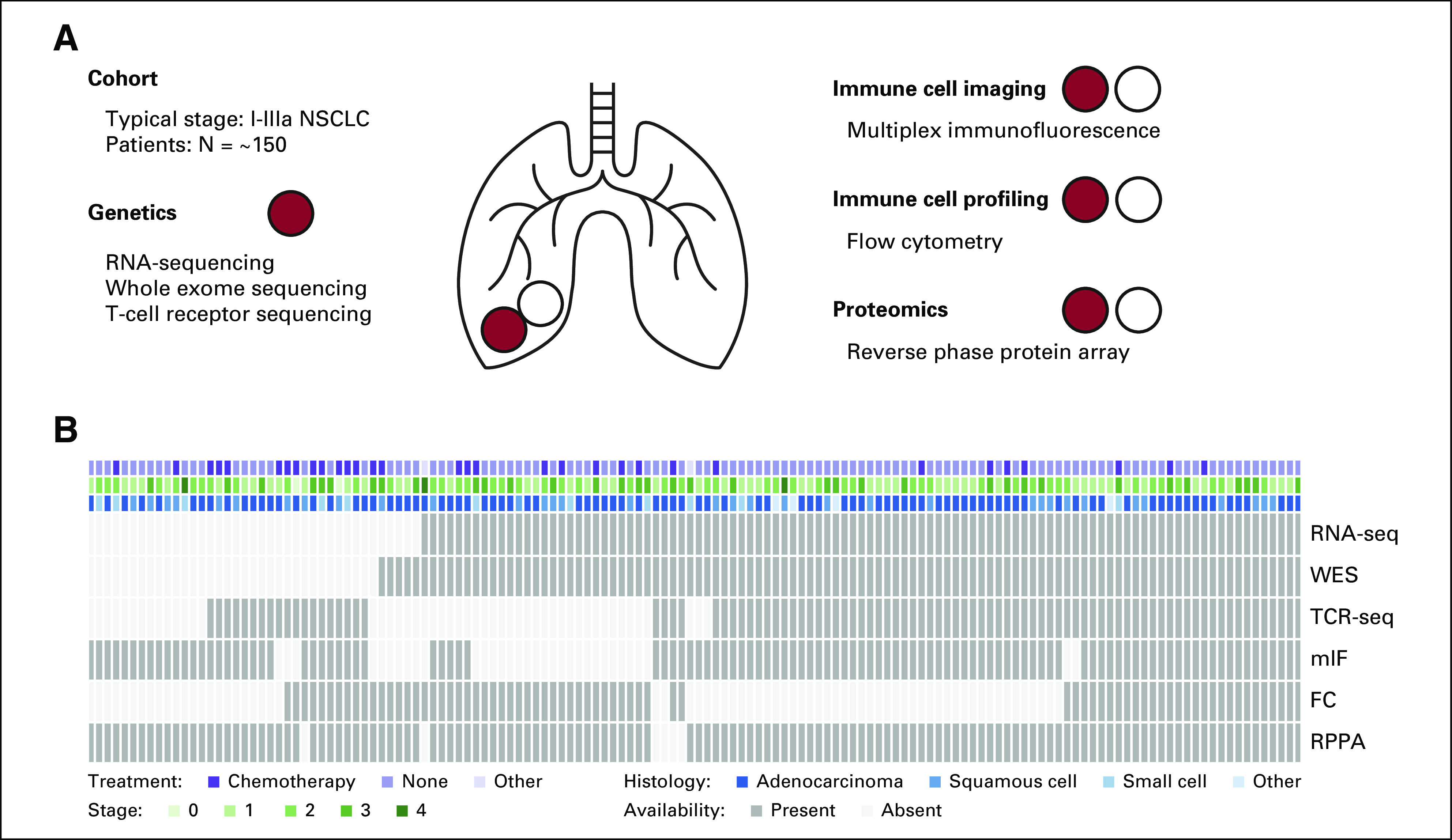
ICON project. (A) Schematic of data collected at the time of surgery through the ICON project. Red denotes tumor samples, and white denotes adjacent uninvolved tissue samples. (B) Heatmap of measurements available in ICON for each platform (rows) across patients (columns). Colors indicate treatment received, stage, histology, and data availability. FC, flow cytometry; ICON, immunogenomic profiling of non–small-cell lung cancer; mIF, multiplex immunofluorescence; NSCLC, non–small-cell lung cancer; RNA-Seq, RNA sequencing; RPPA, reverse phase protein array; TCR-seq, T-cell receptor sequencing; WES, whole-exome sequencing.
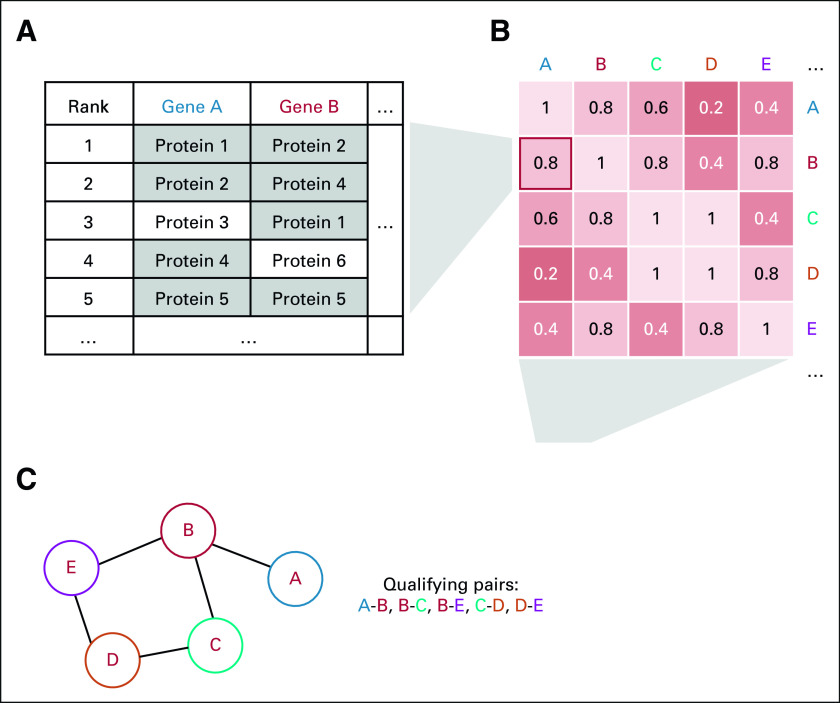
Given their depth, we focused on using RNA-seq, RPPA, and FC data in network building. Measurements from a selected orthogonal platform (eg, RPPA or FC) were ranked by their correlation with a given gene in the RNA-seq data, and for each gene pair, the number of shared top k correlates from the orthogonal platform determined the overlap score of that gene pair (Fig 2A). This process was performed across all gene pairs to compile a SNN matrix for a given orthogonal platform (Fig 2B). From these matrices, gene pairs whose overlap scores met the thresholds selected through LLS benchmarking were included in the network, where each gene is a node and each pair meeting the threshold forms an edge (Fig 2C).
FIG 2.
SNN drives integrated ICON data network. (A) Overlap score is determined by the fraction of shared top k correlates from RPPA, FC. (B) SNN matrix is compiled by performing scoring across all gene pairs for RPPA, FC. (C) Gene pairs meeting score threshold are used to construct the overall network. FC, flow cytometry; ICON, immunogenomic profiling of non–small-cell lung cancer; RPPA, reverse phase protein array; SNN, shared nearest neighbors.
Network Parameter Selection and Benchmarking
We used LLS,10,11 a metric capturing recovery of validated gene pairs, to perform a parameter sweep to select k and the overlap score threshold (Data Supplement). First, we collected annotated gene pairs from a reported set of GO biologic process terms20 and examined LLS of different k and overlap score threshold combinations to select the finalized SNN-based network configuration. Next, for comparison, we used the same annotated gene collection to calculate the LLS of a similarly sized network based solely on correlations between measurements from RNA-seq (Fig 3A); we found that the SNN-based network had a higher LLS than the RNA correlation network, 7.22 versus 7.11, respectively. This increase in the benchmarking metric is notable as it is on the logarithmic scale; in addition, the validated gene pairs used in its calculation are largely derived from expression-based experiments,20 representing a potential tendency away from relationships observed through other measurement platforms. Taken together, these points underscore the strength of the SNN-based approach that we developed.
FIG 3.
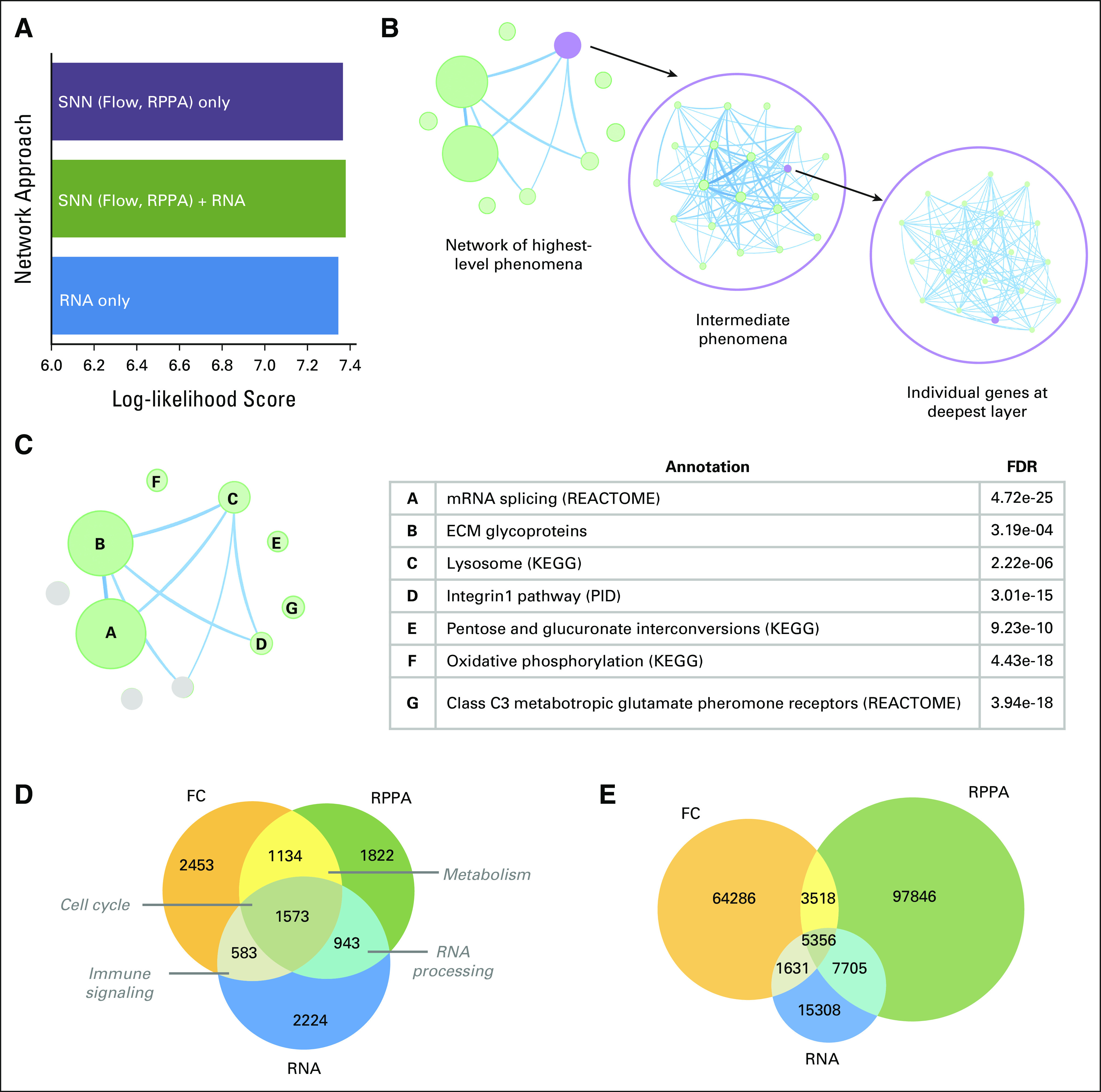
ICON data network combines SNN with RNA-based correlation edges. (A) Selection of SNN with RNA-based edges on the basis of improved performance over SNN, RNA alone. (B) Community detection via Infomap identifies 10 top-level modules and 93 mid-level modules with at least 10 genes. The modules can be examined in tandem with orthogonal data modalities and patient characteristics. (C) Overview of the ICON data network composition. For each top-level module, the most significant annotation (with specific database source in parentheses where applicable) of its mid-level modules is shown. Modules in gray are without annotations meeting FDR < 0.05. (D) Nodes balanced across source modalities. Overlaps in gene nodes from each modality shown along with representative summary annotations. (E) SNN modalities provide more edges per node. Overlaps in network edges from each modality shown. ECM, extracellular matrix; FC, flow cytometry; FDR, false discovery rate; ICON, immunogenomic profiling of non–small-cell lung cancer; KEGG, Kyoto Encyclopedia of Genes and Genomes; PID, Pathway Interaction Database; RPPA, reverse phase protein array; SNN, shared nearest neighbors.
Finally, we considered a network combining gene pairs from the SNN-based approach with additional gene pairs from the RNA correlation network and found that this network outperformed both the RNA correlation network and the SNN-based network with a LLS of 7.29 (Fig 3A). We used this blended strategy to build the ICON data network, which integrates orthogonal measurements from FC, RPPA, and RNA-seq.
ICON Data Network Connects More Than 10,000 Measurements
The ICON data network is composed of 10,732 transcriptomic measurements connected by 195,560 edges, offering a comprehensive view of the ICON data. Given its size, we used Infomap to perform community detection and extract more interpretable subnetworks,12 termed modules. The ICON data network can accordingly be framed as a multilayered network of modules (Fig 3B). We restricted our consideration to modules containing at least 10 genes, leading to 10 top-level modules and 93 mid-level modules within them.
In developing our network-building approach, we connected transcriptomic measurements to each other so that in mining the resulting network, we could leverage pathway databases13,21 to annotate each module's genes. In this manner, we identified annotations with a false discovery rate (FDR) < 0.05 for seven of 10 top-level modules and 23 of 93 mid-level modules (Fig 3C). Transcriptomic measurements were not restricted to protein-coding genes, which can partially explain modules without annotations. Concomitantly, that the network contains modules with and without annotations underscores the recapitulation of known gene relationships along with the identification of new ones through our approach.
Examining the ICON data network by platforms from which nodes and edges were derived, we found that each source platform, that is, FC (SNN approach), RPPA (SNN approach), or RNA-seq (RNA correlation approach), contributed similar numbers of nodes (Fig 3D). We annotated the collections of nodes shared between at least two source platforms and found that each platform captures different biologic phenomena, underscoring the new, more complete perspective gained by integrating across platforms. We noted that more edges were derived from the SNN-based approach developed (Fig 3E), highlighting the richness of information provided.
Network Mining Facilitated by ICON Data Browser
We explored the ICON data network alongside patient metadata and data from additional orthogonal platforms available from ICON. To do so, for each identified module, we tabulated ssGSEA scores,15 which provided a metric of upregulation or downregulation of a module's constituent genes. We then used these scores across patients to contrast modules with measurements from orthogonal platforms and to test modules via multivariate analysis for association with features of interest. To facilitate these operations, we built an interactive Python Dash application16 for data visualization and analysis. This application contains two complementary tools for mining the ICON data network: the first permits browsing of the network, whereas the second enables plotting and modeling of selected measurements, module scores, and patient metadata.
In browsing the ICON data network, the user may examine the relationship between modules and search for specific transcriptomic measurements within the network (Fig 4A). In addition, for a selected module, the user can review its pathway annotation results and determine which underlying measurements from SNN-derived edges drive the module's formation. A selected module's ssGSEA scores or any other measurement across the full ICON data set can be analyzed with the plotting and modeling tool (Fig 4B). In it, the user can plot a selected measurement against individual or combinations of categorical variables from patient metadata. These categorical variables are then used to determine via linear modeling how well they predict the measurement's distribution and to identify other measurements that they predict well. The user can also select a secondary measurement, plot the relationship between the pair of selected measurements, and determine the Spearman correlation between them. With this tool, the user can additionally rank secondary measurements by their correlation with the primary selected measurement. Taken together, the interactive data browser that we developed enables facile exploration of the ICON data network to pave the way for new insights, which we demonstrate through the following vignettes about recurrence and TP53 oncogenotype.
FIG 4.
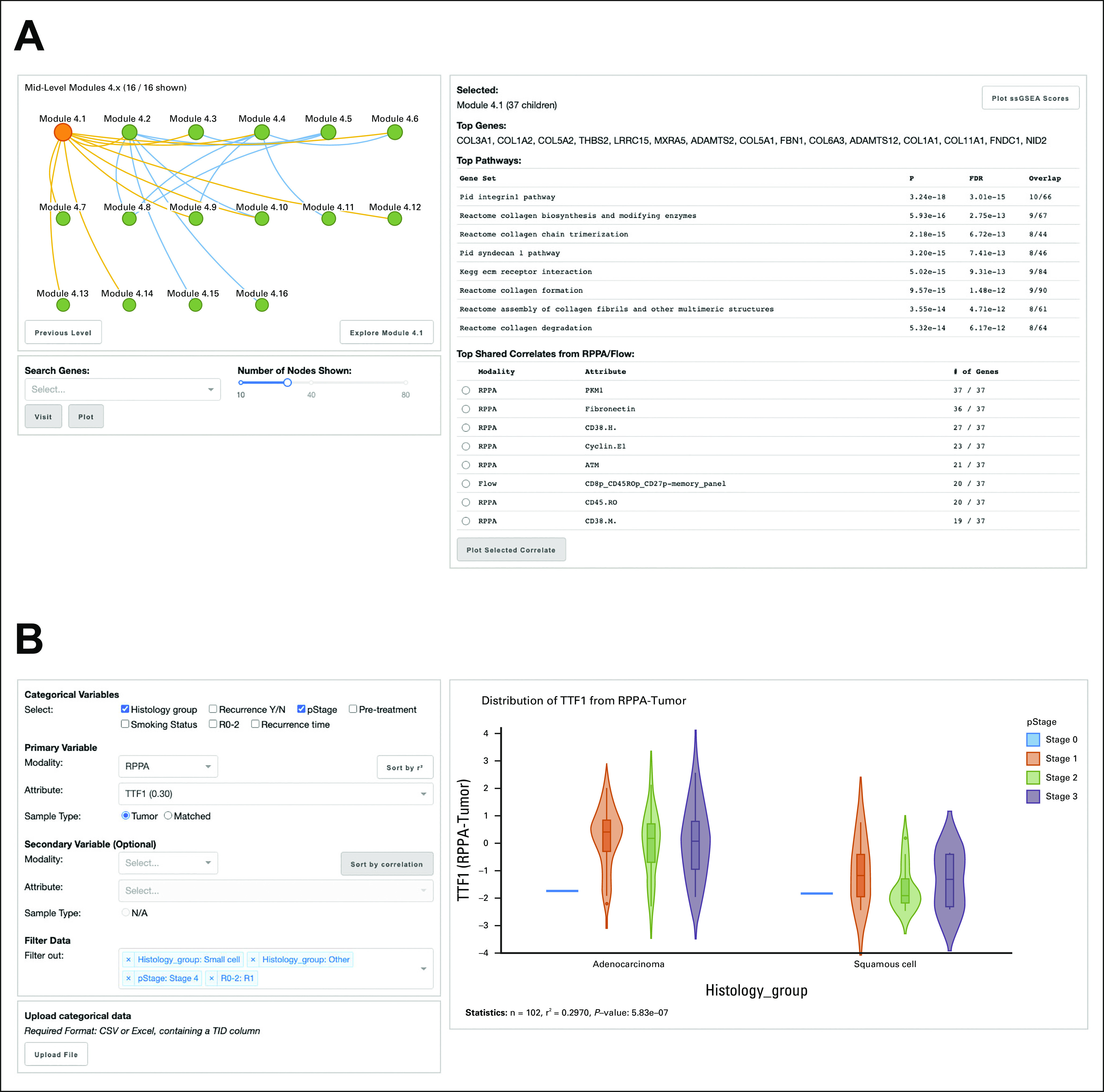
ICON data browser enables exploration of ICON data and network. (A) The network panel offers a view into the multilayered structures of constituent modules, pathway annotation results, and top correlates from the shared nearest neighbors-based network-building approach tied to module formation. (B) The plotting and modeling panel allows for a selected module's ssGSEA scores or any other measurement across the full ICON data set to be compared against categorical metadata or against a secondary continuous measurement. FDR, false discovery rate; ICON, immunogenomic profiling of non–small-cell lung cancer; KEGG, Kyoto Encyclopedia of Genes and Genomes; N/A, not available; PID, Pathway Interaction Database; RPPA, reverse phase protein array; ssGSEA, single sample gene set enrichment; TID, tumor identifer.
Recurrence-Focused Exploration Highlights Metabolic Module
Despite improvements in treatments, approximately half of patients with resectable NSCLC will relapse after curative-intent treatment,22 motivating our inquiry in this area. We leveraged the ICON data network to delve into disease recurrence, focusing on patients who had not received neoadjuvant therapy (Data Supplement). In considering associations between patient characteristics and disease relapse, we found no significant differences in recurrence status of patients who had undergone upfront surgical resection except for the variable of pathologic stage (P = 2.2e–02; Data Supplement).
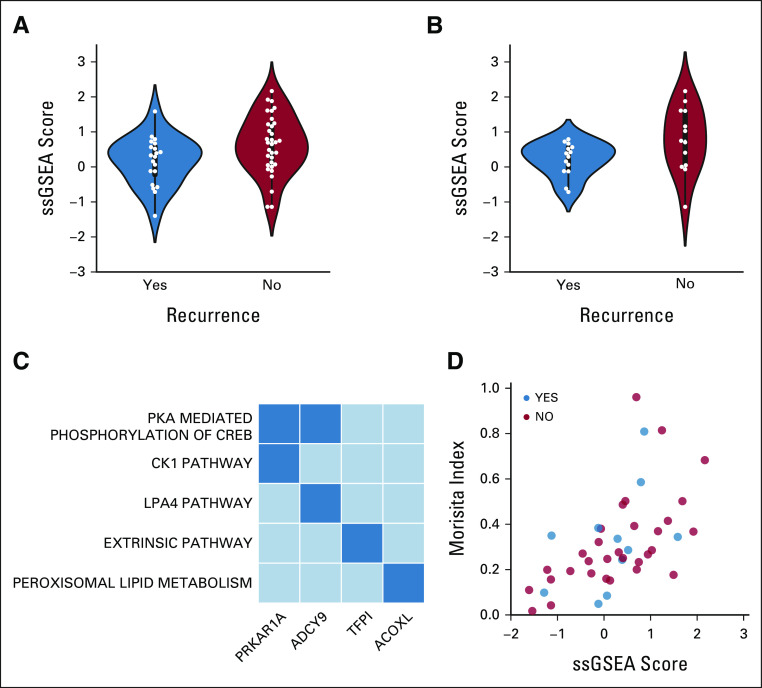
We tested module signatures in this cohort and controlled for stage and histology. This led to the identification of several modules linked to disease recurrence, and we considered in greater detail the most closely associated one (P = 2.14e–06) with pathway annotations meeting FDR < 0.05. The signature score of this module tends to be lower in patients who ultimately relapse (Fig 5A), particularly in patients with stage II-III nonsquamous NSCLC (Fig 5B), suggesting its potential impact in prognosis. We used pathway annotation to further examine this module and found that the genes within it strongly associate with several metabolic pathways (Fig 5C), underscoring the mechanistic insights provided by our approach and highlighting areas for future exploration.
FIG 5.
Network mining identifies module strongly tied to patient recurrence. (A) Distribution of ssGSEA scores by patient response for the neoadjuvant-free cohort. (B) Distribution of ssGSEA scores by patient response for a subset of cohort with nonsquamous, stage II+ tumors. (C) Top pathways for genes driving annotation of the selected module with the false discovery rate < 0.25. (D) Correlation of module scores with the Morisita overlap index from T-cell receptor sequencing (ϱ = 0.53, P = 1.98e–05). ssGSEA, single sample gene set enrichment.
We also considered which measurements from orthogonal platforms not included in the construction of the ICON data network correlated strongly with the distribution of ssGSEA scores from the highlighted module. We found that the correlation of the Morisita Overlap Index (MOI) from TCR sequencing was pronounced (ρ = 0.53, P = 1.98e–05; Fig 5D). As MOI describes the homology between TCRs in the tumor and uninvolved lung,23 we observe that patients who recur tend to have lower MOI, which indicates less similarity between TCRs in the tumor and uninvolved lung. This could suggest that the immune system of patients who eventually relapse do not have widespread presence of a TCR that recognizes the tumor, thus impeding an effective immune response.24
TP53 Oncogenotype in Squamous NSCLC Tied to Transfer RNA
The ICON data network can also be mined from the perspective of patient oncogenotype by incorporating WES data relevant to a query of interest. Given that it is often mutated in NSCLC, we turned our attention to TP53.25,26 TP53 mutations have been reported to associate with therapeutic resistance,27 but further study can help better elucidate the potential mechanisms of action responsible for this association. We focused our inquiry on patients with lung squamous cell carcinoma (LUSC), which although comprising 30% of all new lung cancer diagnoses,28,29 has historically been less studied than lung adenocarcinoma.30
We used WES data to annotate patients with LUSC in ICON by whether they had a functionally relevant mutation in TP53 as determined by the annotation of those mutations in the International Agency for Research on Cancer TP53 database.31 We then performed linear modeling to identify modules associated with the presence or absence of a functionally relevant TP53 mutation in patients with LUSC; we included pathologic stage, whether the patient had received neoadjuvant chemotherapy, and smoking status as covariates. Furthermore, we confirmed that no comutations in KRAS or EGFR were observed in the population of interest, as expected.30
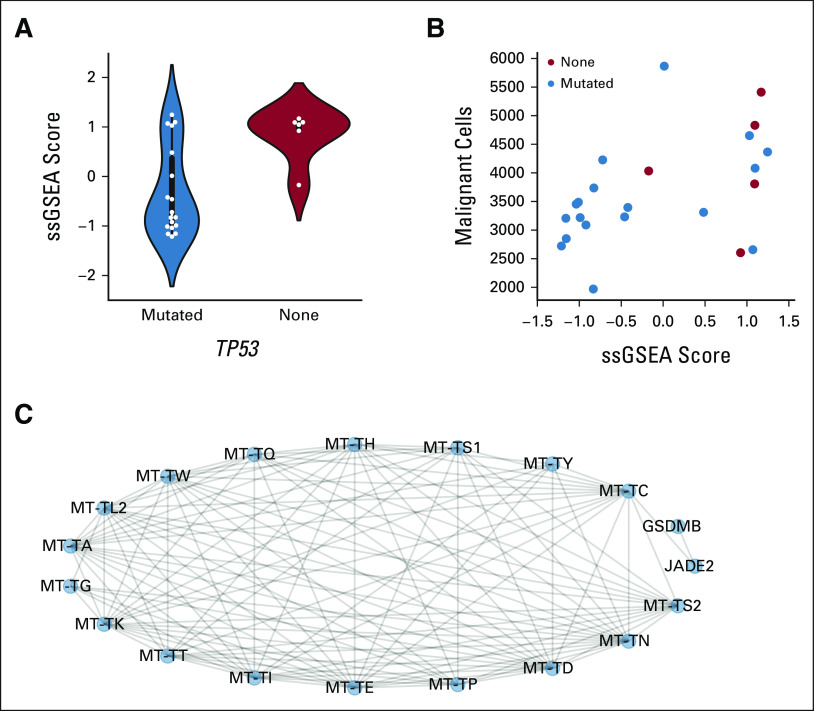
Through this approach, we identified a module linked to TP53 mutation status in patients with LUSC (P = 4.72e–02), and its ssGSEA scores tend to be higher in patients without functionally relevant TP53 mutations (Fig 6A). We performed pathway annotation on the transcriptomic measurements contained within the module; however, we did not find any associated pathways meeting FDR < 0.05. Next, we compared which measurements from orthogonal platforms not included in constructing the ICON data network correlated strongly with the distribution of ssGSEA scores from the highlighted module. We found that the correlation of malignant cell presence in the tumor and stroma (count/mm2) as measured by mIF was robust (ρ = 0.55, P = 6.39e–03; Fig 6B).
FIG 6.
Network mining identifies the transfer RNA–focused module strongly associated with functionally significant TP53 mutations in LUSC. (A) Distribution of ssGSEA scores by TP53 status for LUSC tumors. (B) Correlation of module scores with the presence of malignant cells (count/mm2) in the tumor and stroma via multiplex immunofluorescence (ϱ = 0.55, P = 6.39e–03). (C) Components of the identified module highlight strength of interaction with transfer RNA transcripts. LUSC, lung squamous cell carcinoma; ssGSEA, single sample gene set enrichment.
To delve into this module, we examined its constituents (Fig 6C). We found that the module was primarily composed of transfer RNA transcripts, and most nodes were connected by edges derived from RNA-RNA correlations. Taken together, the generally lower module ssGSEA scores exhibited by TP53-mutant tumors indicate diminished transcription of transfer RNA, which suggests translational dysregulation compared with TP53 wild-type tumors. Moreover, the correlation between the module's ssGSEA scores and the presence of malignant cells attributes generally lower malignant cell presence in the tumors of patients with functionally relevant TP53 mutations. This may be explained as reflecting the dysregulation in tumors with TP53 mutations driving a more diverse tumor microenvironment in which additional cell types have been recruited.32-35
DISCUSSION
We report a SNN-based network approach for integrating multiplatform data and its application to the ICON data set to create a unique resource for the study of resected NSCLC. By harnessing similarities in top correlates between measurements, our approach offers a flexible and generalizable method for holistically analyzing sample data from diverse platforms, thus providing a more complete perspective on underlying biology than possible by considering data from different platforms separately. We show that the network we developed contains established connections between genes in addition to new ones, underscoring the power of the integrative approach presented. Our approach can be used to comprehensively examine multiplatform cohort data sets to better understand diverse patient attributes and explore the interaction between biological phenomena at play. Furthermore, the ICON data browser enables future researchers to explore unique questions with relative ease by making the ICON network accessible in an interactive manner.
We demonstrate that our approach enables the facile identification of collections of measurements that distinguish recurrence and TP53 mutation status in ICON; moreover, these measurements can be further contextualized within the overall ICON data set to learn more about mechanisms linking them. Through our network browser, we show that networks like ours can be explored by pathways associated with collections of measurements, enabling the examination of interactions between pathways. Thus, SNN-based networks can be probed in both a top-down approach, that is, by querying modules, and a bottom-up approach, that is, by querying individual measurements.
Our approach builds upon rich work to date for constructing biologic networks. We drew inspiration from the study by Price et al,36 who directly linked highly correlated measurements from different platforms to create personal, dense, and dynamic data clouds. However, we noted that RNA-seq coexpression networks37 lend themselves well to annotation using established expression-focused resources and approaches like gene set enrichment analysis.13 To achieve multiplatform integration while being able to use existing annotation tools, we developed our SNN-based approach to create a network of transcriptomic measurements informed by measurements from complementary orthogonal platforms (FC, RPPA, and RNA-seq), thus providing a more complete perspective into mechanisms associated with tumor biology.
Our work also fits into a growing collection of interactive interfaces for multiplatform data sets; we highlight one such complementary project, the PanCancer Immunogenomics (PCIG) tool.38 PCIG provides a web platform for mining the Pan-Cancer Analysis of Whole-Genomes study's clinical, genomic, and transcriptomic data from 2,658 samples across 40 indications.39 The PCIG tool presents immune signatures calculated from transcriptomic data to offer perspective on tumor immune phenotype, which can be viewed alongside tabulated copy number, structural variant, and mutation load profiles for a selected indication. Although our work focuses on a smaller cohort of patients from a single broad indication, that is, NSCLC, we provide direct immunophenotype measurements via FC and mIF along with measurements from other platforms for interactive visualization within the ICON data browser; moreover, these data can be viewed in the context of the integrated SNN-based network that we developed. Both the PCIG tool and our project examine cancer from an immune perspective but differ with respect to cohort sizes and data types for exploration. Together, the growing availability of integrated and interactive projects such as these will spur future advances in cancer research.
Our approach is not without limitations. As with other network-building approaches, parameter selection can be somewhat subjective. We attempt to mitigate this by performing benchmarking using LLS and acknowledge that the resulting network can vary on the basis of the combination of k and overlap score cutoffs selected. In mining our network, we recognize that false discoveries can persist despite stringent multiple hypothesis correction. We anticipate that leads from SNN-based networks will require validation through analysis of complementary data sets and experiments in preclinical models.
Taken together, the SNN-based network approach that we developed enables integration of multiplatform data sets, which can help realize the promise of large-scale patient cohort studies across cancer types. Our application of this method to ICON allowed us to a create a rich and deep integrated network that can easily be mined around patient characteristics of interest. We demonstrate that mining the network identifies modules associated with phenomena such as recurrence and TP53 oncogenotype, which provide insights toward a prognostic biomarker and better understanding of disease biology. Ultimately, our SNN network–building approach and accompanying ICON data browser enable integration and exploration of patient data from diverse omics platforms, providing a more complete view of phenomena driving disease and thus unlocking insights for therapeutic targets, biomarkers, and treatment plans.
ACKNOWLEDGMENT
We are grateful to all patients and their families for participating in the ICON project. The authors thank Emily Roarty, Amy Spelman, and May Celestino and all the dedicated research nurses, consent team members, operating room nurses and anesthesiologists, and administrative staff without whom this work would not have been possible.
Alexandre Reuben
Honoraria: Adaptive Biotechnologies
Consulting or Advisory Role: Adaptive Biotechnologies
Natalie Vokes
Honoraria: Sanofi
Consulting or Advisory Role: Sanofi/Regeneron, OncoCyte, Lilly
Research Funding: OncoCyte
Junya Fujimoto
Consulting or Advisory Role: Sakura Finetek Japan, ReasonWhy
Lauren A. Byers
Honoraria: UpToDate, Clinical Care Options
Consulting or Advisory Role: Merck Sharp & Dohme Corp, Arrowhead Pharmaceuticals, Chugai Pharma, AstraZeneca, Genetech Inc, AbbVie, BeiGene, Jazz Pharmaceuticals
Research Funding: AstraZeneca (Inst), Amgen (Inst), Jazz Pharmaceuticals (Inst)
Patents, Royalties, Other Intellectual Property: Pending patents on biomarkers for lung cancer (eg, methods of selection of patients for treatment and novel biomarker tests)—recipient would be my institution and self
Lorenzo Federico
Research Funding: Obsidian Therapeutics (Inst)
Patents, Royalties, Other Intellectual Property: Charles River Laboratories. Agreement for the commercialization of transplantable tumor lines
Marie-Andree Forget
Consulting or Advisory Role: Proteios Technology
Patents, Royalties, Other Intellectual Property: License to Obsidian Therapeutics
Daniel J. McGrail
Patents, Royalties, Other Intellectual Property: Pending patents on biomarkers for sensitivity to cancer therapeutics
Younghee Lee
Employment: MD Anderson Cancer Center
Jeffrey J. Kovacs
Employment: Aktis Oncology
Stock and Other Ownership Interests: Aktis Oncology
Patents, Royalties, Other Intellectual Property: I was the recipient of a royalty payment from an agreement between Ipsen and the MD Anderson Cancer Center pertaining to the licensing and development of IACS-6274/IPN60090
Ignacio I. Wistuba
Consulting or Advisory Role: Genentech/Roche, Bristol Myers Squibb, HTG Molecular Diagnostics, Asuragen, Pfizer, AstraZeneca/MedImmune, GlaxoSmithKline, Guardant Health, Merck, MSD Oncology, Bayer, OncoCyte, Flame Biosciences, Amgen, Novartis, Daiichi Sankyo/Lilly, Daiichi Sankyo/Astra Zeneca, Sanofi
Speakers' Bureau: Pfizer, MSD Oncology, Roche, Merck, AstraZeneca, Daiichi Sankyo/Lilly
Research Funding: Genentech (Inst), Merck (Inst), HTG Molecular Diagnostics (Inst), Adaptimmune (Inst), EMD Serono (Inst), Pfizer (Inst), MedImmune (Inst), Takeda (Inst), Karus Therapeutics (Inst), Amgen (Inst), 4D Molecular Therapeutics (Inst), Bayer (Inst), Novartis (Inst), Guardant Health (Inst), Adaptive Biotechnologies (Inst), Johnson & Johnson (Inst), Iovance Biotherapeutics (Inst), Akoya Biosciences (Inst), Sanofi (Inst)
Andrew Futreal
Stock and Other Ownership Interests: Scorpion Therapeutics
Ara Vaporciyan
Travel, Accommodations, Expenses: AstraZeneca
Boris Sepesi
Consulting or Advisory Role: Bristol Myers Squibb/Medarex, AstraZeneca, Medscape
Speakers' Bureau: AstraZeneca, PeerView
Travel, Accommodations, Expenses: PeerView
John V. Heymach
Stock and Other Ownership Interests: Cardinal Spine, Bio-Tree
Consulting or Advisory Role: AstraZeneca, Bristol Myers Squibb, Spectrum Pharmaceuticals, Guardant Health, Hengrui Pharmaceutical, GlaxoSmithKline, EMD Serono, Takeda, Sanofi/Aventis, Genentech/Roche, Boehringer Ingelheim, Mirati Therapeutics, Janssen, Nexus Health Systems, Pneuma Respiratory, Lilly (Inst)
Speakers' Bureau: IDEOlogy Health, MJH Life Sciences
Research Funding: AstraZeneca (Inst), Spectrum Pharmaceuticals, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Licensing agreement between Spectrum and MD Anderson (including myself) regarding intellectual property for treatment of EGFR and HER2 exon 20 mutations
Chantale Bernatchez
Employment: Cell Therapy Manufacturing Center
Stock and Other Ownership Interests: Myst Therapeutics
Consulting or Advisory Role: Myst Therapeutics
Research Funding: Iovance Biotherapeutics, Obsidian Therapeutics (Inst)
Patents, Royalties, Other Intellectual Property: Patent pending on BTLA as a marker for better CD8 T cells for adoptive immunotherapy, Patent pending—Methods for expansion of Tumor-Infiltrating Lymphocytes and use thereof, Patent pending—Human 4-1BB agonist antibodies and methods of use thereof
Expert Testimony: Allogene Therapeutics
Cara Haymaker
Stock and Other Ownership Interests: BriaCell
Consulting or Advisory Role: Nanobiotix
Research Funding: Idera (Inst), Iovance Biotherapeutics (Inst), Dragonfly Therapeutics (Inst), Sanofi (Inst), BTG (Inst)
Patents, Royalties, Other Intellectual Property: Patent Appl No. 62/977, 672 (Inst)
Tina Cascone
Honoraria: Society for Immunotherapy of Cancer, Roche, Bristol Myers Squibb, Medscape, PeerView
Consulting or Advisory Role: MedImmune, Bristol Myers Squibb, Genentech, EMD Serono, AstraZeneca, Merck, Arrowhead Pharmaceuticals
Research Funding: Bristol Myers Squibb (Inst), Boehringer Ingelheim (Inst), EMD Serono (Inst), MedImmune (Inst), AstraZeneca (Inst)
Travel, Accommodations, Expenses: Parker Institute for Cancer Immunotherapy
Jianjun Zhang
Honoraria: Roche, Sino-USA Biomedical Platform, Geneplus, OrigiMed, Innovent Biologics, CancerNet, Zhejiang Cancer Hospital, Innovent Biologics, Zhejiang Cancer Hospital, Suzhou Medical Association, Hengrui Medicine
Consulting or Advisory Role: AstraZeneca, Geneplus, Capital Medical University, Johnson & Johnson/Janssen, Novartis, Hunan Cancer Hospital
Research Funding: Merck, Novartis
Travel, Accommodations, Expenses: Innovent Biologics, Zhejiang Cancer Hospital
Timothy P. Heffernan
Stock and Other Ownership Interests: Cullgen, Roivant Discovery
Consulting or Advisory Role: Cullgen, Roivant Discovery, Silicon Therapeutics
Research Funding: Boehringer Ingelheim (Inst), Taiho Pharmaceutical (Inst), Blueprint Medicines (Inst), Schrodinger (Inst)
Patents, Royalties, Other Intellectual Property: I am a coinventor on material and method of use patents for therapeutics developed within MD Anderson. I may personally benefit from milestone and royalty agreements through MDA alliances and out-license agreements
Travel, Accommodations, Expenses: Cullgen, Roivant
Marcelo V. Negrao
Consulting or Advisory Role: Mirati Therapeutics, Merck
Research Funding: Mirati Therapeutics, AstraZeneca, Novartis, Pfizer, ZIOPHARM Oncology, Checkmate Pharmaceuticals, Genentech, Alaunos Therapeutics
Other Relationship: ZIOPHARM Oncology, Apothecom, Ashfield Healthcare
Don L. Gibbons
Stock and Other Ownership Interests: Exact Sciences, Nektar
Consulting or Advisory Role: Sanofi, GlaxoSmithKline, Janssen Research & Development, Ribon Therapeutics, Mitobridge, Lilly, Menarini, Napa Therapeutics
Research Funding: Janssen Research & Development, Takeda, AstraZeneca, Mitobridge, Ribon Therapeutics, Boehringer Ingelheim (Inst), Mirati Therapeutics (Inst), NGM Biopharmaceuticals
Travel, Accommodations, Expenses: AstraZeneca/MedImmune, BerGenBio, Takeda
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented as posters at AACR 2021, virtual, April 10-15 and May 17-21, 2021, and AACR-NCI-EORTC 2021, virtual, October 7-10, 2021.
SUPPORT
Supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shots Program, the Gil and Dody Weaver Foundation and Bill and Katie Weaver Charitable Trust, Rexanna's Foundation for Fighting Lung Cancer and grant support from The University of Texas Lung Cancer SPORE NCI P50 (Grant No. CA070907), the MD Anderson Cancer Center Support (Grant No. P30 CA016672), and CPRIT (Grant No. RP200235). Additional support came from the NIH CCSG Award (CA016672 [Institutional Tissue Bank (ITB)] and Research Histology Core Laboratory [RHCL]) and the Translational Molecular Pathology-Immunoprofiling Laboratory (TMP-IL) at the Department Translational Molecular Pathology, The University of Texas MD Anderson Cancer.
M.V.N. and D.L.G. contributed equally to this work.
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/CCI.22.00040.
AUTHOR CONTRIBUTIONS
Conception and design: Stephanie T. Schmidt, Neal Akhave, Natalie Vokes, Andrew Futreal, Ara Vaporciyan, Tina Cascone, Jianjun Zhang, Christopher A. Bristow, Timothy P. Heffernan, Marcelo V. Negrao, Don L. Gibbons
Financial support: Timothy P. Heffernan
Administrative support: Beatriz Sanchez-Espiridion, Ignacio I. Wistuba, Timothy P. Heffernan
Provision of study materials or patients: Jun Li, Lauren A. Byers, Beatriz Sanchez-Espiridion, Ignacio I. Wistuba, Boris Sepesi, John V. Heymach, Marcelo V. Negrao
Collection and assembly of data: Stephanie T. Schmidt, Neal Akhave, Ryan E. Knightly, Alexandre Reuben, Natalie Vokes, Jianhua Zhang, Junya Fujimoto, Lauren A. Byers, Beatriz Sanchez-Espiridion, Lorenzo Federico, Annikka Weissferdt, Younghee Lee, Carmen Behrens, Ignacio I. Wistuba, Andrew Futreal, Boris Sepesi, John V. Heymach, Chantale Bernatchez, Cara Haymaker, Jianjun Zhang, Christopher A. Bristow, Marcelo V. Negrao, Don L. Gibbons
Data analysis and interpretation: Stephanie T. Schmidt, Neal Akhave, Alexandre Reuben, Natalie Vokes, Jun Li, Lauren A. Byers, Lixia Diao, Jing Wang, Marie-Andree Forget, Daniel J. McGrail, Shiaw-Yih Lin, Erika Suzuki, Jeffrey J. Kovacs, Ignacio I. Wistuba, Andrew Futreal, Boris Sepesi, Cara Haymaker, Tina Cascone, Jianjun Zhang, Christopher A. Bristow, Marcelo V. Negrao, Don L. Gibbons
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Alexandre Reuben
Honoraria: Adaptive Biotechnologies
Consulting or Advisory Role: Adaptive Biotechnologies
Natalie Vokes
Honoraria: Sanofi
Consulting or Advisory Role: Sanofi/Regeneron, OncoCyte, Lilly
Research Funding: OncoCyte
Junya Fujimoto
Consulting or Advisory Role: Sakura Finetek Japan, ReasonWhy
Lauren A. Byers
Honoraria: UpToDate, Clinical Care Options
Consulting or Advisory Role: Merck Sharp & Dohme Corp, Arrowhead Pharmaceuticals, Chugai Pharma, AstraZeneca, Genetech Inc, AbbVie, BeiGene, Jazz Pharmaceuticals
Research Funding: AstraZeneca (Inst), Amgen (Inst), Jazz Pharmaceuticals (Inst)
Patents, Royalties, Other Intellectual Property: Pending patents on biomarkers for lung cancer (eg, methods of selection of patients for treatment and novel biomarker tests)—recipient would be my institution and self
Lorenzo Federico
Research Funding: Obsidian Therapeutics (Inst)
Patents, Royalties, Other Intellectual Property: Charles River Laboratories. Agreement for the commercialization of transplantable tumor lines
Marie-Andree Forget
Consulting or Advisory Role: Proteios Technology
Patents, Royalties, Other Intellectual Property: License to Obsidian Therapeutics
Daniel J. McGrail
Patents, Royalties, Other Intellectual Property: Pending patents on biomarkers for sensitivity to cancer therapeutics
Younghee Lee
Employment: MD Anderson Cancer Center
Jeffrey J. Kovacs
Employment: Aktis Oncology
Stock and Other Ownership Interests: Aktis Oncology
Patents, Royalties, Other Intellectual Property: I was the recipient of a royalty payment from an agreement between Ipsen and the MD Anderson Cancer Center pertaining to the licensing and development of IACS-6274/IPN60090
Ignacio I. Wistuba
Consulting or Advisory Role: Genentech/Roche, Bristol Myers Squibb, HTG Molecular Diagnostics, Asuragen, Pfizer, AstraZeneca/MedImmune, GlaxoSmithKline, Guardant Health, Merck, MSD Oncology, Bayer, OncoCyte, Flame Biosciences, Amgen, Novartis, Daiichi Sankyo/Lilly, Daiichi Sankyo/Astra Zeneca, Sanofi
Speakers' Bureau: Pfizer, MSD Oncology, Roche, Merck, AstraZeneca, Daiichi Sankyo/Lilly
Research Funding: Genentech (Inst), Merck (Inst), HTG Molecular Diagnostics (Inst), Adaptimmune (Inst), EMD Serono (Inst), Pfizer (Inst), MedImmune (Inst), Takeda (Inst), Karus Therapeutics (Inst), Amgen (Inst), 4D Molecular Therapeutics (Inst), Bayer (Inst), Novartis (Inst), Guardant Health (Inst), Adaptive Biotechnologies (Inst), Johnson & Johnson (Inst), Iovance Biotherapeutics (Inst), Akoya Biosciences (Inst), Sanofi (Inst)
Andrew Futreal
Stock and Other Ownership Interests: Scorpion Therapeutics
Ara Vaporciyan
Travel, Accommodations, Expenses: AstraZeneca
Boris Sepesi
Consulting or Advisory Role: Bristol Myers Squibb/Medarex, AstraZeneca, Medscape
Speakers' Bureau: AstraZeneca, PeerView
Travel, Accommodations, Expenses: PeerView
John V. Heymach
Stock and Other Ownership Interests: Cardinal Spine, Bio-Tree
Consulting or Advisory Role: AstraZeneca, Bristol Myers Squibb, Spectrum Pharmaceuticals, Guardant Health, Hengrui Pharmaceutical, GlaxoSmithKline, EMD Serono, Takeda, Sanofi/Aventis, Genentech/Roche, Boehringer Ingelheim, Mirati Therapeutics, Janssen, Nexus Health Systems, Pneuma Respiratory, Lilly (Inst)
Speakers' Bureau: IDEOlogy Health, MJH Life Sciences
Research Funding: AstraZeneca (Inst), Spectrum Pharmaceuticals, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Licensing agreement between Spectrum and MD Anderson (including myself) regarding intellectual property for treatment of EGFR and HER2 exon 20 mutations
Chantale Bernatchez
Employment: Cell Therapy Manufacturing Center
Stock and Other Ownership Interests: Myst Therapeutics
Consulting or Advisory Role: Myst Therapeutics
Research Funding: Iovance Biotherapeutics, Obsidian Therapeutics (Inst)
Patents, Royalties, Other Intellectual Property: Patent pending on BTLA as a marker for better CD8 T cells for adoptive immunotherapy, Patent pending—Methods for expansion of Tumor-Infiltrating Lymphocytes and use thereof, Patent pending—Human 4-1BB agonist antibodies and methods of use thereof
Expert Testimony: Allogene Therapeutics
Cara Haymaker
Stock and Other Ownership Interests: BriaCell
Consulting or Advisory Role: Nanobiotix
Research Funding: Idera (Inst), Iovance Biotherapeutics (Inst), Dragonfly Therapeutics (Inst), Sanofi (Inst), BTG (Inst)
Patents, Royalties, Other Intellectual Property: Patent Appl No. 62/977, 672 (Inst)
Tina Cascone
Honoraria: Society for Immunotherapy of Cancer, Roche, Bristol Myers Squibb, Medscape, PeerView
Consulting or Advisory Role: MedImmune, Bristol Myers Squibb, Genentech, EMD Serono, AstraZeneca, Merck, Arrowhead Pharmaceuticals
Research Funding: Bristol Myers Squibb (Inst), Boehringer Ingelheim (Inst), EMD Serono (Inst), MedImmune (Inst), AstraZeneca (Inst)
Travel, Accommodations, Expenses: Parker Institute for Cancer Immunotherapy
Jianjun Zhang
Honoraria: Roche, Sino-USA Biomedical Platform, Geneplus, OrigiMed, Innovent Biologics, CancerNet, Zhejiang Cancer Hospital, Innovent Biologics, Zhejiang Cancer Hospital, Suzhou Medical Association, Hengrui Medicine
Consulting or Advisory Role: AstraZeneca, Geneplus, Capital Medical University, Johnson & Johnson/Janssen, Novartis, Hunan Cancer Hospital
Research Funding: Merck, Novartis
Travel, Accommodations, Expenses: Innovent Biologics, Zhejiang Cancer Hospital
Timothy P. Heffernan
Stock and Other Ownership Interests: Cullgen, Roivant Discovery
Consulting or Advisory Role: Cullgen, Roivant Discovery, Silicon Therapeutics
Research Funding: Boehringer Ingelheim (Inst), Taiho Pharmaceutical (Inst), Blueprint Medicines (Inst), Schrodinger (Inst)
Patents, Royalties, Other Intellectual Property: I am a coinventor on material and method of use patents for therapeutics developed within MD Anderson. I may personally benefit from milestone and royalty agreements through MDA alliances and out-license agreements
Travel, Accommodations, Expenses: Cullgen, Roivant
Marcelo V. Negrao
Consulting or Advisory Role: Mirati Therapeutics, Merck
Research Funding: Mirati Therapeutics, AstraZeneca, Novartis, Pfizer, ZIOPHARM Oncology, Checkmate Pharmaceuticals, Genentech, Alaunos Therapeutics
Other Relationship: ZIOPHARM Oncology, Apothecom, Ashfield Healthcare
Don L. Gibbons
Stock and Other Ownership Interests: Exact Sciences, Nektar
Consulting or Advisory Role: Sanofi, GlaxoSmithKline, Janssen Research & Development, Ribon Therapeutics, Mitobridge, Lilly, Menarini, Napa Therapeutics
Research Funding: Janssen Research & Development, Takeda, AstraZeneca, Mitobridge, Ribon Therapeutics, Boehringer Ingelheim (Inst), Mirati Therapeutics (Inst), NGM Biopharmaceuticals
Travel, Accommodations, Expenses: AstraZeneca/MedImmune, BerGenBio, Takeda
No other potential conflicts of interest were reported.
REFERENCES
- 1.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. : The cancer genome atlas Pan-cancer analysis project. Nat Genet 45:1113-1120, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AACR Project GENIE Consortium : AACR project GENIE: Powering precision medicine through an International Consortium. Cancer Discov 7:818-831, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401-404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez-Cabrero D, Abugessaisa I, Maier D, et al. : Data integration in the era of omics: Current and future challenges. BMC Syst Biol 8:I1, 2014. (suppl 2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang S, Chaudhary K, Garmire LX: More is better: Recent progress in multi-omics data integration methods. Front Genet 8:84, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasin Y, Seldin M, Lusis A: Multi-omics approaches to disease. Genome Biol 18:83, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarvis RA, Patrick EA: Clustering using a similarity measure based on shared near neighbors. IEEE Transact Comput C-22:1025-1034, 1973 [Google Scholar]
- 8.Mitchell KG, Diao L, Karpinets T, et al. : Neutrophil expansion defines an immunoinhibitory peripheral and intratumoral inflammatory milieu in resected non-small cell lung cancer: A descriptive analysis of a prospectively immunoprofiled cohort. J Immunother Cancer 8:e000405, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Federico L, McGrail DJ, Bentebibel SE, et al. : Distinct tumor-infiltrating lymphocyte landscapes are associated with clinical outcomes in localized non-small-cell lung cancer. Ann Oncol 33:42-56, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee I, Date SV, Adai AT, et al. : A probabilistic functional network of yeast genes. Science 306:1555-1558, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Kim E, Dede M, Lenoir WF, et al. : A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci Alliance 2:e201800278, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edler D, Bohlin L, Rosvall M: Mapping higher-order network flows in memory and multilayer networks with Infomap. Algorithms 10:112, 2017 [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, et al. : Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, :15545-15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson GN, Rogers CE: Symbolic description of factorial models for analysis of variance. J R Stat Soc Ser C Appl Stat 22:392-399, 1973 [Google Scholar]
- 15.Barbie DA, Tamayo P, Boehm JS, et al. : Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462:108-112, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hossain S, Calloway C, Lippa D, et al. : Proceedings of the 18th Python in Science Conference. Austin, TX, SciPy Organizers, 2019, pp 126-133 [Google Scholar]
- 17.Gaudreau PO, Negrao MV, Mitchell KG, et al. : Neoadjuvant chemotherapy increases cytotoxic T cell, tissue resident memory T cell, and B cell infiltration in resectable NSCLC. J Thorac Oncol 16:127-139, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reuben A, Zhang J, Chiou SH, et al. : Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat Commun 11:603, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parra ER, Uraoka N, Jiang M, et al. : Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep 7:13380, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Q, Wong AK, Krishnan A, et al. : Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat Methods 12:211-214, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A, Subramanian A, Pinchback R, et al. : Molecular signatures database (MSigDB) 3.0. Bioinformatics 27:1739-1740, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uramoto H, Tanaka F: Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res 3:242-249, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiffelle J, Genolet R, Perez MA, et al. : T-cell repertoire analysis and metrics of diversity and clonality. Curr Opin Biotechnol 65:284-295, 2020 [DOI] [PubMed] [Google Scholar]
- 24.Brambilla E, Le Teuff G, Marguet S, et al. : Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol 34:1223-1230, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcel V, Catez F, Diaz JJ: p53, a translational regulator: contribution to its tumour-suppressor activity. Oncogene 34:5513-5523, 2015 [DOI] [PubMed] [Google Scholar]
- 26.Loayza-Puch F, Drost J, Rooijers K, et al. : p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol 14:R32, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hientz K, Mohr A, Bhakta-Guha D, et al. : The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 8:8921-8946, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bender E: Epidemiology: The dominant malignancy. Nature 513:S2-S3, 2014 [DOI] [PubMed] [Google Scholar]
- 29.Ganti AK, Klein AB, Cotarla I, et al. : Update of incidence, prevalence, survival, and initial treatment in patients with non-small cell lung cancer in the US. JAMA Oncol 7:1824-1832, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network : Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519-525, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouaoun L, Sonkin D, Ardin M, et al. : TP53 Variations in human cancers: New lessons from the IARC TP53 database and genomics data. Hum Mutat 37:865-876, 2016 [DOI] [PubMed] [Google Scholar]
- 32.Uehara I, Tanaka N: Role of p53 in the regulation of the inflammatory tumor microenvironment and tumor suppression. Cancers (Basel) 10:219, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natrajan R, Sailem H, Mardakheh FK, et al. : Microenvironmental heterogeneity parallels breast cancer progression: A histology-genomic integration analysis. PLoS Med 13:e1001961, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palumbo A Jr, Da Costa Nde O, Bonamino MH, et al. : Genetic instability in the tumor microenvironment: A new look at an old neighbor. Mol Cancer 14:145, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mantovani F, Collavin L, Del Sal G: Mutant p53 as a guardian of the cancer cell. Cell Death Differ 26:199-212, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price ND, Magis AT, Earls JC, et al. : A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol 35:747-756, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Horvath S: A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4:17, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Pedrola A, Franch-Expósito S, Lahoz S, et al. : PCIG: A web-based application to explore immune-genomics interactions across cancer types. Bioinformatics 38:2374-2376, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium : Pan-cancer analysis of whole genomes. Nature 578:82-93, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/CCI.22.00040.