Abstract
Myxococcus xanthus social (S) gliding motility has been previously reported by us to require the chemotaxis homologues encoded by the dif genes. In addition, two cell surface structures, type IV pili and extracellular matrix fibrils, are also critical to M. xanthus S motility. We have demonstrated here that M. xanthus dif genes are required for the biogenesis of fibrils but not for that of type IV pili. Furthermore, the developmental defects of dif mutants can be partially rescued by the addition of isolated fibril materials. Along with the chemotaxis genes of various swarming bacteria and the pilGHIJ genes of the twitching bacterium Pseudomonas aeruginosa, the M. xanthus dif genes belong to a unique class of bacterial chemotaxis genes or homologues implicated in the biogenesis of structures required for bacterial surface locomotion. Genetic studies indicate that the dif genes are linked to the M. xanthus dsp region, a locus known to be crucial for M. xanthus fibril biogenesis and S gliding.
Myxococcus xanthus is a gliding bacterium that exhibits complex multicellular development (14). When deprived of nutrients on an appropriate surface, M. xanthus cells use gliding motility to aggregate into multicellular structures referred to as fruiting bodies. Although the mechanism of M. xanthus gliding is not known, studies have indicated that M. xanthus gliding motility is regulated by two genetically separable systems, the A (adventurous) and S (social) motility systems (18, 19). Mutations in either the A- or S-motility genes inactivate the corresponding systems; however, the cells are still motile by means of the remaining system. Whereas A motility is described as motility of well-isolated cells or small cell groups, S motility requires cell proximity or social interaction to function (18, 19, 21, 32). S motility appears more crucial for development than A motility because all of the known M. xanthus S-motility mutants are defective to various degrees in fruiting body development (19, 23).
Two M. xanthus cell surface appendages, pili and fibrils, are required for S motility. M. xanthus pili belong to the bacterial type IV pilus family (39). Numerous experiments and reports have demonstrated the absolute requirement of the polar type IV pili for M. xanthus S motility. The removal of pili either by genetic mutations or by mechanical shearing leads to S-motility defects (20, 27, 39). M. xanthus fibrils are thought to be peritrichous, filamentous structures 10 to 30 nm in diameter and can be many times the length of the cells (13, 22). They have been observed to link neighboring cells or to link cells to the substratum they glide over (2, 5, 26). The fibril-defective dsp mutants are deficient in S motility (2, 3, 29). Chemical disruption of M. xanthus fibrils also results in S-motility defects (3). M. xanthus fibrils consist of approximately equal amounts of proteins and carbohydrates (4). The presence of fibrils correlates well with the specific carbohydrate content and phosphoenolpyruvate carboxykinase activity (22, 26). Recent observations with new techniques indicate that the presence of fibrils on M. xanthus is probably more extensive than previously thought (22). Readily apparent on wild-type cells is a surface layer of fibrillar materials that are evenly distributed over the cell surface (22).
Our previous study showed that a new genetic locus, dif, which encodes a new set of chemotaxis homologues, is required for S motility (43). Here we report that the dif locus is required for the biogenesis of extracellular matrix fibrils which are crucial for S motility. Through various assays, we demonstrate that dif mutants are defective in fibril biogenesis. The developmental defects of the dif mutants can be partially rescued by adding isolated fibril materials. Furthermore, the dif genes were linked to the previously known dsp mutations that result in similar defects in cellular cohesion, S motility, and development (2, 3, 29, 30).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genetic manipulation.
The M. xanthus strains used in this study are listed in Table 1. For maintenance, M. xanthus strains were grown at 32°C on CYE medium (9) plates for 48 h and then stored at room temperature for up to 10 days. Liquid cultures were grown at 32°C in CYE medium on a rotary shaker at 250 rpm. Genetic crosses were conducted by generalized transduction mediated by M. xanthus phage Mx4 (25).
TABLE 1.
M. xanthus strains used in this study
| Strain | Description | Reference |
|---|---|---|
| DK1622 | Wild type | 20 |
| DK1253 | tgl-1 | 19 |
| DK1300 | sglG1 | 19 |
| DK3470 | dsp-1693 Tn5ΩDK1407 | 30 |
| LS300 | dsp-2105 Tn5-132ΩDK1407 | 30 |
| SW501 | difE::Kanr | 43 |
| SW504 | ΔdifA | 43 |
| SW505 | difA::Tn5–kan-903Ω101 | 43 |
Agglutination assays and rescue of cohesion and development.
The cohesion of M. xanthus cells was initially measured by an agglutination assay developed by Shimkets (29) and later modified by Wu et al. (41). Cells were grown and analyzed in CYE medium. The optical density (OD) at 600 nm was measured with a Shimadzu BioSpec-1601 spectrophotometer. The agglutination index was expressed as relative absorbance equal to the OD reading at a given time normalized against the initial OD (41).
Fibril materials were isolated from wild-type strain DK1622 and quantitated as described previously (6, 10). For analysis of the rescue of cohesion by isolated fibrils, the method of Chang and Dworkin (10) was used with the following modification. One milliliter of a cell suspension at 2.5 × 108 cells per ml in cohesion buffer (10 mM morpholine propanesulfonic acid [MOPS; pH 6.8], 1 mM CaCl2, 1 mM MgCl2) with isolated fibril materials at carbohydrate concentrations of 30 to 80 μg/ml was mixed and transferred to a semimicrocuvette. The cuvette was sealed with Parafilm, and the samples were incubated at room temperature in the dark. The OD at 600 nm was measured at different time points. To increase the sensitivity of this assay, the optical surfaces of the cuvette were covered with black paper so that the light beam went through only the upper one-quarter of the cuvette.
Developmental rescue of the dif mutants was performed as described previously (10). Briefly, cells were harvested, washed once with 10 mM MOPS, and then resuspended in cohesion buffer with fibril carbohydrates at 1.0 or 1.5 mg/ml. The final cell density in the mixture was adjusted to 2.5 × 109 cells per ml. A 50-μl volume of the cell mixture was spotted onto TPM agar (10 mM Tris-HCl [pH 7.6], 1 mM potassium phosphate buffer, 8 mM MgSO4). After 48 h of incubation at 32°C, development of the M. xanthus strains was observed and documented. Cells were scraped from the plates after photography and treated with 1% sodium dodecyl sulfate (SDS). The refractile and SDS-resistant spores were examined qualitatively under a phase-contrast microscope.
Detection of pili and fibrils.
Samples containing M. xanthus cell surface pili were prepared by vortexing and differential precipitation as described previously (36). These samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed for the presence of pilin by immunoblotting with polyclonal anti-PilA antibodies (40). Negative staining and transmission electron microscopy (TEM) were used to examine the presence of pili as described previously (39). For the detection of fibril-specific proteins, whole-cell lysates were prepared and analyzed by immunoblotting using MAb2105, a monoclonal antibody against fibril-specific proteins (5, 6). For fluorescence-activated cell sorter (FACS) analysis of fibril proteins on cell surfaces, whole-cell immunofluorescent labeling was performed according to Current Protocols in Molecular Biology (38). MAb2105 was used as the primary antibody, and a goat anti-mouse immunoglobulin G (Fab-specific) fluorescein isothiocyanate conjugate (Sigma, St. Louis, Mo.) was used as the secondary antibody. FACS analysis was performed with a Coulter Epics Elite ESP flow cytometer. The carbohydrate moiety of fibrils was examined by the binding of calcofluor white as previously described (26). Physical examination of fibrils was performed by scanning electron microscopy (SEM) of M. xanthus cells deposited on glass chips as previously described (5).
RESULTS
dif mutants are defective in cellular cohesion.
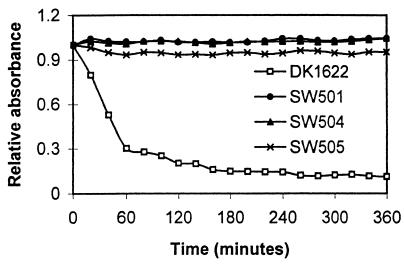
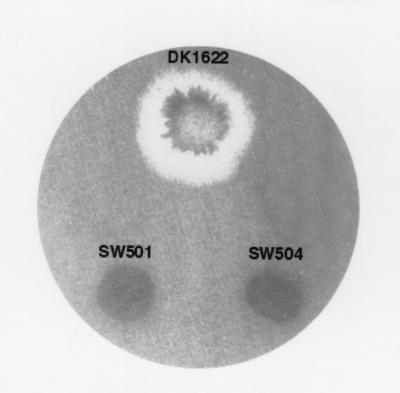
Cellular cohesion is closely correlated with M. xanthus S motility. The cohesion of wild-type M. xanthus strains and S-motility-defective dif mutants (43) was examined using the agglutination assay (41). As shown in Fig. 1, wild-type cells agglutinated as expected. In contrast, the dif mutants (SW501, SW504, and SW505) did not agglutinate over a 3-h period. Even after 24 h, the mutants showed no obvious signs of agglutination. Because cell cohesion measured by this assay depends on the presence of both pili and fibrils (41), the lack of cohesion exhibited by the dif mutants could result from defects in the biosynthesis or assembly of pili, fibrils, or both of these cell surface elements.
FIG. 1.
Agglutination defects of dif mutants. Cells were grown overnight in CYE medium, and the OD at 600 nm was adjusted to about 0.5. The agglutination assay was then conducted as described in Materials and Methods.
Pili are present on the dif mutant cell surface.
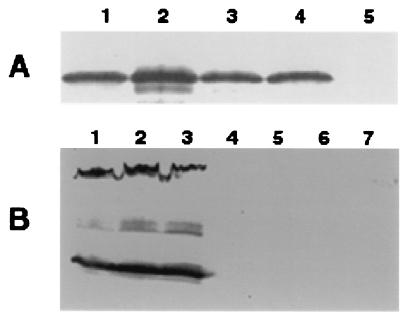
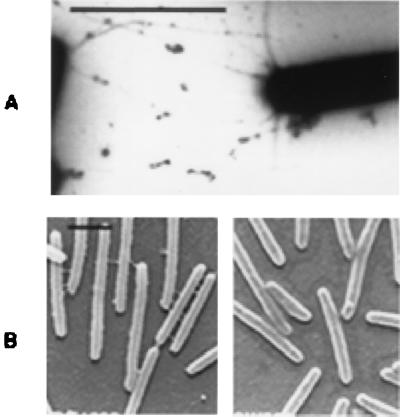
The presence of pili on M. xanthus cells was first examined by a procedure described previously (36). Samples containing cell surface pilin from various M. xanthus strains were prepared and analyzed by immunoblotting with anti-PilA serum (Fig. 2A). A band of about 25 kDa was detected in both wild-type strain DK1622 (lane 4) and dif mutant strains SW501, SW504, and SW505 (lanes 1 to 3), indicating the presence of pili on the cell surfaces (36, 40). Samples from DK1253, an S-motility mutant known to be defective in piliation (20), did not react to the antiserum as expected (lane 5). The samples from the dif (Fig. 2) and dsp (data not shown) mutants consistently reacted more strongly to the anti-PilA serum, even though samples from equal amounts of cells were loaded onto the gel. The reasons for such increases have not been fully investigated. To confirm the piliation of these dif mutants, they were further examined by negative staining and TEM. Figure 3A shows the piliation of SW501, the difE mutant. Both difA mutants (SW504 and SW505) were piliated to an extent similar to that of the difE mutant (data not shown). Therefore, the cohesion and S-motility defects of these dif mutants are not due to the lack of pili.
FIG. 2.
Immunoblot analysis of pilus- and fibril-specific proteins. All samples were separated by SDS–10% PAGE (28). The presence of pilus- and fibril-specific proteins was analyzed using anti-PilA serum (A) and monoclonal antibody MAb2105 (B) as primary antibodies, respectively. In panel A, samples from 5 × 108 cells were loaded on each lane. Whole-cell lysate from 5 × 107 cells was loaded on each lane in panel B. Panel A lanes: 1, SW505; 2, SW501; 3, SW504; 4, DK1622; 5, DK1253. Panel B lanes: 1, DK1622; 2, DK1253; 3, DK1300; 4, SW505; 5, SW501; 6, SW504; 7, DK3470.
FIG. 3.
Examination of pili and fibrils by electron microscopy. The presence of pili on SW501 cells was examined by negative staining and TEM (A). SEM was used to examine the presence of fibrils on cells of both wild-type (DK1622) and difE mutant (SW501) strains (B). Bars, 2 μm.
dif mutants are defective in the synthesis or assembly of extracellular matrix fibrils.
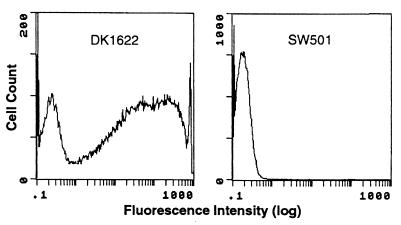
The presence of fibrils on the difE and difA mutant cell surface was first examined by FACS analysis. Monoclonal antibody MAb2105, which reacts with fibril-specific protein antigens (5, 6), was used as the primary antibody for the immunolabeling. Figure 4 shows the results of such analyses, in which difE mutant cells were compared with wild-type DK1622 cells. Whereas the majority of wild-type cells reacted with MAb2105, very few of the difE mutant cells were labeled under the same conditions. The difA mutant cells, similar to those of the difE mutant, did not react to MAb2105 (data not shown). Thus, the fibril-specific proteins recognized by MAb2105 are presumed to be absent from the surface of dif mutant cells.
FIG. 4.
FACS analysis with monoclonal antibody MAb2105. Histograms from FACS analyses of DK1622 and SW501 cells are displayed. Total numbers of cells analyzed: DK1622, 155,831; SW501, 122,003.
The major nonprotein component of the M. xanthus fibrils is the carbohydrates or the extracellular polysaccharides which are believed to form the backbone of fibrils (4). The presence of these polysaccharides on the M. xanthus cell surface was visually examined by the binding of a fluorescent dye, calcofluor white (Fig. 5). DK1622 cells bound calcofluor white and showed fluorescence under UV light as a result. The mutant cells showed no detectable binding of the dye by this assay. The lack of calcofluor white binding by the mutant cells indicates that the dif mutants are also defective in the biosynthesis or export of exopolysaccharides.
FIG. 5.
Calcofluor white (fluorescent dye) binding of M. xanthus. A 10-μl volume of a cell suspension (5 × 107 cells/ml) was spotted onto CYE medium plates containing calcofluor white (50 μg/ml). After 7 days of incubation at 32°C, dye binding was examined and documented using a handheld long-wavelength UV light source and photography.
SEM was used to examine the wild-type M. xanthus strain and the dif mutants for the presence of M. xanthus fibrils. Figure 3B compares the SEM images of difE mutant SW501 and wild-type DK1622. Fibrils are present on DK1622 cells and absent from SW501 cells. Similarly, no extracellular fibrils were detected on the surfaces of difA mutants SW504 and SW505. These results demonstrate that dif mutations affect the biosynthesis and/or assembly of both the carbohydrate and protein components of M. xanthus fibrils.
The absence of the fibril-specific protein antigens from the cell surface could result from defects in the export of proteins containing these antigens to the cell surface. In this case, it might be possible to detect the antigens within the cell. To address this possibility, whole-cell lysates were separated by SDS-PAGE and analyzed by immunoblotting (Fig. 2B) using MAb2105 as the primary antibody. Multiple bands of the wild-type cell lysate (lane 1) were detected as previously described (5). The cell lysate of a dsp mutant (DK3470) which is defective in fibril production did not react to MAb2105 (lane 7). The reaction of cell lysates of two pilus-defecfive S-motility mutants, DK1253 and DK1300 (lanes 2 and 3) (20), was similar to that of the wild type. These data indicate that defects in S motility and piliation per se do not necessarily result in defects in the biosynthesis of fibril protein antigens. In contrast, the dif mutant cell lysates (lanes 4 to 6) did not react to MAb2105, indicating that there are no detectable fibril-specific protein antigens associated with the mutant cells. The data may suggest that the dif mutants are defective in the biosynthesis of these fibril protein antigens or that there is a feedback inhibition preventing the accumulation of these proteins inside cells. Another possibility is that the proteins are synthesized and transported but not maintained on the cell surface. It is also possible that if these mutants are blocked in fibril protein transport or assembly, the proteins inside cells are not stable enough to be detected.
Isolated fibril materials can partially restore cohesion and development to dif mutants.
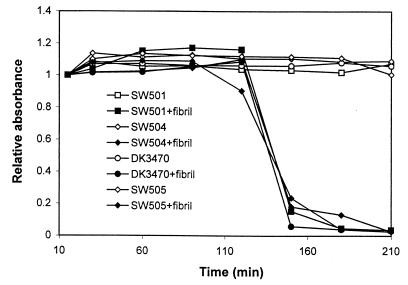
We examined whether the dif mutants could be rescued by the addition of isolated fibrils. Different concentrations of isolated fibril materials were added to mutant cell suspensions, and cohesion was measured by the agglutination assay. The data in Fig. 6 show that cohesion, although delayed compared with that of wild-type cells, was restored to all of the dif mutants when fibrils were added. At fibril carbohydrate concentrations of 40, 60, and 80 μg/ml, cohesion of the mutant cells occurred essentially at the same rate. The rate of cohesion with fibrils at 30 μg/ml or below decreased in a dosage-dependent manner (data not shown). This observed dosage-dependent response is consistent with previous reports (10).
FIG. 6.
Effects of isolated fibrils on cellular adhesion of M. xanthus dif mutants. Cell suspensions (2.5 × 108 cells/ml) with or without fibrils (carbohydrate-equivalent concentration, 40 μg/ml) were analyzed by agglutination. Relative absorbance at 600 nm was used as the cohesion index.
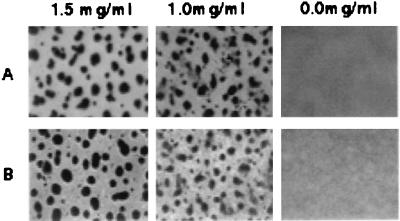
To determine if the addition of fibrils could rescue the ability of the dif mutants to form fruiting bodies, fibrils were added to dif mutants at fibril carbohydrate concentrations of 1.0 and 1.5 mg/ml and the cells were subjected to development-inducing conditions. The results for SW501 and SW505 are shown in Fig. 7. At 1.5 mg/ml, all three dif mutants (SW501, SW504, and SW505) formed fruiting bodies. However, the rescue at 1.0 mg/ml was less complete; loose and irregular fruiting bodies or aggregates were observed. Nevertheless, at both concentrations, the fruiting bodies and the aggregates darkened after 48 h and contained spherical, refractile, SDS-resistant myxospores, as indicated by examination of an SDS-treated sample by phase-contrast microscopy. DK3470, the dsp mutant, could also be rescued in a similar fashion, as expected (data not shown).
FIG. 7.
Developmental rescue of dif mutants by isolated fibrils. The final fibril concentration in cell suspension (2.5 × 109 cells/ml) is indicated at the top as the equivalent of fibril carbohydrates. The experiments were conducted under development-inducing condition as described in Materials and Methods. Shown are the rescues of SW501 (A) and SW505 (B).
dif genes are linked to the dsp region.
Our characterization of the dif mutant phenotypes indicates that they are identical or similar to the previously isolated dsp mutants. This has driven us to examine if dif mutations are linked to the dsp region. dsp mutations are linked to the Tn5 insertion ΩDK1407 (30). To examine the possible linkage between dif genes and the dsp region, SW501 (containing difE::Kanr) was used as the donor and LS300 (containing ΩDK1407 Tn5-132) was used as the recipient in an Mx4-mediated transduction. Of the 249 kanamycin-resistant transductants obtained, 160 had lost the tetracycline resistance marker (Tn5-132), indicating 64% cotransduction of the wild-type locus of the ΩDK1407 insertion and the difE insertion mutation. According to the Wu equation (31, 42), this cotransduction frequency represents a physical distance of about 7.7 kb between difE and ΩDK1407, indicating that the dif genes are at or near the dsp region (30).
DISCUSSION
There are three major mechanisms of active bacterial surface translocation: gliding, twitching, and swarming (17). As described in the introduction to this report, M. xanthus is one of the best-studied gliding bacteria. Swarming motility is widespread among eubacteria (16, 17) and has been extensively studied in Vibrio parahaemolyticus, Proteus mirabilis (1, 7, 24), and recently in Serratia liquefaciens, Escherichia coli, and Salmonella typhimurium (8, 15, 16). On appropriate surfaces, these bacteria undergo a series of changes and transform from “swimmer” cells to differentiated “swarmer” cells. During this differentiation, cell septation ceases and the cell body elongates. Generally, swarmer cells are more flagellated than liquid-grown swimmer cells. In addition, extracellular polysaccharides or slimes often encapsulate swarmer cells. Twitching motility has been best studied in the opportunistic pathogen Pseudomonas aeruginosa (12). Although the mechanism of twitching motility is not yet understood, experimental evidence clearly indicates that type IV pili are indispensable. All of the P. aeruginosa pil mutants defective in type IV pilus biogenesis are unable to display twitching motility (12). Suggestions have been made that the bacterial type IV pilus is a novel type of motor for surface translocation (32, 35).
M. xanthus S motility shares similarities with both twitching and swarming. Type IV pili are required for both M. xanthus S motility (20, 39) and P. aeruginosa twitching motility (12). Swarming motility is strictly a multicellular phenomenon, as single cells emerging from the moving mass are stranded and cease to move (34). Likewise, S motility is a multicellular behavior requiring cell proximity. Consequently, S-motility flares and advancing rafts of swarming bacteria appear strikingly similar to each other (16, 29). In addition, chemotaxis genes or homologues are required for swarming, twitching, and M. xanthus S motility. Studies on a few swarming bacteria have demonstrated that chemotaxis systems play critical roles in swarming motility (16). The P. aeruginosa pilGHIJ cluster encodes a set of chemotaxis homologues required for twitching motility (11). In both twitching and swarming, these chemotaxis genes or homologues appear to be essential for the biogenesis of cell surface structures instead of directed cell movements. For example, the chemotaxis genes in E. coli are required for the differentiation and hyperflagellation of swarmer cells (8) and the P. aeruginosa pilGHIJ genes are essential for the biogenesis of type IV pili (11). The M. xanthus frz genes (37), encoding chemotaxis homologues (43), are required for directed cell movement but not for S motility (37). Prior to this report, there has been no evidence indicating the involvement of chemotaxis genes or homologues in the biogenesis of cell surface structures required for M. xanthus gliding motility. In this study, we have determined that dif genes are required for the biogenesis of fibrils in M. xanthus. Thus, the dif genes are members of a small group of chemotaxis genes or homologues essential for the biogenesis of structures required for bacterial surface translocation. These findings appear to suggest a common theme among twitching, swarming, and M. xanthus S motility; that is, certain chemotaxis genes or homologues have essential functions in all three forms of bacterial surface translocation by regulating the biogenesis of surface structures.
How these chemotaxis genes or homologues regulate a biogenesis process is not clear. The regulation of the biogenesis of these various surface structures could be distinct for each motility system. The regulation of M. xanthus fibril biogenesis by dif genes may occur by two alternate mechanisms which are not mutually exclusive. The dif genes may regulate the biosynthesis of fibrils, or they may regulate their assembly. We do not have evidence to support either mechanism. The chemical composition of M. xanthus fibrils makes distinguishing between these two mechanisms complicated. M. xanthus fibrils contain about equal amounts of protein and carbohydrate, which together account for approximately 85% of the dry weight of fibrils (4). The whole-cell lysates from mutant cells contained no protein antigens reactive with MAb2105 (Fig. 2), which may suggest that such protein antigens are not synthesized in dif mutants. However, the association between fibril proteins and carbohydrates is not yet understood. Since the carbohydrates appear to form the backbone of M. xanthus fibrils (4), the protein materials could be anchored to cell surfaces by the fibril carbohydrates. Defects in the transport or assembly of the carbohydrate backbone may lead to the loss of such proteins at cell surfaces even if they are produced. Alternatively, if the proteins cannot be transported correctly, they may be degraded within the cells or their synthesis may be inhibited. The development of new assays and further biochemical characterization of the fibrils should facilitate the understanding of the associations and interactions between the carbohydrate and protein components of fibrils and help to elucidate the mechanism by which dif genes regulate fibril biogenesis.
What roles do the fibrils play in M. xanthus social motility? It has been suggested that M. xanthus fibrils provide the major force for cell cohesion and, as a result, maintain the integrity of M. xanthus multicellular or S swarms (2). The roles of fibrils in S motility probably go beyond simple physical cohesion required for S motility. Genetic studies (18, 19) clearly indicated that S motility is not a collective movement of cell with A motility by physical attachment because A-motility-defective mutants retain their S motility. Although there have been suggestions that M. xanthus type IV pili are the motor or part of a motor specific for S motility (39, 41), it is not clear how and by what structure the mechanical force is generated for S gliding. It is clear, however, that the S-gliding mechanism needs activation via cell proximity-related signals (18, 19, 32). When cells with S motility (A− S+) are isolated from one another, they show no net translocation. However, when they are in proximity to one another, motility is apparent (33). The S-motility defects of dif mutants are distinct from those of pil mutants. Similar to A− dsp double mutants (41), A− dif double mutants show a fringe around their colonies after prolonged incubation (data not shown), indicating that the dif mutants retain some residual S motility. The residual S motility of dif and dsp mutants suggests that the components of the S-gliding-specific motor have not been entirely eliminated in these mutants. It is possible, instead, that it is the activation of the S-gliding mechanism that is impaired in dif and dsp mutants. Interestingly, cell proximity is also required for bacterial swarming motility (16, 17). Perhaps, the requirement for cell proximity reflects a common activation mechanism in M. xanthus S motility and the swarming motility of other bacteria.
If we consider the possibility of an activation defect in dif and dsp mutants, these genes may affect such an activation process in two different ways. First, the effects of dif and dsp genes on activation could be indirect. Since both dif and dsp mutants lack fibrils, it may be the lack of fibrils, rather than the lack of dif and dsp gene products themselves, that results in the motility defects. Fibrils can either be the activation signal or facilitate the transmission of such a signal. It is conceivable that fibril materials, which encircle and link M. xanthus cells, provide a suitable and managed microenvironment for the transmission of signals of cell proximity. Interestingly, differentiated swarmer cells also produce extracellular slimes composed of polysaccharides. Second, dif and dsp genes may play a more direct role in the activation process. The dif gene products are expected to form a chemotaxis-like signal transduction pathway that may potentially interact with the S-gliding-specific motor, analogous to the way enteric bacterial chemotaxis genes interact with components of the bacterial flagella. Further studies are necessary to determine how dif and dsp regulate S motility.
ACKNOWLEDGMENTS
We are grateful to Martin Dworkin and Dale Kaiser for providing antibodies and strains. We thank David Zusman and Martin Dworkin for their enthusiasm and for very helpful discussions. We owe debts of thanks to Alice Thompson and Birgitta Sjostrand for expert help with electron microscopy, to Anahid Jewett for help with the flow cytometer, and to Sharon Hunt Gerardo for critical reading and careful editing of the manuscript.
This work was supported by NIH grants GM54666 to W. Shi and GM47444 to H. Kaplan, NSF grant MCB9601077 to L. Shimkets, and NIH training grants 5-T32-AI-07323 and 5-T32-DE-07296 to Z. Yang.
REFERENCES
- 1.Allison C, Hughes C. Bacterial swarming: an example of prokaryotic differentiation and multicellular behaviour. Sci Prog. 1991;75:403–422. [PubMed] [Google Scholar]
- 2.Arnold J W, Shimkets L J. Cell surface properties correlated with cohesion in Myxococcus xanthus. J Bacteriol. 1988;170:5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold J W, Shimkets L J. Inhibition of cell-cell interactions in Myxococcus xanthus by Congo red. J Bacteriol. 1988;170:5765–5770. doi: 10.1128/jb.170.12.5765-5770.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behmlander R M, Dworkin M. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J Bacteriol. 1994;176:6295–6303. doi: 10.1128/jb.176.20.6295-6303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behmlander R M, Dworkin M. Extracellular fibrils and contact-mediated cell interactions in Myxococcus xanthus. J Bacteriol. 1991;173:7810–7821. doi: 10.1128/jb.173.24.7810-7820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behmlander R M, Dworkin M. Integral proteins of the extracellular matrix fibrils of Myxococcus xanthus. J Bacteriol. 1994;176:6304–6311. doi: 10.1128/jb.176.20.6304-6311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belas R. Proteus mirabilis and other swarming bacteria. In: Shopiro J, Dworkin M, editors. Bacteria as multicellular organisms. New York, N.Y: Oxford University Press; 1997. pp. 183–219. [Google Scholar]
- 8.Burkart M, Toguchi A, Harshey R M. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos J M, Zusman D R. Regulation of development in Myxococcus xanthus: effect of 3′:5′-cyclic AMP, ADP, and nutrition. Proc Natl Acad Sci USA. 1975;72:518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang B Y, Dworkin M. Isolated fibrils rescue cohesion and development in the dsp mutant of Myxococcus xanthus. J Bacteriol. 1994;176:7190–7196. doi: 10.1128/jb.176.23.7190-7196.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 12.Darzins A, Russell M A. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system—a review. Gene. 1997;192:109–115. doi: 10.1016/s0378-1119(97)00037-1. [DOI] [PubMed] [Google Scholar]
- 13.Dworkin M. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. Bioessays. 1999;21:590–595. doi: 10.1002/(SICI)1521-1878(199907)21:7<590::AID-BIES7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 14.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciens MG1. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harshey R M. Bees aren't the only ones: swarming in gram-negative bacteria. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 17.Henrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol Gen Genet. 1979;171:167–176. [Google Scholar]
- 19.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 20.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser D, Crosby C. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 1983;3:227–245. [Google Scholar]
- 22.Kim S-H, Ramaswamy S, Downard J. Regulated exopolysaccharide production in Myxococcus xanthus. J Bacteriol. 1999;181:1496–1507. doi: 10.1128/jb.181.5.1496-1507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacNeil S D, Mouzeyan A, Hartzell P L. Genes required for both gliding motility and development in Myxococcus xanthus. Mol Microbiol. 1994;14:785–795. doi: 10.1111/j.1365-2958.1994.tb01315.x. [DOI] [PubMed] [Google Scholar]
- 24.McCarter L, Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990;4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor K A, Zusman D R. Genetic analysis of Myxococcus xanthus and isolation of gene replacements after transduction under conditions of limited homology. J Bacteriol. 1986;167:744–748. doi: 10.1128/jb.167.2.744-748.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramaswamy S, Dworkin M, Downard J. Identification and characterization of Myxococcus xanthus mutants deficient in calcofluor white binding. J Bacteriol. 1997;179:2863–2871. doi: 10.1128/jb.179.9.2863-2871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbluh A, Eisenbach M. Effect of mechanical removal of pili on gliding motility of Myxococcus xanthus. J Bacteriol. 1992;174:5406–5413. doi: 10.1128/jb.174.16.5406-5413.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Shimkets L J. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J Bacteriol. 1986;166:837–841. doi: 10.1128/jb.166.3.837-841.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimkets L J. Role of cell cohesion in Myxococcus xanthus fruiting body formation. J Bacteriol. 1986;166:842–848. doi: 10.1128/jb.166.3.842-848.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sodergren E, Cheng Y, Avery L, Kaiser D. Recombination in the vicinity of insertions of transposon Tn5 in Myxococcus xanthus. Genetics. 1983;105:281–291. doi: 10.1093/genetics/105.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spormann A M. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spormann A M, Kaiser D. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J Bacteriol. 1999;181:2593–2601. doi: 10.1128/jb.181.8.2593-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturdza S A. La reaction d'immobilisation des filaments de Proteus sur les milieux geloses. Arch Roum Pathol Exp Microbiol. 1973;32:575–580. [PubMed] [Google Scholar]
- 35.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 36.Wall D, Wu S S, Kaiser D. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J Bacteriol. 1998;180:759–761. doi: 10.1128/jb.180.3.759-761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 38.Watkins S. Immunohistochemistry. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 14.6.1–14.6.13. [Google Scholar]
- 39.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 40.Wu S S, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S S, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 42.Wu T T. A model for three-point analysis of random general transduction. Genetics. 1966;54:405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Geng Y, Xu D, Kaplan H B, Shi W. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol Microbiol. 1998;30:1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]