Keywords: O-GlcNAcylation rhythm, metabolic input, circadian input, biological rhythms, phosphorylation, hexosamine biosynthetic pathway
Abstract
O-linked-N-acetylglucosaminylation (O-GlcNAcylation) is a nutrient-sensitive protein modification that alters the structure and function of a wide range of proteins involved in diverse cellular processes. Similar to phosphorylation, another protein modification that targets serine and threonine residues, O-GlcNAcylation occupancy on cellular proteins exhibits daily rhythmicity and has been shown to play critical roles in regulating daily rhythms in biology by modifying circadian clock proteins and downstream effectors. We recently reported that daily rhythm in global O-GlcNAcylation observed in Drosophila tissues is regulated via the integration of circadian and metabolic signals. Significantly, mistimed feeding, which disrupts coordination of these signals, is sufficient to dampen daily O-GlcNAcylation rhythm and is predicted to negatively impact animal biological rhythms and health span. In this review, we provide an overview of published and potential mechanisms by which metabolic and circadian signals regulate hexosamine biosynthetic pathway metabolites and enzymes, as well as O-GlcNAc processing enzymes to shape daily O-GlcNAcylation rhythms. We also discuss the significance of functional interactions between O-GlcNAcylation and other post-translational modifications in regulating biological rhythms. Finally, we highlight organ/tissue-specific cellular processes and molecular pathways that could be modulated by rhythmic O-GlcNAcylation to regulate time-of-day-specific biology.
1. Introduction
Organisms from all domains of life exhibit daily biological rhythms to adapt to changes in their environment over the 24 h day–night cycle. In animals, daily rhythms of physiology, metabolism and behaviour are strongly regulated by the circadian clock, an endogenous biological timer that enables animals to anticipate predictable changes in biotic and abiotic factors [1,2]. The circadian clock is a molecular oscillator that relies on transcriptional–translational feedback mechanisms operated by key clock transcription factors to generate daily oscillations in gene expression. In coordination with processes that are regulated by post-transcriptional mechanisms, clock-regulated rhythmic gene expression programs that are often tissue- and cell-specific produce daily rhythms in clock outputs. The outputs of animal circadian clocks are all-encompassing and include rhythmic processes such as sleep–wake cycles, feeding–fasting cycles, metabolism, hormone production and secretion, immune response, neuronal excitability and even permeability of the blood–brain barrier [3–10]. There is growing evidence that some clock outputs are themselves zeitgebers (i.e. time-givers) and can feedback to the molecular oscillator to reinforce and/or modulate daily biological rhythms. The feeding–fasting cycle is one such clock output and studies have shown that key clock transcription factors that form the core of the molecular oscillator can be regulated by metabolites or nutrient-sensitive hormones, such as heme, NAD/NADH (nicotinamide adenine dinucleotide/reduced form of nicotinamide adenine dinucleotide), AMP/ATP (adenosine monophosphate/adenosine triphosphate), acetyl coenzyme A, glucocorticoids and glucagon (reviewed in [11]).
Besides impacting daily biological rhythms by modulating the activities of key clock transcription factors, metabolic feedback from feeding–fasting cycles can also regulate daily rhythms through other mechanisms beyond the circadian clock. For example, feeding–fasting cycles can drive rhythmic production of NAD+, which serves as coenzyme for histone deacetylases class III, also known as sirtuins, to regulate daily rhythmicity in epigenomic landscape and global gene expression [12,13]. Feeding activity also contributes to daily oscillation of protein translation [14]. This was shown to be mediated by the nutrient-sensitive mTOR pathway, amino acid sensing pathways and metabolic modification of mRNA.
We recently established that integration of circadian signals and rhythmic metabolic input can regulate daily cellular physiology through rhythmic protein O-linked-N-acetylglucosaminylation (O-GlcNAcylation) [15]. O-GlcNAcylation has the potential to modify the function of thousands of proteins [16–19], and has been shown to play a critical role in maintaining animal circadian rhythms [20–23]. Furthermore, since both O-GlcNAcylation and phosphorylation modify serine and threonine residues [16,17,24], rhythmic O-GlcNAcylation may contribute to robust oscillation of the 24 h phosphoproteome and regulate its time-of-day specific function [25–28]. These post-translational mechanisms could bypass regulation at the transcriptional level to directly modulate protein function in a time-specific and nutrient-sensitive manner. Interestingly, our studies showed that the amplitude of daily protein O-GlcNAcylation rhythm is severely dampened if animals are fed at an unnatural time window (i.e. time of day at which they are normally fasting [15]). This suggests that rhythmic functions of cellular proteins could be impaired by mistimed meals, a common occurrence in modern society. Our findings point to the likelihood that the beneficial effects of time-restricted feeding [29–35], a practice that limits food consumption to 8–12 h during an individual's natural active period and has been shown to maintain robust circadian rhythms, enhance health span and alleviate metabolic diseases, may be partially mediated via daily O-GlcNAcylation rhythm.
In the remainder of the review, we will summarize the regulation of daily rhythmicity in O-GlcNAcylation by metabolic and circadian signals, outline interactions between O-GlcNAcylation and other post-translational modifications (PTMs), and highlight cellular processes that are potentially regulated by rhythmic O-GlcNAcylation.
2. Regulation of O-GlcNAcylation in the context of daily biological rhythm
Protein O-GlcNAcylation is nutrient-sensitive and is tightly linked to cellular metabolic status. For this reason, the regulation and function of O-GlcNAcylation have been extensively studied in the context of metabolic diseases, specifically diabetes and cancers [17,36]. On the contrary, although metabolism and energy status are highly rhythmic over the day–night cycle, the number of studies on rhythmic O-GlcNAcylation and the consequences of its disruption dwarfs in comparison. The cycling of O-GlcNAc groups on proteins is regulated by the level of UDP-GlcNAc (the substrate) and the activities of two O-GlcNAc processing enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) (figure 1). UDP-GlcNAc is produced from the hexosamine biosynthetic pathway (HBP), which integrates metabolites from glucose metabolism (glucose), amino acid metabolism (glutamine), lipid metabolism (acetyl-CoA) and nucleotide metabolism. In this section, we discuss current findings on the regulation of O-GlcNAcylation under the framework of rhythmic biology over a 24 h day–night cycle.
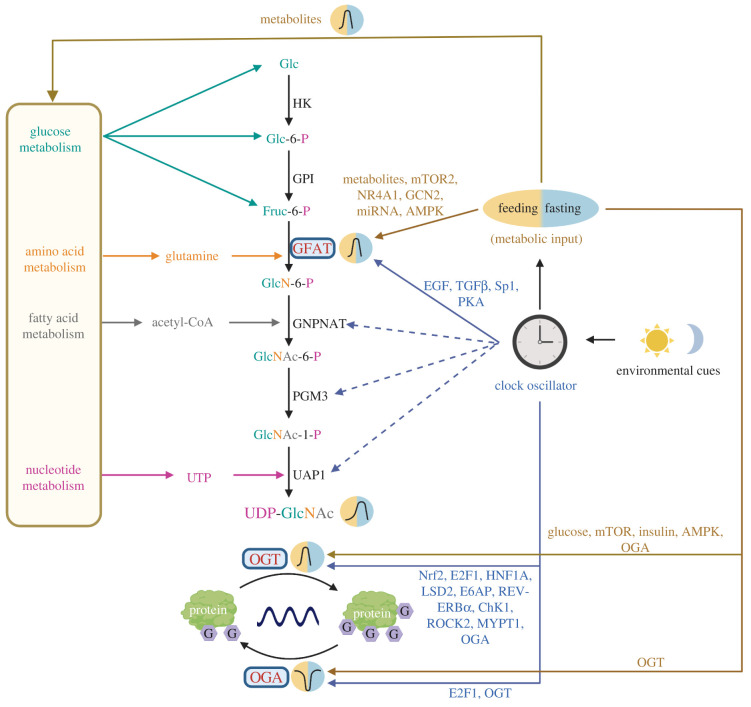
Figure 1.
Schematic illustrating metabolic and circadian regulation of rhythmic protein O-linked-N-acetylglucosaminylation (O-GlcNAcylation). The circadian clock oscillator receives environmental signals and regulates daily feeding–fasting cycles. Feeding–fasting cycles rhythmically provide input to hexosamine biosynthetic pathway (HBP), which contributes to rhythmic production of UDP-GlcNAc [15]. O-GlcNAc transferase (OGT) takes UDP-GlcNAc as a substrate and transfers GlcNAc onto serine and threonine residues of proteins. This process is recognized as O-GlcNAcylation (O-GlcNAc is depicted as G on protein molecules). Metabolic input can also regulate the O-GlcNAcylation rhythm through modifying the activities of glutamine:fructose-6-phosphate amidotransferase (GFAT) [37–49], OGT [21,50–63] and O-GlcNAcase (OGA) [64]. Additionally, the clock oscillator not only regulates feeding–fasting cycles, but also regulates the expression or enzymatic activities of all the HBP enzymes [39,47,65–76] and O-GlcNAc processing enzymes [62,63,71,72,77–93]. The potential mediating factors of metabolic and circadian inputs are illustrated in the schematic diagram; metabolic inputs are depicted in brown and circadian inputs are depicted in blue. The dashed arrows indicate potential regulation without known mechanisms. HK, Hexokinase; GPI, phosphoglucose isomerase; GFAT, glutamine–fructose-6-phosphate aminotransferase; GNPNAT, glucosamine-phosphate N-acetyltransferase; PGM3, phosphoacetylglucosamine mutase; UAP1, UDP-N-acetyl glucosamine pyrophosphorylase 1; OGT, O-GlcNAc transferase; OGA, O-GlcNAcase; Glc, glucose; Glc-6-P, glucose-6-phosphate; Fruc-6-P, fructose-6-phosphate; GlcN-6-P, glucosamine-6-phosphate; GlcNAc-6-P, N-acetylglucosamine-6-phosphate; GlcNAc-1-P, N-acetylglucosamine-1-phosphate; UTP , uridine triphosphate; UDP-GlcNAc, uridine diphosphate N-acetylglucosamine; mTOR, mammalian target of rapamycin; NR4A1, nuclear subfamily 4 group A member 1; GCN2, general control nonderepressible2; miRNA, microRNA; AMPK, AMP-activated protein kinase; EGF, epidermal growth factor; TGFβ, transforming growth factorβ; Sp1, specificity protein 1; PKA, protein kinase A; Nrf2, nuclear factor E2-related factor-2; E2F1, transcription factor E2F1; HNF1A, hepatocyte nuclear factor 1 homologue A; LSD2, lysine-specific histone demethylase 1B; E6AP, ubiquitin ligase E6AP; ChK1, checkpoint kinase 1; ROCK2, Rho-associated coiled-coil forming protein kinase 2; MYPT1, myosin phosphatase target subunit 1.
2.1. Regulation of HBP pathway by daily rhythm of nutrient availability
Nutrient availability directly determines the level of building blocks for producing UDP-GlcNAc. Given that there is strong support from metabolomics studies showing that nutrient input correlates with feeding activity [15,94–96], metabolic influx into the HBP is expected to be highly rhythmic over the day–night cycle and probably contributes to daily rhythmicity in O-GlcNAcylation. In studies conducted using cultured cells or tissues, elevated production of UDP-GlcNAc has been shown to correlate with higher nutrient concentration in cell media, including glucose, glutamine, glucosamine (GlcN), acetylglucosamine (GlcNAc), free fatty acids and uridine [37,50,97–105]. However, some glucose starvation studies showed contradictory results; glucose starvation was observed to result in elevation of O-GlcNAcylation level [38,50,51]. These conflicting observations could be explained by divergent properties of cell lines or tissue types. For example, Pham et al. [106] showed that O-GlcNAcylation levels in different subtypes of diffuse large B-cell lymphoma cell lines respond differently to glucose deprivation. Additionally, increased O-GlcNAcylation upon glucose starvation could be due to altered levels of OGT, OGA or glutamine:fructose-6-phosphate amidotransferase (GFAT) [38,50,51].
Although cell culture and ex vivo studies have firmly established the importance of nutritional regulation of HBP and O-GlcNAcylation, in vivo studies especially ones that take into account daily rhythmic biology and feeding–fasting cycles are still limited. In the 1990 s, Hawkins et al. [107] showed that continuously infusing lipid emulsion, uridine or GlcN for 7 h increases UDP-GlcNAc levels in rat skeletal muscles. To establish the relationship between feeding activity and levels of HBP metabolites including UDP-GlcNAc, we recently monitored feeding rhythm and HBP metabolites in Drosophila flies over a 24 h day–night cycle [15]. We observed strong correlation between fly feeding rhythm and daily rhythms in HBP metabolites in fly body tissues. Significantly, we found that shifting the time of food consumption significantly altered the peak time of HBP metabolite rhythm. In summary, we conclude that HBP metabolites and UDP-GlcNAc level are strongly regulated by clock-controlled feeding–fasting cycle and metabolic input. Whether this phenomenon is consistent in other animals, including nocturnal animals, will need to be explored in future studies.
2.2. Daily regulation of HBP enzymes
Besides rhythmic metabolic input into the HBP facilitated by clock-controlled feeding–fasting cycles, rhythmic expression and activity of HBP enzymes could also contribute to daily rhythms in protein O-GlcNAcylation. Searching through CirGRDB, a mammalian circadian transcriptomic database [108] and published Drosophila transcriptomic datasets [27,109], we found that transcripts encoding all HBP enzymes oscillate in at least one study. Additionally, data mining in circadian proteomic and phosphoproteomic datasets [25,27,110] revealed that the majority of the HBP enzymes have oscillating protein levels and/or phosphorylation. Our recent study reported that GFAT enzyme activity oscillates over a 24 h cycle in flies and rhythmic GFAT activity is regulated by the integration of metabolic and circadian signals [15]. In this section, we will elaborate on our findings and discuss potential mechanisms mediating daily regulation of HBP enzymes. In particular, we will focus on the regulation of GFAT, the rate-limiting enzyme of HBP and the most well studied of all HBP enzymes (figure 1).
The expression of gfat mRNA is highly regulated by nutrient availability and nutrient-sensing pathways. gfat has two isoforms in animals, gfat1 and gfat2. Both isoforms encode GFAT enzymes that perform the same catalytic function but have distinct tissue-specific distribution (reviewed in [111]). We showed that the expression of gfat2 mRNA in fly body tissues is strongly induced by food consumption in Drosophila [15], although we did not explore the mechanisms that mediate the observed induction. In tissue culture, expression of gfat mRNA has been shown to be stimulated by various nutrients [37,39–42] and mediated by multiple molecular pathways, including mammalian target of rapamycin2 (mTOR2) [41,43], nuclear subfamily 4 group A member 1 (NR4A1) [40], microRNA (miR)-27b-3p [42] and general control nonderepressible2-activating transcription factor 4 pathway [38]. Expression of gfat mRNA can potentially be regulated by clock-controlled factors/processes in addition to feeding–fasting cycles and rhythmic metabolic input. These include angiotensin II [112–114], epidermal growth factor (EGF) [39,65,66], transforming growth factorβ (TGFβ) [67–69] and Specificity protein 1 [70–72], all of which are known to influence gfat expression.
Beyond transcriptional regulation, GFAT enzyme activity is known to be influenced by PTMs and feedback regulation from HBP metabolites. We reported that the circadian clock strongly regulates daily GFAT activity through unknown post-transcriptional and/or post-translational mechanism(s) [15]. Interestingly, the kinases that have previously been identified to regulate GFAT activities are also known effectors of circadian signals or clock-controlled metabolic signals. These include AMP-activated protein kinase (AMPK) [44,45], mTOR2 [46] and protein kinase A (PKA) [47,73–76]. In particular, PKA-directed phosphorylate site at GFAT1 S235 [74] is shown to oscillate over a circadian cycle in mouse liver [25]. However, the function of GFAT1 pS235 is currently unclear. Finally, glucosamine-6-phosphate (GlcN-6-P) and UDP-GlcNAc, the direct product from the GFAT-catalysed reaction and end product of the HBP respectively, can feedback to inhibit GFAT activity [47–49]. As our study found that GlcN-6-P and UDP-GlcNAc levels oscillate over the day–night cycle with peak time corresponding to feeding period [15], HBP metabolites likely represent important signals to shape daily GFAT activity.
In summary, as we concluded in our studies in Drosophila [15], the HBP enzyme GFAT represents an important integration hub of circadian and metabolic signals to regulate the production of UDP-GlcNAc and cellular protein O-GlcNAcylation.
2.3. Daily regulation of O-GlcNAc processing enzymes
There is strong evidence showing that OGT and OGA, the two O-GlcNAc processing enzymes that drive the cycling of GlcNAc group on and off proteins, are subjected to control by the circadian clock, but data on direct measurements of OGT and OGA enzyme activities over a daily cycle are still lacking to the best of our knowledge. Circadian transcriptomic and proteomic analyses showed that the oga mRNA and OGA protein oscillate in mouse livers and fly heads [27,71,115–119], while ogt mRNA but not OGT protein was observed to oscillate in mouse livers and fly heads [20,21,27,71,109,110,120–122]. We showed that in Drosophila fly bodies, the transcripts and encoded proteins of the two O-GlcNAc processing enzymes are modulated by both circadian and metabolic input [15]. This section is devoted to review potential molecular mechanisms that mediate metabolic and circadian regulation of OGT and OGA.
ogt mRNA and its encoded protein OGT are regulated by nutrient levels and nutrient-sensing pathways that are expected to be highly rhythmic over the day–night cycle. There are two nutrient-sensing pathways that are known to regulate OGT protein level, mTOR [52–54] and insulin signalling [55], and glucose itself [50,51,56] has also been shown to modulate ogt mRNA expression. Currently, it is unclear how expression of ogt mRNA and their encoded proteins are regulated by the circadian clock. The clock can potentially orchestrate rhythmic ogt mRNA expression by targeting rhythmically active transcription factors. Candidates include nuclear factor E2-related factor-2 (Nrf2) [77–80], E2F1 transcription factor [81,82] and hepatocyte nuclear factor 1 homologue A (HNF1A) [72,83,84], which are known to regulate ogt expression. With regard to OGT protein cycling, lysine-specific histone demethylase 1B (KDM1B or LSD2) and ubiquitin ligase E6AP have been shown to facilitate OGT ubiquitylation and degradation through their ubiquitin ligase activity [85,86]. Interestingly, LSD2 and E6AP transcripts are both observed to oscillate in circadian transcriptome studies [71,72,82]. Finally, the clock protein REV-ERBα directly interacts with OGT and stabilizes OGT in different cellular compartments, as the cellular localization of REV-ERBα oscillates [87]. Whether and how these mechanisms contribute to circadian regulation of OGT levels will need to be explored in future studies. When compared with the regulation of ogt expression, much less is known about pathways that modulate oga expression. Given that E2F1 regulates oga expression in addition to ogt expression [81], it represents a transcription factor candidate [82] that can drive rhythmic oga expression.
At the post-transcriptional level, OGT enzymatic activity is regulated by multiple PTMs, which have been shown to respond to metabolic or circadian signals. Metabolic input has been shown to regulate OGT phosphorylation and thereby enzymatic activity through insulin signalling [21,57,58], as well as AMPK [59] and CAMKII [60,61] phosphorylation. Glycogen synthase kinase 3β (GSK3β), which happens to be an insulin signalling effector and a clock kinase, is shown to phosphorylate OGT at S3 or S4 to increase its enzymatic activity [21]. GSK3β-dependent phosphorylation of OGT can also change substrate selectivity [58]. Moreover, the circadian clock has been shown to rhythmically regulate the kinases and phosphatases that modify OGT, such as Checkpoint kinase 1 (ChK1) [88,89], Rho-associated coiled-coil forming protein kinase 2 (ROCK2) [71,72,90], myosin phosphatase target subunit 1 (MYPT1) [71,72,91,92]. Finally, O-GlcNAcylation of OGT S389 is shown to increase OGT nuclear localization [62]. As O-GlcNAcylation can integrate both metabolic and circadian signals [15], it will be interesting to explore whether OGT exhibits daily oscillation of subcellular localization. OGA can also be modified by phosphorylation and O-GlcNAcylation [123]. However, the functional role of these PTMs on OGA is less defined. Whether the phosphorylation and O-GlcNAcylation status of OGT and OGA is rhythmically regulated over a 24 h day–night cycle and how that modulates their activities needs future investigation.
To conclude the discussion on daily regulation of O-GlcNAc processing enzymes, it is important to point out that OGT and OGA impose reciprocal regulation against one another to maintain O-GlcNAc homeostasis. For example, as OGT protein level decreases, OGT forms a repressor complex with mSin3A and histone deacetylase1 (HDAC1) at the promoter of oga to inhibit its expression [64]. OGA can promote ogt expression by reducing O-GlcNAcylation of CCAAT/enhancer-binding proteinβ (C/EBPβ) recruited to the promoter of ogt [63]. Notably, increasing O-GlcNAcylation level using an OGA inhibitor has been observed to elevate OGA protein while decreasing OGT protein level [93]. The reciprocal regulation of the two O-GlcNAc processing enzymes is expected to contribute to shaping daily rhythmicity of O-GlcNAcylation.
3. Crosstalk between O-GlcNAcylation and other post-translational modifications to regulate daily cellular physiology
Different types of PTMs co-occur on proteins to regulate their functions in response to diverse physiological and environmental signals. O-GlcNAc modifications have been shown to exhibit crosstalk with other PTMs, such as phosphorylation (reviewed in [24]), acetylation [124–126] and ubiquitination [127,128]. The crosstalk between O-GlcNAcylation and phosphorylation has attracted the most attention, as both PTMs target serine and threonine residues. Given that circadian proteomics studies in recent years demonstrated that phospho-occupancy in many cellular proteins exhibit daily rhythmicity to regulate time-of-day protein functions [25–28], it is intriguing to explore how O-GlcNAcylation and phosphorylation (and possibly other PTMs) could work in conjunction to regulate daily rhythmicity in protein functions. Crosstalk between O-GlcNAcylation and other PTMs can present itself in two manners: (i) modify the function of writers and erasers (enzyme level), or (ii) modify the same sites or nearby sites on protein substrates to modulate the level of other PTMs (substrate level) (figure 2). With an emphasis on phosphorylation, we next review the mechanisms by which O-GlcNAcylation could shape daily rhythmicity in phosphoproteome to regulate biological rhythms.
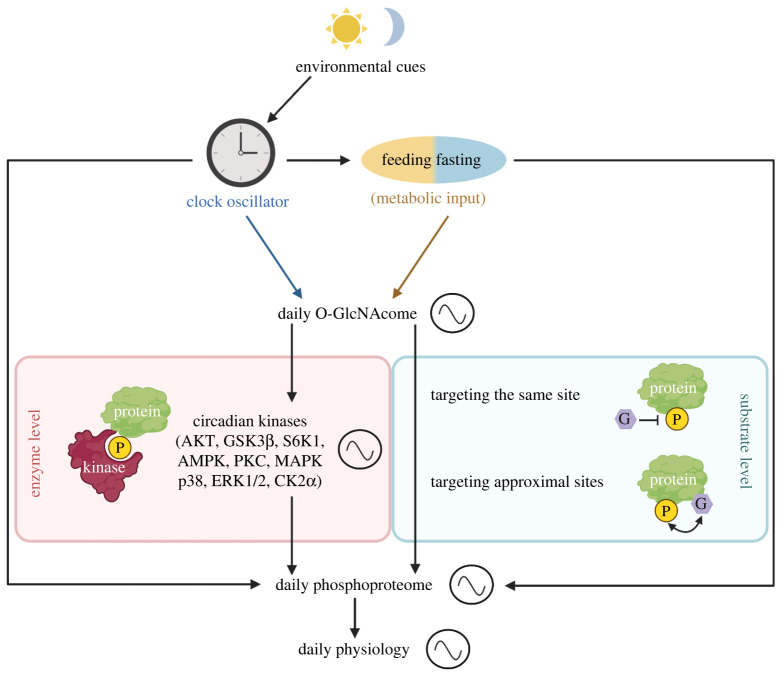
Figure 2.
Daily rhythmicity of the O-GlcNAcome can integrate metabolic and circadian signals to modulate rhythmicity of the phosphoproteome. O-GlcNAcylation can modulate rhythmic activities of ‘circadian kinases’, which have been previously identified by analysing circadian/daily phosphoproteomic datasets [25,27]. We define this as enzyme level regulation. For substrate level regulation, O-GlcNAcylation can directly compete with phosphorylation by targeting the same residue on substrate proteins and/or modulate the protein conformation to promote or inhibit phosphorylation by targeting an approximal site [129–133]. Independently, phosphorylation is also sensitive to environmental and metabolic signals. In sum, interplay between daily phosphorylation and O-GlcNAcylation regulates time-of-day functions of cellular proteins and daily physiological rhythms. GSK3β, Glycogen synthase kinase3β; S6K1, ribosomal protein S6 kinase 1; AMPK, AMP-activated protein kinase; PKC, protein kinase C; MAPK p38, mitogen-activated protein kinase p38, ERK1/2, extracellular signal-regulated kinase1/2; CK2α casein kinase2α.
3.1. Daily O-GlcNAcylation-phosphorylation crosstalk (O-P crosstalk) at the enzyme level
Since global O-GlcNAcylation level oscillates over a 24 h cycle [15] and O-GlcNAcylation has been shown to regulate the function of a myriad of kinases and phosphatases [24,134,135], time-of-day specific O-GlcNAcylation of kinases and phosphatases could represent an important mechanism to remodel daily rhythmicity in the phosphoproteome. In 2012, Dias et al. [136] systematically analysed O-GlcNAcylation of kinases using an in vitro OGT assay. They screened through 152 full-length human kinases and identified 42 O-GlcNAcylated kinases. More recently, Schwein & Woo [135] reviewed the O-GlcNAcomic datasets, and found more than 100 O-GlcNAcylated kinases, which covers all six major kinase families (AGC, CMGC, CAMK, STE, CK1 and TK/TKL) and some atypical protein kinases. Not surprisingly, a number of phosphatases are also found to be O-GlcNAcylated, including MYPT1, PPFIA2−4, PPP6R2, PTPN6, PTPN7, PTPRC, TNS2 and SIRPA [135].
Much progress has been made in characterizing the function of O-GlcNAcylation on many kinases. In table 1, we highlight the O-GlcNAcylated ‘circadian kinases', which we identified by analysing the circadian phosphoproteome [25,27]. Nevertheless, proteomic studies on how O-GlcNAc sites of certain kinases could modulate the phosphoproteome are still rather limited. Schwein et al. [146] recently used nanobody-OGT and nanobody-split OGA to specifically modify O-GlcNAc S347 on CK2α and analysed the phosphoproteome in HEK293 cells. They observed that increased CK2α O-GlcNAcylation promotes the phosphorylation of 39 proteins, enriched for chromatin modification, metabolism and ribosome, while decreasing the phosphorylation of 12 proteins. In conclusion, O-GlcNAcylation could regulate cellular physiology through modifying the kinome and thereby the phosphoproteome (figure 2). Future investigation is warranted to reveal the O-GlcNAcylation-kinome crosstalk under the framework of the 24 h day–night cycle.
Table 1.
O-GlcNAcylation of kinases known to regulate circadian rhythm. GSK3β, glycogen synthase kinase3β; S6K1, ribosomal protein S6 kinase 1; AMPK, AMP-activated protein kinase; PKC, protein kinase C; MAPK p38, mitogen-activated protein kinase p38; ERK1/2, extracellular signal-regulated kinase1/2, CK2α casein kinase2α.
| kinases | O-GlcNAc sites | function of O-GlcNAcylation | references |
|---|---|---|---|
| AKT | T308, S473 (characterized by mutagenesis) | inhibit AKT phosphorylation at T308 and S473, and inhibit AKT activity | [137,138] |
| GSK3β | n.a. | promote GSK3β phosphorylation at S9, and inhibit GSK3 activity | [137,139,140] |
| S6K1 | S489 (characterized by mutagenesis) | inhibit S6K1 phosphorylation at S418 and T229, and inhibit S6K1 activity | [141] |
| AMPK | n.a. | inhibit AMPKα phosphorylation at T174, and inhibit AMPK activity | [59,142] |
| PKCζ | T408, T410 (characterized by mutagenesis) | inhibit PKCζ phosphorylation at T410, and inhibit PKCζ activity | [143] |
| MAPK p38 | n.a. | promote p38 phosphorylation, and activate p38 activity | [144] |
| ERK1/2 | n.a. | promote ERK1/2 phosphorylation, and activate ERK1/2 activity | [144] |
| CK2α | S347 (validated by Edman sequencing) | inhibit CK2α phosphorylation at T344, reduce the interaction between CK2α and PIN1, promote CK2α degradation, and alter substrate selectivity | [145] |
3.2. Daily O-P crosstalk at the substrate level
Since the discovery that O-GlcNAcylation and phosphorylation can modify the same amino acid residue on the same protein [147], crosstalk between O-GlcNAcylation and phosphorylation on substrates is recognized as an important mechanism for regulating protein function. Early study investigating O-P crosstalk used OGA inhibitors to elevate the global O-GlcNAcylation level and assayed the phosphoproteome in cell culture [129]. Out of the 711 phosphopeptides detected, 148 phosphopeptides increased and 280 decreased upon OGA inhibition. The phosphoproteins identified by Wang et al. [129] were enriched for cytoskeleton, cytoskeleton binding and RNA/DNA binding proteins. In addition to using inhibitors to globally alter protein O-GlcNAcylation status, other approaches have also been used to characterize physiologically relevant O-P crosstalks. Trinidad et al. [130] studied O-P crosstalks at murine synapses, as both OGT and OGA are enriched at synapses [148,149]. They sequentially enriched for O-GlcNAc and phosphopeptides and successfully detected O-GlcNAcylation and phosphorylation on the same peptides [130]. Among the 1750 O-GlcNAc sites and 16 500 phosphosites detected, 135 sites can be modified by both O-GlcNAcylation and phosphorylation. More recently, Fan et al. [131] developed a HILIC enrichment method to simultaneously enrich for O-GlcNAc and phosphopeptides. They assayed O-P crosstalk on 1115 RNA binding proteins (RBPs) and found that 213 RBPs (25%) can be both O-GlcNAcylated and phosphorylated. Taken together, direct competition between O-GlcNAcylation and phosphorylation may not be the dominant mechanism for O-P crosstalk, and O-GlcNAcylation and phosphorylation are more likely to regulate each other through modifying approximal sites (figure 2).
To elucidate the mechanisms for O-P crosstalk, researchers have started to analyse potential consensus sequence of O-P crosstalk. Although sites modified by both O-GlcNAcylation and phosphorylation occur at a relatively low frequency, Yao et al. [132] extracted three motifs (Pxx[S], Txxx[S] and [T]xxxxxxxxxP) that are overrepresented in S/T exhibiting O-P crosstalk at the same residue. For O-P crosstalk at approximal sites, Leney et al. [133] performed a systematic analysis using a MS-based in vitro kinetic assay and identified a motif with four amino acids: N-[S/T]P(V/A/T)[S/T]-C. Phosphorylation occurs at the N terminal S/T and O-GlcNAcylation modifies the C terminal S/T, and the two PTMs tend to reciprocally inhibit one other [133]. Data mining in PhosphoSite Plus showed that 1048 proteins could be regulated by this potential mechanism, and previous studies support that O-P crosstalk on proteins such as eukaryotic initiation factor 4 (eIF4) and Sin3A could be mediated by this motif [133,150,151]. Kinase prediction shows that extracellular signal-regulated kinase1 (ERK1), ERK2, CK1 and GSK3β are likely to modify the motifs mentioned above [132,133]. Interestingly, these kinases overlap with ‘circadian kinases' that can rhythmically phosphorylate proteins over a 24 h cycle [25,27], suggesting that daily cycling of O-GlcNAcylation could potentially regulate daily rhythmicity in the phosphoproteome at the substrate level (figure 2). Nevertheless, this hypothesis needs to be tested with the advances of O-P peptide enrichment methods and MS proteomics.
4. O-GlcNAcylation is an important mechanism that regulates daily cellular physiology
A large body of work contributed to our understanding of diverse mechanisms by which metabolic input interacts with the endogenous circadian clock to regulate daily biological rhythms (reviewed in [11,13,152]). Our recent study highlights protein O-GlcNAcylation as a key post-translational mechanism that integrates metabolic and circadian signals to regulate rhythmic physiology [15]. At the molecular level, there are two ways O-GlcNAcylation can regulate daily rhythms of cellular physiology: (i) O-GlcNAcylation can modulate core clock proteins and the pace of the molecular clock, which in turn alters rhythmicity of diverse cellular processes; (ii) O-GlcNAcylation can rhythmically modify cellular proteins outside of the molecular oscillator to regulate their time-of-day specific functions (figure 3). In this section, we review the impact of O-GlcNAcylation on clock protein functions and outline other rhythmic cellular processes that can be modified by O-GlcNAcylation beyond the core clock.
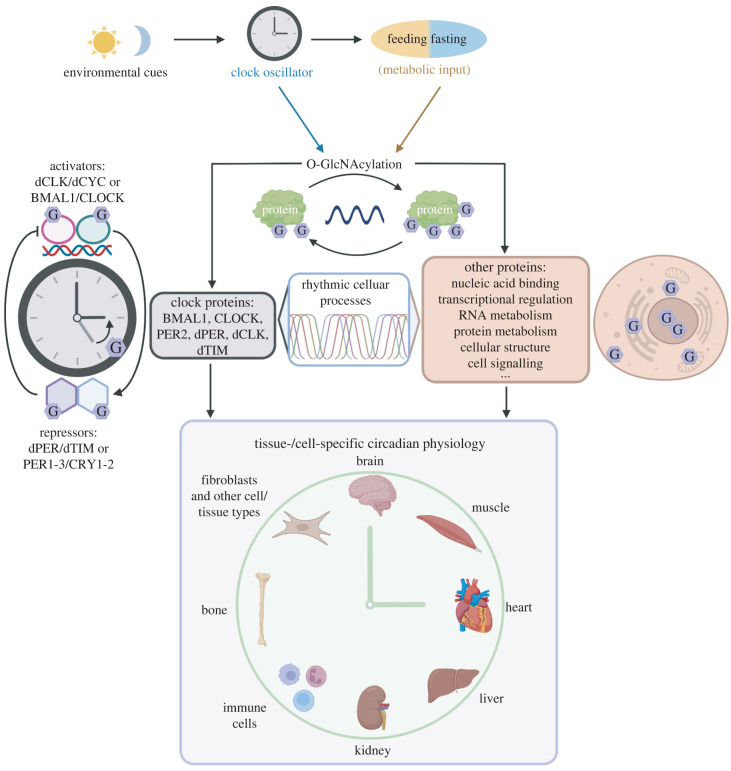
Figure 3.
O-GlcNAcylation regulates daily biological rhythms from cellular to organismal level. O-GlcNAcylation rhythmically modifies circadian clock proteins, key components of the molecular oscillator [21–23]. Global increase in cellular O-GlcNAcylation slows down the pace of circadian clocks, which in turn alters timing of rhythmic cellular processes [20–22]. In addition to clock proteins, thousands of other cellular proteins are also O-GlcNAcylated. O-GlcNAcylation is directly involved in regulating basic cellular functions, such as transcriptional regulation, RNA metabolism, translation, protein metabolism [16–19]. Furthermore, O-GlcNAcylation can also modify activities of organ-, tissue- or cell-specific processes [153–173]. In summary, rhythmic O-GlcNAcylation ranging from subcellular to organ levels manifest into robust daily biological rhythms at the organismal level. dCLK, Drosophila CLOCK; dCYC Drosophila CYCLE; BMAL1, brain and muscle Arnt-like protein-1; CLOCK, circadian locomotor output cycles kaput; dPER, Drosophila PERIOD; dTIM, Drosophila TIMELESS; PER1-3, PERIOD1-3; CRY1-2, CRYPTOCHROME1-2.
4.1. Regulation of clock proteins within the core molecular oscillator by O-GlcNAcylation
The molecular oscillator of the animal circadian timing system relies on transcription-translation feedback mechanisms to maintain approximately 24 h biological rhythms (reviewed in [1,2]) (figure 3). Brain and muscle Arnt-like protein-1 (BMAL1) and circadian locomotor output cycles kaput (CLOCK) are the key transcriptional activators of the mammalian clock (Drosophila homologues are dCYCLE and dCLOCK (dCLK)), and as heterodimers, they drive the expression of thousands of clock-controlled genes including genes that encode their own transcriptional repressors, PERIOD1-3 (PER1-3) and CRYPTOCHROME1-2 (CRY1-2) (dPER and dTIMELESS (dTIM) in Drosophila) (figure 3). The molecular oscillator is critical for generating daily rhythmicity of gene expression that manifest into a range of rhythmic biological processes (reviewed in [1–10]).
PTMs, especially phosphorylation, have been established as essential mechanisms for maintaining the pace of the molecular oscillator [174–176]. Phosphorylation of the clock protein dPER was first characterized by Edery et al. [177] and subsequent studies continue to highlight diverse properties of clock proteins that are regulated by phosphorylation (reviewed in [178,179]). O-GlcNAcylation was first introduced as a mechanism to regulate the molecular clock and circadian rhythms by Kim et al. [20], Kaasik et al. [21] and Li et al. [22]. These pioneering studies showed that in both fly and mammalian models, increasing global O-GlcNAcylation slows down the pace of the clock and results in period lengthening of behavioural rhythms, while reducing O-GlcNAcylation has the opposite effect. Furthermore, these studies and subsequent studies [23,180] showed that many clock proteins, including dCLK, dPER, dTIM, BMAL1, CLOCK, are O-GlcNAcylated and period-altering effects are mediated by disrupting clock protein O-GlcNAcylation.
Interestingly but perhaps not surprisingly, some clock proteins even display daily rhythms of O-GlcNAcylation that are sensitive to feeding and nutrient input [21–23]. In Drosophila, O-GlcNAcylation of dPER promotes its stability and inhibits nuclear entry [20]. Transcriptional reporter assays in Drosophila S2 tissue culture showed that manipulating O-GlcNAcylation levels by overexpressing OGT or OGA also changes dPER and dCLK transcriptional activities [21]. To begin to dissect site-specific O-GlcNAc regulation, our group mapped dPER and dTIM O-GlcNAc sites in fly heads using MS proteomics [23,180]. We found that O-GlcNAcylation of dPER S942 inhibits the interaction of dPER and dCLK and reduces dPER repressor activity [23]. The function of O-GlcNAcylation on dTIM however remains to be determined. In mammalian clock, O-GlcNAcylation is shown to stabilize BMAL1 and CLOCK by inhibiting their ubiquitination [22]. In HEK293 cells, O-GlcNAcylation and phosphorylation compete at PER2 S662, and O-GlcNAcylation of S662 increases PER2 repressor activity [21]. In summary, O-GlcNAcylation is highly involved in the regulation of molecular oscillators (figure 3). Future site-specific characterization of clock proteins is needed to further understand the mechanisms by which metabolic input regulates molecular oscillators and biological rhythms through O-GlcNAcylation.
4.2. Regulation of rhythmic cellular processes beyond the molecular oscillator by O-GlcNAcylation
O-GlcNAcylation not only occurs on clock proteins, but also regulates the function of a large part of the proteome (figure 3). By cataloging results from over 1700 articles, O-GlcNAcome Database listed 7789 human O-GlcNAc proteins and 3503 mouse O-GlcNAc proteins [18]. Meta analysis on the human O-GlcNAcylated proteins from Wulff-Fuentes et al. [18] and combing through published O-GlcNAcomic papers [153–173] suggest that O-GlcNAc proteins are heavily involved in nucleic acid binding/transcriptional regulation/RNA metabolism, metabolism of proteins, cellular structure and cell signalling (figure 3). Many elegant reviews have summarized published functional investigations of O-GlcNAcylation on cellular proteins (e.g. [16,17,19,181,182]). To date, although circadian/daily rhythm of the O-GlcNAcome has yet to be conducted, our study showing robust daily rhythmicity in global protein O-GlcNAcylation in Drosophila tissues suggests that diverse cellular processes and molecular pathways could potentially be rhythmically regulated by daily cycling O-GlcNAcylation that is sensitive to metabolic input [15].
In table 2, we summarize published efforts to identify O-GlcNAcylated proteins in different cell types or tissues, which could provide insights into tissue- or cell-specific daily O-GlcNAc regulation on biological rhythms. In particular, we highlight pathways with O-GlcNAcylated factors that are critical for performing tissue- or cell-specific functions. We excluded O-GlcNAcomic studies conducted using cancer cell lines, as cellular O-GlcNAc status is known to be altered in cancer cells compared to cells under physiological conditions [183–185]. Finally, we also excluded studies conducted using whole organisms, such as D. melanogaster and Caenorhabditis elegans [186–188], since our focus in table 2 is on tissue-specific characterization.
Table 2.
O-GlcNAcomic studies in animal tissues and cell lines.
| tissue or cell type | organism | number of O-GlcNAc proteins | number of O-GlcNAc sites | tissue- or cell-specific function of O-GlcNAcylation | references |
|---|---|---|---|---|---|
| nervous system | |||||
| forebrain | rat | 25 | n.a. | cellular communication/signal transduction; intracellular transport | [153,154] |
| hippocampus | mouse | 14 | n.a. | neuronal structure; glucose metabolism | [155] |
| cerebral cortical tissue | mouse | 274 | n.a. | neurogenesis; synaptic transmission; learning and memory; cytoskeleton | [156] |
| cortex | mouse | 278 | n.a. | synaptic trafficking; notch/Wnt signalling; circadian clock proteins | [157] |
| brain | rat | 30 | n.a. | signal transduction; cytoskeleton and vesicle trafficking | [158] |
| brain | human | 530 | 1094 | receptor signalling; substrate-adhesion dependent cell spreading; cell projection assembly | [159] |
| muscular system | |||||
| gastrocnemius muscle | rat | 14 | n.a. | glycolytic pathway and energetic metabolism; contractile protein | [160] |
| C2C12 myotubes | mouse | 342 | n.a. | cytoskeleton and chaperones; transporter and binding proteins; cell adhesion molecules | [161] |
| right ventricle | rat | 500 | n.a. | oxidation–reduction process; intracellular transport; metabolism; cellular respiration and energy | [162] |
| excretory system | |||||
| embryonic kidney cells (HEK293) | human | 1500 | 180 | cell death; molecular transport; cellular assembly and organization; cell cycle, growth and proliferation; cell morphology; PTM | [163] |
| embryonic kidney cells (HEK293) | human | 75 | n.a. | cell-cell adhesion; cell cycle; molecular transport; Purine ribonucleoside monophosphate biosynthetic process; cellular response to heat; viral process | [164] |
| embryonic kidney cells (HEK293) | human | 215 | n.a. | Metabolism; Signal transduction; Translation; Transport | [165] |
| urine | human | 457 | n.a. | organelle organization; cell cycle; cellular localization; heterocycle metabolic processes; DNA repair; cellular response to stress; developmental processes; transport | [166] |
| immune system | |||||
| T cell | mouse | 116 | n.a. | metabolic process; cellular component organization/biogenesis; DNA packing | [167] |
| T cell | human | 133 | n.a. | nucleotide, nucleic acid transport | [168] |
| T cell | human | 1045 | n.a. | viral process; cell-cell adhesion; cell cycle; cellular transport; protein sumoylation | [169] |
| embryonic macrophage-like cells (S2 cells) | fruit fly | 51 | n.a. | metabolism; stress response; cell cycle | [170] |
| other tissues or cell types | |||||
| liver | rat | 68 | n.a. | metabolism; transport; signal transduction | [165] |
| osteoblasts (MC3T3E1) | mouse | 20 | n.a. | post-translational regulation; systemic nutrient homeostasis | [171] |
| placental trophoblasts (BeWo) | human | 829 | n.a. | translational initiation; viral transcription; SRP-dependent co-translational protein targeting to membrane | [172] |
| fibroblasts (NIH3T3) | mouse | 374 | n.a. | metabolism; intracellular transport | [173] |
In addition to tissue- or cell-specific studies, there are other in depth O-GlcNAcomic studies at the organelle level, such as mitochondria from rat heart [189,190] and rat liver [191], cardiac myofilament from rat [192], synapses from mouse brain [130,193,194], ribosome from rat liver [195] and nuclei from mouse embryonic stem cells [196]. Additionally, comparative O-GlcNAcomic studies have been carried out in cell culture systems to investigate the role of O-GlcNAcylation under different conditions [197–199] or during the progression of cell cycles [200,201]. However, comparative O-GlcNAcomic studies over different time points of a 24 h day and in different organs/tissues in vivo are warranted to reveal the role of O-GlcNAcylation in regulating daily rhythmicity in organ- and tissue-specific physiology and potential differential regulation by metabolic and circadian signals.
5. Conclusion
Significant progress has been made to elucidate the regulation of metabolic signals on daily rhythms in physiology and behaviour. Metabolic input regulates gene expression at specific times of day through nutrient-sensing pathways influenced by feeding–fasting cycles. This is accomplished through functional modification of core clock proteins and/or epigenetic regulation of the genomic landscape. Our recent study showed that O-GlcNAcylation is also an important mechanism that integrates metabolic and circadian signals to regulate the daily biological rhythms [15]. In this review, we summarize published and potential mechanisms by which metabolic and circadian signals can shape daily O-GlcNAc oscillation, discuss crosstalk between O-GlcNAcylation and the phosphoproteome to regulate rhythmic protein functions, and highlight cellular pathways that may be regulated by oscillating O-GlcNAcylation in different tissues or cell types. As time-restricted feeding/eating is emerging as a non-invasive therapeutic strategy to alleviate metabolic syndromes [30,35,202,203], our review provides mechanistic insight into the significance of properly aligning our eating time with biological rhythm. Since we showed in Drosophila that food consumption at unnatural feeding time of the day–night cycle can dampen the oscillation of protein O-GlcNAcylation [15], it is likely that rhythmic functions of O-GlcNAc proteins/pathways would be disrupted with mistimed eating. This may contribute to deleterious effects of mistimed eating and high-fat diet, which has been shown to impair feeding–fasting rhythms and rhythmic metabolic input [204,205].
It is important to note that different organs or tissues are likely differentially regulated by metabolic input versus circadian input. For example, despite that the blood–brain barrier is expected to render brain tissues less sensitive to daily oscillation of metabolites, O-GlcNAcylation has been detected in brain tissues and shown to oscillate on clock proteins in fly heads [20,21,23]. Our understanding of the similarities and divergence among O-GlcNAcomes in multiple organs or tissues and how organisms coordinate organ/tissue-specific O-GlcNAcomic rhythms to properly maintain time-of-day physiology at the organismal level will improve with continued development of comparative O-GlcNAcomic methods [198,201].
In this review, we largely focused on O-GlcNAcylation as an intracellular mechanism that underlies metabolic regulation of daily biological rhythms. We briefly mentioned a few intercellular signals, such as insulin, EGF, TGFβ, which could contribute to regulation of protein O-GlcNAcylation. However, there are many other intercellular signals, including neuronal signals, hormones (melatonin, adrenal cortex hormones, thyroid hormones etc.) and gut microbiota, which can relay time-of-day specific metabolic signals to influence cellular protein functions. How O-GlcNAcylation responds to these intercellular signals is unknown and beyond the scope of this review.
Finally, it is important to note that O-GlcNAcylation is only one of many nutrient-sensitive PTMs. Feeding–fasting cycles likely regulate daily cellular physiology through other metabolite-driven PTMs. Figlia et al. [206] reviewed over 20 different types of PTM using metabolites, such as lipids, amino acids, Coenzyme-A, acetate, malonate and lactate. Future studies are warranted to determine whether these nutrient-sensitive PTMs are also involved in regulation of daily biological rhythms. Recently, Bludau et al. [207] developed an exciting tool to predict protein structure with PTMs. In combination with site-specific information of these metabolite-driven PTMs, the metabolic regulation of protein functions could be computationally predicted, which could provide a more comprehensive view on the metabolic effect on daily biological rhythms.
Acknowledgements
We thank Yao D. Cai for critical reading of the manuscript. Figures are created with BioRender.com. We apologize to any colleagues whose works were not included in this review owing to space limitations.
Data accessibility
This article has no additional data.
Authors' contributions
X.L.: conceptualization, data curation, formal analysis, investigation, methodology, writing—original draft, writing—review and editing; J.C.C.: conceptualization, funding acquisition, project administration, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was funded by NIH R01 DK124068 to J.C.C.
References
- 1.Cox KH, Takahashi JS. 2019. Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 63, R93-R102. ( 10.1530/JME-19-0153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patke A, Young MW, Axelrod S. 2020. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67-84. ( 10.1038/s41580-019-0179-2) [DOI] [PubMed] [Google Scholar]
- 3.Deota S, Panda S. 2021. New horizons: circadian control of metabolism offers novel insight into the cause and treatment of metabolic diseases. J. Clin. Endocrinol. Metab. 106, e1488-e1493. ( 10.1210/clinem/dgaa691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono D. 2022. Neural circuits in the central circadian clock and their regulation of sleep and wakefulness in mammals. Neurosci. Res. 182, 1-6. ( 10.1016/j.neures.2022.05.005) [DOI] [PubMed] [Google Scholar]
- 5.Yildirim E, Curtis R, Hwangbo D. 2022. Roles of peripheral clocks: lessons from the fly. FEBS Lett. 596, 263-293. ( 10.1002/1873-3468.14251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng F, Li X, Xiao F, Zhao R, Sun Z. 2022. Circadian clock, diurnal glucose metabolic rhythm, and dawn phenomenon. Trends Neurosci. 45, 471-482. ( 10.1016/j.tins.2022.03.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Y, Silver R. 2022. Mutual shaping of circadian body-wide synchronization by the suprachiasmatic nucleus and circulating steroids. Front. Behav. Neurosci. 16, 877256. ( 10.3389/fnbeh.2022.877256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Lutes LK, Barnoud C, Scheiermann C. 2022. The circadian immune system. Sci. Immunol. 7, eabm2465. ( 10.1126/sciimmunol.abm2465) [DOI] [PubMed] [Google Scholar]
- 9.Tabuchi M, Coates KE, Bautista OB, Zukowski LH. 2021. Light/clock influences membrane potential dynamics to regulate sleep states. Front. Neurol. 12, 625369. ( 10.3389/fneur.2021.625369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuddapah VA, Zhang SL, Sehgal A. 2019. Regulation of the blood–brain barrier by circadian rhythms and sleep. Trends Neurosci. 42, 500-510. ( 10.1016/j.tins.2019.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panda S. 2016. Circadian physiology of metabolism. Science 354, 1008-1015. ( 10.1126/science.aah4967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mauvoisin D, et al. 2017. Circadian and feeding rhythms orchestrate the diurnal liver acetylome. Cell Rep. 20, 1729-1743. ( 10.1016/j.celrep.2017.07.065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato T, Sassone-Corsi P. 2022. Nutrition, metabolism, and epigenetics: pathways of circadian reprogramming. EMBO Rep. 23, e52412. ( 10.15252/embr.202152412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu XE, Swanda RV, Qian S-B. 2020. Nutrient control of mRNA Translation. Annu. Rev. Nutr. 40, 51-75. ( 10.1146/annurev-nutr-120919-041411) [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Blaženović I, Contreras AJ, Pham TM, Tabuloc CA, Li YH, Ji J, Fiehn O, Chiu JC. 2021. Hexosamine biosynthetic pathway and O-GlcNAc-processing enzymes regulate daily rhythms in protein O-GlcNAcylation. Nat. Commun. 12, 4173. ( 10.1038/s41467-021-24301-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart GW. 2019. Nutrient regulation of signaling and transcription. J. Biol. Chem. 294, 2211-2231. ( 10.1074/jbc.AW119.003226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatham JC, Zhang J, Wende AR. 2021. Role of O-linked N-acetylglucosamine protein modification in cellular (patho)physiology. Physiol. Rev. 101, 427-493. ( 10.1152/physrev.00043.2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J, Kahsay R, Olivier-Van Stichelen S. 2021. The human O-GlcNAcome database and meta-analysis. Sci. Data 8, 25. ( 10.1038/s41597-021-00810-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma J, Hou C, Wu C. In press. Demystifying the O-GlcNAc code: a systems view. Chem. Rev. 10.1021/acs.chemrev.1c01006) [DOI] [PubMed] [Google Scholar]
- 20.Kim EY, Jeong EH, Park S, Jeong H-J, Edery I, Cho JW. 2012. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26, 490-502. ( 10.1101/gad.182378.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaasik K, et al. 2013. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291-302. ( 10.1016/j.cmet.2012.12.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M-D, Ruan H-B, Hughes ME, Lee J-S, Singh JP, Jones SP, Nitabach MN, Yang X. 2013. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 17, 303-310. ( 10.1016/j.cmet.2012.12.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YH, Liu X, Vanselow JT, Zheng H, Schlosser A, Chiu JC. 2019. O-GlcNAcylation of PERIOD regulates its interaction with CLOCK and timing of circadian transcriptional repression. PLoS Genet. 15, e1007953. ( 10.1371/journal.pgen.1007953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laarse SAM, Leney AC, Heck AJR. 2018. Crosstalk between phosphorylation and O-GlcNAcylation: friend or foe. FEBS J. 285, 3152-3167. ( 10.1111/febs.14491) [DOI] [PubMed] [Google Scholar]
- 25.Robles MS, Humphrey SJ, Mann M. 2017. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 25, 118-127. ( 10.1016/j.cmet.2016.10.004) [DOI] [PubMed] [Google Scholar]
- 26.Horta MAC, et al. 2019. Broad substrate-specific phosphorylation events are associated with the initial stage of plant cell wall recognition in Neurospora crassa. Front. Microbiol. 10, 2317. ( 10.3389/fmicb.2019.02317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, et al. 2020. Integrated omics in Drosophila uncover a circadian kinome. Nat. Commun. 11, 2710. ( 10.1038/s41467-020-16514-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krahmer J, Hindle M, Perby LK, Mogensen HK, Nielsen TH, Halliday KJ, van Ooijen G, Le Bihan T, Millar AJ. 2022. The circadian clock gene circuit controls protein and phosphoprotein rhythms in Arabidopsis thaliana. Mol. Cell. Proteomics 21, 100172. ( 10.1016/j.mcpro.2021.100172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acosta-Rodríguez V, Rijo-Ferreira F, Izumo M, Xu P, Wight-Carter M, Green CB, Takahashi JS. 2022. Circadian alignment of early onset caloric restriction promotes longevity in male C57BL/6 J mice. Science 376, 1192-1202. ( 10.1126/science.abk0297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaix A, Manoogian ENC, Melkani GC, Panda S. 2019. Time-restricted eating to prevent and manage chronic metabolic diseases. Annu. Rev. Nutr. 39, 291-315. ( 10.1146/annurev-nutr-082018-124320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaix A, Lin T, Le HD, Chang MW, Panda S. 2019. Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 29, 303-319.e4. ( 10.1016/j.cmet.2018.08.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaix A, Zarrinpar A, Miu P, Panda S. 2014. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 20, 991-1005. ( 10.1016/j.cmet.2014.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaix A, Deota S, Bhardwaj R, Lin T, Panda S. 2021. Sex- and age-dependent outcomes of 9-hour time-restricted feeding of a Western high-fat high-sucrose diet in C57BL/6 J mice. Cell Rep. 36, 109543. ( 10.1016/j.celrep.2021.109543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gill S, Le HD, Melkani GC, Panda S. 2015. Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science 347, 1265-1269. ( 10.1126/science.1256682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatori M, et al. 2012. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848-860. ( 10.1016/j.cmet.2012.04.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Very N, Vercoutter-Edouart A-S, Lefebvre T, Hardivillé S, El Yazidi-Belkoura I. 2018. Cross-dysregulation of O-GlcNAcylation and PI3 K/AKT/mTOR axis in human chronic diseases. Front. Endocrinol. 9, 602. ( 10.3389/fendo.2018.00602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weigert C, Klopfer K, Kausch C, Brodbeck K, Stumvoll M, Häring HU, Schleicher ED. 2003. Palmitate-induced activation of the hexosamine pathway in human myotubes. Diabetes 52, 650-656. ( 10.2337/diabetes.52.3.650) [DOI] [PubMed] [Google Scholar]
- 38.Chaveroux C, et al. 2016. Nutrient shortage triggers the hexosamine biosynthetic pathway via the GCN2-ATF4 signalling pathway. Sci. Rep. 6, 27278. ( 10.1038/srep27278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson AJ, Kudlow JE. 1995. Regulation of glutamine:fructose-6-phosphate amidotransferase gene transcription by epidermal growth factor and glucose. Endocrinology 136, 2809-2816. ( 10.1210/endo.136.7.7789306) [DOI] [PubMed] [Google Scholar]
- 40.Dai W, Dierschke SK, Toro AL, Dennis MD. 2018. Consumption of a high fat diet promotes protein O-GlcNAcylation in mouse retina via NR4A1-dependent GFAT2 expression. Biochim. Biophys. Acta 1864, 3568-3576. ( 10.1016/j.bbadis.2018.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B, Huang Z-B, Chen X, See Y-X, Chen Z-K, Yao H-K. 2019. Mammalian target of rapamycin 2 (MTOR2) and C-MYC modulate glucosamine-6-phosphate synthesis in glioblastoma (GBM) cells through glutamine:fructose-6-phosphate aminotransferase 1 (GFAT1). Cell. Mol. Neurobiol. 39, 415-434. ( 10.1007/s10571-019-00659-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng H, Huang J, Zhang M, Zhao H-J, Chen P, Zeng Z-H. 2022. miR-27b-3p improved high glucose-induced spermatogenic cell damage via regulating Gfpt1/HBP signaling. Eur. Surg. Res. 63, 64-76. ( 10.1159/000518960) [DOI] [PubMed] [Google Scholar]
- 43.Moloughney JG, et al. 2016. mTORC2 responds to glutamine catabolite levels to modulate the hexosamine biosynthesis enzyme GFAT1. Mol. Cell 63, 811-826. ( 10.1016/j.molcel.2016.07.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eguchi S, Oshiro N, Miyamoto T, Yoshino K, Okamoto S, Ono T, Kikkawa U, Yonezawa K. 2009. AMP-activated protein kinase phosphorylates glutamine:fructose-6-phosphate amidotransferase 1 at Ser243 to modulate its enzymatic activity. Genes Cells 14, 179-189. ( 10.1111/j.1365-2443.2008.01260.x) [DOI] [PubMed] [Google Scholar]
- 45.Zibrova D, et al. 2017. GFAT1 phosphorylation by AMPK promotes VEGF-induced angiogenesis. Biochem. J. 474, 983-1001. ( 10.1042/BCJ20160980) [DOI] [PubMed] [Google Scholar]
- 46.Moloughney JG, et al. 2018. mTORC2 modulates the amplitude and duration of GFAT1 Ser-243 phosphorylation to maintain flux through the hexosamine pathway during starvation. J. Biol. Chem. 293, 16 464-16 478. ( 10.1074/jbc.RA118.003991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graack HR, Cinque U, Kress H. 2001. Functional regulation of glutamine:fructose-6-phosphate aminotransferase 1 (GFAT1) of Drosophila melanogaster in a UDP-N-acetylglucosamine and cAMP-dependent manner. Biochem. J. 360, 401-412. ( 10.1042/0264-6021:3600401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broschat KO, Gorka C, Page JD, Martin-Berger CL, Davies MS, Huang H, Gulve EA, Salsgiver WJ, Kasten TP. 2002. Kinetic characterization of human glutamine-fructose-6-phosphate amidotransferase I. J. Biol. Chem. 277, 14 764-14 770. ( 10.1074/jbc.M201056200) [DOI] [PubMed] [Google Scholar]
- 49.Ruegenberg S, Horn M, Pichlo C, Allmeroth K, Baumann U, Denzel MS. 2020. Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis. Nat. Commun. 11, 687. ( 10.1038/s41467-020-14524-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor RP, Geisler TS, Chambers JH, McClain DA. 2009. Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. J. Biol. Chem. 284, 3425-3432. ( 10.1074/jbc.M803198200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou L, Zhu-Mauldin X, Marchase RB, Paterson AJ, Liu J, Yang Q, Chatham JC. 2012. Glucose deprivation-induced increase in protein O-GlcNAcylation in cardiomyocytes is calcium-dependent. J. Biol. Chem. 287, 34 419-34 431. ( 10.1074/jbc.M112.393207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F, Snead CM, Catravas JD. 2012. Hsp90 regulates O-linked β-N-acetylglucosamine transferase: a novel mechanism of modulation of protein O-linked β-N-acetylglucosamine modification in endothelial cells. Am. J. Physiol.-Cell Physiol. 302, C1786-C1796. ( 10.1152/ajpcell.00004.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park S, Pak J, Jang I, Cho J. 2014. Inhibition of mTOR affects protein stability of OGT. Biochem. Biophys. Res. Commun. 453, 208-212. ( 10.1016/j.bbrc.2014.05.047) [DOI] [PubMed] [Google Scholar]
- 54.Sodi VL, Khaku S, Krutilina R, Schwab LP, Vocadlo DJ, Seagroves TN, Reginato MJ. 2015. mTOR/MYC axis regulates O-GlcNAc transferase expression and O-GlcNAcylation in breast cancer. Mol. Cancer Res. 13, 923-933. ( 10.1158/1541-7786.MCR-14-0536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez-Cervera Y, Dehennaut V, Gil MA, Guedri K, Mata CJS, Stichelen SO, Michalski J, Foulquier F, Lefebvre T. 2013. Insulin signaling controls the expression of O-GlcNAc transferase and its interaction with lipid microdomains. FASEB J. 27, 3478-3486. ( 10.1096/fj.12-217984) [DOI] [PubMed] [Google Scholar]
- 56.Lo W-Y, Yang W-K, Peng C-T, Pai W-Y, Wang H-J. 2018. MicroRNA-200a/200b modulate high glucose-induced endothelial inflammation by targeting O-linked N-acetylglucosamine transferase expression. Front. Physiol. 9, 355. ( 10.3389/fphys.2018.00355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whelan SA, Lane MD, Hart GW. 2008. Regulation of the O-linked β-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 283, 21 411-21 417. ( 10.1074/jbc.M800677200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Pandey A, Hart GW. 2007. Dynamic interplay between O-Linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell. Proteomics 6, 1365-1379. ( 10.1074/mcp.M600453-MCP200) [DOI] [PubMed] [Google Scholar]
- 59.Bullen JW, Balsbaugh JL, Chanda D, Shabanowitz J, Hunt DF, Neumann D, Hart GW. 2014. Cross-talk between two essential nutrient-sensitive enzymes. J. Biol. Chem. 289, 10 592-10 606. ( 10.1074/jbc.M113.523068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huttlin EL, et al. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174-1189. ( 10.1016/j.cell.2010.12.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruan H-B, et al. 2017. Calcium-dependent O-GlcNAc signaling drives liver autophagy in adaptation to starvation. Genes Dev. 31, 1655-1665. ( 10.1101/gad.305441.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seo HG, Kim HB, Kang MJ, Ryum JH, Yi EC, Cho JW. 2016. Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci. Rep. 6, 34614. ( 10.1038/srep34614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian K, et al. 2018. Transcriptional regulation of O-GlcNAc homeostasis is disrupted in pancreatic cancer. J. Biol. Chem. 293, 13 989-14 000. ( 10.1074/jbc.RA118.004709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaidyanathan K, et al. 2017. Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 292, 8948-8963. ( 10.1074/jbc.M116.771030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tuomela T, Viinikka L, Perheentupa J. 1990. Epidermal growth factor in mice: changes during circadian and female reproductive cycles. Acta Endocrinol. (Copenh.) 123, 643-648. ( 10.1530/acta.0.1230643) [DOI] [PubMed] [Google Scholar]
- 66.Haus E, Haus E, Dumitriu L, Nicolau GY, Bologa S, Sackett-Lundeen L. 2001. Circadian rhythms of basic fibroblast growth factor (bFGF), epidermal growth factor (EGF), insulin-like growth factor-1 (IGF-1), insulin-like growth factor binding protein-3 (IGFBP-3), cortisol, and melatonin in women with breast cancer. Chronobiol. Int. 18, 709-727. ( 10.1081/CBI-100106083) [DOI] [PubMed] [Google Scholar]
- 67.Lucena MC, et al. 2016. Epithelial mesenchymal transition induces aberrant glycosylation through hexosamine biosynthetic pathway activation. J. Biol. Chem. 291, 12 917-12 929. ( 10.1074/jbc.M116.729236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaghefi SSE, Mousavi F, Khaksari M, Asadikaram G, Soltani Z. 2021. Sex-related changes in circadian rhythm of inflammatory and oxidative stress markers in CKD. Iran J. Kidney Dis. 15, 351-363. ( 10.52547/ijkd.6242) [DOI] [PubMed] [Google Scholar]
- 69.Yang H, Yang L-T, Liu J, Tang S, Zhao X, Wang Q, Zhang S, Pan W, Yang P-C. 2018. Circadian protein CLK suppresses transforming growth factor-β expression in peripheral B cells of nurses with day-night shift rotation. Am. J. Transl. Res. 10, 4331-4337. [PMC free article] [PubMed] [Google Scholar]
- 70.Yamazaki K, Mizui Y, Oki T, Okada M, Tanaka I. 2000. Cloning and characterization of mouse glutamine:fructose-6-phosphate amidotransferase 2 gene promoter. Gene 261, 329-336. ( 10.1016/s0378-1119(00)00497-2) [DOI] [PubMed] [Google Scholar]
- 71.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl Acad. Sci. USA 111, 16 219-16 224. ( 10.1073/pnas.1408886111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Atger F, et al. 2015. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl Acad. Sci. USA 112, E6579-E6588. ( 10.1073/pnas.1515308112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou J, Huynh QK, Hoffman RT, Crook ED, Daniels MC, Gulve EA, McClain DA. 1998. Regulation of glutamine:fructose-6-phosphate amidotransferase by cAMP-dependent protein kinase. Diabetes 47, 1836-1840. ( 10.2337/diabetes.47.12.1836) [DOI] [PubMed] [Google Scholar]
- 74.Chang Q, Su K, Baker JR, Yang X, Paterson AJ, Kudlow JE. 2000. Phosphorylation of human glutamine:fructose-6-phosphate amidotransferase by cAMP-dependent protein kinase at serine 205 blocks the enzyme activity. J. Biol. Chem. 275, 21 981-21 987. ( 10.1074/jbc.M001049200) [DOI] [PubMed] [Google Scholar]
- 75.Hu Y, Riesland L, Paterson AJ, Kudlow JE. 2004. Phosphorylation of mouse glutamine-fructose-6-phosphate amidotransferase 2 (GFAT2) by cAMP-dependent protein kinase increases the enzyme activity. J. Biol. Chem. 279, 29 988-29 993. ( 10.1074/jbc.M401547200) [DOI] [PubMed] [Google Scholar]
- 76.Ruegenberg S, Mayr FAMC, Atanassov I, Baumann U, Denzel MS. 2021. Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1. Nat. Commun. 12, 2176. ( 10.1038/s41467-021-22320-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X, et al. 2017. Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J. Exp. Med. 214, 1093-1109. ( 10.1084/jem.20161105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ishii T, Warabi E, Mann GE. 2019. Circadian control of BDNF-mediated Nrf2 activation in astrocytes protects dopaminergic neurons from ferroptosis. Free Radic. Biol. Med. 133, 169-178. ( 10.1016/j.freeradbiomed.2018.09.002) [DOI] [PubMed] [Google Scholar]
- 79.Joshi A, Upadhyay KK, Vohra A, Shirsath K, Devkar R. 2021. Melatonin induces Nrf2-HO-1 reprogramming and corrections in hepatic core clock oscillations in non-alcoholic fatty liver disease. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 35, e21803. ( 10.1096/fj.202002556RRR) [DOI] [PubMed] [Google Scholar]
- 80.Wang J, et al. 2021. Circadian clock gene BMAL1 reduces urinary calcium oxalate stones formation by regulating NRF2/HO-1 pathway. Life Sci. 265, 118853. ( 10.1016/j.lfs.2020.118853) [DOI] [PubMed] [Google Scholar]
- 81.Muthusamy S, Hong KU, Dassanayaka S, Hamid T, Jones SP. 2015. E2F1 transcription factor regulates O-linked N-acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase expression. J. Biol. Chem. 290, 31 013-31 024. ( 10.1074/jbc.M115.677534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schick S, Becker K, Thakurela S, Fournier D, Hampel MH, Legewie S, Tiwari VK. 2016. Identifying novel transcriptional regulators with circadian expression. Mol. Cell. Biol. 36, 545-558. ( 10.1128/MCB.00701-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. 2012. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1, e00011. ( 10.7554/eLife.00011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang C, et al. 2019. Hepatocyte nuclear factor 1 alpha (HNF1A) regulates transcription of O-GlcNAc transferase in a negative feedback mechanism. FEBS Lett. 593, 1050-1060. ( 10.1002/1873-3468.13381) [DOI] [PubMed] [Google Scholar]
- 85.Yang Y, Yin X, Yang H, Xu Y. 2015. Histone demethylase LSD2 Acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. Mol. Cell 58, 47-59. ( 10.1016/j.molcel.2015.01.038) [DOI] [PubMed] [Google Scholar]
- 86.Peng K, et al. 2021. Regulation of O-linked N-acetyl glucosamine transferase (OGT) through E6 stimulation of the ubiquitin ligase activity of E6AP. Int. J. Mol. Sci. 22, 10286. ( 10.3390/ijms221910286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berthier A, et al. 2018. Combinatorial regulation of hepatic cytoplasmic signaling and nuclear transcriptional events by the OGT/REV-ERBα complex. Proc. Natl Acad. Sci. USA 115, E11 033-E11 042. ( 10.1073/pnas.1805397115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kondratov RV, Antoch MP. 2007. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends Cell Biol. 17, 311-317. ( 10.1016/j.tcb.2007.07.001) [DOI] [PubMed] [Google Scholar]
- 89.Li Z, Li X, Nai S, Geng Q, Liao J, Xu X, Li J. 2017. Checkpoint kinase 1–induced phosphorylation of O-linked β-N-acetylglucosamine transferase regulates the intermediate filament network during cytokinesis. J. Biol. Chem. 292, 19 548-19 555. ( 10.1074/jbc.M117.811646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deng X, Yi X, Huang D, Liu P, Chen L, Du Y, Hao L. 2020. ROCK2 mediates osteosarcoma progression and TRAIL resistance by modulating O-GlcNAc transferase degradation. Am. J. Cancer Res. 10, 781-798. [PMC free article] [PubMed] [Google Scholar]
- 91.Janich P, Toufighi K, Solanas G, Luis NM, Minkwitz S, Serrano L, Lehner B, Benitah SA. 2013. Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell 13, 745-753. ( 10.1016/j.stem.2013.09.004) [DOI] [PubMed] [Google Scholar]
- 92.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. 2008. O-linked β-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 283, 33 935-33 941. ( 10.1074/jbc.M806199200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Z, Tan EP, VandenHull NJ, Peterson KR, Slawson C. 2014. O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. 5, 206. ( 10.3389/fendo.2014.00206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petrus P, et al. 2022. The central clock suffices to drive the majority of circulatory metabolic rhythms. Sci. Adv. 8, eabo2896. ( 10.1126/sciadv.abo2896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xin H, et al. 2021. A multi-tissue multi-omics analysis reveals distinct kineztics in entrainment of diurnal transcriptomes by inverted feeding. iScience 24, 102335. ( 10.1016/j.isci.2021.102335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rhoades SD, Nayak K, Zhang SL, Sehgal A, Weljie AM. 2018. Circadian- and light-driven metabolic rhythms in Drosophila melanogaster. J. Biol. Rhythms 33, 126-136. ( 10.1177/0748730417753003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marshall S, Nadeau O, Yamasaki K. 2004. Dynamic actions of glucose and glucosamine on hexosamine biosynthesis in isolated adipocytes. J. Biol. Chem. 279, 35 313-35 319. ( 10.1074/jbc.M404133200) [DOI] [PubMed] [Google Scholar]
- 98.Liu J, Marchase RB, Chatham JC. 2007. Increased O-GlcNAc levels during reperfusion lead to improved functional recovery and reduced calpain proteolysis. Am. J. Physiol.-Heart Circ. Physiol. 293, H1391-H1399. ( 10.1152/ajpheart.00285.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grigorian A, Lee S-U, Tian W, Chen IJ, Gao G, Mendelsohn R, Dennis JW, Demetriou M. 2007. Control of T cell-mediated autoimmunity by metabolite flux to N-glycan biosynthesis. J. Biol. Chem. 282, 20 027-20 035. ( 10.1074/jbc.M701890200) [DOI] [PubMed] [Google Scholar]
- 100.Nakajima K, Kitazume S, Angata T, Fujinawa R, Ohtsubo K, Miyoshi E, Taniguchi N. 2010. Simultaneous determination of nucleotide sugars with ion-pair reversed-phase HPLC. Glycobiology 20, 865-871. ( 10.1093/glycob/cwq044) [DOI] [PubMed] [Google Scholar]
- 101.Wellen KE, Lu C, Mancuso A, Lemons JMS, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. 2010. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 24, 2784-2799. ( 10.1101/gad.1985910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palorini R, Cammarata FP, Balestrieri C, Monestiroli A, Vasso M, Gelfi C, Alberghina L, Chiaradonna F. 2013. Glucose starvation induces cell death in K-ras-transformed cells by interfering with the hexosamine biosynthesis pathway and activating the unfolded protein response. Cell Death Dis. 4, e732. ( 10.1038/cddis.2013.257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abdel Rahman AM, Ryczko M, Pawling J, Dennis JW. 2013. Probing the hexosamine biosynthetic pathway in human tumor cells by multitargeted tandem mass spectrometry. ACS Chem. Biol. 8, 2053-2062. ( 10.1021/cb4004173) [DOI] [PubMed] [Google Scholar]
- 104.Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DMF, Cantrell DA. 2016. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17, 712-720. ( 10.1038/ni.3439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vasconcelos-dos-Santos A, et al. 2017. Hyperglycemia exacerbates colon cancer malignancy through hexosamine biosynthetic pathway. Oncogenesis 6, e306. ( 10.1038/oncsis.2017.2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pham LV, et al. 2016. Targeting the hexosamine biosynthetic pathway and O-linked N-acetylglucosamine cycling for therapeutic and imaging capabilities in diffuse large B-cell lymphoma. Oncotarget 7, 80 599-80 611. ( 10.18632/oncotarget.12413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hawkins M, Barzilai N, Liu R, Hu M, Chen W, Rossetti L. 1997. Role of the glucosamine pathway in fat-induced insulin resistance. J. Clin. Invest. 99, 2173-2182. ( 10.1172/JCI119390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li X, et al. 2018. CirGRDB: a database for the genome-wide deciphering circadian genes and regulators. Nucleic Acids Res. 46, D64-D70. ( 10.1093/nar/gkx944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rodriguez J, Tang C-HA, Khodor YL, Vodala S, Menet JS, Rosbash M. 2013. Nascent-Seq analysis of Drosophila cycling gene expression. Proc. Natl Acad. Sci. USA 110, E275-E284. ( 10.1073/pnas.1219969110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F. 2014. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl Acad. Sci. USA 111, 167-172. ( 10.1073/pnas.1314066111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamazaki K. 2014. Glutamine–Fructose-6-Phosphate Transaminase 1,2 (GFPT1,2). In Handbook of glycosyltransferases and related genes (eds Taniguchi N, Honke K, Fukuda M, Narimatsu H, Yamaguchi Y, Angata T), pp. 1465-1479. Tokyo, Japan: Springer. [Google Scholar]
- 112.Richards AM, Nicholls MG, Espiner EA, Ikram H, Cullens M, Hinton D. 1986. Diurnal patterns of blood pressure, heart rate and vasoactive hormones in normal man. Clin. Exp. Hypertens. A 8, 153-166. ( 10.3109/10641968609074769) [DOI] [PubMed] [Google Scholar]
- 113.James LR, Ingram A, Ly H, Thai K, Cai L, Scholey JW. 2001. Angiotensin II activates the GFAT promoter in mesangial cells. Am. J. Physiol.-Ren. Physiol. 281, F151-F162. ( 10.1152/ajprenal.2001.281.1.F151) [DOI] [PubMed] [Google Scholar]
- 114.Isobe S, et al. 2016. Augmented circadian rhythm of the intrarenal renin–angiotensin systems in anti-thymocyte serum nephritis rats. Hypertens. Res. 39, 312-320. ( 10.1038/hr.2015.151) [DOI] [PubMed] [Google Scholar]
- 115.Hughes ME, Hong H-K, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, Hogenesch JB. 2012. Brain-specific rescue of clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet. 8, e1002835. ( 10.1371/journal.pgen.1002835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442. ( 10.1371/journal.pgen.1000442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Masri S, et al. 2014. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659-672. ( 10.1016/j.cell.2014.06.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. 2015. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 25, 1848-1859. ( 10.1101/gr.195404.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang G, et al. 2016. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 8, 324ra16. ( 10.1126/scitranslmed.aad3305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. 2013. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455. ( 10.1371/journal.pbio.1001455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kuintzle RC, Chow ES, Westby TN, Gvakharia BO, Giebultowicz JM, Hendrix DA. 2017. Circadian deep sequencing reveals stress-response genes that adopt robust rhythmic expression during aging. Nat. Commun. 8, 14529. ( 10.1038/ncomms14529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Terajima H, Yoshitane H, Ozaki H, Suzuki Y, Shimba S, Kuroda S, Iwasaki W, Fukada Y. 2017. ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat. Genet. 49, 146-151. ( 10.1038/ng.3731) [DOI] [PubMed] [Google Scholar]
- 123.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512-D520. ( 10.1093/nar/gku1267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Allison DF, Wamsley JJ, Kumar M, Li D, Gray LG, Hart GW, Jones DR, Mayo MW. 2012. Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-κB acetylation and transcription. Proc. Natl Acad. Sci. USA 109, 16 888-16 893. ( 10.1073/pnas.1208468109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ma Z, Chalkley RJ, Vosseller K. 2017. Hyper-O-GlcNAcylation activates nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) signaling through interplay with phosphorylation and acetylation. J. Biol. Chem. 292, 9150-9163. ( 10.1074/jbc.M116.766568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kronlage M, et al. 2019. O-GlcNAcylation of histone deacetylase 4 protects the diabetic heart from failure. Circulation 140, 580-594. ( 10.1161/CIRCULATIONAHA.117.031942) [DOI] [PubMed] [Google Scholar]
- 127.Fujiki R, et al. 2011. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, 557-560. ( 10.1038/nature10656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ruan H-B, Nie Y, Yang X. 2013. Regulation of protein degradation by O-GlcNAcylation: crosstalk with ubiquitination. Mol. Cell. Proteomics MCP 12, 3489-3497. ( 10.1074/mcp.R113.029751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Z, Gucek M, Hart GW. 2008. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl Acad. Sci. USA 105, 13 793-13 798. ( 10.1073/pnas.0806216105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trinidad JC, Barkan DT, Thalhammer A, Sali A. 2012. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215-229. ( 10.1074/mcp.O112.018366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fan Z, Li J, Liu T, Zhang Z, Qin W, Qian X. 2021. A new tandem enrichment strategy for the simultaneous profiling of O-GlcNAcylation and phosphorylation in RNA-binding proteome. Analyst 146, 1188-1197. ( 10.1039/D0AN02305A) [DOI] [PubMed] [Google Scholar]
- 132.Yao H, Li A, Wang M. 2015. Systematic analysis and prediction of In Situ cross talk of O-GlcNAcylation and phosphorylation. BioMed Res. Int. 2015, 279823. ( 10.1155/2015/279823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Leney AC, El Atmioui D, Wu W, Ovaa H, Heck AJR. 2017. Elucidating crosstalk mechanisms between phosphorylation and O-GlcNAcylation. Proc. Natl Acad. Sci. USA 114, E7255-E7261. ( 10.1073/pnas.1620529114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Butkinaree C, Park K, Hart GW. 2010. O-linked β-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96-106. ( 10.1016/j.bbagen.2009.07.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schwein PA, Woo CM. 2020. The O-GlcNAc modification on kinases. ACS Chem. Biol. 15, 602-617. ( 10.1021/acschembio.9b01015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dias WB, Cheung WD, Hart GW. 2012. O-GlcNAcylation of kinases. Biochem. Biophys. Res. Commun. 422, 224-228. ( 10.1016/j.bbrc.2012.04.124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shi J, Wu S, Dai C, Li Y, Grundke-Iqbal I, Iqbal K, Liu F, Gong C-X. 2012. Diverse regulation of AKT and GSK-3β by O-GlcNAcylation in various types of cells. FEBS Lett. 586, 2443-2450. ( 10.1016/j.febslet.2012.05.063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shi J, Gu J, Dai C, Gu J, Jin X, Sun J, Iqbal K, Liu F, Gong C-X. 2015. O-GlcNAcylation regulates ischemia-induced neuronal apoptosis through AKT signaling. Sci. Rep. 5, 14500. ( 10.1038/srep14500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kazemi Z, Chang H, Haserodt S, McKen C, Zachara NE. 2010. O-Linked β-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3β-dependent manner. J. Biol. Chem. 285, 39 096-39 107. ( 10.1074/jbc.M110.131102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Inoue Y, Moriwaki K, Ueda Y, Takeuchi T, Higuchi K, Asahi M. 2018. Elevated O-GlcNAcylation stabilizes FOXM1 by its reduced degradation through GSK-3β inactivation in a human gastric carcinoma cell line, MKN45 cells. Biochem. Biophys. Res. Commun. 495, 1681-1687. ( 10.1016/j.bbrc.2017.11.179) [DOI] [PubMed] [Google Scholar]
- 141.Yang Y, et al. 2020. OGT suppresses S6K1-mediated macrophage inflammation and metabolic disturbance. Proc. Natl Acad. Sci. USA 117, 16 616-16 625. ( 10.1073/pnas.1916121117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pang Y, et al. 2021. High fat activates O-GlcNAcylation and affects AMPK/ACC pathway to regulate lipid metabolism. Nutrients 13, 1740. ( 10.3390/nu13061740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miura T, Kume M, Kawamura T, Yamamoto K, Hamakubo T, Nishihara S. 2018. O-GlcNAc on PKCζ Inhibits the FGF4-PKCζ-MEK-ERK1/2 pathway via inhibition of PKCζ phosphorylation in mouse embryonic stem cells. Stem Cell Rep. 10, 272-286. ( 10.1016/j.stemcr.2017.11.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guo X, Deng Y, Zhan L, Shang J, Liu H. 2021. O-GlcNAcylation contributes to intermittent hypoxia-associated vascular dysfunction via modulation of MAPKs but not CaMKII pathways. Mol. Med. Rep. 24, 744. ( 10.3892/mmr.2021.12384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tarrant MK, et al. 2012. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol. 8, 262-269. ( 10.1038/nchembio.771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schwein PA, Ge Y, Yang B, D'Souza A, Mody A, Shen D, Woo CM. 2022. Writing and erasing O-GlcNAc on casein kinase 2 alpha alters the phosphoproteome. ACS Chem. Biol. 17, 1111-1121. ( 10.1021/acschembio.1c00987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dong DL, Xu ZS, Chevrier MR, Cotter RJ, Cleveland DW, Hart GW. 1993. Glycosylation of mammalian neurofilaments. Localization of multiple O-linked N-acetylglucosamine moieties on neurofilament polypeptides L and M. J. Biol. Chem. 268, 16 679-16 687. ( 10.1016/S0021-9258(19)85471-6) [DOI] [PubMed] [Google Scholar]
- 148.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. 2001. Dynamic O-Glycosylation of nuclear and cytosolic proteins. J. Biol. Chem. 276, 9838-9845. ( 10.1074/jbc.M010420200) [DOI] [PubMed] [Google Scholar]
- 149.Akimoto Y, Comer FI, Cole RN, Kudo A, Kawakami H, Hirano H, Hart GW. 2003. Localization of the O-GlcNAc transferase and O-GlcNAc-modified proteins in rat cerebellar cortex. Brain Res. 966, 194-205. ( 10.1016/S0006-8993(02)04158-6) [DOI] [PubMed] [Google Scholar]
- 150.Yang X, Zhang F, Kudlow JE. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69-80. ( 10.1016/s0092-8674(02)00810-3) [DOI] [PubMed] [Google Scholar]
- 151.Hahne H, Gholami AM, Kuster B. 2012. Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol. Cell. Proteomics MCP 11, 843-850. ( 10.1074/mcp.M112.019463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reinke H, Asher G. 2019. Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20, 227-241. ( 10.1038/s41580-018-0096-9) [DOI] [PubMed] [Google Scholar]
- 153.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. 2004. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl Acad. Sci. USA 101, 13 132-13 137. ( 10.1073/pnas.0403471101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khidekel N, et al. 2007. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339-348. ( 10.1038/nchembio881) [DOI] [PubMed] [Google Scholar]
- 155.Tramutola A, et al. 2018. Proteomic identification of altered protein O-GlcNAcylation in a triple transgenic mouse model of Alzheimer's disease. Biochim. Biophys. Acta 1864, 3309-3321. ( 10.1016/j.bbadis.2018.07.017) [DOI] [PubMed] [Google Scholar]
- 156.Alfaro JF, et al. 2012. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl Acad. Sci. USA 109, 7280-7285. ( 10.1073/pnas.1200425109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huynh VN, et al. 2021. Defining the dynamic regulation of O-GlcNAc proteome in the mouse cortex—the O-GlcNAcylation of synaptic and trafficking proteins related to neurodegenerative diseases. Front. Aging 2, 757801. ( 10.3389/fragi.2021.757801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. 2002. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791-804. ( 10.1074/mcp.M200048-MCP200) [DOI] [PubMed] [Google Scholar]
- 159.Wang S, et al. 2017. Quantitative proteomics identifies altered O-GlcNAcylation of structural, synaptic and memory-associated proteins in Alzheimer's disease. J. Pathol. 243, 78-88. ( 10.1002/path.4929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cieniewski-Bernard C, Bastide B, Lefebvre T, Lemoine J, Mounier Y, Michalski J-C. 2004. Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 3, 577-585. ( 10.1074/mcp.M400024-MCP200) [DOI] [PubMed] [Google Scholar]
- 161.Deracinois B, Camoin L, Lambert M, Boyer J-B, Dupont E, Bastide B, Cieniewski-Bernard C. 2018. O-GlcNAcylation site mapping by (azide-alkyne) click chemistry and mass spectrometry following intensive fractionation of skeletal muscle cells proteins. J. Proteomics 186, 83-97. ( 10.1016/j.jprot.2018.07.005) [DOI] [PubMed] [Google Scholar]
- 162.Prisco SZ, et al. 2020. Excess protein O-GlcNAcylation links metabolic derangements to right ventricular dysfunction in pulmonary arterial hypertension. Int. J. Mol. Sci. 21, 7278. ( 10.3390/ijms21197278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DMF, Agnew B, Kuster B. 2013. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J. Proteome Res. 12, 927-936. ( 10.1021/pr300967y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhu Y, Willems LI, Salas D, Cecioni S, Wu WB, Foster LJ, Vocadlo DJ. 2020. Tandem bioorthogonal labeling uncovers endogenous cotranslationally O-GlcNAc modified nascent proteins. J. Am. Chem. Soc. 142, 15 729-15 739. ( 10.1021/jacs.0c04121) [DOI] [PubMed] [Google Scholar]
- 165.Teo CF, Ingale S, Wolfert MA, Elsayed GA, Nöt LG, Chatham JC, Wells L, Boons G-J. 2010. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat. Chem. Biol. 6, 338-343. ( 10.1038/nchembio.338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shen B, Zhang W, Shi Z, Tian F, Deng Y, Sun C, Wang G, Qin W, Qian X. 2017. A novel strategy for global mapping of O-GlcNAc proteins and peptides using selective enzymatic deglycosylation, HILIC enrichment and mass spectrometry identification. Talanta 169, 195-202. ( 10.1016/j.talanta.2017.03.049) [DOI] [PubMed] [Google Scholar]
- 167.Lopez Aguilar A, Gao Y, Hou X, Lauvau G, Yates JR, Wu P. 2017. Profiling of protein O-GlcNAcylation in Murine CD8 + effector- and memory-like T cells. ACS Chem. Biol. 12, 3031-3038. ( 10.1021/acschembio.7b00869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lund PJ, Elias JE, Davis MM. 2016. Global analysis of O-GlcNAc glycoproteins in activated human T cells. J. Immunol. 197, 3086-3098. ( 10.4049/jimmunol.1502031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, Pitteri SJ. 2018. Mapping and quantification of over 2000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (Isotag). Mol. Cell. Proteomics 17, 764-775. ( 10.1074/mcp.RA117.000261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sprung R, Nandi A, Chen Y, Kim SC, Barma D, Falck JR, Zhao Y. 2005. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res. 4, 950-957. ( 10.1021/pr050033j) [DOI] [PubMed] [Google Scholar]
- 171.Nagel AK, Schilling M, Comte-Walters S, Berkaw MN, Ball LE. 2013. Identification of O-linked N-acetylglucosamine (O-GlcNAc)-modified osteoblast proteins by electron transfer dissociation tandem mass spectrometry reveals proteins critical for bone formation. Mol. Cell. Proteomics 12, 945-955. ( 10.1074/mcp.M112.026633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu J, et al. 2021. Quantitative chemoproteomics reveals O-GlcNAcylation of cystathionine γ-lyase (CSE) represses trophoblast syncytialization. Cell Chem. Biol. 28, 788-801.e5. ( 10.1016/j.chembiol.2021.01.024) [DOI] [PubMed] [Google Scholar]
- 173.Zaro BW, Yang Y-Y, Hang HC, Pratt MR. 2011. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc. Natl Acad. Sci. USA 108, 8146-8151. ( 10.1073/pnas.1102458108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Okamoto-Uchida Y, Izawa J, Nishimura A, Hattori A, Suzuki N, Hirayama J. 2019. Post-translational modifications are required for circadian clock regulation in vertebrates. Curr. Genomics 20, 332-339. ( 10.2174/1389202919666191014094349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mauvoisin D. 2019. Circadian rhythms and proteomics: it's all about posttranslational modifications! Wiley Interdiscip. Rev. Syst. Biol. Med. 11, e1450. ( 10.1002/wsbm.1450) [DOI] [PubMed] [Google Scholar]
- 176.Srikanta SB, Cermakian N. 2021. To Ub or not to Ub: regulation of circadian clocks by ubiquitination and deubiquitination. J. Neurochem. 157, 11-30. ( 10.1111/jnc.15132) [DOI] [PubMed] [Google Scholar]
- 177.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. 1994. Temporal phosphorylation of the Drosophila period protein. Proc. Natl Acad. Sci. USA 91, 2260-2264. ( 10.1073/pnas.91.6.2260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Narasimamurthy R, Virshup DM. 2021. The phosphorylation switch that regulates ticking of the circadian clock. Mol. Cell 81, 1133-1146. ( 10.1016/j.molcel.2021.01.006) [DOI] [PubMed] [Google Scholar]
- 179.Mendoza-Viveros L, Bouchard-Cannon P, Hegazi S, Cheng AH, Pastore S, Cheng H-YM. 2017. Molecular modulators of the circadian clock: lessons from flies and mice. Cell. Mol. Life Sci. 74, 1035-1059. ( 10.1007/s00018-016-2378-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cai YD, et al. 2021. CK2 inhibits TIMELESS nuclear export and modulates CLOCK transcriptional activity to regulate circadian rhythms. Curr. Biol. 31, 502-514.e7. ( 10.1016/j.cub.2020.10.061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang X, Qian K. 2017. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452-465. ( 10.1038/nrm.2017.22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gonzalez-Rellan MJ, Fondevila MF, Dieguez C, Nogueiras R. 2022. O-GlcNAcylation: a sweet hub in the regulation of glucose metabolism in health and disease. Front. Endocrinol. 13, 873513. ( 10.3389/fendo.2022.873513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lee JB, Pyo K-H, Kim HR. 2021. Role and function of O-GlcNAcylation in cancer. Cancers 13, 5365. ( 10.3390/cancers13215365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sun L, Lv S, Song T. 2021. O-GlcNAcylation links oncogenic signals and cancer epigenetics. Discov. Oncol. 12, 54. ( 10.1007/s12672-021-00450-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yang R, Wang L, Wu Z, Yin Y, Jiang S-W. 2022. How nanotechniques could vitalize the O-GlcNAcylation-targeting approach for cancer therapy. Int. J. Nanomedicine 17, 1829-1841. ( 10.2147/IJN.S360488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Selvan N, et al. 2017. A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat. Chem. Biol. 13, 882-887. ( 10.1038/nchembio.2404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Qin W, Xie Z, Wang J, Ou G, Wang C, Chen X. 2020. Chemoproteomic profiling of O-GlcNAcylation in Caenorhabditis elegans. Biochemistry 59, 3129-3134. ( 10.1021/acs.biochem.9b00622) [DOI] [PubMed] [Google Scholar]
- 188.Akan I, Halim A, Vakhrushev SY, Clausen H, Hanover JA. 2021. Drosophila O-GlcNAcase mutants reveal an expanded glycoproteome and novel growth and longevity phenotypes. Cells 10, 1026. ( 10.3390/cells10051026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ma J, Liu T, Wei A-C, Banerjee P, O'Rourke B, Hart GW. 2015. O-GlcNAcomic profiling identifies widespread O-linked β-N-acetylglucosamine modification (O-GlcNAcylation) in oxidative phosphorylation system regulating cardiac mitochondrial function. J. Biol. Chem. 290, 29 141-29 153. ( 10.1074/jbc.M115.691741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ma J, et al. 2016. Comparative proteomics reveals dysregulated mitochondrial O-GlcNAcylation in diabetic hearts. J. Proteome Res. 15, 2254-2264. ( 10.1021/acs.jproteome.6b00250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cao W, Cao J, Huang J, Yao J, Yan G, Xu H, Yang P. 2013. Discovery and confirmation of O-glcnacylated proteins in rat liver mitochondria by combination of mass spectrometry and immunological methods. PLoS ONE 8, e76399. ( 10.1371/journal.pone.0076399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ramirez-Correa GA, Jin W, Wang Z, Zhong X, Gao WD, Dias WB, Vecoli C, Hart GW, Murphy AM. 2008. O-Linked GlcNAc modification of cardiac myofilament proteins: a novel regulator of myocardial contractile function. Circ. Res. 103, 1354-1358. ( 10.1161/CIRCRESAHA.108.184978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vosseller K, et al. 2006. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923-934. ( 10.1074/mcp.T500040-MCP200) [DOI] [PubMed] [Google Scholar]
- 194.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. 2009. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl Acad. Sci. USA 106, 8894-8899. ( 10.1073/pnas.0900288106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zeidan Q, Wang Z, De Maio A, Hart GW. 2010. O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol. Biol. Cell 21, 1922-1936. ( 10.1091/mbc.e09-11-0941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Myers SA, Panning B, Burlingame AL. 2011. Polycomb repressive complex2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 108, 9490-9495. ( 10.1073/pnas.1019289108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lee A, Miller D, Henry R, Paruchuri VDP, O'Meally RN, Boronina T, Cole RN, Zachara NE. 2016. Combined antibody/lectin enrichment identifies extensive changes in the O-GlcNAc sub-proteome upon oxidative stress. J. Proteome Res. 15, 4318-4336. ( 10.1021/acs.jproteome.6b00369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li J, et al. 2019. An isotope-coded photocleavable probe for quantitative profiling of protein O-GlcNAcylation. ACS Chem. Biol. 14, 4-10. ( 10.1021/acschembio.8b01052) [DOI] [PubMed] [Google Scholar]
- 199.Lin C-H, Liao C-C, Wang S-Y, Peng C-Y, Yeh Y-C, Chen M-Y, Chou T-Y. 2022. Comparative O-GlcNAc proteomic analysis reveals a role of O-GlcNAcylated SAM68 in lung cancer aggressiveness. Cancers 14, 243. ( 10.3390/cancers14010243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Drougat L, Olivier-Van Stichelen S, Mortuaire M, Foulquier F, Lacoste A-S, Michalski J-C, Lefebvre T, Vercoutter-Edouart A-S. 2012. Characterization of O-GlcNAc cycling and proteomic identification of differentially O-GlcNAcylated proteins during G1/S transition. Biochim. Biophys. Acta 1820, 1839-1848. ( 10.1016/j.bbagen.2012.08.024) [DOI] [PubMed] [Google Scholar]
- 201.Liu J, Hao Y, He Y, Li X, Sun D, Zhang Y, Yang P-Y, Chen X. 2021. Quantitative and site-specific chemoproteomic profiling of protein O-GlcNAcylation in the cell cycle. ACS Chem. Biol. 16, 1917-1923. ( 10.1021/acschembio.1c00301) [DOI] [PubMed] [Google Scholar]
- 202.Fleischer JG, Das SK, Bhapkar M, Manoogian ENC, Panda S. 2022. Associations between the timing of eating and weight-loss in calorically restricted healthy adults: findings from the CALERIE study. Exp. Gerontol. 165, 111837. ( 10.1016/j.exger.2022.111837) [DOI] [PubMed] [Google Scholar]
- 203.Wilkinson MJ, et al. 2020. Ten-hour time-restricted eating reduces weight, blood pressure, and atherogenic lipids in patients with metabolic syndrome. Cell Metab. 31, 92-104.e5. ( 10.1016/j.cmet.2019.11.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pendergast JS, Branecky KL, Yang W, Ellacott KLJ, Niswender KD, Yamazaki S. 2013. High-fat diet acutely affects circadian organisation and eating behavior. Eur. J. Neurosci. 37, 1350-1356. ( 10.1111/ejn.12133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Branecky KL, Niswender KD, Pendergast JS. 2015. Disruption of daily rhythms by high-fat diet is reversible. PLoS ONE 10, e0137970. ( 10.1371/journal.pone.0137970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Figlia G, Willnow P, Teleman AA. 2020. Metabolites regulate cell signaling and growth via covalent modification of proteins. Dev. Cell 54, 156-170. ( 10.1016/j.devcel.2020.06.036) [DOI] [PubMed] [Google Scholar]
- 207.Bludau I, Willems S, Zeng W-F, Strauss MT, Hansen FM, Tanzer MC, Karayel O, Schulman BA, Mann M. 2022. The structural context of posttranslational modifications at a proteome-wide scale. PLoS Biol. 20, e3001636. ( 10.1371/journal.pbio.3001636) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.